Abstract
There are nearly 400 modern domestic dog breeds with a unique histories and genetic profiles. To track the genetic signatures of breed development, we have assembled the most diverse dataset of dog breeds, reflecting their extensive phenotypic variation and heritage. Combining genetic distance, migration, and genome-wide haplotype sharing analyses, we uncover geographic patterns of development and independent origins of common traits. Our analyses reveal the hybrid history of breeds and elucidate the effects of immigration, revealing for the first time a suggestion of New World dog within some modern breeds. Finally, we used cladistics and haplotype sharing to show that some common traits have arisen more than once in the history of the dog. These analyses characterize the complexities of breed development resolving long standing questions regarding individual breed origination, the effect of migration on geographically distinct breeds, and by inference, transfer of trait and disease alleles among dog breeds.
Keywords: canine, domestication, population, migration, morphology, behavior
Graphical abstract
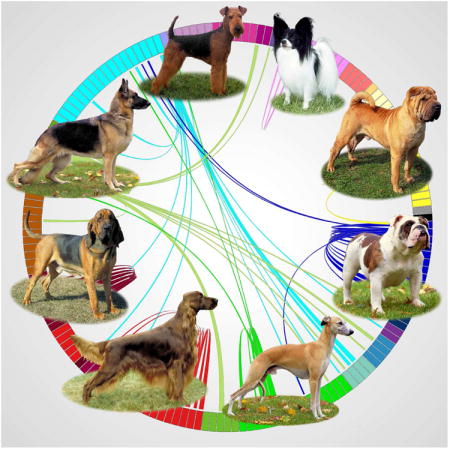
Introduction
The dog, Canis familiaris, is the first domesticate earning a place within nearly every society across the globe for thousands of years (Druzhkova et al., 2013; Thalmann et al., 2013; Vila et al., 1999; Vila et al., 1997). Over the millennia dogs have assisted humans with hunting and livestock management, guarding house and field, and played crucial roles in major wars (Moody et al., 2006). Providing a range of services from companionship to production of fur and meat (Wilcox and Walkowicz, 1995), the diversity of talents and phenotypes combined with an unequalled emotional connection between dog and man has led to the creation of more than 350 distinct breeds, each of which is a closed breeding population that reflects a collage of defining traits (www.akc.org).
Previous studies have addressed the genomic makeup of a limited number of breeds, demonstrating that dogs from the same breed share common alleles and can be grouped using measures of population structure (Irion et al., 2003; Koskinen, 2003; Parker et al., 2004), and breeds that possess similar form and function often share similar allelic patterns (Parker et al., 2004; Parker et al., 2007; Vonholdt et al., 2010). However, none of these studies have effectively accounted for the variety of mechanisms through which modern breeds may have developed, such as geographic separation and immigration; the role of hybridization in the history of the breeds; and the time-line of the formation of breeds. In this study we overcome these barriers by presenting an expansive dataset including pure-breeds sampled from multiple sections of the globe and genotyped on a dense scale. By applying both phylogenetic methods as well as a genome-wide analysis of recent haplotype sharing, we have unraveled common population confounders for many breeds leading us to propose a two-step process of breed creation beginning with ancient separation by functional employment followed by recent selection for physical attributes. These data and analyses provide a basis for understanding which and why numerous, sometimes deleterious, mutations are shared across seemingly unrelated breeds.
Results
We examined genomic data from the largest and most diverse group of breeds studied to date, amassing a dataset of 1346 dogs representing 161 breeds. Included are populations with vastly different breed histories, originating from all continents except Antarctica, and sampled from North America, Europe, Africa, and Asia. We have specifically included breeds that represent the full range of phenotypic variation present amongst modern dogs, as well as three breeds sampled from both the US and their country of origin. Samples from 938 dogs representing 127 breeds and nine wild canids were genotyped using the Illumina CanineHD bead array following standard protocols (Illumina, San Diego, CA). Data were combined with publically available information from 405 dogs genotyped using the same chip (Hayward et al., 2016; Vaysse et al., 2011). For three dogs from one breed, genotypes were retrieved from publically available sequence files and all were merged into a single dataset (Table S1). After pruning for low quality or genotyping rate 150,067 informative SNPs were retained.
Ascertainment bias has been shown to skew population genetic calculations that require estimation of allele frequencies and diversity measures (Lachance and Tishkoff, 2013). It has also been shown that ascertainment based on a single individual provides less bias than a mixed group (Patterson et al., 2012). The SNPs used in this study were identified primarily within the boxer or from boxer compared to another genome (Vaysse et al., 2011) which has exaggerated the boxer minor allele frequency (0.351 in boxer compared to 0.260 overall) but has little affect the other breeds (MAF range 0.247-0.284). To minimize the effect this might have, we have chosen to use distance measures based on allele sharing rather than frequency and to enhance these analyses with unbiased haplotype sharing for a robust assessment of canine population structure.
A bootstrapped cladogram was obtained using an identity-by-state distance matrix and a neighbor joining tree algorithm (Methods in the Supplement). After 100 bootstraps, 91% of breeds (146/161) formed single, breed-specific nodes with 100% bootstrap support (Figure 1). Of the 15 breeds that did not meet these criteria, seven (Belgian Tervuren, Belgian sheepdog, Cane Corso, Bull terrier, Miniature Bull terrier, Rat terrier, American Hairless terrier) were part of two- or three-breed clades that were supported at 98% or greater, and two breeds (Lhasa Apso, and Saluki) formed single-breed clades that were supported at 50 and 78%, respectively. Four breeds (Redbone Coonhound, Sloughi, Cane Paratore, Jack Russell terrier) were split within single multi-breed clades and the last two breeds, Xoloitzcuintli and Peruvian Hairless dog, were split between divergent clades. Nine of the breeds that were not monophyletic were either newly recognized by the American Kennel Club (AKC) or not recognized at the time of sample collection and likely represent a breed under development. Two other non-monophyletic breeds are comprised of dogs collected in two countries; the Cane Corsos collected in Italy form a fully supported, single clade, as do the Salukis collected in the United States (U.S.). However, the Cane Corsos collected in the U.S. form a paraphyletic clade near the Neapolitan Mastiffs and the Salukis collected in the Middle East form multiple paraphyletic groups within a clade that includes the U.S. Salukis and Afghan hounds.
Figure 1.
