Abstract
Purpose
Current Lynch syndrome (LS) prediction models quantify the risk to an individual of carrying a pathogenic germline mutation in three mismatch repair (MMR) genes: MLH1, MSH2, and MSH6. We developed a new prediction model, PREMM5, that incorporates the genes PMS2 and EPCAM to provide comprehensive LS risk assessment.
Patients and Methods
PREMM5 was developed to predict the likelihood of a mutation in any of the LS genes by using polytomous logistic regression analysis of clinical and germline data from 18,734 individuals who were tested for all five genes. Predictors of mutation status included sex, age at genetic testing, and proband and family cancer histories. Discrimination was evaluated by the area under the receiver operating characteristic curve (AUC), and clinical impact was determined by decision curve analysis; comparisons were made to the existing PREMM1,2,6 model. External validation of PREMM5 was performed in a clinic-based cohort of 1,058 patients with colorectal cancer.
Results
Pathogenic mutations were detected in 1,000 (5%) of 18,734 patients in the development cohort; mutations included MLH1 (n = 306), MSH2 (n = 354), MSH6 (n = 177), PMS2 (n = 141), and EPCAM (n = 22). PREMM5 distinguished carriers from noncarriers with an AUC of 0.81 (95% CI, 0.79 to 0.82), and performance was similar in the validation cohort (AUC, 0.83; 95% CI, 0.75 to 0.92). Prediction was more difficult for PMS2 mutations (AUC, 0.64; 95% CI, 0.60 to 0.68) than for other genes. Performance characteristics of PREMM5 exceeded those of PREMM1,2,6. Decision curve analysis supported germline LS testing for PREMM5 scores ≥ 2.5%.
Conclusion
PREMM5 provides comprehensive risk estimation of all five LS genes and supports LS genetic testing for individuals with scores ≥ 2.5%. At this threshold, PREMM5 provides performance that is superior to the existing PREMM1,2,6 model in the identification of carriers of LS, including those with weaker phenotypes and individuals unaffected by cancer.
INTRODUCTION
Nearly 1 million individuals in the United States have Lynch syndrome (LS) but most are unaware of their diagnosis.1 LS is caused by germline alterations in DNA mismatch repair (MMR) genes (MLH1, MSH2, MSH6, or PMS2) or EPCAM (which causes epigenetic silencing of MSH2) and confers a 40% to 80% lifetime risk of colorectal cancer (CRC).2-4 In addition to CRC, mutation carriers are at increased risk for cancers of the endometrium, ovaries, stomach, small intestine, pancreas, urinary tract, brain, and cutaneous sebaceous glands.5-8 Personal and family histories of these component cancers can guide genetic testing to identify mutation carriers. If unidentified, these individuals miss the opportunity to pursue interventions known to effectively reduce the risk of Lynch-associated cancers, such as frequent colonoscopies, prophylactic surgeries, and chemoprevention.9-14
Prediction models, such as PREMM1,2,6, are evidence-based tools that can help identify carriers of LS by using the personal and family history of Lynch-associated malignancies in an individual to quantify the likelihood of carrying a germline mutation in the MLH1, MSH2, and MSH6 genes.15 Clinical practice guidelines from various national organizations recommend that individuals with ≥5% likelihood of LS by PREMM1,2,6 undergo genetic testing.2,3,16 However, current LS prediction models do not assess for PMS2 or EPCAM mutations.17,18 Our aim was to develop and validate a new prediction model to quantify the risk of an individual carrying a pathogenic mutation in any of the five genes associated with LS (MLH1, MSH2, MSH6, PMS2, and EPCAM).
PATIENTS AND METHODS
Patients
Development cohort.
We analyzed data from 18,734 patients (probands) who underwent germline testing of the MLH1, MSH2, MSH6, PMS2, and EPCAM genes by Myriad Genetic Laboratories (Salt Lake City, UT) after May 2011. Clinical data were obtained from the test order form completed by health care professionals who ordered germline testing, as previously described,15,19 including proband age at genetic testing, sex, personal and family cancer histories (including ages at diagnoses), and germline testing results. Cancer history included Lynch-associated malignancies of the colorectum, endometrium, stomach, ovaries, urinary tract, small intestine, pancreas, bile ducts, brain, or sebaceous glands. Probands without personal history of these cancers were defined as unaffected. Family history was limited to first-degree relatives (FDRs) and second-degree relatives (SDRs) on the affected side.
Validation cohort.
Validation of the model was performed in an independent cohort of 1,058 patients with CRC who were recruited to an institutional sample registry at Dana-Farber Cancer Institute from 2008 to 2014, without preselection for high-risk features of LS (eg, age at diagnosis, personal or family cancer histories, or tumor microsatellite instability/MMR deficiency). All participants underwent germline testing for multiple genes, including MLH1, MSH2, MSH6, PMS2, and EPCAM.
Laboratory Methods
Germline analysis was uniform for probands in both cohorts and was performed by Myriad Genetics. MLH1, MSH2, and MSH6 analyses were performed as previously described.15 PMS2 testing involved sequencing of all exons and adjacent intronic regions as well as large rearrangement testing by multiplex ligation-dependent probe amplification. EPCAM analysis involved large rearrangement testing by microarray comparative genomic hybridization for the 3-prime region. Individuals with deleterious/suspected deleterious mutations were named mutation positive. Patients with polymorphisms, unclassified variants, no alterations, or missense mutations for which clinical significance is not established were named mutation-negative.15
Statistical Methods
Variables in the previous PREMM1,2,6 and PREMM1,2 models were considered for the new PREMM5 model.15,19,20 We examined the association of proband sex with mutation presence, because male sex was a predictor of mutations in previous analyses.15 Proband age at testing was considered for inclusion because of the high proportion of unaffected individuals who underwent testing. Summary statistics for each variable were catalogued by gene type for probands and relatives, and ages at diagnosis of CRC and endometrial cancer were truncated at the lower and upper one percentile to stabilize the resulting model. Patients were excluded if all cancer-related data were missing, if sex was unreported (n = 11), or if two pathogenic mutations (other than EPCAM and MSH2) were detected (n = 5). Multiple imputation was applied for missing values; five completed data sets were analyzed, and results were combined by Rubin’s rules.21 The imputation model included personal and family cancer characteristics, and the outcome was presence of a specific gene mutation. Among the 18,734 probands, missing data were imputed for ages at CRC (n = 28), endometrial cancer (n = 28), other cancers (n = 50), and genetic testing (n = 14). Among the 49,237 relatives, missing ages of CRC (n = 1,995), endometrial cancer (n = 407), and other cancers (n = 4,581) was imputed. Associations of overall and gene-specific mutation statuses were analyzed with F tests (continuous variables) and χ2 tests (categoric variables). A two-sided P value of less than .05 indicated statistical significance.
Development of the PREMM5 model.
PREMM5 predictions of any pathogenic mutation were based on a logistic regression model derived from the full cohort. The associations of predictors with mutation status were reported as odds ratios (ORs) with 95% CIs. Interaction terms were added to test for sex or age-specific effects. Different models were explored and adjusted for unaffected individuals and age at testing. MSH2 and EPCAM were combined into one category (MSH2), because there were few EPCAM mutations and because these mutations induce epigenetic inactivation of MSH2.22 Polytomous logistic regression was used to assess the associations of clinical features with mutation status by the specific gene, in which the response variable was a categoric variable with five levels: (1) MLH1, (2) MSH2 or EPCAM, (3) MSH6, (4) PMS2, and (5) mutation-negative status. For each individual, the PREMM5 score was calculated as the predicted probability of a mutation within the resulting multivariable polytomous logistic regression model. Internal validation was performed by bootstrap resampling that used 200 random samples drawn with replacement. Predictive models were developed in each bootstrap sample and evaluated in the entire cohort to quantify the optimism in the estimated apparent performance.23
Assessment of Model Performance.
