Abstract
The induction of autophagy by nutrient deprivation leads to a rapid increase in the formation of autophagosomes, unique organelles that replenish the cellular pool of nutrients by sequestering cytoplasmic material for degradation. The urgent need for membrane to form autophagosomes during starvation, to maintain homeostasis, leads to a dramatic rearrangement of intracellular membranes. Here we discuss recent findings that have begun to uncover how different parts of the secretory pathway directly and indirectly contribute to autophagosome formation during starvation.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a highly conserved bulk degradation process that is rapidly upregulated to maintain homeostasis when cells are starved for nutrients. During autophagy, large double-membrane vesicles called autophagosomes, non-selectively deliver cytoplasmic components to the vacuole or lysosome for degradation (Mizushima et al., 2011). The source of the membrane used to form autophagosomes has remained elusive, and many membranes including the endoplasmic reticulum (ER), Golgi, recycling endosomes, plasma membrane and mitochondra have been implicated (Lamb et al., 2013). Key events in autophagosome formation require the activation and recruitment of both specific autophagy-related (Atg) proteins and membrane trafficking machinery from the secretory pathway. The secretory pathway traffics membrane-bound and secreted proteins through a complex network of intracellular compartments (Palade, 1975). Recent findings have highlighted the early secretory pathway as a key player in autophagosome biogenesis. In this review we will focus on the role of the secretory pathway and its associated trafficking machinery in the early stages of autophagosome formation during starvation.
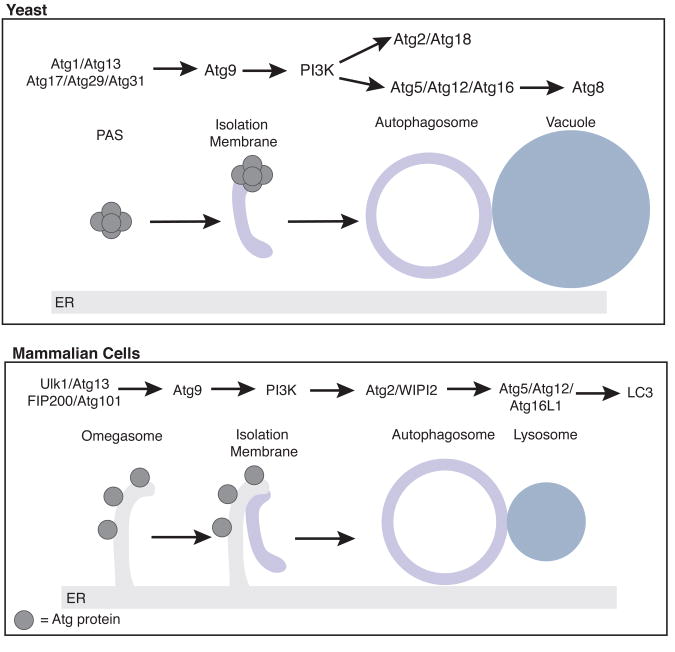
In higher eukaryotes, autophagy plays an important role in the pathophysiology of various diseases and is essential for proper development and immune responses (Hale et al., 2013; Schneider and Cuervo, 2014). There are four major stages of autophagosome formation that have been characterized in both yeast and mammalian cells; initiation, expansion, closure and fusion with the endolysomal system (Lamb et al., 2013) (Figure 1). Upon autophagy induction, autophagosome formation is initiated adjacent to a pre-autophagosomal structure (PAS). The PAS was originally identified in the yeast Saccharomyces cerevisiae as a location where Atg proteins are recruited (Nakatogawa et al., 2009). A double track membrane, called the phagophore, or isolation membrane, extends from the PAS engulfing cytoplasmic materials and organelles. In mammalian cells, there is not a single PAS, rather there are multiple sites of autophagosome formation. Additionally, Atg proteins in mammalian cells are recruited to a domain of the ER termed the omegasome, which cradles the developing phagophore and appears to be functionally similar to the PAS (Axe et al., 2008; Hayashi-Nishino et al., 2009). After sealing, the outer membrane of the fully formed autophagosome fuses with the vacuole or lysosome where hydrolases degrade the delivered cargo. This catabolic process is upregulated during nutrient deprivation, resulting in a dramatic increase in autophagosome formation. .
Figure 1. Overview of autophagosome formation.
The Atg proteins, which form 6 major groups, are recruited in a hierarchical manner to the PAS in yeast (top) or the omegasome in mammals (bottom). A double membrane track called the isolation membrane forms. The isolation membrane expands and then seals to form an autophagosome before it fuses with the vacuole in yeast or lysosome in mammals to release its contents for degradation.
Genetic screens in yeast, reviewed in (Mizushima et al., 2011; Nakatogawa et al., 2009), first identified Atg proteins that are necessary for autophagosome formation. The Atgs can be grouped into six major complexes that are recruited in a hierarchical manner to sites of autophagosome formation (Suzuki et al., 2007). These complexes are well conserved and mammalian homologues have been identified for all key Atg proteins. The most upstream complex is the Atg1/Ulk1 initiating complex, which contains the serine/threonine kinase Atg1/Ulk1. In yeast Atg1 forms a complex with Atg13, Atg17, Atg29 and Atg31 (Kawamata et al., 2008; Ragusa et al., 2012), while mammalian Ulk1 complexes with Atg13, FIP200 (mammalian Atg17) and Atg101, which is absent in budding yeast (Hosokawa et al., 2009; Qi et al., 2015). When autophagy is induced, the initiating complex activates Atg1/Ulk1 kinase activity and recruits downstream Atg complexes, including the multispanning transmembrane protein Atg9, followed by the autophagy-specific class III phosphatidylinositol 3-kinase (PI3K) complex (Rao et al., 2016; Suzuki et al., 2015). The PI3K complex generates phosphatidylinositol-3-phosphate (PI(3)P) at the PAS by phosphorylating phosphatidylinositol (PI) at the 3-position hydroxyl group. PI(3)P is needed for the recruitment of the PI(3)P-binding protein Atg18 (WIPI2 in mammals) and its partner Atg2, which are involved in Atg9 recycling (Obara et al., 2008; Reggiori et al., 2004).