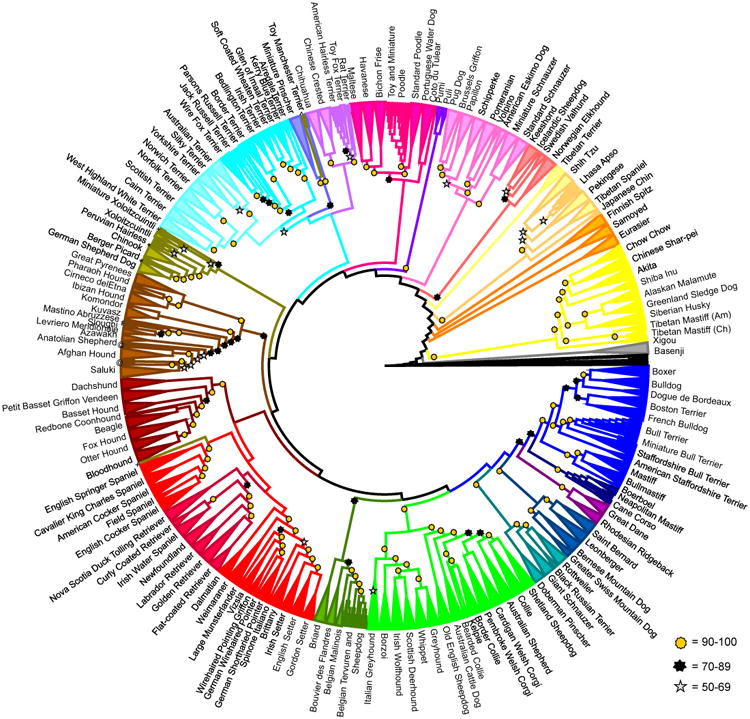
Cladogram of 161 domestic dog breeds. Breeds that form unique clades supported by 100% of bootstraps are combined into triangles. For all other branches the gold star =90% or better, Black star = 70% -89%, Silver star = 50% -69% bootstrap support. Breeds are listed on the perimeter of the circle. A small number of dogs do not cluster with the rest of their breed indicated as follows: * - Cane paratore; + - Peruvian Hairless dog; # - Sloughi; @ - Country of origin Salukis; ^ - Miniature Xoloitzcuintli
Not including those that are breed-specific, this study defined 105 phylogenetic nodes supported by ≥ 90% of bootstrap replicates, 133 by ≥70%, and 150 supported by ≥ 50% of replicates. We identify 29 multi-breed clades that are supported at ≥90%. Each of these clades includes two to 16 breeds and together account for 78% of breeds in the dataset. One hundred and fifty breeds, or 93% of the dataset, can be divided into 23 clades of two to 18 breeds each, supported at >50%. These multi-breed clades reflect common behaviors, physical appearance, and/or related geographic origin (Figure 2).
Figure 2.

Representatives from each of the 23 clades of breeds. Breeds and clades are listed for each picture from left to right, top to bottom: a)Akita – Asian Spitz, b) Shih Tzu – Asian Toy (by Mary Bloom), c) Icelandic sheepdog – Nordic Spitz (by Veronica Druk) d) Miniature Schnauzer - Schnauzer, e) Pomeranian – Small Spitz, f) Brussels Griffon – Toy Spitz (by Mary Bloom), g) Puli - Hungarian, h) Standard Poodle - Poodle, i) Chihuahua – American Toy, j) Rat Terrier – American Terrier (by Stacy Zimmerman), k) Miniature Pinscher - Pinscher, l) Irish Terrier -Terrier, m) German Shepherd Dog - New World (by Mary Bloom), n) Saluki -Mediterranean (by Mary bloom), o) Basset Hound – Scent Hound (by Mary Bloom), p) American Cocker Spaniel – Spaniel (by Mary Bloom), q) Golden Retriever – Retriever (by Mary Bloom), r) German Shorthaired Pointer – Pointer Setter (by Mary Bloom), s)Briard – Continental Herder (by Mary Bloom), t)Shetland Sheepdog – U.K. Rural, u) Rottweiler - Drover, v) Saint Bernard - Alpine, w) English Mastiff – European Mastiff (by Mary Bloom).
Eleven breeds did not group with significance to any other breeds. Five breeds form independent clades and six others are paraphyletic to established clades with <50% bootstrap support (Table S2). The lack of grouping may indicate that we have not sampled the closest relatives of these breeds or that these breeds comprise outcrossings that are not shared by similar breeds.
To assess hybridization across the clades, identical-by-decent (IBD) haplotype sharing was calculated between all pairs of dogs from the 161 breeds. Haplotypes were phased using the program Beagle (Browning and Browning, 2013) in 100 SNP windows, resulting in a minimum haplotype size of 232kb, well above the shared background level established in previous studies (Lindblad-Toh et al., 2005; Sutter et al., 2004). The large haplotypes specifically target admixture resulting from breed formation rather than domestication, which previous studies have not addressed. The total length of the shared haplotypes was summed for each pair of dogs. Individuals from within the same breed-clade share nearly four times more of their genome within large IBD haplotype blocks than dogs in different breed clades [median shared haplotype lengths of 9,742,000bp and 2,184,000bp, respectively, P(K-S and Wilcox) < 2.2e-16 (Figure 3a)]. Only 5% of the across-breed pairings have a median greater than 9,744,974bp. These exceptions argue for recent admixture events between breeds, as evidenced by the example of the Eurasier breed, created in the 1970's by mixing Chow Chow with other spitz-type breeds (Fogle, 2000) (Figure 3b). The data reveals not only the components of the breed but also the explanation for its placement on the cladogram. The Eurasier (unclustered) shows significant haplotype sharing with the Samoyed (unclustered), Keeshond (Nordic Spitz) and Chow Chow (Asian Spitz)(Figure 3b). Because all three breeds are located in different clades, unrelated to each other, the Eurasier falls between the component breeds and forms its own single-breed clade. Haplotype-sharing bar graphs for each of 161 breeds, including 152 AKC breeds, are available in the supplemental material (Data File S1). This provides a long term resource for identifying populations that likely share rare and common traits that will be invaluable for mapping the origins of deleterious and beneficial mutations.
Figure 3.
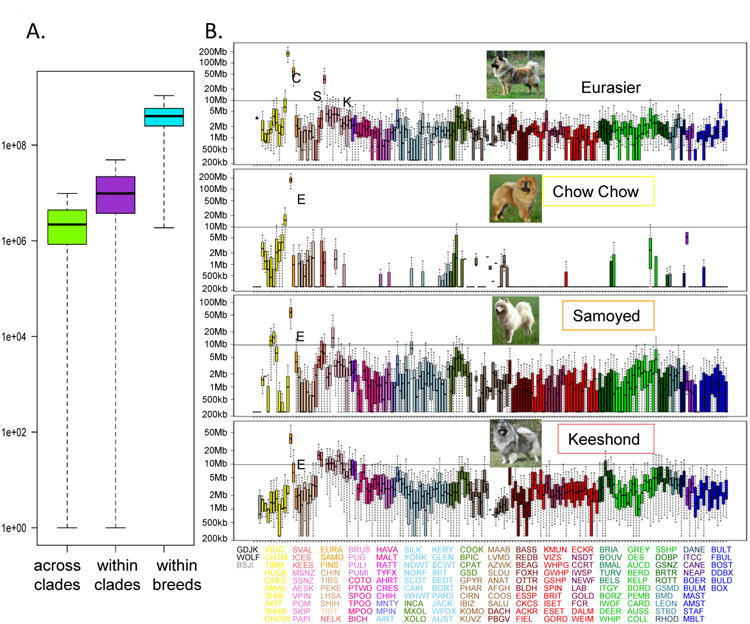
Gross haplotype sharing across breeds. A) Boxplot of total haplotype sharing between all pairs of dogs from breeds within the same clade, across different clades and within the same breed. The difference between the distributions is highly significant, p<2e-16. B) Example of haplotype sharing between three breeds (Samoyed, Chow Chow and Keeshond) and a fourth (Eurasier) that was created as a composite of the other three. Combined haplotype length is displayed on the y-axis, 169 breeds and populations are listed on the x-axis in the order they appear on the cladogram starting with the jackal and continuing counter-clockwise. Haplotype sharing of zero is set at 250,000 for graphing, a value just below what is detected in this analysis. Breeds are colored by clade. 95% significance level is indicated by the horizontal line. Breed abbreviations are listed under the graph, in the order they appear and colored by clade.