We quantified the overall ability of the PREMM5 model to discriminate carriers from noncarriers by the area under the receiver operating characteristic curve (AUC), which also was used for each specific gene in the development cohort.24 Decision curve analysis was used to determine the clinical usefulness of the model, and the true-positive (TP) and false-positive (FP) classifications were considered at increasing decision thresholds. This methodology evaluates prediction models for their potential to improve clinical decision making24-28 and, in this case, to refer individuals for genetic testing. A decision curve shows the net benefit (NB) of using a model at different thresholds. The NB sums the TPs minus a weighted number of FPs: NB = (TP − wFP) / n, in which n is the total sample size and w is the relative weight of the harm of unnecessary testing versus the benefit of identification of a carrier. The relative weight—w—is defined by the threshold probability to define at-risk patients who need genetic testing. The NB of PREMM5 and two reference strategies—test none or test all—was calculated. Thresholds between 0% (test all) and 10% (test high-risk probands) were considered, and 5% was the focus, as recommended by national guidelines.2,3,16 The number needed to test (NNT), which represents the number of patients who have PREMM5 scores greater than a given threshold who should undergo testing to identify one mutation carrier, was calculated. The same performance metrics were analyzed with the validation cohort.
Comparison of the PREMM5 and PREMM1,2,6 Models.
PREMM1,2,6 predictions were calculated for all patients and were compared with PREMM5 predictions by using receiver operating characteristic curves and reclassification analysis. Under- or overprediction of each model was quantified by the calibration intercept and was converted to a ratio of observed to expected (O/E) results: O/E = exp (intercept).29 Statistical analysis was performed using SAS (version 9.4) for data management and univariable analysis, and R software (version 3.1.2) was used for multivariable analysis, bootstrap and external validation, and decision curve and reclassification analyses.
The study was investigator-initiated for model development; data collection and germline analyses occurred at Myriad Genetics, and anonymized data sets were provided to investigators at Dana-Farber Cancer Institute and Columbia University. Statistical analyses were conducted by researchers and independent statisticians, all of whom were unaffiliated with Myriad. Investigational review board approval was obtained for this study.
RESULTS
Pathogenic mutations were found in 1,000 (5%) of 18,734 probands in the development cohort: MLH1 (n = 306), MSH2 (n = 354), MSH6 (n = 177), PMS2 (n = 141), and EPCAM (n = 2; Table 1). A total of 15,363 (82%) of 18,734 participants were women. Pathogenic mutations were more frequent among men (370 [11%] of 3,371) than women (630 [4%] of 15,363; P < .001).
Table 1.
Clinical Characteristics of Gene Mutation Carriers Stratified by Gene
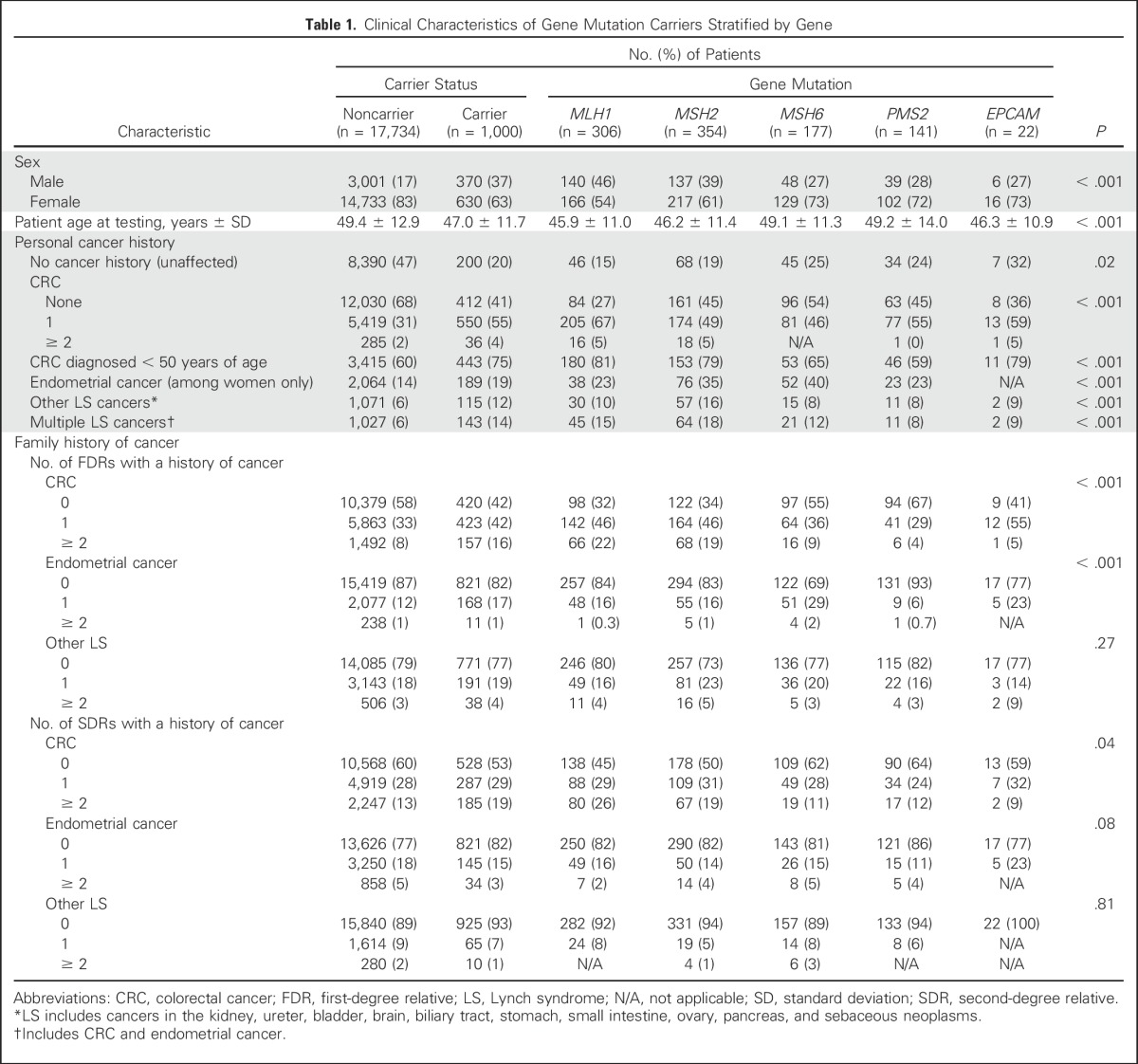
Carriers of gene mutations were younger at the time of genetic testing than noncarriers (47.0 v 49.4 years of age), and carriers of MLH1, MSH2, and EPCAM were younger at the time of testing (45.9 v 46.2 v 46.3 years of age, respectively) than carriers of MSH6 and PMS2 (49 years of age for both; P < .001). Forty-six percent of probands (8,590 of 18,734) were unaffected by cancer, including 200 (20%) of 1,000 mutation carriers.
Multiple CRCs and other LS-associated malignancies were more frequent among carriers of MLH1, MSH2, and EPCAM than carriers of MSH6 and PMS2. Carriers of MSH6 and PMS2 were older than other carriers diagnosed with CRC (P < .001) or endometrial cancer (P = .034) (Appendix Table A3, online only). Carriers of PMS2 had fewer relatives with CRC (P < .001) and fewer FDRs with endometrial cancer (P < .001) compared with other carriers.
In the validation cohort, the mean CRC age was 55.7 years (standard deviation, 12.6 years), and 587 (55.5%) of 1,058 were men (Appendix Table A4, online only). Pathogenic gene mutations were detected in 33 (3.1%) of 1,058 individuals: 13 with MLH1, seven with MSH2, six with MSH6, seven with PMS2, and none with EPCAM.