The most downstream Atg complexes are the two ubiquitin-like conjugation systems, Atg12 and Atg8. The Atg12 system results in the formation of the Atg16/Atg12/Atg5 complex, which acts as an E3 ligase for the conjugation of Atg8 (LC3 in mammals) to phosphatidylethanolamine (PE) (Hanada et al., 2007; Noda et al., 2013). The Atg16/Atg5/Atg12 complex localizes to the outer membrane of the autophagosome and is released before vacuole fusion (Kuma et al., 2002). Conversely, Atg8-PE is embedded in both the inner and outer autophagosomal membrane and some Atg8 is delivered with the autophagosome to the vacuole (Kirisako et al., 1999). As a result, Atg8 is a commonly used marker for both the phagophore and mature autophagosome. While there is one Atg8 family member in yeast, mammals contain several homologues that form three subfamilies (LC3, GABARAP and GATE-16; (Slobodkin and Elazar, 2013). Yeast Atg8 has been reported to regulate the later stages of autophagosome formation including expansion and closure by an unknown mechanism (Nakatogawa et al., 2007; Xie et al., 2008). Additionally, while the recruitment of the Atg complexes to sites of autophagosome formation is well defined, how these complexes work with trafficking machinery to coordinate the membrane rearrangements required for autophagosome formation is only beginning to emerge.
Early secretory pathway
The ER appears to play a critical role in the early stages of autophagosome formation. In mammalian cells, Atg proteins are recruited to the ER upon autophagy induction and the growing phagophore or isolation membrane becomes cradled between sheets of ER-associated membrane called the omegasome (Axe et al., 2008; Hayashi-Nishino et al., 2009). The omegasome, named for its Ω-shape, is a PI(3)P enriched region of the ER that was originally identified by localization studies with the PI(3)P binding protein DFCP1. Formation of the omegasome occurs de novo during starvation and is dependent on the PI3K complex (Axe et al., 2008). This PI(3)P enriched subdomain of the ER enwraps the phagophore as it expands, with occasional connections between the two membranes (Hayashi-Nishino et al., 2009). Before full maturation, the autophagosome is released and the omegasome contracts back to the ER. Recently, specific subdomains of the ER, ER Exit Sites (ERES) and ER-mitochondria contact sites, have been implicated as sites of autophagosome biogenesis.
ER Exit Sites
ERES are domains of the ER where COPII vesicles, that transport cargo between the ER and the Golgi, are formed (Budnik and Stephens, 2009). Assembly of the COPII coat is initiated at ERES by the GTPase Sar1 (Figure 2, Table 1). Activated Sar1 couples membrane curvature with the recruitment of the inner COPII coat (Sec23/Sec24), or coat adaptor, to capture cargo into the vesicle. The coat adaptor recruits the outer shell (Sec13/Sec31), which leads to membrane deformation and vesicle budding (Lord et al., 2013; Zanetti et al., 2011). After vesicle budding, Sar1 is released from the vesicle (Barlowe et al., 1994), however, the COPII adaptor and outer shell remain on the vesicle until it docks with its target membrane (Cai et al., 2007; Lord et al., 2011). The coat adaptor then plays an important role in targeting the vesicle to its acceptor membrane (Cai et al., 2007; Lord et al., 2011).
Figure 2. COPII vesicles, tethers and other factors in autophagy.
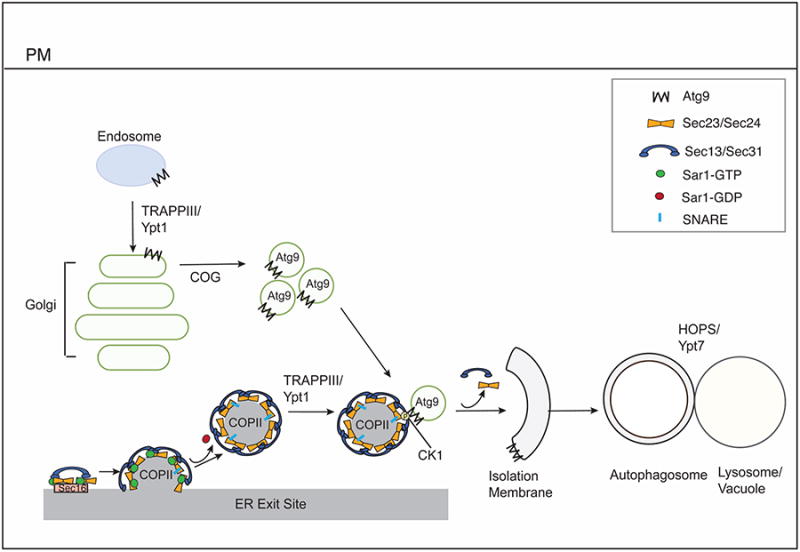
Formation of the COPII coat is initiated at ERES by the GEF Sec12. Sec16 at ERES facilitates coat assembly. Sec12 activates Sar1 and recruits the Sec23/Sec24 complex, which is followed by recruitment of the Sec13/Sec31 complex. After the coat has formed, Sar1 is released from the vesicle, and the coat remains on the vesicle to aid in intracellular targeting. CK1 phosphorylates Sec24, which enhances its interaction with Atg9. Atg9 vesicles are found at the growing edge of the isolation membrane that faces the ERES.
Constitutive Atg9 trafficking requires the TRAPPIII complex, for transport from the early endosome to Golgi in yeast or endocytic to Golgi traffic in mammalian cells. Intra Golgi transport requires COG, which is needed for the trafficking of Atg9 vesicles to sites of autophagosome formation. TRAPPIII also binds the COPII coat and activates the GTPase Rab1, that participates in autophagosome formation. HOPS and Rab7 act after autophagosome formation in autophagosome-lysosome fusion.
Table 1. Secretory factors required for macroautophagy.
Abbreviations: ERES, ER exit sites; PAS, pre-autophagosomal structure; ERGIC, ER-Golgi intermediate compartment.