Strong evidence of admixture across the clades was found in 117 breeds (Figure 4). A small number of these were identified in previous studies using migration analysis (Pickrell and Pritchard, 2012; Shannon et al., 2015) Thirty percent of these breeds share with only one breed outside their clade. Therefore, more than half (54%) of the breeds that make up the 23 established clades share large haplotypes with one or zero breeds outside their clade indicating breed creation by selection based on the initial founder population rather than recent admixture. Only six of the 161 breeds share extensive haplotypes with many (>8) different groups suggesting recent creation of these breeds from multiple others or that they provide a popular modern breed component. The overall low level of sharing across diverse breeds suggests that interclade crosses are done thoughtfully and for specific reasons, such as the introduction of a new trait or the immigration of a breed to a new geographic region.
Figure 4.
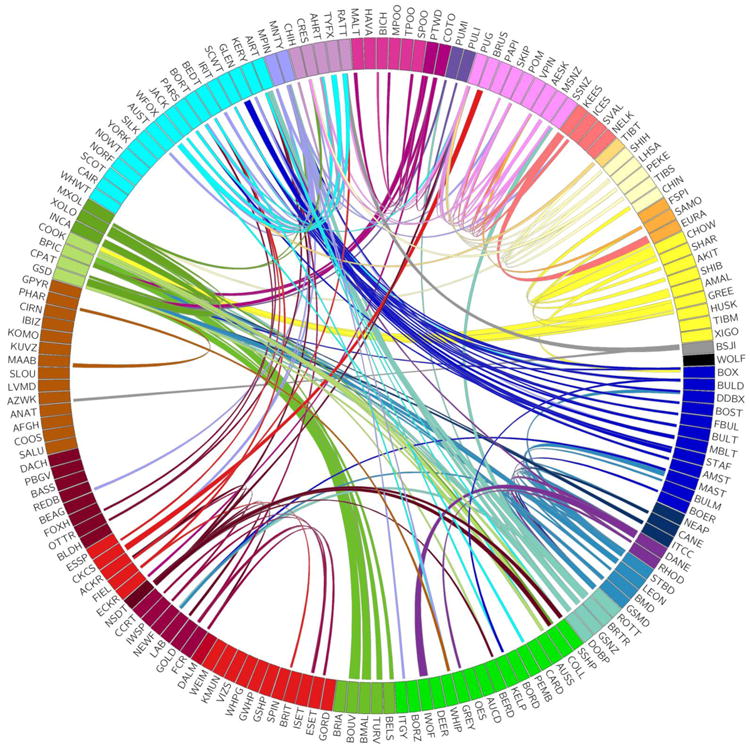
Haplotype sharing between breeds from different phylogenetic clades. The circos plot is ordered and colored to match the tree in Figure 1. Ribbons connecting breeds indicate a median haplotype sharing between all dogs of each breed in excess of 95% of all haplotype sharing across clades.
As importation and establishment in a new country has been shown to have a measurable effect on breed structure (Quignon et al., 2007), we chose three breeds, the Tibetan mastiff, Saluki, and Cane Corso, for inclusion in the study, with each collected in the country of origin as well as from established populations in the U.S. In each case there is division of the breed based on collection location. The split between the U.S. and Chinese Tibetan mastiffs is likely due to independent lineage formation stemming from an importation bottleneck, as is evident from estimations of inbreeding coefficients (Chinese Tibetan mastiffs average F = 0.07, and U.S. Tibetan mastiffs average F = 0.15). Similarly, the average inbreeding coefficient of Salukis collected in the U.S. is twice as high as those sampled from the countries of origin, (F = 0.21 and 0.10 respectively). Since the U.S. Salukis form a more strongly bootstrapped clade than the country of origin dogs, we suggesting that there is a less diverse gene pool in the U.S. In comparison, the Cane Corsos from Italy form a single clade, while the Cane Corsos from the U.S. cluster with the Neapolitan mastiffs, also collected in the U.S. Significant shared haplotypes are observed between the U.S. Cane Corsos and the Rottweiler that are not evident in the Italian Cane Corsos, as well as increased shared haplotypes with the other mastiffs. Cane Corsos have been in the U.S. for less than 30 years (American, 1998).
Our analyses were designed to detect recent admixture, therefore we were able to identify hybridization events that are described in written breed histories and stud-book records. Using the most reliably-dated crosses that produced modern breeds, we established a linear relationship between the total length of haplotype sharing and the age of an admixture event, occurring between 35 and 160 years before present (ybp)(Figure 5a). Applying this equation to the total shared haplotypes calculated from the genotyping data, we have validated this relationship on a second set of recently created breeds arriving at historically accurate time estimations (Figure 5b). Using the relationship equation, y = -1,613,084.67x + 262,137,843.89, where y is the total shared haplotype length and x is the number of years, we can estimate the time at which undocumented crosses or divisions from older breeds took place. For example, based on a median haplotype sharing value of 66,993,738 the Golden Retriever was separated from the Flat-coated Retriever in 1895 and the written history of the Golden retriever dates to crosses between multiple breeds taking place between 1868 and 1890 (Figure 5b), a near perfect match.
Figure 5.
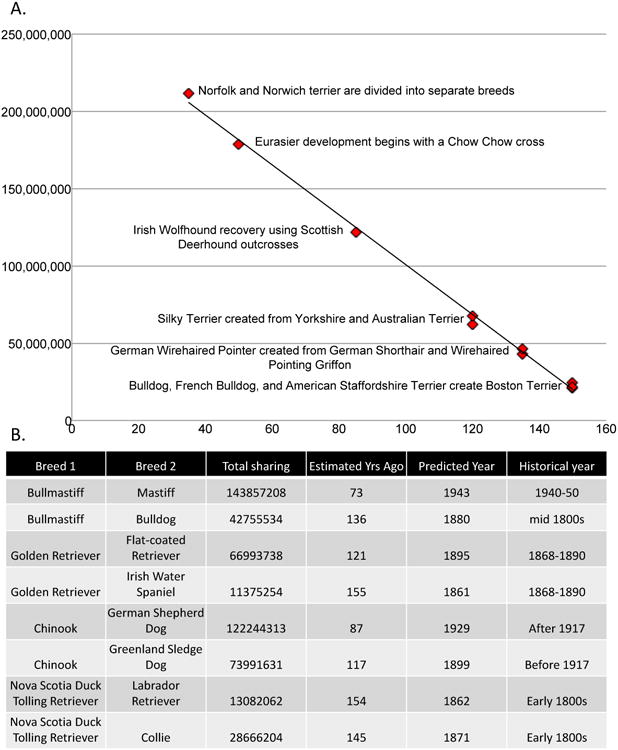
Total haplotype sharing is inversely correlated with the time of hybridization between breeds that have developed within the last 200 years. A) The time of hybridization in years-before-present is graphed on the X-axis and the median total haplotype sharing on the Y-axis for six breeds of dog with reliable recent histories of admixture in breed formation or recovery. The trendline shows a linear correlation with r2=1. B) The slope and intercept of the trendline from A was applied to the median haplotype sharing values from the data for four additional breeds with reliable breed creation dates to establish accuracy of estimated hybridization dates.