Development, Validation, and Performance of PREMM5
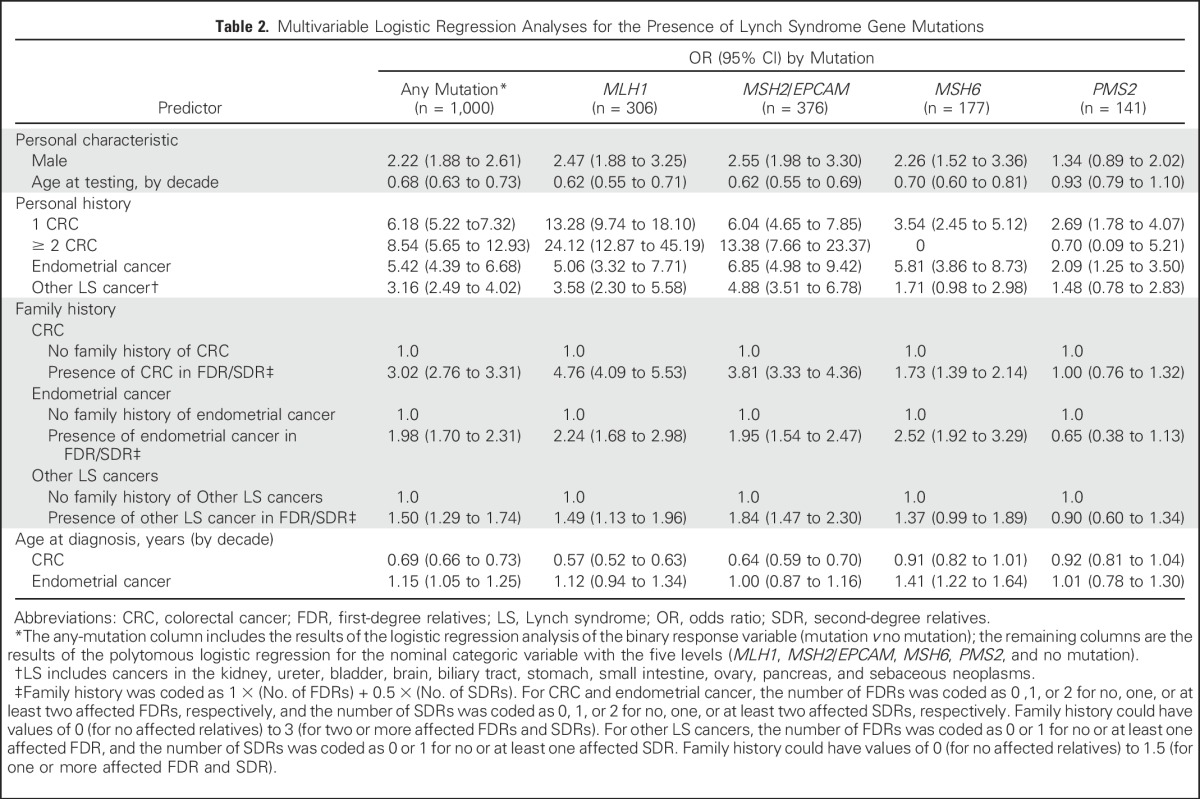
In multivariable analysis (Table 2), younger age at testing was associated with the presence of any gene mutation (OR, 0.69 per decade; 95% CI, 0.66 to 0.73) and added into PREMM5 as a new predictor. After analysis was adjusted for all other predictors, male sex was associated with the presence of any mutation (OR, 2.22; 95% CI, 1.88 to 2.61); other predictors included personal history of CRC with one occurrence (OR, 6.18, 95% CI, 5.22 to 7.32), or multiple occurrences (OR, 8.54; 95% CI, 5.65 to 12.93), and relatives with CRC (OR, 3.02; 95% CI, 2.76 to 3.31). Personal and family CRC diagnoses and corresponding ages were less predictive of PMS2 mutations (OR, 2.69; 95% CI, 1.78 to 4.07 and OR, 1.00; 95% CI, 0.76 to 1.32, respectively) compared with other genes. Personal history of endometrial cancer was associated with any mutation (OR, 5.42; 95% CI, 4.39 to 6.68), but age was only predictive of MSH6 mutations. Personal history of other LS-associated cancers had an OR of 3.16 (95% CI, 2.49 to 4.02) for any mutation but was not predictive of MSH6 or PMS2 mutations. Family history of endometrial cancer was predictive of all gene mutations except PMS2. Family history of other LS-associated cancers was weakly predictive of any mutation (OR, 1.50; 95% CI, 1.29 to 1.74) but not of MSH6 or PMS2. The PREMM5 prediction model is available online30 and in Appendix Tables A1 and A2 (online only).
Table 2.
Multivariable Logistic Regression Analyses for the Presence of Lynch Syndrome Gene Mutations
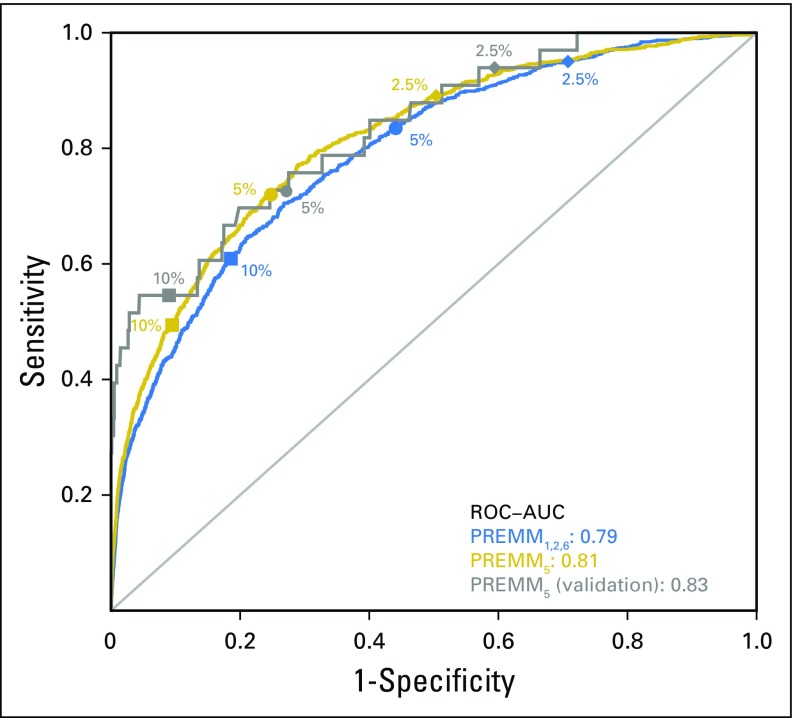
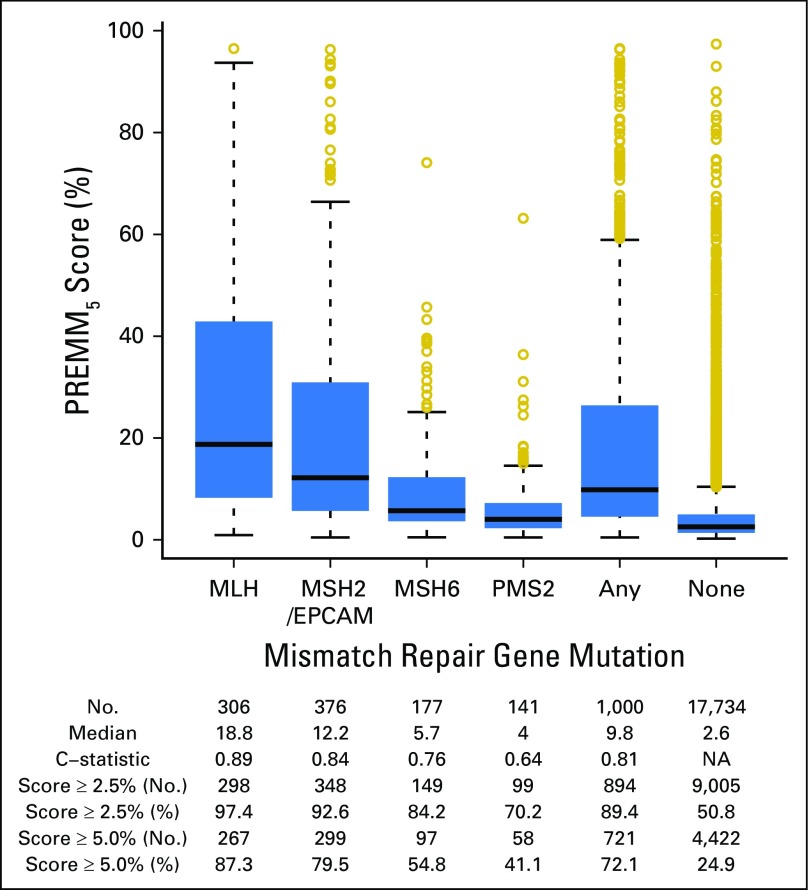
PREMM5 performed well in discriminating carriers from noncarriers; the optimism-corrected AUC was 0.81 (95% CI, 0.79 to 0.82; Fig 1). The polytomous multivariable model showed good discrimination for MLH1 (AUC, 0.89; 95% CI, 0.87 to 0.91), MSH2/EPCAM (AUC, 0.84; 95% CI, 0.82 to 0.86), and MSH6 (AUC, 0.76; 95% CI, 0.73 to 0.79) but less discrimination for PMS2 (AUC, 0.64; 95% CI, 0.60 to 0.68; Appendix Table A5, online only). PREMM5 had similar discrimination on external validation (AUC, 0.83; 95% CI, 0.75 to 0.92; Fig 1).
