Abstract
19F MRI is valuable for in vivo imaging due to the only trace amounts of fluorine in biological systems. Because of the low sensitivity of MRI however, designing new fluorochemicals remains a significant challenge for achieving sufficient 19F signal. Here, we describe a new class of high-signal, water-soluble fluorochemicals as 19F MRI imaging agents. A polyamide backbone is used for tuning the proteolytic stability to avoid retention within the body, which is a limitation of current state-of-the-art perfluorochemicals. We show that unstructured peptides containing alternating N-ε-trifluoroacetyllysine and lysine provide a degenerate 19F NMR signal. 19F MRI phantom images provide sufficient contrast at micromolar concentrations, showing promise for eventual clinical applications. Finally, the degenerate high signal characteristics were retained when conjugated to a large protein, indicating potential for in vivo targeting applications, including molecular imaging and cell tracking.
Keywords: 19F NMR spectroscopy, fluorochemical, molecular imaging, MRI, peptides
Molecular and cellular therapies have the potential to treat numerous diseases, but there is a significant need for noninvasive techniques to monitor the biodistribution of these therapeutics in vivo. The trace amounts of organic fluorine in biological settings enables monitoring of fluorine-tagged molecules in whole organisms by magnetic resonance imaging (MRI).[1] MRI using the 19F nucleus holds great potential for diagnostic molecular imaging with applications for attaching fluorinated molecules to targeting domains,[2] cell tracking,[3] and oxygen sensing.[4] 19F MRI is less developed than 1H MRI, in part due to the need for high signal, biocompatible 19F probe molecules.[5] Synthesis of high-symmetry fluorochemicals, such as perfluorocrownethers,[6] fluoropolymers,[7] and fluorinated dendrimers[8] (e.g., Figure 1) leads to an increase in the number of magnetically equivalent fluorine nuclei for improving signal intensity. Emulsions of these fluorochemicals represents the current state-of-the-art for 19F MRI probes which are reaching the limit of detection threshold at clinical field strengths.[7],[2] However, these emulsions are susceptible to stability problems, poor physicochemical properties, and retention in the body.[9],[10] Environmental persistence and health concerns are also prompting legal regulations of some perfluorochemicals and their degradation products.[11],[12]
Figure 1.
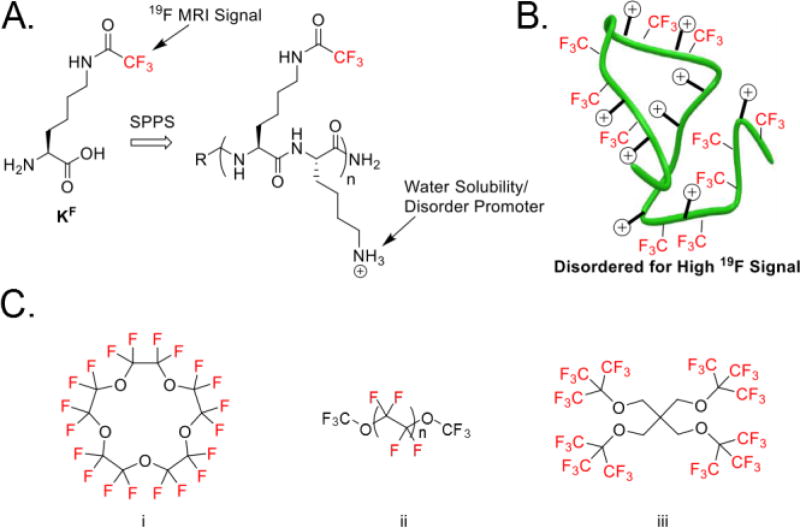
Disordered 19F peptide design constructed by solid-phase peptide synthesis (SPPS) and current state-of-the-art molecules. A) Generic structure of disordered 19F peptides with key features highlighted. A complete list of peptides used in this study can be found in Figure S1 B) Disordered peptide design showing 19F sources alternating with positive charge causing a random-coil through electrostatic repulsion. C) Select fluorinated molecules used for 19F MRI: i) Perfluoro-15-crown-5 ether[6] ii) Perfluoro(polyethylene glycol) methyl ether[13] and iii) PERFECTA.[14]