To determine if the multi-breed clades are formed through recent admixture rather than through common ancestral sources, we examined migration in 18 groups of four or more breeds. These include 16 of the clades established on the tree including nearby unclustered breeds, and two groups of small clades (American Terrier/American Toy and Small Spitz/Toy Spitz/Schnauzer) that are monophyletic, but not well supported. Using the program Treemix (Pickrell and Pritchard, 2012), and allowing zero to 12 predicted migration events, we determined the effect of admixture on clade formation by calculating the increase in maximum likelihood score over a zero migration tree (Figure 6A). Only two of the 18 clades, New World and Asian Toy (Figure 6B-C), showed evidence of extensive hybridization between the breeds. Thus the modern breeds were likely created through selection for unique traits within an ancient breed type with possible admixture from unrelated breeds to enhance the trait.
Figure 6.
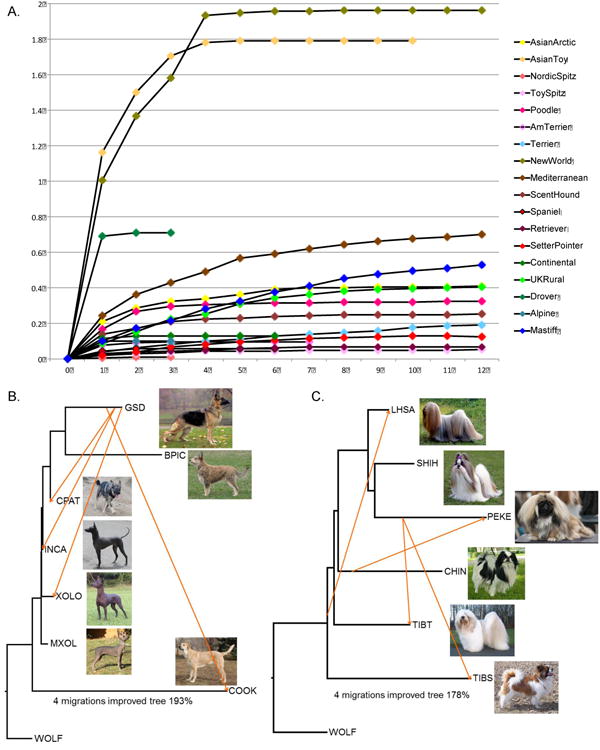
Assessment of migration between breeds within clades. Admixture was measured in Treemix for 18 groups of breeds representing clades or combinations of small clades. A) Improvement to the maximum likelihood tree of each group as the result of admixture. They-axis shows fold improvement over the zero admixture tree. B) Cladogram of the New World breeds with European herders allowing four migration events. Arrows show estimated migration between breeds colored by weight (yellow to red = 0 - 0.5). C) Cladogram showing migration within the Asian Toy clade including a neighboring breed, the Tibetan Terrier. Pictures by Yuri Hooker (INCA), Mary Bloom (GSD and SHIH), Maurizio Marziali (CPAT), Mary Malkiel (COOK) and John & Debbie Caponetto (large and small XOLO/MXOL)
Our hybridization analysis reveals evidence for disease sharing across the clades. For instance, Collie eye anomaly (CEA) is a disease that affects the development of the choroid in several herding breeds including the Collie, Border collie, Shetland sheepdog, and Australian shepherd, all members of the U.K. Rural clade (Lowe et al., 2003). The mutation and haplotype pattern displayed IBD across all affected breeds and we speculated that all share a common obviously affected ancestor (Parker et al., 2007). We were unable to explain, however the presence of the disease in the Nova Scotia duck tolling retriever, a sporting dog developed in Canada from an unknown mixture of local breeds, which also shares the same haplotype. This perplexing observation can now be explained, as this analysis shows that Collie and/or Shetland sheepdog were strong, undocumented, contributors to the formation of the Nova Scotia duck tolling retriever and, therefore, the likely source of the CEA mutation within that breed (Figure 7a).
Figure 7.
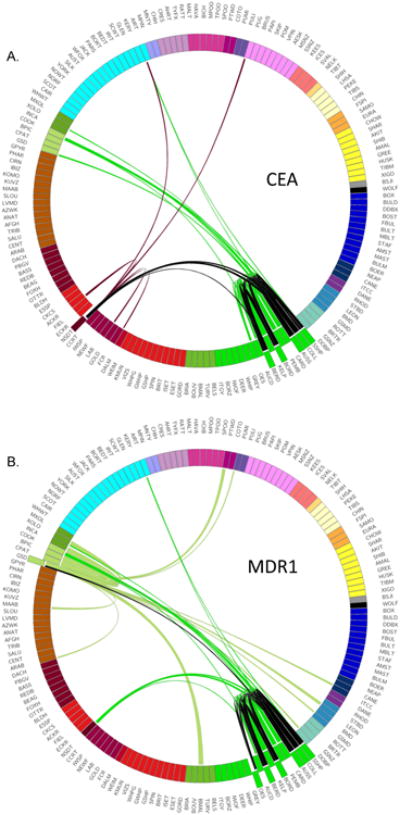
Haplotypes shared with breeds that carry known deleterious mutations. Breeds are connected if the median shared haplotype size exceeds the 95% threshold for interclade sharing. Sharing between breeds that are known to carry the mutation are colored black, sharing with other breeds are colored according to the breed that carries the mutation. A) Collie eye anomaly is found in a number of herding breeds developed in the UK and some sporting breeds developed in the US. B) Multi-drug resistance 1 mutation is carried by many UK herding breeds as well as the German Shepherd.
Similarly, a mutation in the MDR1 gene (multi-drug resistance 1), which causes life threatening reactions to multiple drugs in many of the U.K. Rural breeds, has been reported in 10% of German shepherd dogs (Mealey and Meurs, 2008). These data display a link between the German shepherd dog and U.K. Rural breeds through the Australian shepherd, highlighting the unexpected role the Australian shepherd or its predecessor played in the development of the modern German shepherd dog (Figure 6b). Earlier this year the MDR1 mutation was identified in the Chinook at a frequency of 15% (Donner et al., 2016). Our analysis reveals recent admixture between this breed and the German shepherd dog as well as previously unknown addition of Collie, both carriers of the MDR1 mutation. Haplotype sharing with the same affected breeds is found in the Xoloitzcuintli, which allows us to predict that this rare breed may also carry the deleterious allele but has yet to be tested.