Fig 1.
Receiver operating characteristic (ROC) curves to discriminate mismatch repair mutation carriers from noncarriers for the PREMM5 and the PREMM1,2,6 models. AUC, area under the ROC curve.
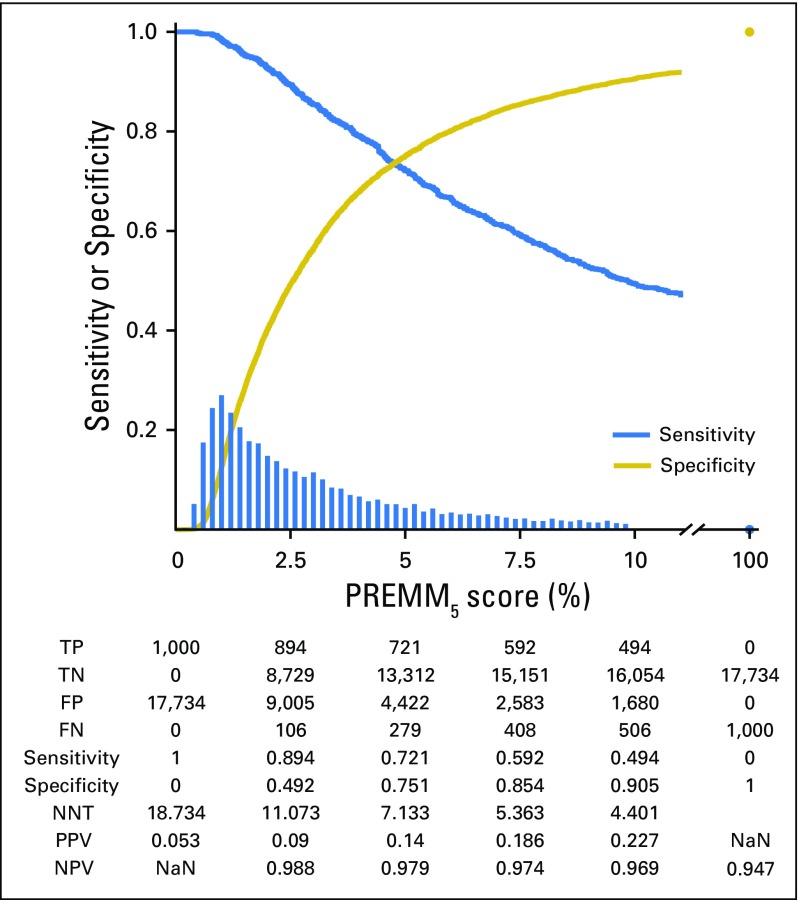
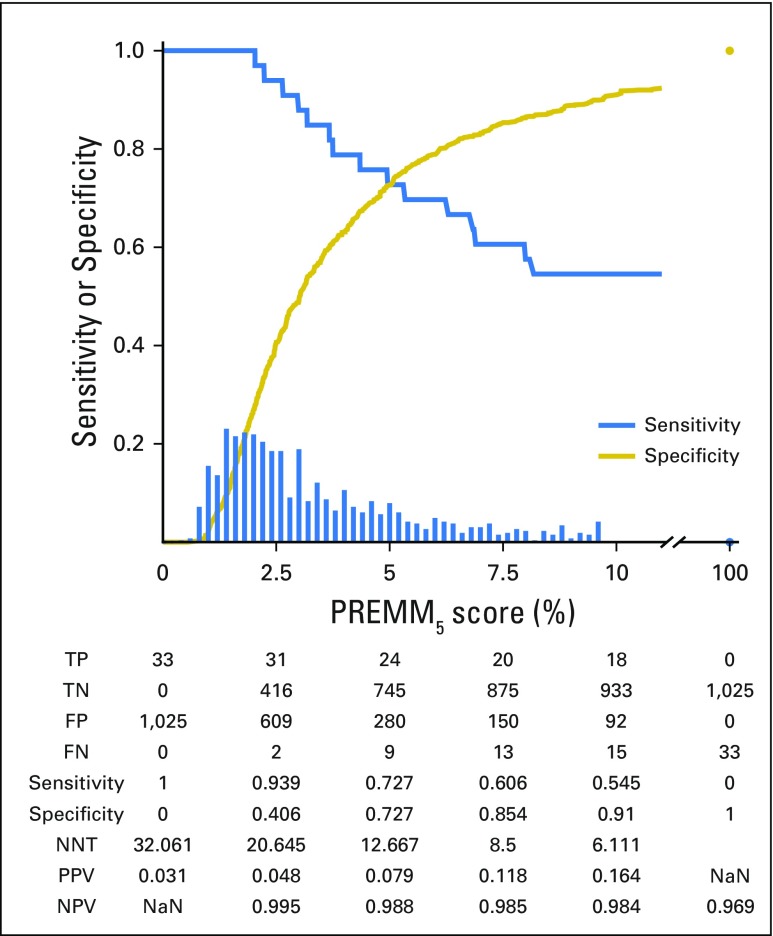
The median PREMM5 score for carriers was 9.8% versus 2.6% for noncarriers. At the recommended ≥ of 5% threshold, 721 of 1,000 carriers were identified (Fig 2) with variability by specific gene (Fig 3). A threshold ≥ 2.5% identified 894 of 1,000 carriers, but with lower specificity. The NNT to identify one carrier at 5% and 2.5% was seven and 11 individuals, respectively; a decrease from 19 needed to test if PREMM5 was not used. Between 2.5% and 10%, the negative predictive value was 97% to 99% of individuals correctly identified as mutation negative. Similar sensitivity, specificity, and NNT data were observed at external validation with 2.5% and 5.0% thresholds (Appendix Fig A1, online only).
Fig 2.
Performance characteristics of PREMM5. Decreasing sensitivity and increasing specificity are shown for increasing risk thresholds for the PREMM5 score, with a histogram for the distribution of predicted risks. FN, false negative; FP, false positive; NaN, not a number; NNT, number needed to test; NPV, negative predictive value; PPV, positive predictive value; TN, true negative; TP, true positive. FN, FP, TN, and TP are reported for a given cutoff value on the PREMM5 score. NNT= (TP + FP) / TP.