| Protein or complex | Role in Secretory Pathway | Role in Macroautophagy |
|---|---|---|
| ERES and COPII vesicle-related proteins | ||
| Sar1 | Initiates COPII vesicle budding | Regulates autophagosome formation |
| Sec16 | Scaffold required for COPII vesicle formation | Regulates autophagosome formation |
| Sec24 | COPII coat protein | Regulates autophagosome abundance |
| Sec23 | COPII coat protein | Defect in autophagy if the Sec23-TRAPPIII interaction is disrupted |
| TRAPP | Ypt1/Rab1 GEFs in early secretory pathway (TRAPPI and TRAPPII) | TRAPPIII interacts with Atg17 and Sec23, recruits and activates Ypt1/RAB1 in autophagy |
| TECPR2 | Stabilizes COPII coat protein SEC24D and maintains functional ERES | Regulates autophagosome formation |
| Hrr25 | COPII coat phoshorylation | Regulates autophagosome abundance |
| SNAREs | ||
| Sso1/Sso 2 | Mediates fusion of secretory vesicles at plasma membrane | Regulates Atg9 traffic to the PAS |
| Sec9 | Mediates fusion of secretory vesicles at plasma membrane | Regulates Atg9 traffic to the PAS |
| Tlg2 | Mediates fusion of endosome-derived vesicles with late Golgi | Regulates Atg9 traffic to the PAS |
| Sec 22 | Mediates ER-Golgi traffic | May regulate Atg9 traffic to the PAS |
| Ykt6 | Mediates Golgi traffic and vacuole fusion | May regulate Atg9 traffic to the PAS |
| Ufe1 | Mediates fusion of Golgi-derived vesicles at ER | Regulates autophagosome formation |
| STX17 | Maintains the architecture of ERGIC and Golgi | Recruits autophagy machinery to ER-mitochondria contact site and regulates autophagosome-lysosome fusion |
| SNX18 | Modulates endocytic traffic | Recruits ATG16L1 and RAB11 to recycling endosomes |
| VAMP3 | Mediates transport from endosome to the trans-Golgi network | Regulates heterotypic fusion of mATG9- and ATG16L1-containing vesicles |
| VAMP7 | Mediates fusion of endosomes with lysosomes | Regulates homotypic fusion of ATG16L1-positive vesicles |
| Tethers | ||
| COG | Regulates intra-Golgi traffic via COPI coated vesicles | Regulates anterograde trafficking of Atg9 to the PAS and autophagosome formation |
| Exocyst | Regulates the tethering of post-Golgi secretory vesicles to the plasma membrane | Role in autophagy is controversial |
| HOPS | Regulates tethering and fusion events at the vacuole/lysosome | Regulates autophagosome fusion with the vacuole/lysosome |
| CORVET | Regulates early to late endosome maturation | CORVET-specific subunits required for autophagy |
Proteomics has recently linked the ERES to the autophagy machinery (Graef et al., 2013), and Atg proteins were found to assemble adjacent to the ERES (Graef et al., 2013; Suzuki et al., 2013). In particular, (Graef et al., 2013) have found that the growing edge of the phagophore, where Atg9 and Atg2 localize, was shown to face the ERES (Suzuki et al., 2013). In mammalian cells, approximately 54% of the LC3 puncta colocalize with the ERES protein Sec16 (Graef et al., 2013), a scaffold protein that interacts with multiple COPII coat subunits to regulate coat assembly (Budnik and Stephens, 2009). Similar observations were also made by others (Karanasios et al., 2016). Additionally, loss of function temperature-sensitive mutations in genes that encode secretory machinery such as SEC16 or SEC12, the guanine nucleotide exchange factor (GEF) for Sar1 (Barlowe and Schekman, 1993), inhibit autophagosome formation (Graef et al., 2013; Ishihara et al., 2001). Disruption of ERES by knockdown of Sar1 or TECPR2, a protein required for the stability of SEC24D in mammalian cells, also inhibits autophagosome formation at an early stage (Stadel et al., 2015; Zoppino et al., 2010). ERES have been proposed to function in autophagy through the production of COPII vesicles, which may directly provide membrane for the growing phagophore. Alternatively, ERES may act as a platform on which Atg proteins and the phagophore assemble (Sanchez-Wandelmer et al., 2015). Although these models are not entirely mutually exclusive, a growing body of evidence discussed above and below favors the former model.
COPII vesicles
Almost 15 years ago, mutants that block the early secretory pathway were also shown to block autophagy (Ishihara et al., 2001). However, it was argued that these mutants were inhibiting autophagy indirectly by disrupting the trafficking of Atg machinery to the PAS. Recent work has begun to separate out the function of COPII vesicles in autophagy from their role in secretion, demonstrating that COPII vesicles directly act in autophagosome formation.
Supporting a direct role for COPII vesicles in autophagy, COPII coated structures were found to accumulate at the PAS when autophagy was blocked, and a coat subunit (Sec23) was shown to bind directly to machinery (TRAPPIII, described below) that acts in autophagy (Tan et al., 2013). Interestingly, although mutations in the COPII vesicle trafficking machinery broadly inhibit autophagy, a notable exception is the tether Uso1, which links COPII vesicles to the Golgi (Tan et al., 2013). Thus, current findings indicate that ER-Golgi traffic is not directly relevant to autophagy, however, certain components of the ER-Golgi trafficking machinery are needed. ER-Golgi SNAREs, type II membrane proteins required for COPII vesicle fusion (Jahn and Scheller, 2006), are also required for autophagy (Nair et al., 2011; Tan et al., 2013), as is the ER localized SNARE, Ufe1. A recent study suggested that Ufe1 may traffic to sites of autophagosome formation in a special class of COPII vesicles (Lemus et al., 2016).
While the findings described above are consistent with a role for COPII vesicles in autophagosome formation, how the vesicle recognizes the autophagy machinery has been unclear. In line with previous findings showing that the COPII coat plays an active role in targeting the vesicle to its final intracellular destination (Cai et al., 2007; Lord et al., 2011), yeast Sec24 was recently shown to have a central role in engaging the vesicle with the Atg machinery during nutrient deprivation (Davis et al., 2016). In particular, phosphorylation of a cluster of amino acids on the membrane distal surface of Sec24 was found to commit COPII vesicles to autophagy. Mutating these phosphorylation sites to alanine specifically inhibited autophagy, but not ER-Golgi transport. Failure to phosphorylate the Sec24 membrane distal surface disrupted the interaction of Sec24 with Atg9, which was shown to be needed for autophagy (Davis et al., 2016). Casein kinase 1, Hrr25, is a key kinase that phosphorylates this Sec24 surface, and both Sec24 phosphorylation and Hrr25 modulate autophagosome abundance during starvation. These phosphorylation events were found to be independent of the assembly of the Atg hierarchy, indicating that Sec24 phosphorylation does not indirectly disrupt autophagy by blocking the delivery of the Atg machinery to sites of autophagosome formation (Davis et al., 2016). These findings show that phosphorylation regulates the membrane rearrangements that take place during starvation induced autophagy and the COPII coat is a target of this regulation. As the Sec24 phosphosites are partially conserved in some isoforms and the role of the CK1 kinase Hrr25 (CK1δ in mammals) is highly conserved, these events are likely to occur in mammals. Additional experiments are needed to confirm this.