Discussion
Phylogenetic analyses have often been applied to determine the relationships between dog breeds with the understanding that a tree structure cannot fully explain the development of breeds. Prior studies have shown that single mutations produce recognizable traits that are shared across breeds from diverse clades, suggesting that admixture across clades is a notable source of morphologic diversity (Cadieu et al., 2009; Parker et al., 2009; Sutter et al., 2007). Studies of linkage disequilibrium and haplotype sharing suggest, further, that within regions of ∼10-15kb, there exist a small number of haplotypes that are shared by the majority of breeds, while breed specificity is revealed only in large haplotypes (Lindblad-Toh et al., 2005; Sutter et al., 2004).
In this study we observe that the majority of dog breeds either do not share large haplotypes outside their clade or share with only one remote breed. The small number of breeds that share excessively outside their assigned clade could be recently created from multiple diverse breeds or may have been popular contributors to other breeds. For example, the Pug dog groups closely with the European toy breed, Brussels Griffon (Figure 2f), in the Toy Spitz clade but also shares extensive haplotypes with the Asian Toy breeds (Figure 2b) as well as many small dog breeds from multiple other clades. This likely indicates the Pug's early exportation from Asia and subsequent contribution to many small breeds (Watson, 1906). Consider also the extensive cross-clade haplotype sharing in the Chinook, a recently created breed with multiple ancestors from different breeds. Our data both recapitulates and enhances the written history of this breed (http://www.chinook.org/history.html) (Data File S1). Extreme examples such as these underscore the complications implicit in relying on phylogeny alone to describe breed relationships. Overall our data shows that admixture has played an important role in the development of many breeds and, as new hybrids are added to phylogenetic analyses, the topology of the cladogram will likely rearrange to accommodate.
The ability to determine a time of hybridization for recent admixture events can refine sparse historical accounts of breed formation. For example, when dog fighting was a popular form of entertainment, many combinations of terriers and mastiff or bully-type breeds were crossed to create dogs that would excel in that sport. In this analysis, all of the bull and terrier crosses map to the terriers of Ireland and date to 1860-1870. This coincides perfectly with the historical descriptions that, though they do not clearly identify all breeds involved, report the popularity of dog contests in Ireland and the lack of stud book veracity, hence undocumented crosses, during this era of breed creation (Lee, 1894).
The dates estimated from these data are approximations, as selection for or against traits that accompanied each cross, as well as the size of the population at the time of the cross, would have affected retention of the haplotypes within the genome. Based on these estimates the excess haplotype sharing that we have identified represents the creation of breeds since the Victorian era breed explosion. Most breeds within each clade share haplotypes at this level (<200 ybp), however, the lack of sharing across the clades, outside of very specific crosses, suggests the clades were developed much earlier than the breed registries. Dividing the data by clade, the median haplotype sharing is lowest in the Asian Spitz (median = 0) and the Mediterranean clades (median = 516,900) (median range across all clades = 0-3,459,000), indicating that these clades are most divergent and possibly older than the rest. This fits well with previous studies that suggest the earliest dogs came from Central and East Asia (Pang et al., 2009; Shannon et al., 2015). Interestingly, the mean haplotype sharing is slightly higher in the Asian Spitz clade than it is in the Mediterranean clade (mean = 1,596,000 and 1,317,000, respectively) (Figure S1), implying that the Asian Spitz breeds have been used in recent crosses while the Mediterranean breeds are currently more segregated. These data describe a staggered pattern of dog breed creation starting with separation by type based on required function, and the form necessary to carry out that function. This would have taken place as the need arose during early human progression from hunter-gather to pastoral, agricultural and finally urban lifestyles. During the last 200 years, these breed-types were refined into very specific breeds by dividing the original functional dog into morphotypes based on small changes in appearance and with occasional outcrosses to enhance appearance or alter behavior (i.e. reduce aggression, increase biddability).
Though most breeds within a clade appear to be the result of descent from a common ancestor, the New World dogs and the Asian Toys showed nearly 200% improvement in the maximum likelihood score by allowing for admixture between the breeds within the clade. Based on this analysis, the Asian Toy dogs were likely not considered separate breeds when first exported from their country of origin resulting in multiple admixture events (Figure 6C). Unexpectedly, the New World clade admixture events center exclusively on the German shepherd dog which informs both the development of this breed as well as immigration of dog breeds to the New World (Figure 6B). The inclusion of German shepherd dog with Cane Paratore, an Italian working farm dog, likely indicates a recent common ancestor among these breeds as the German shepherd dog was derived from a herding dog of unknown ancestry in the late 1800's (gsdca.org). However, the hybridization of the German shepherd dog with the Peruvian Hairless dog and the Xoloitzcuintli, also a hairless breed, is unexpected and could be the result of recent admixture to enhance the larger varieties of these breeds or could indicate admixture of generic herding dogs from Southern Europe into South America during the Columbian Exchange.
Dogs have been in the Americas for more than 10,000 years, likely travelling from East Asia with the first humans (Wang et al., 2016). However, studies of mitochondrial DNA suggest that the original New World dogs were almost entirely replaced through European contact (Castroviejo-Fisher et al., 2011; Wayne and Ostrander, 1999; Witt et al., 2014) and additional Asian migrations (Brown et al., 2015). As colonists came to the Americas from the 16th to the 19th centuries, they brought Old World livestock, and therefore the dogs required to manage and tend the livestock, to the New World (Crosby, 1972). Many of the newly introduced animals outcompeted the native animals, which may explain the surprising and very strong herding dog signature in the native hairless breeds of South and Central America that were not developed to herd. In this analysis we observe that the ancient hairless breeds show extensive hybridization with herding dogs from Europe and, to a lesser extent, with each other. We also identify two additional clades of New World breeds, the American Terriers and the American Toys (Figure 2i and 2j), two monophyletic clades of small-sized breeds from North/Central America, which include a set of related terriers, and the Chihuahua and Chinese Crested. Written records state that the terriers trace their ancestry to the feists, a North American landrace dog bred for hunting, (http://www.americantreeingfeist.com,akc.org). The Chihuahua and Chinese Crested are both believed to have originated in Central America (American, 1998; Parker et al., 2017), despite the nomenclature of the latter, which implies Asian ancestry. In contrast, most new breeds developed in the Americas were created from crosses of European breeds and cluster accordingly (i.e. Boston terrier (European Mastiff), Nova Scotia duck tolling retriever (Retriever), Australian shepherd (UK Rural)). The separation of the older American breeds on the cladogram, despite recent European admixture suggests that both clades may retain the aboriginal New World dog genomic signatures intermixed with the European breed haplotypes, similar to the admixture between European, African, and Native American genomes that can be found in modern South American human populations (Mathias et al., 2016; Ruiz-Linares et al., 2014). This is the first indication that the New World dog signature may not be entirely extinct in modern dog breeds, as has been previously suggested (Leonard et al., 2002).