Fig 3.
Box plots of PREMM5 scores by specific gene mutation at model development. C-statistic: area under the receiver operating characteristic curve. Median values are represented by the horizontal lines in the boxes; 25th and 75th percentiles are represented by the lower and upper lines of the boxes, respectively. NA, not available.
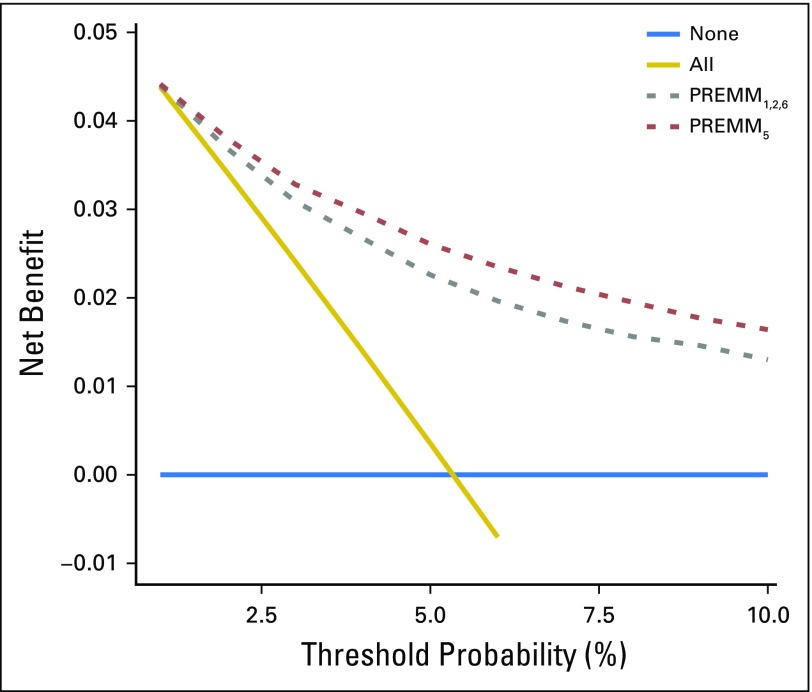
By decision curve analysis, the clinical impact of PREMM5 to identify individuals for germline testing was observed at thresholds ≥ 2.5%; maximal utility occurred at 5%. A threshold ≥ 2.5% to guide testing was superior compared with testing of all patients (Fig 4). PREMM5 had minimal clinical impact at less than 2.5%, because few individuals would be excluded from genetic testing at this threshold, so it was similar to a test-all approach.
Fig 4.
Net benefit curves for PREMM5 compared with PREMM1,2,6. The y-axis measures net benefit, which is calculated by summing the benefits (true positives) and subtracting the harms (false positives), in which the latter are weighted by a factor related to the relative harm of a missed mutation carrier compared with the harm of unnecessary genetic testing. A model is considered of clinical value if it has the highest net benefit compared with other models and simple strategies, such as performing genetic testing in all patients (gold line) or no patients (horizontal blue line) across the full range of threshold probabilities at which a patient would undergo genetic testing. For example, the net benefit of using PREMM5 to selectively test for mutation carriers exceeds that of PREMM1,2,6, and testing all at risk threshold ≥ 2.5%.
Performance Comparison of PREMM5 and PREMM1,2,6
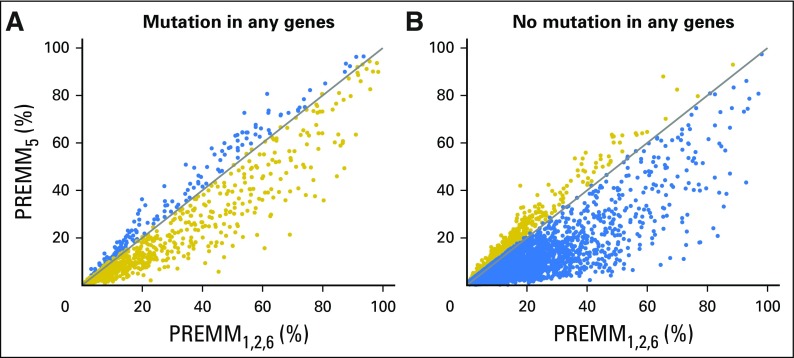
PREMM1,2,6 had an AUC of 0.83 (95% CI, 0.82 to 0.84) for the identification of carriers of MLH1, MSH2, or MSH6 among the 18,734 probands. Extending prediction to all five genes decreased discrimination to 0.79 (95% CI, 0.78 to 0.81), whereas PREMM5 prediction yielded an AUC of 0.81 (95% CI, 0.79 to 0.82). PREMM1,2,6 overpredicted mutation-positive status (ie, the observed mutation fraction for carriers of MLH1, MSH2, and MSH6 was 4.5% but the average PREMM1,2,6 prediction was 8.0% [O/E ratio, 0.557]). Reclassification plots confirmed overprediction by PREMM1,2,6 and showed considerable differences between PREMM5 and PREMM1,2,6 predictions (Appendix Fig A2, online only).
DISCUSSION
To improve the identification of carriers of LS mutations, we developed and validated the PREMM5 model, a risk assessment tool that uses readily ascertained clinical features (age, sex, and personal and family cancer history) to provide accurate and comprehensive risk estimation for all five associated genes. PREMM5 is a simple and efficient online tool for a diverse array of health care providers (eg, oncologists, gynecologists, gastroenterologists, and primary care practitioners) to rapidly identify individuals for germline testing for LS in routine clinical practice. The performance of the model is robust in quantifying an individual’s overall risk of carrying any pathogenic gene mutation, and its performance exceeds that of the previous model, PREMM1,2,6, which only predicted MLH1, MSH2, and MSH6 gene mutation status. We propose that PREMM5 should replace PREMM1,2,6 and that individuals with a ≥ 2.5% likelihood of LS undergo genetic testing.
Beyond the use of prediction models such as PREMM5, the primary strategy to identify individuals with LS involves screening CRC and endometrial cancer tumor specimens for microsatellite instability or deficient MMR protein expression, followed by germline testing of individuals who have suggestive tumor testing results.2,3 The limitation of tumor testing is that it is only relevant to patients with cancer, so the opportunity for cancer prevention in the proband has inherently been lost. Prior versions of PREMM and other LS models (eg, MMRpro, MMRpredict) were developed and validated in cohorts predominated by patients with cancer,17,18,31-35 and the performance of these models in unaffected patients is poorly understood. In this study, 46% of the development cohort (including 20% of LS mutation carriers) were unaffected but had a family history of LS-associated cancer, which supports the potential use of PREMM5 as a risk assessment tool for unaffected individuals.
Despite existing recommendations for universal molecular tumor testing of all colorectal and endometrial cancers to screen for LS, the majority of the nearly 1 million carriers in the United States remain unidentified.1 Use of the PREMM5 model can ameliorate the gap that exists between those identified through tumor testing and the vast majority of undiagnosed carriers of LS, many of whom are unaffected by cancer. Therefore, systematic implementation of PREMM5 can be considered by providers in routine clinical care, including primary and preventive health care, for individuals with a personal or family history of LS-associated cancers. Also, the PREMM5 model can be used in individuals who have colorectal and endometrial cancer when molecular tumor testing is unavailable or when resources are limited and universal tumor testing cannot be adopted.