Fluorinated biopolymers are an untapped material for 19F MRI applications. Fluorinated proteins characteristically lead to well-resolved fluorine resonances for which the same fluorine-labeled amino acids can produce resonances that span over 10 ppm.[15], Such dispersion is due to the high sensitivity of the fluorine chemical shift to distinct protein environments.[16] To overcome this, we were inspired from spectral studies of intrinsically disordered proteins that result in limited resonance dispersion.[17] Therefore, our current approach seeks to override the high environmental sensitivity of the 19F nucleus through synthesis of highly disordered peptide chains lacking secondary or tertiary structure. Here, we describe the design of a series of high-signal 19F MRI agents for future molecular imaging and cell tracking applications using sequence-defined peptide-based materials that are emulsion-free while possessing tunable biological and environmental stability. As an initial evaluative step towards molecular imaging, we show that protein bioconjugation of the fluorinated polypeptide is able to maintain both a narrow linewidth and high 19F magnetic resonance signal for future applications.
Our initial designs focused on the solid-phase synthesis of peptide oligomers containing an alternating pattern of N-ε-trifluoroacetylated lysine (TFA-lysine) and lysine. These amino acids were chosen to favor high solubility, while disfavoring both aggregation as well as secondary structure formation due to electrostatic repulsion (Figure 1a, b). TFA-lysine, was chosen based on three magnetically equivalent fluorine nuclei yielding high intensity narrow resonances that are devoid of scalar couplings. Ye et al. also showed that the chemical shift sensitivity of CF3 groups next to carbonyls are significantly less responsive to environmental changes relative to benzylic CF3 groups.[18] Regarding faster spectral acquisition time, the T1 relaxation of CF3 groups tend to be shorter than those found on aromatic rings.[19] Grage et al. showed by solid state 19F NMR that amino acids with aliphatic CF3 groups have T1 relaxation times between 0.32 and 1.86 s, whereas fluorinated aromatic residues had T1 times ranging from 14.9 to 352 s.[19],[20] As a final consideration, the trifluoroacetamide group can also hydrolyze under alkaline conditions for eventual degradation in the environment.
Our first peptide series consisted of a 17 residue peptide 1 containing seven TFA-lysines (Figure 2). Encouragingly, when the peptide was dissolved in aqueous solution and studied by 19F NMR, a single resonance was observed with only a slight shoulder, indicating that the TFA-lysine residues were experiencing a similar chemical environment. The resonance spanned 15 Hz, with a 7 Hz measurement at half-height. Surprisingly, when the N-terminus was left unacetylated a slight decrease in resonance degeneracy was observed as indicated by the appearance of a set of non-overlapping resonances. We therefore focused on peptides of alternating TFA-lysine and lysine with acylated N-termini.
Figure 2.