In addition to the effects on the native population, our analysis of geographically distinct subsets of the same breeds shows that some degree of admixture also occurs in the imported breeds when first introduced into a new country. These data suggest two outcomes of breed immigration that mirrors human immigration into a new region; the immigrant population is less diverse than the founding population and there is often admixture with the native population in early generations (Baharian et al., 2016; Zhai et al., 2016).
We observe further evidence of the role geography plays in the distribution of breeds within the clades. For instance, both the U.K. Rural and the Mediterranean clades include both sighthound and working dog breeds, two highly divergent groups in terms of physical and behavioral phenotype. Sighthounds are lithe and leggy hunters, built to run fast and have a strong prey drive. Working dogs include both the tall and heavy flock guards that are bred to live among herds without human interaction, preventing predator attacks, and mid-sized herders (Figure 2t), which are agile and bred to work closely with humans to control the movement of the flock without harming them. Yet despite the opposing phenotypes under selection, both breed types form single clades stemming from distinct geographical regions. Haplotype analysis shows no recent admixture between the geographically distinct clades, suggesting that these groups arose independently (Figure S2a-b). Archeological depictions show sighthound-type hunting dogs that date back 4000ybp (Alderton, 2002; Fogle, 2000), and one of the earliest known writings regarding segregation of dogs based by type clearly delineates hunting dogs from working dogs (Columella, 70). The new cladogram presented herein suggests that the switch from hunting to agricultural pursuits may have initiated early breed formation and that this occurred in multiple regions. These data show that geographical region can define a foundational canid population within which selection for universally relevant behaviors occurred independently, separating the regional groups also by function long ago.
The lack of admixture across clades that appear to share a common trait suggests that these traits may have arisen independently, multiple times. For example, these data show no recent haplotype sharing between the giant flock guards of the Mediterranean and the European mastiffs (Figure S2d). These breed types required large size for guarding, however each used that size in a different way, a fact that was recognized at least 2000 years ago (Columella, 70). The flock guards use their size to defeat animal predators while the mastiffs use their size to keep human predators at bay, often through fierce countenance rather than action. The phylogenetic placement of these breeds and lack of recent admixture suggests that giant size developed independently in the different clades, and that it may have been one of the earliest traits by which breeds were segregated thousands of years ago.
The cladogram of 161 breeds presented here represents the most diverse dataset of domestic dog breeds analyzed to date, displaying 23 well-supported clades of breeds representing breed-types that existed before the advent of breed clubs and registries. While the addition of more rare or niche breeds will produce a denser tree, the results here address many unanswered questions regarding the origins of breeds. We show that many traits such as herding, coursing, and intimidating size, which are associated with specific canine occupations have likely been developed more than once in different geographical locals during the history of modern dog. These data also show that extensive haplotype sharing across clades is a likely indicator of recent admixture that took place in the time since the advent of breed registries, thus leading to the creation of most of the modern breeds. However, the primary breed types were developed well before this time indicating selection and segregation of dog populations in the absence of formal breed recognition. Breed prototypes have been forming through selective pressures since ancient times depending on the job they were most required to perform. A second round of hybridization and selection has been applied within the last 200 years to create the many unique combinations of traits that modern breeds display. By combining genetic distance relationships with patterns of haplotype sharing, we can now elucidate the complex makeup of modern dogs breeds and guide the search for genetic variants important to canine breed development, morphology, behavior, and disease.
Experimental Procedures
Contact for Reagent and Resource Sharing
Further information and requests for data may be directed to, and will be fulfilled by the first author Heidi G. Parker; hgparker@mail.nih.gov
Experimental Model and Subject Details
Samples from adult, domestic dogs were collected with owners signed consent in accordance with standard protocols approved by the NHGRI IACUC committee, protocol #GFS-05-1. Of these samples, 48% were male, 51% were female and 1% did not report a sex. No experimental models were used
Method Details
Sample collection
Samples from domestic dogs were collected through dog events and mail-in submission with owners signed consent in accordance with standard protocols approved by the NHGRI IACUC committee. Blood draws were performed by licensed veterinarians or veterinary technicians and DNA extracted using the cell lysis protocol described by Bell et al. (Bell et al., 1981) followed by phenol/chloroform extraction with phase separation performed in 15 mL Phase-lock tubes (5-Prime, Inc. Gaithersburg, MD, USA). Saliva samples were collected by the owner or by a member of the Ostrander lab using the Performagene (PG-100) saliva collection kit and extracted following standard protocols (DNA Genotek, Ottowa, ON, Canada). DNA was resuspended in 10 mMolar Tris with 0.01 mM EDTA, pH 8.0 and stored at -80°C.
SNP genotyping
DNA samples from 947 canids were run on the Illumina Canine HD SNP chip (Illumina, San Diego, CA) using standard protocols. Genotype calls were made with Genome Studio V2011.1 with genotyping module v1.9.4 (Illumina). Subsets of these data were described in Dreger et al. (Dreger et al., 2016a; Dreger et al., 2016b). The dataset consists of 938 dogs from 127 breeds and nine wild canids. Genotyping data from 195 dogs from 30 breeds and 210 dogs from 39 breeds were added to the dataset from Vaysse et al. and Hayward et al., respectively(Hayward et al., 2016; Vaysse et al., 2011). One additional breed was included by pulling genotypes for the relevant SNP positions from published whole genome sequence data aligned to the Canfam3.1 reference sequence. Basic statistics place this breed on par with the others in the dataset (MAF= 0.25, breed average MAF=0.26; Observed homozygosity=0.81, breed range homozygosity= 0.64 to 0.84), therefore it was included in all analyses. The final dataset consisted of 1346 dogs from 161 breeds, three country of origin populations, three breed varieties, seven wolves, and two golden jackals genotyped at 150112 SNPs. Where possible, dogs from multiple datasets were included in a breed to increase the amount of variation captured and to assure that there was no source bias in the datasets. A list of breeds and genotype sources can be found in Table S1.
Quantification and Statistical Analysis
Distance matrix and dendrogram
A pairwise identity-by-state (IBS) distance matrix was computed using plink v1.07 and the --genome command followed by the --cluster command (Purcell et al., 2007). Matrices were fed into phylip using the neighbor program to build neighbor-joining cladograms (Felsenstein, 1989). Bootstrapping was performed using 100 datasets resampled with replacement from the original. The consense program in phylip was used to combine the bootstrap results and build a consensus tree by majority rule(Felsenstein, 1989). Cladograms were visualized using FigTree v1.4.2 (Rambaut, 2006) http://tree.bio.ed.ac.uk/software/figtree/. Tree file can be found in the supplement, Data File S2.
Population admixture analyses
The breeds that make up individual clades were analyzed using Treemix(Pickrell and Pritchard, 2012). The wolf was used as the outgroup in each run. Allele frequencies for input data were calculated in plink. Each clade was analyzed allowing 0-12 migration events. The effect of recent admixture on the clade was assessed based on the improvement in likelihood measurement of the tree, from a zero migration tree, with each additional migration. Predicted migration events were viewed using plotting_functions.R, an R script provided with the Treemix package. The trees shown were drawn in FigTree v1.4.2 (Rambaut, 2006) with migration lines added based on the R figures.