Membrane fractionation also identified the mammalian ER-Golgi intermediate compartment (ERGIC) as required for LC3 lipidation in an in vitro reaction (Ge et al., 2013). The ERGIC is a site of both COPII vesicle fusion and COPI vesicle budding. COPI vesicles mediate multiple trafficking events, including retrograde traffic from the ERGIC to the ER (Lorente-Rodriguez and Barlowe, 2011). While the relationship of the omegasome to the ERGIC is unclear, the ERES were found to move to the ERGIC fraction when autophagy was induced. Interestingly, it was shown that budding of COPII, but not COPI vesicles, from this ERGIC fraction is required for LC3 lipidation (Ge et al., 2014). COPII vesicles were also found to relocalize to the ERGIC during starvation in a PI3K-dependent manner (Ge et al., 2014). The collective findings discussed above suggest that only those translocated COPII vesicles with phosphorylated Sec24 engage the autophagy machinery and contribute to autophagosome formation (Figure 2).
ER-mitochondria Contact Sites
Non-vesicular lipid transfer takes place at ER-mitochondria contact sites (Phillips and Voeltz, 2014), which have also been proposed to be sites of autophagosome biogenesis. Key autophagy machinery such as the PI3K complex component Atg14, and Atg5 localize to ER-mitochondria contact sites during starvation in a Stx17 dependent manner (Hamasaki et al., 2013). Stx17 is a SNARE required late in autophagy for autophagosome-lysosome fusion (Diao et al., 2015). Disruption of ER-mitochondria contact sites by knockdown of PACS2, or the ER-mitochondria tether mitofusion-2, disrupts autophagosome formation at an early stage. However, live cell imaging revealed that the ER is the dominant platform for autophagosome biogenesis, with the mitochondria interacting more dynamically (Hamasaki et al., 2013). Interestingly, a significant fraction of the COPII coat subunit Sec31 was also reported to colocalize with ER-mitochondria markers during starvation, raising the possibility that COPII vesicles are a source of membrane for autophagosomes at the ER-mitochondria contact sites (Tan et al., 2013).
As the basic mechanism of autophagy is well conserved from yeast to man, an essential role for mitochondria in the biogenesis of the autophagosome should be conserved. In yeast, however, a role for ER-mitochondria contact sites in autophagy has not been substantiated. ERMES, a critical tether for maintaining ER-mitochondria contact sites, is required for selective autophagy of the mitochondria (mitophagy), but not general autophagy (Bockler and Westermann, 2014). Additionally, mitochondria were not found to colocalize significantly with the PAS, phagophores or autophagosomes (Graef et al., 2013).
Golgi/Endosome/Plasma Membrane
The Golgi, endosome and plasma membrane have all been implicated in autophagosome formation. Recent findings suggest the contribution of each of these organelles is largely through the regulation of Atg9 and Atg16 trafficking. In this section we will discuss the role that each of these organelles plays in the constitutive trafficking of Atg9 and Atg16 as well as the delivery of these Atg proteins to sites of autophagosome formation.
Atg9
As mentioned above, the multispanning Atg9 transmembrane protein is necessary for the recruitment of downstream Atg proteins, including the Atg2/Atg18 (WIPI2) complex and the PI3K complex (Suzuki et al., 2007; Suzuki et al., 2015). In yeast, Atg9 is incorporated into the autophagosomal membrane, but is recycled before the autophagosome fuses with the vacuole (Yamamoto et al., 2012). Although mammalian Atg9 (mATG9) is required for formation of the omegasome (Orsi et al., 2012), it does not appear to incorporate into the autophagosomal membrane, rather it localizes to tubular-vesicular clusters adjacent to sites of autophagosome formation (Orsi et al., 2012). More recent studies suggest that while mATG9 has a complex trafficking route, it associates with ATG16L1 in RAB11 positive recycling endosomes, which may function as an autophagosome precursor (Puri et al., 2013).
Yeast Atg9 traffics through the early secretory pathway before it localizes to late-Golgi/endosomal compartments (Ohashi and Munro, 2010). Atg9 also traffics from the early endosome to the late Golgi to provide Atg9 reservoirs for autophagosome formation (Figure 3, left). Atg9 trafficking from the early endosome to the Golgi (Shirahama-Noda et al., 2013) is mediated by the sorting nexin Atg24 and the TRAPPIII complex that activates the Rab GTPase Ypt1 (Lynch-Day et al., 2010). During starvation, late endosome-Golgi bypass trafficking pathways may also add to the reservoirs to ensure sufficient Atg9 is transported to sites of autophagosome formation during periods of stress (Ohashi and Munro, 2010; Shirahama-Noda et al., 2013) (Figure 3, left).
Figure 3. The Golgi and endosomal system in autophagy.
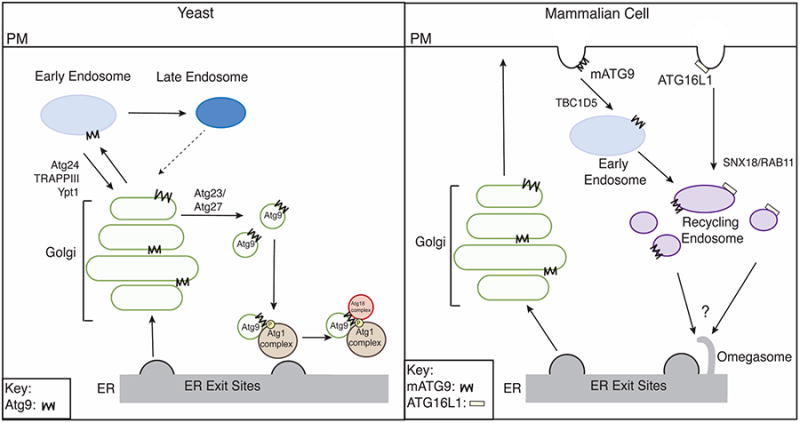
Left, in yeast, Atg9 traffics through the secretory pathway before it localizes to the early endosome. During starvation, TRAPPII traffics Atg9 from the early endosome to the late Golgi where Atg23 and Atg27 mediate the biogenesis of Atg9 vesicles that translocate to the PAS. Late endosome trafficking bypass pathways may also exist during starvation (dotted arrow). Right, at the plasma membrane (PM) in mammals, mATG9 and ATG16L1 are packaged into separate clathrin-coated vesicles. The interaction of mATG9 with RAPGAP TBC1D5 and AP2 facilitates its transport from the PM to the early endosome before it traffics to the recycling endosome, where it meets ATG16L1. ATG16L1 is recruited to the recycling endosome by SNX18/RAB11. The perinuclear recycling endosome is tubulated during starvation and ATG16L1 and mATG9 are subsequently recruited to the omegasome.