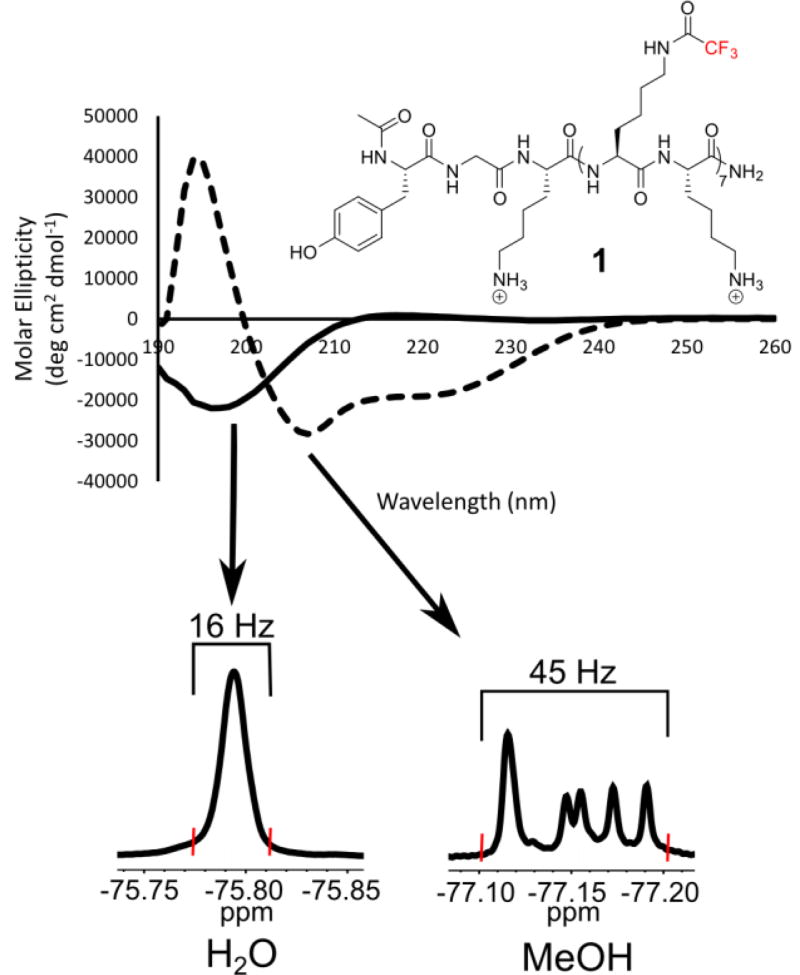
Top) Far-UV circular dichroism spectra of 100 µM 1 in water (solid lines) and methanol (dashed lines). Bottom) A random-coil is observed in aqueous environment resulting in a degenerate 19F NMR resonance spanning 16 Hz. However in methanol, an α-helix is observed resulting in resolved 19F NMR resonances spanning 45 Hz (See Figure S51). Linewidths were measured from baseline to baseline at the red hatch marks for comparison.
In order to discern the secondary structure of the peptide sequence and support our disordered peptide designs, we performed circular dichroism on 1 in both aqueous and organic media (Figure 2). The far-UV CD spectra of 1 display a strong solvent dependence. In methanol, a structure-promoting solvent, a characteristic α-helical signature was observed with double minima at 208 and 222 nm. This peptide was determined to be 55% α-helical based on the molar ellipticity at 222 nm. Alternatively, when 1 was dissolved in water, a CD spectrum consistent with a random-coil was observed. Because of the low resolution of CD, we cannot rule out additional population of a polyproline II helix (PPII) based on the small positive absorption at 215 nm, which has been observed for polylysine containing peptides in the PPII conformation[21]. We measured 1H NMR 3JHNCα coupling constants from the backbone amides of 1 which range from 6.11–6.80 Hz (Figure S51). These values are within the range to slightly below the random coil values calculated by Serrano[22] from Karplus analysis and higher than 5.16 Hz determined by Shi et al for the PPII helix.[23]
Comparing the 19F NMR spectra from these two solutions also supports the structural design. Whereas all 19F resonances remain degenerate when the peptide is dissolved in water, when methanol is used, the 19F resonances resolve with minimal overlap, consistent with the hyperresponsive character of the 19F nucleus to distinct chemical environments.[16] This observation is indicative of the 19F-labeled side chains being in several distinct environments within the α-helix, yielding resolved NMR resonances. (Figure 2, Fig. S52)
Peptides tend to adopt increased α-helical structure at long chain lengths. To evaluate a length dependence on our designs, peptides S1a–i were synthesized containing 5–21 amino acids and up to ten TFA-lysine residues. In many cases a similar degeneracy was observed in the NMR spectra with a slight shoulder in some cases (Figure S4–21). Therefore, even up to 30 equivalent fluorine resonances can easily be obtained using our initial designs. Typically, when symmetrical molecules such as perfluorocrownethers or dendrimers are used to maximize the number of magnetically equivalent 19F resonances, synthesis of these large molecules can become challenging.[24] Additionally, these molecules have a dramatic loss of water solubility.[9] Our peptide constructs however can be readily obtained using solid phase synthesis and readily dissolve in water at millimolar concentrations.