Haplotype sharing
Genome wide haplotype sharing was assessed using Beagle identity-by-decent sharing without imputation on 100 SNP windows with 25 SNP overlap and 10 SNP end trimming(Browning and Browning, 2013). Shared haplotypes were summed between all pairs of dogs that did not belong to the same breed. The distribution of haplotype sharing among dogs from breeds in the same clade versus dogs from breeds in different clades was assessed using a Kolmogorov-Smirnov test as implemented in R. Haplotype sharing across clades was determined to be significant between two breeds if greater than the 95the percentile boundary from all pairs of dogs from different clades. Boxplots of haplotype sharing distributions between a single breed and all other breeds were performed using R boxplot. Circle plots showing haplotype sharing between breeds from different clades with an average above the 95th percentile were created using Circos(Krzywinski et al., 2009). We assessed the amount of sharing identified in groups of two to ten dogs per breed both by Wilcoxon rank sum test between two and ten, and the Pearson's correlations between number of individuals representing a breed and number of breeds with significant sharing and found no significant difference (P(Wilcox)=0.62; Pearson's Correlation = -0.086, P=0.28). The average haplotype sharing between nine pairs of breeds was plotted against the age of historical admixture events reported in credible breed histories and/or current pedigree databases. A linear relationship was fitted to this plot; y = -1,613,084.67x + 262,137,843.89, where y is the total shared haplotype length and x is the number of years. This equation was applied to the average haplotype sharing between eight additional breed pairs and results compared to reported dates of admixture to show the relative accuracy of the estimations.
Data Availability
Raw data files for the SNP genotype arrays have been deposited in the NCBI Gene Expression Omnibus under accession numbers GEO: GSE90441, GSE83160, GSE70454, and GSE96736.
Supplementary Material
Key Resources Table.
| Reagent Or Resource | Source | Identifier |
|---|---|---|
| Deposited Data | ||
| Raw Data for Illumina Canine HD SNP genotypes | NCBI Gene Expression Omnibus (GEO) | GEO: GSE90441, GSE83160, GSE70454, GSE96736 |
| Published SNP Genotypes | LUPA | http://dogs.genouest.org/SWEEP.dir/Supplemental.html |
| Published SNP Genotypes | DRYAD | datadryad.org, doi:10.5061/dryad.266k4 |
| Whole Genome Sequences | Short Read Archive (SRA) | BioProject PRJNA263947 |
| Software and Algorithms | ||
| Plink v1.07 | Purcell et al., 2007 | http://pngu.mgh.harvard.edu/∼purcell/plink/ |
| Phylip | Felsenstein, 1989 | http://evolution.genetics.washington.edu/phylip.html |
| Treemix | Pickrell and Pritchard, 2012 | http://pritchardlab.stanford.edu/software.html |
| Beagle | Browning and Browning, 2013 | https://faculty.washington.edu/browning/beagle/beagle.html |
| Other | ||
| Illumina Canine HD SNP arrays | Illumina Corp | Cat# WG-440-1001 |
Acknowledgments
We gratefully acknowledge support from the Intramural Program of the National Human Genome Research Institute. We thank Sir Terence Clark for collecting DNA samples from multiple breeds of sighthounds from their countries of origin in Africa and Asia, Mauricio Lima, Flavio Bruno and Robert Gennari for collecting samples from native Italian breeds, and Lei Song for collecting samples from native Tibetan Mastiffs.
Footnotes
Author Contributions: HGP conceived of project, performed analyses, created figures, prepared manuscript; DLD created figures, assisted in manuscript preparation; MR ran SNP chips, worked on early analysis; BWD and AB performed experiments; GC-R performed sample collection and DNA isolation; EAO organized and directed study, contributed to manuscript preparation
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alderton D. Dogs. New York: Dorling Kindersley Ltd; 2002. [Google Scholar]
- American Kennel Club. The Complete Dog Book, 19th Edition Revised. New York, NY: Howell Book House; 1998. [Google Scholar]
- Baharian S, Barakatt M, Gignoux CR, Shringarpure S, Errington J, Blot WJ, Bustamante CD, Kenny EE, Williams SM, Aldrich MC, et al. The Great Migration and African-American Genomic Diversity. PLoS Genet. 2016;12:e1006059. doi: 10.1371/journal.pgen.1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI, Karam JH, Rutter WJ. Polymorphic DNA region adjacent to the 5′ end of the human insulin gene. Proc Natl Acad Sci U S A. 1981;78:5759–5763. doi: 10.1073/pnas.78.9.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SK, Darwent CM, Wictum EJ, Sacks BN. Using multiple markers to elucidate the ancient, historical and modern relationships among North American Arctic dog breeds. Heredity (Edinb) 2015;115:488–495. doi: 10.1038/hdy.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194:459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, Parker HG, Vonholdt BM, Rhue A, Boyko A, Byers A, et al. Coat variation in the domestic dog is governed by variants in three genes. Science. 2009;326:150–153. doi: 10.1126/science.1177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castroviejo-Fisher S, Skoglund P, Valadez R, Vila C, Leonard JA. Vanishing native American dog lineages. BMC Evol Biol. 2011;11:73. doi: 10.1186/1471-2148-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columella LJM. On Agriculture (De Re Rustica), Vol Books 5-9. Cambridge, MA: Harvard University Press; 70. [Google Scholar]
- Crosby AW., Jr . The Columbian Exchange. Westport, CT, USA: Greenwood Publishing Company; 1972. [Google Scholar]
- Donner J, Kaukonen M, Anderson H, Moller F, Kyostila K, Sankari S, Hytonen M, Giger U, Lohi H. Genetic Panel Screening of Nearly 100 Mutations Reveals New Insights into the Breed Distribution of Risk Variants for Canine Hereditary Disorders. PLoS One. 2016;11:e0161005. doi: 10.1371/journal.pone.0161005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger DL, Davis BW, Cocco R, Sechi S, Di Cerbo A, Parker HG, Polli M, Marelli SP, Crepaldi P, Ostrander EA. Commonalities in Development of Pure Breeds and Population Isolates Revealed in the Genome of the Sardinian Fonni's Dog. Genetics. 2016a;204:737–755. doi: 10.1534/genetics.116.192427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger DL, Rimbault M, Davis BW, Bhatnagar A, Parker HG, Ostrander EA. Whole-genome sequence, SNP chips and pedigree structure: building demographic profiles in domestic dog breeds to optimize genetic-trait mapping. Dis Model Mech. 2016b;9:1445–1460. doi: 10.1242/dmm.027037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhkova AS, Thalmann O, Trifonov VA, Leonard JA, Vorobieva NV, Ovodov ND, Graphodatsky AS, Wayne RK. Ancient DNA analysis affirms the canid from Altai as a primitive dog. PLoS One. 2013;8:e57754. doi: 10.1371/journal.pone.0057754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP -- Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fogle B. The New Encyclopedia of the Dog. second. New York: Dorling Kindersley Publishing, Inc; 2000. [Google Scholar]