Upon autophagy induction, Atg9 vesicles form at the late-Golgi (Figure 3, left) in an Atg23 and Atg27 dependent manner (Yamamoto et al., 2012). Atg23 is a peripheral membrane protein and Atg27 is a single-pass transmembrane protein (Tucker et al., 2003; Yen and Klionsky, 2007). Atg23 and Atg27 form a complex with Atg9 (Legakis et al., 2007) and regulate the amount of Atg9 that is incorporated into vesicles (Backues et al., 2015). Atg9 vesicles are recruited to the PAS through the Atg1 initiating complex. Specifically, the N-terminal cytoplasmic domain of Atg9 was shown to interact with the HORMA domain of Atg13, which is required for Atg9 PAS recruitment (Suzuki et al., 2015). Additionally, the Atg9 core was reported to interact with Atg17 (Rao et al., 2016). The Atg9 binding region of Atg17 is inhibited by Atg29-Atg31, but incorporation of Atg1-Atg13 into the complex relieves this inhibition, allowing Atg17 to interact with Atg9 (Rao et al., 2016). At the PAS, Atg1 phosphorylates the C-terminus of Atg9, which facilitates the recruitment of Atg18 (Papinski et al., 2014). Atg18 and its binding partner Atg2 regulate Atg9 recycling from the autophagosome through an unknown mechanism (Reggiori et al., 2004).
Multiple SNAREs have been implicated in Atg9 trafficking and autophagy (Table 1). The exocytic SNARES Sso1/Sso2 and Sec9 as well as the late Golgi SNARE Tlg2 have been proposed to regulate Atg9 trafficking to the PAS (Nair et al., 2011). Tlg2, which is required for endosome to Golgi traffic, only displays a severe autophagy defect when combined with other endosome-Golgi mutants, consistent with the observation that Atg9 uses multiple trafficking pathways during nutrient deprivation (Ohashi and Munro, 2010). Interestingly, the functionally redundant exocytic SNAREs Snc1/2 are not required for autophagy (Nair et al., 2011). Sec22 and Ykt6 have also been implicated in autophagy (Nair et al., 2011), potentially through regulating Atg9 traffic, although the precise role of these SNARES in autophagy is unclear. Currently, it is unknown which SNAREs are present on Atg9 vesicles and which are required to traffic Atg9 to the PAS. In the future it will be important to address this point.
mATG9 follows a similar, albeit slightly more complex, trafficking network (Figure 3, right) as it has been reported to localize to the trans-Golgi network (TGN) as well as early, late and recycling endosomes (Young et al., 2006). When autophagy is induced, mATG9 disperses from the TGN to peripheral endosome compartments (Young et al., 2006). mATG9 is thought to traffic to the plasma membrane where it is endocytosed in AP2 vesicles and then transported through the early endosome to the recycling endosome (Popovic and Dikic, 2014; Puri et al., 2013). The RabGAP TBC1D5 interacts with mATG9 and AP2 during starvation to facilitate mATG9 traffic from the plasma membrane (Popovic and Dikic, 2014). mATG9 has been reported to form tubular-vesicular clusters from perinuclear RAB11-positive recycling endosomes. Tubulation of RAB11 recycling endosomes is induced upon nutrient deprivation and is dependent on the curvature inducing protein Bif-1 and dynamin 2 (Takahashi et al., 2016). mATG9 tubular-vesicular clusters are ultimately recruited adjacent to sites of autophagosome formation (Orsi et al., 2012), and this recruitment is independent of Ulk1 (Orsi et al., 2012) (Figure 3, right). Like yeast Atg9, mATG9 colocalizes with ATG13 adjacent to ER subdomains (Karanasios et al., 2016). Although yeast and mammalian ATG9 have somewhat different trafficking routes, this conserved protein is likely to play the same role in autophagy in both species.
Atg16
Atg16 (ATG16L1 in mammals) relies on trafficking machinery for its proper localization and function (Figure 3, right). Atg16 is a peripheral membrane protein that forms a complex with Atg5-Atg12 (Romanov et al., 2012). Together Atg5-Atg12-Atg16 act as an E3-like enzyme catalyzing the conjugation of Atg8/LC3 to PE. Thus, Atg16 has been proposed to be the key determinant for the location of Atg8/LC3 lipidation (Fujita et al., 2008). In mammalian cells, ATG16L1 associates with clathrin coated pits and is endocytosed from the plasma membrane into distinct vesicles from mATG9 (Puri et al., 2013; Ravikumar et al., 2010). After endocytosis, ATG16L1 containing vesicles have been reported to undergo homotypic fusion, which is dependent on the SNARE VAMP7 (Moreau et al., 2011). ATG16L1 then coalesces with mATG9 in RAB11-positive recycling endosomes. This fusion event is directed by another SNARE, VAMP3 (Puri et al., 2013). Recruitment of ATG16L1 to recycling endosomes requires SNX18, a sorting nexin that binds PtdIns(4,5)P2 (Knaevelsrud et al., 2013). During starvation, SNX18 recruits RAB11 to perinuclear recycling endosomes where together they recruit ATG16L1 (Knaevelsrud et al., 2013) (Figure 3, right). It is unknown if Atg16 is trafficked by a similar mechanism in lower eukaryotes. Additionally, the precise relationship between the recycling endosome and the omegasome is unclear, as additional studies have suggested the recycling endosome may act as a site of autophagosome formation (Puri et al., 2013). Further work is needed to establish the exact role of the recycling endosome in autophagy.
During starvation, ATG16L1 is recruited to the site of autophagosome formation. Atg16 recruitment to the PAS in yeast is dependent on its complex partner Atg5 and PI(3)P (Suzuki et al., 2007). PI(3)P helps recruit Atg16 in part through Atg21, which interacts with both PI(3)P and Atg16. Additional factors are also likely to be needed (Juris et al., 2015). In mammalian cells, after trafficking to the recycling endosome, ATG16L1 is recruited to the omegasome by the PI(3)P binding protein WIPI2b (Dooley et al., 2014). ATG16L1 also interacts with FIP200 (Nishimura et al., 2013), but this interaction does not appear to be essential for autophagy and it cannot bypass the dependence of ATG16L1 membrane recruitment on PI(3)P (Dooley et al., 2014).