Sequential addition of TFA-lysine to the peptide sequence only minimally affects the hydrophobicity of the peptide chain which we assessed based on reverse-phase HPLC retention time using acetonitrile as the elution solvent, in which case peptides S1a–i differ in retention by only 11 min (23–34% acetonitrile). We conclude from these first studies that a highly disordered peptide sequence that dissolves readily in water, can be maintained at long chain lengths by simple alternation of TFA-lysine and lysine, giving rise to high intensity degenerate 19F NMR spectra.
In addition to a signal sensitivity requirement, a second priority for the design of our fluorinated peptides is the ability to tune their degradation susceptibility in vivo to avoid persistence within the body, as well as in the environment. To probe this, we subjected our lysine-rich peptides to trypsin to monitor the rate of degradation. An 11 residue model peptide, 2, was used for these experiments, which was then modified via incorporation of D-lysine to tune the stability of the molecule. As we expected a 100% D-amino acid containing peptide to be proteolytically stable,[25] we investigated decreased percentages of D-amino acids to allow us to assess a range of stabilities from these diastereomeric peptides, as well as any protection from the fluorinated side-chain.
We monitored the hydrolysis of three peptides over time. In all cases presented, the L-variant of TFA-lysine was used. The all L-lysine containing peptide 2a was degraded very rapidly with complete hydrolysis of the peptide backbone observed approximately one minute after addition of trypsin at 25 °C, and pH 8.1. Alternatively, peptide 2b, containing 30% D-amino acids, showed a 10-fold increase in protease stability. The final peptide studied contained 50% D-amino acids, 2c, and showed 72% remaining after 3 hours. After 24 h, 12% of the peptide remained, which matches approximately the behavior of the peptide in buffer alone, indicating the D-lysines provided sufficient protection from trypsin until the hydrolysis of TFA-protecting groups was observed. (Table 1).
Table 1.
Lifetime of model peptide series 2 during tryptic digest (20:1 S/E)
| Peptide | Sequence[a] | Lifetime |
|---|---|---|
| 2a |
|
1 min |
| 2b |
|
10 min |
| 2c |
|
> 24 h[b] |
| 2b pH = 8.1, buffer only |
|
> 24 h[c] |
| 2b pH 7.1, buffer only |
|
> 48 h[d] |
 designates N-ε-trifluoroacetyllysine, and lower-case k refers to D-lysine.
designates N-ε-trifluoroacetyllysine, and lower-case k refers to D-lysine.
12% of 2c remains after 24 h.
11% of 2b remains after 24 h in pH 8.1 buffer without trypsin.
10% of 2b remains after 48 h in pH 7.1 buffer without trypsin.
We tested the buffer conditions of the trypsin assay on the peptide to ensure it would not appreciably interfere with the digest, and found that out to three hours there is an 11% reduction of full length peptide, but near complete hydrolysis of the TFA side chains has occurred by 24 hours. This experiment was also performed at pH 7.1, where the peptide showed no hydrolysis up to five hours, but near complete hydrolysis after 48 hours (Table 1). Although more stable, 19F NMR spectra in water for peptides 2b and 2c showed two resonances, indicating a change in structure (See Supporting Information). Further analysis and design of peptides containing partial incorporation of D amino acids must be performed in order to reestablish degenerate signals.