The cohort used to develop PREMM5 was far larger than those used for older prediction models, including PREMM1,2,6. We examined data on 1,000 mutation carriers, in which 32% carried pathogenic PMS2 and MSH6 mutations, the less penetrant but more prevalent MMR genes associated with LS.36 Although PREMM5 performs well estimating individuals’ overall risk of carrying any MMR gene mutation, variability exists in estimates for each individual gene. Reliable prediction of PMS2 was more difficult than other genes because of a weaker phenotype—carriers of PMS2 were older at CRC diagnoses and had fewer cancers among relatives compared with other families with LS.37 This is in line with studies of carriers of the PMS2 gene mutation, in which retention of mRNA expression resulted in a milder phenotype in patients who were older at the time of their CRC diagnosis and/or had no family history of CRC.38 The performance measures associated with PREMM5 were comparable in the cohort used for external validation. A strength of the validation data set is that it minimized selection bias; patients had CRC but were not selected for genetic testing for LS because of features suggestive of the condition, such as young age of diagnosis, fulfillment of clinical criteria, or results of MMR tumor testing.
The large size of the development cohort allowed us to reassess previous predictors of mutation carrier status, such as sex of the individual tested. The frequency of pathogenic mutations differed between men and women (11% v 4% mutation prevalence, respectively), despite the presence of more women (73%) in the data set. This is consistent with prior reports6 and may be due to unmeasured selection bias in the development cohort. The sex distribution was more even in the validation cohort and did not alter the ability of the model to discern mutation carriers from noncarriers.
Incorporation of age at genetic testing improved risk estimation with PREMM5; with every decade increase in age, the likelihood an individual would carry a pathogenic mutation decreased by 32%. This refinement compared with PREMM1,2,6 is relevant to individuals unaffected by cancer, because the lack of such history in a 75-year-old patient who undergoes genetic evaluation is strong evidence against LS, whereas the absence of personal cancer history is less reassuring in a 25-year-old patient.
At the currently recommended threshold of 5%, the PREMM5 model identifies fewer individuals than PREMM1,2,6 for genetic evaluation and has a higher likelihood than PREMM1,2,6 of mutation carrier detection. Compared with PREMM5, PREMM1,2,6 overpredicts an individual’s need to obtain genetic evaluation. With PREMM5 at a ≥ 5% threshold, seven patients would need to be tested to identify one mutation carrier, with a negative predictive value of 98% (Fig 1). However, a ≥ 2.5% PREMM5 score cutoff to guide LS genetic testing markedly increased the number of identified mutation carriers and preserved a high negative predictive value of 99% compared with the ≥ 5% cutoff endorsed by national guidelines for PREMM1,2,6.
With ongoing advances in next-generation DNA sequencing technologies, many commercial laboratories currently offer multigene hereditary cancer panels that provide simultaneous germline analysis of dozens of cancer risk genes. PREMM5 provides accurate and comprehensive risk assessment specifically for LS, so its performance must be examined when genetic testing includes an expanded panel of genes. Recent data to examine such panels have shown that individuals with PREMM1,2,6 scores ≥ 5% often have mutations in cancer susceptibility genes beyond those linked to LS, including APC, MUTYH, BRCA1, and BRCA2.39 These results suggest that prediction models such as PREMM5 may identify individuals who may have underlying mutations in a wide spectrum of syndromes, rather than just LS.
In conclusion, PREMM5 provides comprehensive LS risk estimation for all five genes, including PMS2 and EPCAM. PREMM5 identifies fewer high-risk individuals for genetic evaluation and testing who have a higher likelihood of carrying a pathogenic mutation compared with PREMM 1,2,6. These analyses support the use of a ≥ 2.5% threshold to identify individuals suitable for genetic evaluation for LS; this threshold optimizes identification of carriers of gene mutations, including those who carry MMR genes associated with a weaker phenotype or who are unaffected by cancer but have a family history suggestive of an inherited CRC syndrome.
Appendix
Equation for the PREMM5 Model
We encourage researchers who use this equation to contact the PREMM investigators for guidance on the appropriate application. Variables are explained in detail in Appendix Table A1. The use of the term “PREMM” is protected by a servicemark.
Predicted probability of any mismatch repair gene mutation: p(any) = predicted probability of MLH1 mutation + predicted probability of MSH2/EPCAM mutation + predicted probability of MSH6 mutation + predicted probability of PMS2 mutation
Predicted probability of a mutation in MLH1: p(MLH1)
Predicted probability of a mutation in MSH2 or EPCAM: p(MSH2/EPCAM)
Predicted probability of a mutation in MSH6: p(MSH6)
Predicted probability of a mutation in PMS2: p(PMS2)
Predicted probability of no mutation: p(none)
p(none) = 1 − [p(MLH1) + p(MSH2/EPCAM) + p(MSH6) + p(PMS2)]
p(MLH1) = exp (lp(MLH1)) / [(1 + exp(lp(MLH1)) + exp(lp(MSH2/EPCAM)) + exp (lp(MSH6)) + exp(lp(PMS2))]
p(MSH2/EPCAM) = exp(lp(MSH2)) / [(1 + exp(lp(MLH1)) + exp(lp(MSH2)) + exp(lp(MSH6) + exp(lp(PMS2))]
p(MSH6) = exp(lp(MSH6)) / [(1 + exp(lp(MLH1)) + exp(lp(MSH2)) + exp(lp(MSH6)) + exp(lp(PMS2))]
p(PMS2) = exp(lp(PMS2)) / [(1 + exp(lp(MLH1)) + exp(lp(MSH2)) + exp(lp(MSH6)) + exp(lp(PMS2))]
lp(MLH1) = −5.325 + (0.904 × V0) + (2.586 × V1) + (3.183 × V2) + (1.621 × V3) + (1.276 × V4) + (1.560 × V5) + (0.804 × V6) + (0.397 × V7) + (−0.0557 × V8) + (0.0115 × V9) + (−0.0476 × V10);
lp(MSH2/EPCAM) = −4.427 + (0.937 × V0) + (1.799 × V1) + (2.593 × V2) + (1.924 × V3) + (1.585 × V4) + (1.337 × V5) + (0.670 × V6) + (0.607 × V7) + (−0.0441 × V8) + (0.0002 × V9) + (−0.0482 × V10);
lp(MSH6) = −4.675 + (0.816 × V0) + (1.265 × V1) + (−53.205 × V2) + (1.759 × V3) + (0.538 × V4) + (0.545 × V5) + (0.923 × V6) + (0.313 × V7) + (−0.0095 × V8) + (0.0344 × V9) + (−0.0363 × V10);
lp(PMS2) = −4.913 + (0.294 × V0) + (0.989 × V1) + (−0.354 × V2) + (0.739 × V3) + (0.395 × V4) + (−0.002 × V5) + (−0.426 × V6) + (−0.105 × V7) + (−0.0086 × V8) + (0.0008 × V9) + (−0.0074 × V10);
Table A1.