- Hayward JJ, Castelhano MG, Oliveira KC, Corey E, Balkman C, Baxter TL, Casal ML, Center SA, Fang M, Garrison SJ, et al. Complex disease and phenotype mapping in the domestic dog. Nature communications. 2016;7:10460. doi: 10.1038/ncomms10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion DN, Schaffer AL, Famula TR, Eggleston ML, Hughes SS, Pedersen NC. Analysis of genetic variation in 28 dog breed populations with 100 microsatellite markers. J Hered. 2003;94:81–87. doi: 10.1093/jhered/esg004. [DOI] [PubMed] [Google Scholar]
- Koskinen MT. Individual assignment using microsatellite DNA reveals unambiguous breed identification in the domestic dog. Anim Genet. 2003;34:297–301. doi: 10.1046/j.1365-2052.2003.01005.x. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance J, Tishkoff SA. SNP ascertainment bias in population genetic analyses: why it is important, and how to correct it. Bioessays. 2013;35:780–786. doi: 10.1002/bies.201300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RB. A history and description of the modern dogs of Great Britain and Ireland. London: Horace Cox; 1894. [Google Scholar]
- Leonard JA, Wayne RK, Wheeler J, Valadez R, Guillen S, Vila C. Ancient DNA evidence for Old World origin of New World dogs. Science. 2002;298:1613–1616. doi: 10.1126/science.1076980. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, 3rd, Zody MC, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lowe JK, Kukekova AV, Kirkness EF, Langlois MC, Aguirre GD, Acland GM, Ostrander EA. Linkage mapping of the primary disease locus for collie eye anomaly. Genomics. 2003;82:86–95. doi: 10.1016/s0888-7543(03)00078-8. [DOI] [PubMed] [Google Scholar]
- Mathias RA, Taub MA, Gignoux CR, Fu W, Musharoff S, O'Connor TD, Vergara C, Torgerson DG, Pino-Yanes M, Shringarpure SS, et al. A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nature communications. 2016;7:12522. doi: 10.1038/ncomms12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealey KL, Meurs KM. Breed distribution of the ABCB1-1Delta (multidrug sensitivity) polymorphism among dogs undergoing ABCB1 genotyping. J Am Vet Med Assoc. 2008;233:921–924. doi: 10.2460/javma.233.6.921. [DOI] [PubMed] [Google Scholar]
- Moody JA, Clark LA, Murphy KE. Canine History and Breed Clubs. In: Ostrander EA, Giger U, Lindblad-Toh K, editors. The Dog and Its Genome. New York, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 1–18. [Google Scholar]
- Pang JF, Kluetsch C, Zou XJ, Zhang AB, Luo LY, Angleby H, Ardalan A, Ekstrom C, Skollermo A, Lundeberg J, et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol. 2009;26:2849–2864. doi: 10.1093/molbev/msp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Harris A, Dreger DL, Davis BW, Ostrander EA. The bald and the beautiful: hairlessness in domestic dog breeds. Philosophical Transactions of the Royal Society B. 2017;372 doi: 10.1098/rstb.2015.0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, Johnson GS, DeFrance HB, Ostrander EA, Kruglyak L. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Parker HG, Kukekova AV, Akey DT, Goldstein O, Kirkness EF, Baysac KC, Mosher DS, Aguirre GD, Acland GM, Ostrander EA. Breed relationships facilitate fine-mapping studies: a 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 2007;17:1562–1571. doi: 10.1101/gr.6772807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon P, Herbin L, Cadieu E, Kirkness EF, Hedan B, Mosher DS, Galibert F, Andre C, Ostrander EA, Hitte C. Canine population structure: assessment and impact of intra-breed stratification on SNP-based association studies. PLoS ONE. 2007;2:e1324. doi: 10.1371/journal.pone.0001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. FigTree 2006 [Google Scholar]
- Ruiz-Linares A, Adhikari K, Acuna-Alonzo V, Quinto-Sanchez M, Jaramillo C, Arias W, Fuentes M, Pizarro M, Everardo P, de Avila F, et al. Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10:e1004572. doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon LM, Boyko RH, Castelhano M, Corey E, Hayward JJ, McLean C, White ME, Abi Said M, Anita BA, Bondjengo NI, et al. Genetic structure in village dogs reveals a Central Asian domestication origin. Proc Natl Acad Sci U S A. 2015;112:13639–13644. doi: 10.1073/pnas.1516215112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Eberle MA, Parker HG, Pullar BJ, Kirkness EF, Kruglyak L, Ostrander EA. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann O, Shapiro B, Cui P, Schuenemann VJ, Sawyer SK, Greenfield DL, Germonpre MB, Sablin MV, Lopez-Giraldez F, Domingo-Roura X, et al. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science. 2013;342:871–874. doi: 10.1126/science.1243650. [DOI] [PubMed] [Google Scholar]
- Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Rosengren Pielberg G, Sigurdsson S, Fall T, Seppala EH, Hansen MS, Lawley CT, et al. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011;7:e1002316. doi: 10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila C, Maldonado JE, Wayne RK. Phylogenetic relationships, evolution, and genetic diversity of the domestic dog. J Hered. 1999;90:71–77. doi: 10.1093/jhered/90.1.71. [DOI] [PubMed] [Google Scholar]
- Vila C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, Honeycutt RL, Crandall KA, Lundeberg J, Wayne RK. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- Vonholdt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GD, Zhai W, Yang HC, Wang L, Zhong L, Liu YH, Fan RX, Yin TT, Zhu CL, Poyarkov AD, et al. Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res. 2016;26:21–33. doi: 10.1038/cr.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. The Dog Book, Vol II. New York, USA: Doubleday; Page & Company; 1906. [Google Scholar]
- Wayne RK, Ostrander EA. Origin, genetic diversity, and genome structure of the domestic dog. Bioessays. 1999;21:247–257. doi: 10.1002/(SICI)1521-1878(199903)21:3<247::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Wilcox B, Walkowicz C. Atlas of Dog Breeds of the World. 5th. Neptune City, NJ: T.F.H. Publications; 1995. [Google Scholar]
- Witt KE, Judd K, Kitchen A, Grier C, Kohler TA, Ortman SG, Kemp BM, Malhi RS. DNA analysis of ancient dogs of the Americas: Identifying possible founding haplotypes and reconstructing population histories. Journal of human evolution. 2014 doi: 10.1016/j.jhevol.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Zhai G, Zhou J, Woods MO, Green JS, Parfrey P, Rahman P, Green RC. Genetic structure of the Newfoundland and Labrador population: founder effects modulate variability. Eur J Hum Genet. 2016;24:1063–1070. doi: 10.1038/ejhg.2015.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data files for the SNP genotype arrays have been deposited in the NCBI Gene Expression Omnibus under accession numbers GEO: GSE90441, GSE83160, GSE70454, and GSE96736.