While peptide 2a maintained overlapping resonances in the 19F NMR spectra in water, in biological systems, local chemical environments can change affecting chemical shift dispersion. Therefore, despite its instability towards proteases, we also measured the 19F NMR spectra of 2a in an E. coli cellular lysate Fig S53). In this case, although the resonance moved upfield 0.07 ppm, the overlap was maintained with an upfield tail, which was also observed in the reference TFA resonance consistent with imperfect shimming.
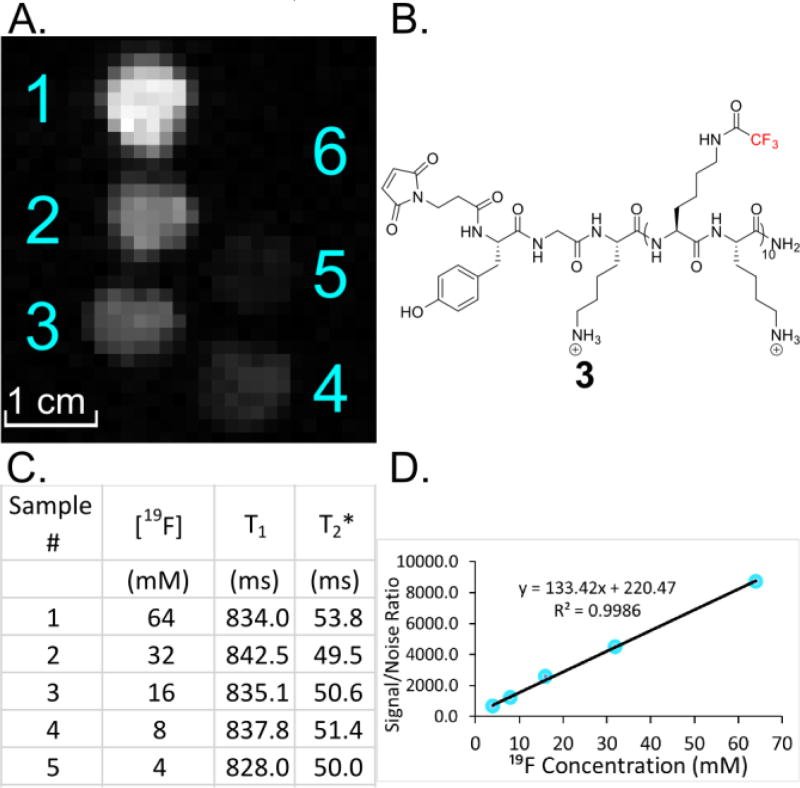
With a tunable peptide system in hand, we turned our attention to peptide 3 containing 30 equivalent fluorine atoms for initial 19F MRI studies. To assess the limit of detection, a 19F MRI phantom image was acquired of 3 at varying concentrations to evaluate the image quality (Figure 3). A 16.4 T Varian MRI instrument was used to scan five samples of 3 with 19F concentrations ranging from 4–64 mM equivalent fluorine (or 0.13 – 2.13 mM peptide). Images were acquired in ten minutes, and a linear dependent intensity gradient can be observed throughout the samples with the lowest concentration still being visible at 4 mM fluorine (0.13 mM peptide) (Figure 3D). Highly fluorinated materials tend to aggregate at high concentrations, which is reflected in short T2 relaxation and results in broadened resonances. Therefore, we also measured the T1 and T2* values of the varying peptide concentrations by 19F NMR (470 MHz, 25 °C). We used T2* as an estimation of T2 due to the importance of the peak width in the resolution of our spectra. We do not observe a significant concentration effect on T1 or T2* consistent with the narrow line widths (5.9–6.4 Hz) and linear concentration dependence on signal to noise (Figure 3C,D) indicating that these peptides do not self-assemble in solution.
Figure 3.
A) 19F MRI phantom image of 3 at various concentrations: 1) 2.13 mM (64 mM 19F), 2) 1.07 mM (32 mM 19F), 3) 0.53 mM (16 mM 19F), 4) 0.27 mM (8 mM 19F), 5) 0.13 mM (4 mM 19F), 6) 0 mM. B) Structure of 3 utilized in MRI experiments. C) NMR relaxation times determined for 3 at various concentrations, showing no appreciable change as a function of concentration. D) Signal to noise ratio of 3 increases linearly with respect to concentration, indicating no concentration dependent higher order structure.