Tethers and Rabs
Tethers are essential membrane trafficking machinery that physically connect vesicles with their target membrane. They also facilitate the transition from vesicle tethering to fusion by binding to SNAREs and promoting SNARE complex assembly (Hong and Lev, 2014). Tethers are typically Rab effectors and can be divided into two main subgroups; multisubunit complexes and long coiled-coil tethers. Long coiled-coil tethers include p115 (Uso1 in yeast) and the Golgins (Whyte and Munro, 2002). The multisubunit complexes include COG (conserved oligomeric Golgi), the exocyst and HOPS (homotypic fusion and vacuole sorting protein), which will be discussed in this section. We will also discuss the TRAPP (transport particle protein) complexes here. While the TRAPP complexes do not directly tether membranes, they participate in membrane tethering events via Ypt1 effectors that act after TRAPP activates the Rab (Lord et al., 2011). Some tethers and Rabs function directly in autophagosome formation and autophagosome-lysosome fusion, while others act indirectly by regulating the trafficking of Atg9 (Figure 2, Table 1).
TRAPP
Rabs are molecular switches that regulate membrane traffic. They are inactive in their GDP-bound state and active when bound to GTP (Mizuno-Yamasaki et al., 2012). The Rab GTPase Ypt1 is activated by the guanine nucleotide exchange factor (GEF) called TRAPP. This GEF was originally identified in yeast where it forms 3 distinct complexes; TRAPPI, TRAPPII and TRAPPIII. TRAPPI is required for ER-Golgi transport, TRAPPII is needed for Golgi traffic, and TRAPPIII functions in autophagy (Barrowman et al., 2010). These three complexes share a core set of 6 subunits. In addition to the core subunits, TRAPPII and TRAPPIII contain adaptor subunits that target each of these complexes to different parts of the cell. Recent work has shown that TRAPPIII, which contains the unique subunit Trs85, is required for autophagy (Lynch-Day et al., 2010). The single-particle EM structure of TRAPPIII revealed that Trs85 caps one end of the elongated core TRAPPI structure (Cai et al., 2008; Tan et al., 2013). The Bet3 subunit, present in all three complexes, interacts with the COPII coat subunit Sec23 (Cai et al., 2007). The structure of TRAPPIII revealed that the addition of Trs85 to the TRAPPI core does not obscure the Sec23 binding site on Bet3, and in vitro binding studies confirmed that TRAPPIII binds to Sec23 (Tan et al., 2013). Therefore, similar to TRAPPI, TRAPPIII likely functions in part by activating Ypt1 on COPII vesicles. In addition to localizing to COPII vesicles, TRAPPIII is recruited to the PAS by Atg17, an event that is needed for the PAS recruitment of Ypt1 (Lynch-Day et al., 2010; Wang et al., 2013). At the PAS Ypt1 recruits effectors that include Atg1 and the CK1 kinase Hrr25 (Wang et al., 2015; Wang et al., 2013).
In addition to recruiting Ypt1 to COPII vesicles and the PAS, TRAPPIII resides on the late Golgi and facilitates endosomal to Golgi transport and Atg9 trafficking (Shirahama-Noda et al., 2013). Supporting a role for TRAPPIII in Atg9 traffic (Figure 3A, left), Trs85 was found on Atg9 vesicles and partially colocalizes with Atg9 (Kakuta et al., 2012; Lynch-Day et al., 2010). Although TRAPPIII is essential for Atg9 trafficking during nutrient rich conditions, late endosome to Golgi trafficking bypass pathways exist during starvation (Figure 3, left) that mask this requirement (Shirahama-Noda et al., 2013). Atg9 was also reported to recruit Trs85 to the PAS (Kakuta et al., 2012), however, others have found no effect on Trs85 recruitment in atg9Δ cells (Lynch-Day et al., 2010; Wang et al., 2013). Currently, it is unclear how TRAPPIII is recruited to the late-Golgi/early endosome to regulate early endosome traffic. Interactions with different vesicle coat complexes (COPII and COPI) have been shown to recruit TRAPPI and TRAPPII to the ER-Golgi (COPII) and Golgi (COPI) trafficking pathways respectively (Cai et al., 2007; Yamasaki et al., 2009). Whether TRAPPIII is engaged by a similar mechanism is unknown. Thus, in yeast, the many roles of TRAPPIII in autophagy can be attributed to its role in activating Ypt1 at multiple sites: COPII vesicles, the PAS and probably the late Golgi where it regulates Atg9 traffic. The activated Rab then recruits Ypt1 effectors (Hrr25 and Atg1) to each of these sites.
In mammalian cells, two forms of TRAPP have been identified: a TRAPPIII-like and TRAPPII-like complex (Bassik et al., 2013; Wang et al., 2013; Yamasaki et al., 2009). The mammalian TRAPPIII-like complex regulates both secretion and autophagy and contains TRAPPC8, the mammalian homologue of Trs85. Knockdown of TRAPPC8 disrupts Golgi integrity and general secretion (Lamb et al., 2016; Scrivens et al., 2011). Additionally, during starvation, knockdown of TRAPPC8 reduces the number of omegasomes, suggesting it is required for autophagy at an early stage (Lamb et al., 2016). Similarly, RAB1a is required for autophagosome formation and in particular omegasome formation (Huang et al., 2011; Zoppino et al., 2010). TRAPPC8 was recently found to directly interact with the RAP GAP TBC1D14. TBC1D14 overexpression was previously shown to inhibit autophagy by inducing the formation of an enlarged, Ulk1-positive tubulated recycling endosome (Longatti et al., 2012). TBC1D14 and TRAPPC8 work together to regulate constitutive mAtg9 trafficking from RAB11-positive recycling endosomes to a RAB1-positive Golgi compartment (Lamb et al., 2016). While TRAPPC8 is critical for constitutive mATG9 trafficking to the Golgi, it did not affect the co-localization of mATG9 with LC3 during starvation (Lamb et al., 2016). Although the roles of the mammalian TRAPP complexes are still being elucidated, thus far the general function of TRAPPIII in autophagosome formation and constitutive Atg9 trafficking appears to be largely conserved.
COG
The conserved oligomeric Golgi complex (COG), which regulates intra-Golgi traffic via COPI coated vesicles (Willett et al., 2013), has been implicated in autophagy in yeast via its ability to regulate Atg9 traffic. COG is comprised of eight subunits, divided into two lobes, lobe A and lobe B. Disruptions in the essential lobe A COG subunits inhibit autophagosome formation (Yen et al., 2010). In particular, COG was found to affect the anterograde trafficking of Atg9 to the PAS. This is consistent with proper Golgi transport needed for Atg9 PAS delivery. COG was also suggested to play additional roles in tethering membranes at the PAS (Yen et al., 2010), although future work will be required to validate this model. The trafficking function of COG is well conserved in mammalian cells (Willett et al., 2013), but a role for COG in mammalian autophagy has not yet been identified.