Definitions for Variables of the PREMM5 Model Equation
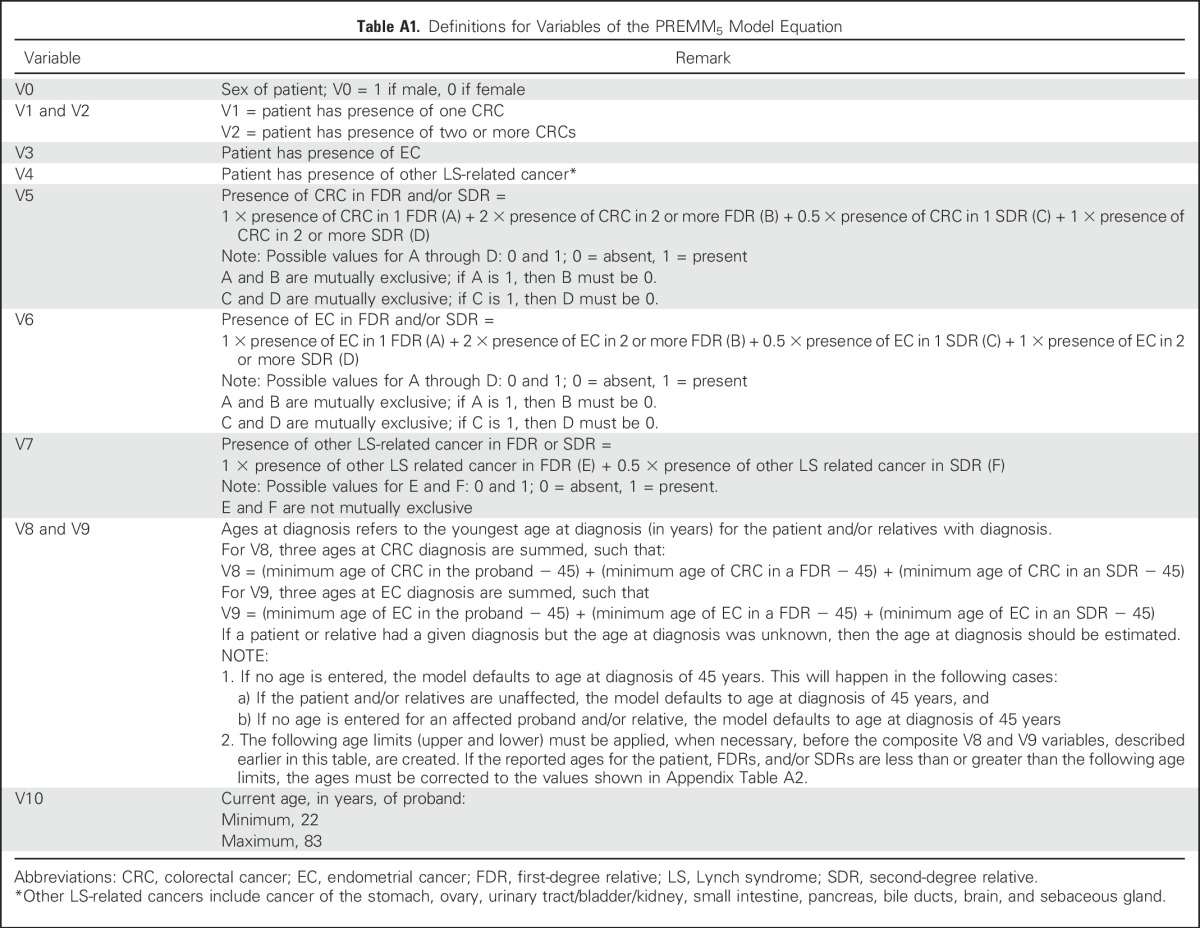
Table A2.
Ages at Diagnoses of CRC and EC in Calculation of Overall Risk Estimate
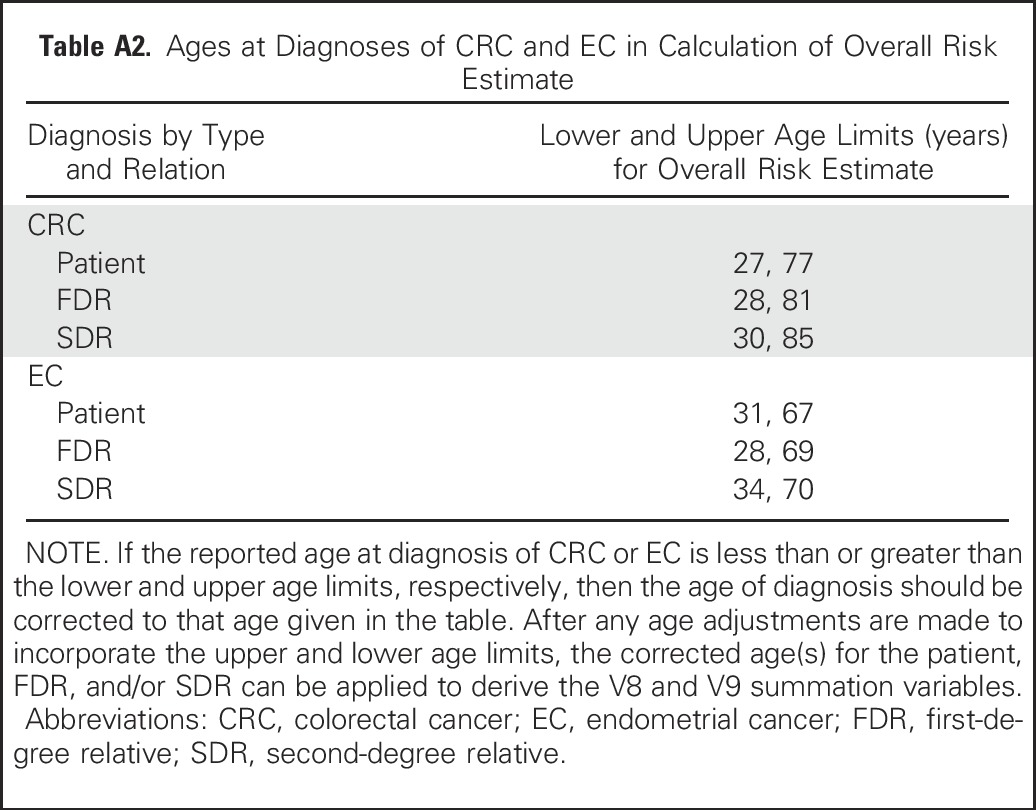
Table A3.
Age of Diagnosis Among Gene Mutation Carriers and Noncarriers in the Development Cohort
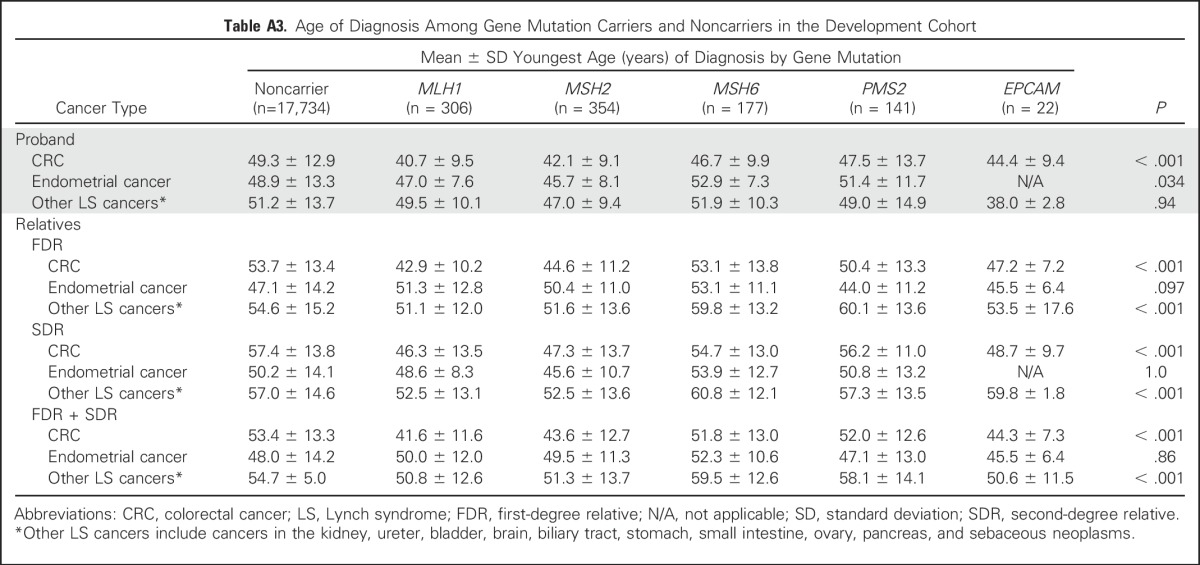
Table A4.
Clinical Characteristics of Individuals With Colorectal Cancer in the External Validation Cohort
Table A5.