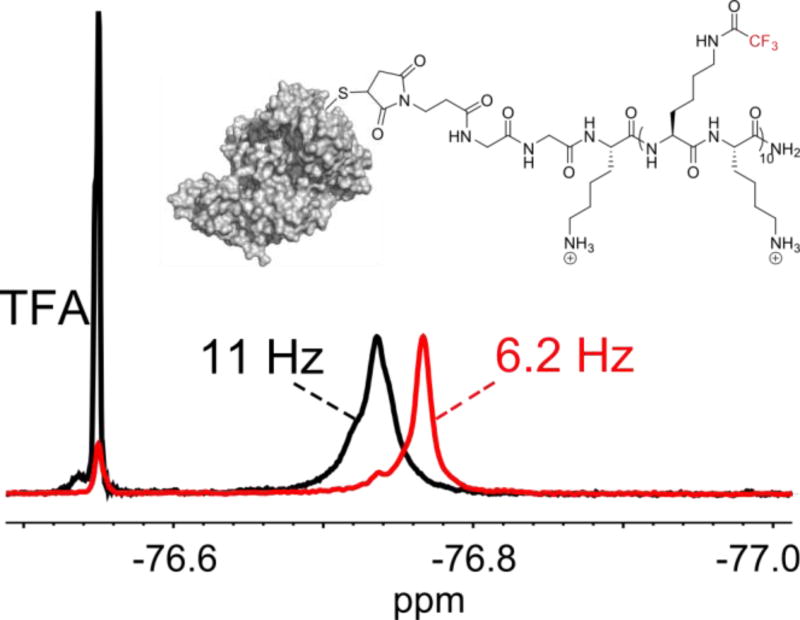
One potential application of this type of peptide construct is for use as a 19F MRI imaging tag conjugated to a larger targeting protein. To investigate if the newly introduced chemical environment of a bioconjugated protein with the peptide affects its NMR resonances, we used a model protein, bovine serum albumin (BSA), to react via the sulfhydryl moiety of its lone free cysteine residue to a maleimide attached to the N-terminus of peptide 3. The bioconjugation reaction was performed by incubating 50 µM BSA with 100 µM 3 for 24 h at 37 °C (0.2 M acetate buffer, pH = 5.0). Protein adduct formation was verified by MALDI-TOF MS (See Supporting Information). After removal of unreacted peptide via dialysis, a 19F NMR was performed on the protein-peptide conjugate (Figure 4). The resulting NMR spectrum showed a singular resonance with a minimal downfield shoulder for the construct. Although still narrow, the resonance of the conjugate was broadened (11 Hz) in comparison to the free peptide (6.2 Hz), consistent with the attachment of a large protein (66 kDa). The T2* of the conjugate was approximately 40% shorter than the peptide alone (29 vs 50 ms). Such a small reduction in T2 suggests that despite being attached to a large protein, resonance broadening from chemical shift anisotropy is mitigated due to significant rotational freedom of the CF3 group, and the fluorinated peptide side-chains maintain a significant amount of conformational dynamics, similar to the behavior of intrinsically disordered proteins.[26]
Figure 4.
19F NMR of BSA conjugated to 3 (black line). A single resonance is obtained, with only minimal broadening observed. The linewidths are compared to unconjugated 3 (red line), and were measured at the dashed line. Trifluoroacetic acid was added as a reference standard.