Exocyst
The exocyst is an octameric complex that regulates the tethering of post-Golgi secretory vesicles to the plasma membrane (Munson and Novick, 2006). In mammalian cells two exocyst subunits, Exo84 and Sec5, are effectors of the Ras-like GTPase RalB (Moskalenko et al., 2002; Moskalenko et al., 2003). RalB was shown to be required for autophagosome formation and its activity was found to be enhanced during starvation. It was hypothesized that RalB functions as a switch during autophagy to regulate the assembly of an Atg initiation complex, which employs an exocyst subcomplex as a platform. The role of the exocyst in autophagy, however, has recently been called into question. Findings in Drosophila show that the Ral GTPase and the exocyst are not needed for starvation-induced autophagy and only act during development in an autophagy pathway associated with cell death (Tracy et al., 2016). In yeast, there have been conflicting reports as to whether exocyst subunits are required for autophagy (Geng et al., 2010; Ishihara et al., 2001; Nair et al., 2011). The GTPase Sec4 and its GEF Sec2, which associate with the exocyst, have been shown to contribute to autophagy by regulating Atg9 trafficking to the PAS.
HOPS
HOPS works with the Rab GTPase Ypt7/Rab7 to regulate multiple tethering and fusion events at the vacuole/lysosome. During autophagy, HOPS and Ypt7/Rab7 are required for autophagosome fusion with the vacuole/lysosome. In yeast, deletion of HOPS subunits and Ypt7 results in the accumulation of autophagosomes (Kirisako et al., 1999; Rieder and Emr, 1997). However, a clear role for HOPS in autophagosome fusion in higher eukaryotes has only recently emerged. Knockdown of multiple HOPS subunits leads to the accumulation of autophagosomes in both mammalian cells and Drosophila (Jiang et al., 2014; Takats et al., 2014). Additionally, HOPS subunits were shown to interact with the autophagosomal SNARE, Stx17, which helps recruit HOPS to the autophagosome (Jiang et al., 2014; Takats et al., 2014). A role for the HOPS-Stx17 interaction in autophagy has subsequently been confirmed in a mouse model (Zhen and Li, 2015). HOPS has also been proposed to be linked to autophagosomes in mammalian cells by another Rab7 effector, Pleckstrin homology domain containing protein family member 1 (PLEKHM1). PLEKHM1 is thought to connect LC3 on autophagosomes with HOPS by binding the C-terminus of the VPS34 subunit (McEwan et al., 2015).
HOPS shares a core set of subunits with another tethering complex called CORVET, that regulates early to late endosome maturation (Balderhaar and Ungermann, 2013). HOPS and CORVET also contain additional unique subunits that interact with different Rabs, Rab7 (HOPS) and Rab5 (CORVET) (Balderhaar and Ungermann, 2013). Subunits only present in CORVET have been implicated in autophagy in yeast (Chen et al., 2014).
Concluding Remarks
Recent studies have highlighted the importance of the secretory pathway in the transport of Atg proteins to sites of autophagosome formation during starvation. Additionally, the early secretory pathway has been shown to play a direct and critical role in contributing membrane to autophagosome formation. The COPII coat, in particular, facilitates the delivery of membrane and SNAREs to sites of autophagosome formation via its ability to recognize the Atg machinery. While it is not clear if the late secretory pathway directly contributes membrane to autophagosome formation, key autophagy proteins, such as Atg9 and Atg16, use the secretory and endocytic pathways during nutrient deprivation. These Atgs have tightly coordinated trafficking itineraries that regulate their delivery to sites of autophagosome formation.
Although significant progress has been made in understanding the initiation of autophagosome formation, the precise mechanism by which the autophagosomal membrane expands and closes has remained elusive. Additional studies will be needed to determine if the secretory pathway also contributes to this event. While this review has focused on autophagosome formation during starvation, recent work has highlighted the importance of selective autophagy pathways that also use autophagosomes (Mizushima et al., 2011). These pathways target a particular cargo, often a damaged organelle or protein aggregate, for degradation. Future work will be required to determine if the secretory pathway also plays a role in forming autophagosomes on these less characterized pathways.
Acknowledgments
Salary and stipend support for S. F-N. and S.D. was provided by NIGMS under award numbers R01GM114111 and R01GM115422 to S.F-N. The BMS graduate program at UCSD also supported S.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backues SK, Orban DP, Bernard A, Singh K, Cao Y, Klionsky DJ. Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae. Traffic. 2015;16:172–190. doi: 10.1111/tra.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Schekman R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat Rev Mol Cell Biol. 2010;11:759–763. doi: 10.1038/nrm2999. [DOI] [PubMed] [Google Scholar]
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockler S, Westermann B. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev Cell. 2014;28:450–458. doi: 10.1016/j.devcel.2014.01.012. [DOI] [PubMed] [Google Scholar]
- Budnik A, Stephens DJ. ER exit sites--localization and control of COPII vesicle formation. FEBS Lett. 2009;583:3796–3803. doi: 10.1016/j.febslet.2009.10.038. [DOI] [PubMed] [Google Scholar]
- Cai H, Yu S, Menon S, Cai Y, Lazarova D, Fu C, Reinisch K, Hay JC, Ferro-Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- Cai Y, Chin HF, Lazarova D, Menon S, Fu C, Cai H, Sclafani A, Rodgers DW, De La Cruz EM, Ferro-Novick S, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhou F, Zou S, Yu S, Li S, Li D, Song J, Li H, He Z, Hu B, et al. A Vps21 endocytic module regulates autophagy. Mol Biol Cell. 2014;25:3166–3177. doi: 10.1091/mbc.E14-04-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Wang J, Zhu M, Stahmer K, Lakshminarayan R, Ghassemian M, Jiang Y, Miller EA, Ferro-Novick S. Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. Elife. 2016;5 doi: 10.7554/eLife.21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife. 2014;3:e04135. doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale AN, Ledbetter DJ, Gawriluk TR, Rucker EB., 3rd Autophagy: regulation and role in development. Autophagy. 2013;9:951–972. doi: 10.4161/auto.24273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014;24:35–43. doi: 10.1016/j.tcb.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Birmingham CL, Shahnazari S, Shiu J, Zheng YT, Smith AC, Campellone KG, Heo WD, Gruenheid S, Meyer T, et al. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy. 2011;7:17–26. doi: 10.4161/auto.7.1.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juris L, Montino M, Rube P, Schlotterhose P, Thumm M, Krick R. PI3P binding by Atg21 organises Atg8 lipidation. EMBO J. 2015;34:955–973. doi: 10.15252/embj.201488957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta S, Yamamoto H, Negishi L, Kondo-Kakuta C, Hayashi N, Ohsumi Y. Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J Biol Chem. 2012;287:44261–44269. doi: 10.1074/jbc.M112.411454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, Ahmed Q, Domart MC, Collinson L, Ktistakis NT. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016;7:12420. doi: 10.1038/ncomms12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaevelsrud H, Soreng K, Raiborg C, Haberg K, Rasmuson F, Brech A, Liestol K, Rusten TE, Stenmark H, Neufeld TP, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol. 2013;202:331–349. doi: 10.1083/jcb.201205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- Lamb CA, Nuhlen S, Judith D, Frith D, Snijders AP, Behrends C, Tooze SA. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 2016;35:281–301. doi: 10.15252/embj.201592695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- Legakis JE, Yen WL, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- Lemus L, Ribas JL, Sikorska N, Goder V. An ER-Localized SNARE Protein Is Exported in Specific COPII Vesicles for Autophagosome Biogenesis. Cell Rep. 2016;14:1710–1722. doi: 10.1016/j.celrep.2016.01.047. [DOI] [PubMed] [Google Scholar]
- Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Bhandari D, Menon S, Ghassemian M, Nycz D, Hay J, Ghosh P, Ferro-Novick S. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 2011;473:181–186. doi: 10.1038/nature09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Ferro-Novick S, Miller EA. The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the golgi. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a013367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente-Rodriguez A, Barlowe C. Entry and exit mechanisms at the cisface of the Golgi complex. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015;57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–659. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, Camonis J, White MA. Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Kaizuka T, Cadwell K, Sahani MH, Saitoh T, Akira S, Virgin HW, Mizushima N. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep. 2013;14:284–291. doi: 10.1038/embor.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 2013;14:206–211. doi: 10.1038/embor.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:867. doi: 10.1126/science.189.4206.867-b. [DOI] [PubMed] [Google Scholar]
- Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2014;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D, Dikic I. TBC1D5 and the AP2 complex regulate ATG9 trafficking and initiation of autophagy. EMBO Rep. 2014;15:392–401. doi: 10.1002/embr.201337995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Kim do J, Stjepanovic G, Hurley JH. Structure of the Human Atg13-Atg101 HORMA Heterodimer: an Interaction Hub within the ULK1 Complex. Structure. 2015;23:1848–1857. doi: 10.1016/j.str.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Perna MG, Hofmann B, Beier V, Wollert T. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat Commun. 2016;7:10338. doi: 10.1038/ncomms10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Wandelmer J, Ktistakis NT, Reggiori F. ERES: sites for autophagosome biogenesis and maturation? J Cell Sci. 2015;128:185–192. doi: 10.1242/jcs.158758. [DOI] [PubMed] [Google Scholar]
- Schneider JL, Cuervo AM. Autophagy and human disease: emerging themes. Curr Opin Genet Dev. 2014;26:16–23. doi: 10.1016/j.gde.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrivens PJ, Noueihed B, Shahrzad N, Hul S, Brunet S, Sacher M. C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol Biol Cell. 2011;22:2083–2093. doi: 10.1091/mbc.E10-11-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahama-Noda K, Kira S, Yoshimori T, Noda T. TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J Cell Sci. 2013;126:4963–4973. doi: 10.1242/jcs.131318. [DOI] [PubMed] [Google Scholar]
- Slobodkin MR, Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 2013;55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- Stadel D, Millarte V, Tillmann KD, Huber J, Tamin-Yecheskel BC, Akutsu M, Demishtein A, Ben-Zeev B, Anikster Y, Perez F, et al. TECPR2 Cooperates with LC3C to Regulate COPII-Dependent ER Export. Mol Cell. 2015;60:89–104. doi: 10.1016/j.molcel.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Suzuki SW, Yamamoto H, Oikawa Y, Kondo-Kakuta C, Kimura Y, Hirano H, Ohsumi Y. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc Natl Acad Sci U S A. 2015;112:3350–3355. doi: 10.1073/pnas.1421092112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Tsotakos N, Liu Y, Young MM, Serfass J, Tang Z, Abraham T, Wang HG. The Bif-1-Dynamin 2 membrane fission machinery regulates Atg9-containing vesicle generation at the Rab11-positive reservoirs. Oncotarget. 2016;7:20855–20868. doi: 10.18632/oncotarget.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Pircs K, Nagy P, Varga A, Karpati M, Hegedus K, Kramer H, Kovacs AL, Sass M, Juhasz G. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25:1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Cai Y, Wang J, Zhang J, Menon S, Chou HT, Ferro-Novick S, Reinisch KM, Walz T. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc Natl Acad Sci U S A. 2013;110:19432–19437. doi: 10.1073/pnas.1316356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy K, Velentzas PD, Baehrecke EH. Ral GTPase and the exocyst regulate autophagy in a tissue-specific manner. EMBO Rep. 2016;17:110–121. doi: 10.15252/embr.201541283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KA, Reggiori F, Dunn WA, Jr, Klionsky DJ. Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J Biol Chem. 2003;278:48445–48452. doi: 10.1074/jbc.M309238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Davis S, Menon S, Zhang J, Ding J, Cervantes S, Miller E, Jiang Y, Ferro-Novick S. Ypt1/Rab1 regulates Hrr25/CK1delta kinase activity in ER-Golgi traffic and macroautophagy. J Cell Biol. 2015;210:273–285. doi: 10.1083/jcb.201408075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Menon S, Yamasaki A, Chou HT, Walz T, Jiang Y, Ferro-Novick S. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci U S A. 2013;110:9800–9805. doi: 10.1073/pnas.1302337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- Willett R, Ungar D, Lupashin V. The Golgi puppet master: COG complex at center stage of membrane trafficking interactions. Histochem Cell Biol. 2013;140:271–283. doi: 10.1007/s00418-013-1117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki A, Menon S, Yu S, Barrowman J, Meerloo T, Oorschot V, Klumperman J, Satoh A, Ferro-Novick S. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol Biol Cell. 2009;20:4205–4215. doi: 10.1091/mbc.E09-05-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WL, Klionsky DJ. Atg27 is a second transmembrane cycling protein. Autophagy. 2007;3:254–256. doi: 10.4161/auto.3823. [DOI] [PubMed] [Google Scholar]
- Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2011;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- Zhen Y, Li W. Impairment of autophagosome-lysosome fusion in the buff mutant mice with the VPS33A(D251E) mutation. Autophagy. 2015;11:1608–1622. doi: 10.1080/15548627.2015.1072669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]