These results are encouraging for future molecular imaging applications, in which such peptides could be conjugated to larger antibodies, or smaller targeting agents such as affibodies[27] and fibronectin domains.[28] Although, we note a single peptide possessing 30 equivalent fluorine atoms may be insufficient for imaging applications at clinical field strengths (≤ 3 T), the possibility of attaching multiple fluorinated peptides to a protein construct such as to a tetracysteine tag, or the surface of a nanoparticle opens up the possibility of highly fluorinated imaging constructs.[29]
In conclusion, these studies lay the groundwork for a molecular design strategy for obtaining high signal 19F MRI agents, based on introducing a high degree of structural disorder. Through careful choice of a peptide backbone and fluorinated side-chain, the stability of the fluorochemical can be rationally tuned for breakdown in vivo or in the environment. Due to the increased interest in new fluorochemicals for imaging applications, and concerns over toxicity and environmental persistence, these new designs will be studied for broader use in 19F MRI-based biomedical applications. We note, that with the increased availability of synthetic methods for more highly fluorinated amino acids,[30],[31] even higher SNR may be achievable. Paramagnetic metal chelates as T1 agents,[32] as well as improvement in data processing such as sparse sampling,[33] may help bring designs of fluorinated materials closer to practical applications in the 19F MRI field.
Supplementary Material
Acknowledgments
This study has been funded via the University of Minnesota McKnight Land Grant Professorship, Research Corporation Cottrell Scholar Award, Heisig-Gleysteen Fellowship, and University of Minnesota Undergraduate Research Scholarship (URS). The MRI work was supported in part by the Minnesota Lions Diabetes Foundation, the Schott Family Foundation, the Carol Olson Memorial Diabetes Research Fund, and the NIH [grants P41 EB015894 and S10 RR025031].
References
- 1.Ahrens ET, Zhong J. NMR Biomed. 2013;26:860–71. doi: 10.1002/nbm.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kok MB, De Vries A, Abdurrachim D, Prompers JJ, Grüll H, Nicolay K, Strijkers GJ. Contrast Media Mol. Imaging. 2011;6:19–27. doi: 10.1002/cmmi.398. [DOI] [PubMed] [Google Scholar]
- 3.Ahrens ET, Flores R, Xu H, a Morel P. Nat. Biotechnol. 2005;23:983–7. doi: 10.1038/nbt1121. [DOI] [PubMed] [Google Scholar]
- 4.Mignion L, Magat J, Schakman O, Marbaix E, Gallez B, Jordan BF. Magn. Reson. Med. 2013;69:248–254. doi: 10.1002/mrm.24245. [DOI] [PubMed] [Google Scholar]
- 5.Schlemmer HP, Becker M, Bachert P, Dietz A, Rudat V, Vanselow B, Wollensack P, Zuna I, Knopp MV, Weidauer H, Wannenmacher M, Van Kaick G. Cancer Res. 1999;59:2363–2369. [PubMed] [Google Scholar]
- 6.Nakamura T, Sugihara F, Matsushita H, Yoshioka Y, Mizukami S, Kikuchi K. Chem. Sci. 2015;6:1986–1990. doi: 10.1039/c4sc03549f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janjic JM, Srinivas M, Kadayakkara DKK, Ahrens ET. J. Am. Chem. Soc. 2008;130:2832–2841. doi: 10.1021/ja077388j. [DOI] [PubMed] [Google Scholar]
- 8.Yu W, Yang Y, Bo S, Li Y, Chen S, Yang Z, Zheng X, Jiang Z-X, Zhou X. J. Org. Chem. 2015;80:4443–4449. doi: 10.1021/acs.joc.5b00294. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz-Cabello J, Barnett BP, Bottomley PA, Bulte JWM. NMR Biomed. 2011;24:114–29. doi: 10.1002/nbm.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfannkuch N, Schnoy F. Anaesthesist. 1979;28:511–516. [PubMed] [Google Scholar]
- 11.Dunster KR, Davies MW, Fraser JF. BioMed Eng OnLine. 2006;5:5–10. doi: 10.1186/1475-925X-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicole W. Environ. Health Perspect. 2013;121:A340–A340. doi: 10.1289/ehp.121-A340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mountain GA, Jelier BJ, Bagia C, Friesen CM, Janjic JM. J. Fluor. Chem. 2014;162:38–44. doi: 10.1016/j.jfluchem.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tirotta I, Mastropietro A, Cordiglieri C, Gazzera L, Baggi F, Baselli G, Grazia Bruzzone M, Zucca I, Cavallo G, Terraneo G, Baldelli Bombelli F, Metrangolo P, Resnati G. J. Am. Chem. Soc. 2014;136:8524–8527. doi: 10.1021/ja503270n. [DOI] [PubMed] [Google Scholar]
- 15.Mishra NK, Urick AK, Ember SWJ, Schönbrunn E, Pomerantz WC. ACS Chem. Biol. 2014;9:2755–2760. doi: 10.1021/cb5007344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitevski-LeBlanc JL, Prosser RS. Prog. Nucl. Magn. Reson. Spectrosc. 2012;62:1–33. doi: 10.1016/j.pnmrs.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Wang GF, Li C, Pielak GJ. Protein Sci. 2010;19:1686–1691. doi: 10.1002/pro.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye L, Larda ST, Frank Li YF, Manglik A, Prosser RS. J. Biomol. NMR. 2015;62:97–103. doi: 10.1007/s10858-015-9922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grage SL, Dürr UHN, Afonin S, Mikhailiuk PK, Komarov IV, Ulrich AS. J. Magn. Reson. 2008;191:16–23. doi: 10.1016/j.jmr.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Dürr UHN, Grage SL, Witter R, Ulrich AS. J. Magn. Reson. 2008;191:7–15. doi: 10.1016/j.jmr.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 21.Rucker AL, Creamer TP. Protein Sci. 2002;11:994–994. doi: 10.1110/ps.4550102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serrano L. J. Mol. Biol. 1995;254:322–333. doi: 10.1006/jmbi.1995.0619. [DOI] [PubMed] [Google Scholar]
- 23.Shi Z, Chen K, Liu Z, Ng A, Bracken WC, Kallenbach NR. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17964–17968. doi: 10.1073/pnas.0507124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu YB. Wiley Interdiscip. Rev. Nanomedicine Nanobiotechnology. 2013;5:646–661. doi: 10.1002/wnan.1239. [DOI] [PubMed] [Google Scholar]
- 25.Tsuyuki E, Tsuyuki H, Stahmann MA. J. Biol. Chem. 1956;222:479–485. [PubMed] [Google Scholar]
- 26.Li C, Lutz EA, Slade KM, Ruf RAS, Wang GF, Pielak GJ. Biochemistry. 2009;48:8578–8584. doi: 10.1021/bi900872p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antaris AL, Chen H, Cheng K, Sun Y, Hong G, Qu C, Diao S, Deng Z, Hu X, Zhang B, Zhang X, Yaghi OK, Alamparambil ZR, Hong X, Cheng Z, Dai H. Nat. Mater. 2015;15:235–242. doi: 10.1038/nmat4476. [DOI] [PubMed] [Google Scholar]
- 28.Hackel BJ, Kimura RH, Gambhir SS. Radiology. 2012;263:179–188. doi: 10.1148/radiol.12111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu G, Tang J, Bai X, Xu S, Wang L. Nano Research. 2016;9:1–9. [Google Scholar]
- 30.Tressler CM, Zondlo NJ. Org. Lett. 2016;18:6240–6243. doi: 10.1021/acs.orglett.6b02858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buer BC, Levin BJ, Marsh ENG. J. Pept. Sci. 2013;19:308–314. doi: 10.1002/psc.2501. [DOI] [PubMed] [Google Scholar]
- 32.Harvey P, Kuprov I, Parker D. Eur. J. Inorg. Chem. 2012:2015–2022. [Google Scholar]
- 33.Zhong J, Mills PH, Hitchens TK, Ahrens ET. Magn. Reson. Med. 2013;69:1683–1690. doi: 10.1002/mrm.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.