Discrimination of PREMM5 for Any MMR Gene and by Specific Gene
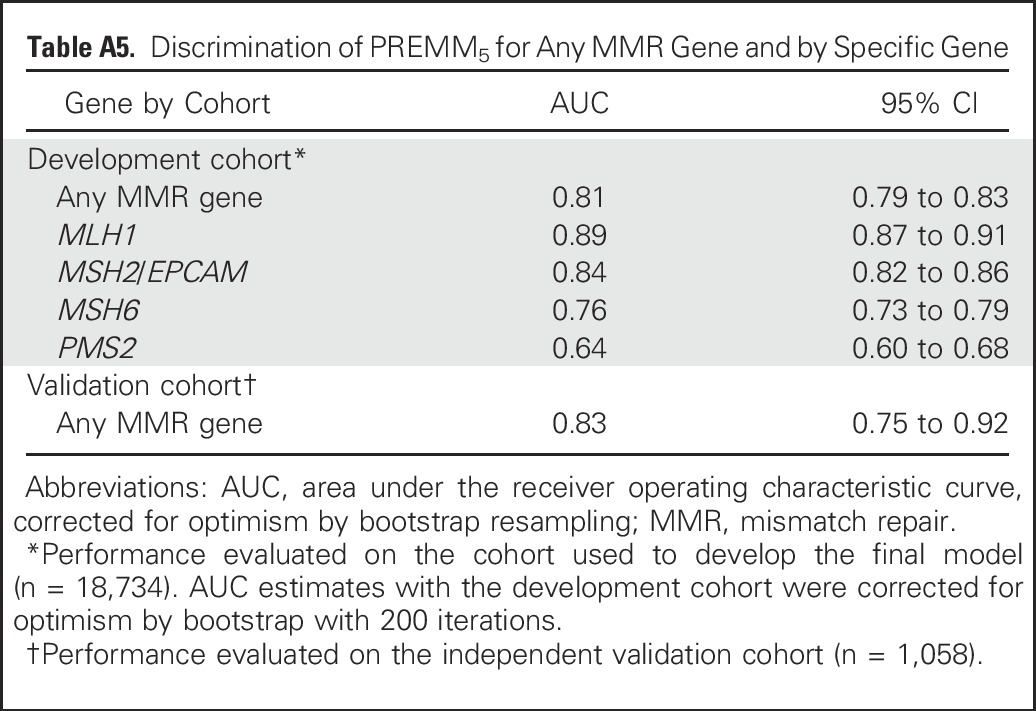
Fig A1.
Performance characteristics of PREMM5: validation cohort. FN, false negative; FP, false positive; NNT, number needed to test; NPV, negative predictive value; PPV, positive predictive value; TN, true negative; TP, true positive.
Fig A2.
Reclassification plots for the comparison of PREMM1,2,6 with PREMM5. The scatterplot diagrams show the added predictive value provided by the PREMM5 model in identification of (A) mutation carriers (mutation in any gene) and (B) noncarriers (no mutation in any gene) on the basis of the data from this study. (A) PREMM5 predicts mutation carriers, as shown by patients (blue circles) above the line. This is regardless of the mutation carrier status of the patient. (B) Compared with PREMM1,2,6, the extended PREMM5 model better predicts noncarriers, as shown by more patients (blue circles) below the line of identity (diagonal line). Gold circles indicate reclassification by PREMM5, in which, on average, the extended model provided lower risk estimates than PREMM1,2,6.
Footnotes
Supported by Grants No. R01CA132829 and K24CA113433 (to S.S.), and K07CA151769 (to F.K.) and by the Louis V. Gerstner Jr. Scholars Program (F.K.).
AUTHOR CONTRIBUTIONS
Conception and design: Fay Kastrinos, Matthew H. Kulke, Deborah Schrag, Jeffrey A. Meyerhardt, Robert J. Mayer, Kimmie Ng, Sapna Syngal
Provision of study materials or patients: Charles S. Fuchs
Collection and assembly of data: Fay Kastrinos, Chinedu Ukaegbu, Carmelita Alvero, Ashley McFarland, Matthew H. Kulke, Deborah Schrag, Jeffrey A. Meyerhardt, Charles S. Fuchs, Robert J. Mayer, Kimmie Ng
Data analysis and interpretation: Fay Kastrinos, Hajime Uno, Chinedu Ukaegbu, Carmelita Alvero, Ashley McFarland, Matthew B. Yurgelun, Ewout W. Steyerberg, Sapna Syngal
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Development and Validation of the PREMM5 Model for Comprehensive Risk Assessment of Lynch Syndrome
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Fay Kastrinos
No relationship to disclose
Hajime Uno
Consulting or Advisory Role: Ion Pharma
Chinedu Ukaegbu
No relationship to disclose
Carmelita Alvero
No relationship to disclose
Ashley McFarland
No relationship to disclose
Matthew B. Yurgelun
Research Funding: Myriad Genetics
Matthew H. Kulke
No relationship to disclose
Deborah Schrag
Consulting or Advisory Role: New Century Health, Ohio State University
Other Relationship: Journal of the American Medical Association
Jeffrey A. Meyerhardt
Consulting or Advisory Role: Genentech
Charles S. Fuchs
Consulting or Advisory Role: Genentech (a Roche group), Lilly, Sanofi, Bayer, Celgene, Merck, Bristol-Myers Squibb, Entrinic Health, Five Prime Therapuetics, Agios
Robert J. Mayer
Honoraria: Taiho Pharmaceutical
Consulting or Advisory Role: CASI Pharmaceuticals
Kimmie Ng
Consulting or Advisory Role: Genentech, Lilly, Tarrex
Research Funding: Gilead Sciences, Travegene, Tarrex
Ewout W. Steyerberg
Patents, Royalties, Other Intellectual Property: Royalties from Springer for book on prediction models
Sapna Syngal
No relationship to disclose
REFERENCES
- 1.Blue Ribbon Panel Cancer Moonshot Blue Ribbon Panel Report 2016. https://www.cancer.gov/research/key-initiatives/moonshot-cancer-initiative/blue-ribbon-panel-report-2016.pdf
- 2.Giardiello FM, Allen JI, Axilbund JE, et al. Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US Multi-Society Task Force on colorectal cancer. Gastroenterology. 2014;147:502–526. doi: 10.1053/j.gastro.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Syngal S, Brand RE, Church JM, et al. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol. 2015;110:223–262, quiz 263. doi: 10.1038/ajg.2014.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jasperson KW, Tuohy TM, Neklason DW, et al. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenkins MA, Baglietto L, Dowty JG, et al. Cancer risks for mismatch repair gene mutation carriers: A population-based early onset case-family study. Clin Gastroenterol Hepatol. 2006;4:489–498. doi: 10.1016/j.cgh.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Stoffel E, Mukherjee B, Raymond VM, et al. Calculation of risk of colorectal and endometrial cancer among patients with Lynch syndrome. Gastroenterology. 2009;137:1621–1627. doi: 10.1053/j.gastro.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engel C, Loeffler M, Steinke V, et al. Risks of less common cancers in proven mutation carriers with Lynch syndrome. J Clin Oncol. 2012;30:4409–4415. doi: 10.1200/JCO.2012.43.2278. [DOI] [PubMed] [Google Scholar]
- 8.Win AK, Young JP, Lindor NM, et al. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: A prospective cohort study. J Clin Oncol. 2012;30:958–964. doi: 10.1200/JCO.2011.39.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Järvinen HJ, Aarnio M, Mustonen H, et al. Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 2000;118:829–834. doi: 10.1016/s0016-5085(00)70168-5. [DOI] [PubMed] [Google Scholar]
- 10.de Jong AE, Hendriks YM, Kleibeuker JH, et al. Decrease in mortality in Lynch syndrome families because of surveillance. Gastroenterology. 2006;130:665–671. doi: 10.1053/j.gastro.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 11.Vasen HF, Abdirahman M, Brohet R, et al. One to 2-year surveillance intervals reduce risk of colorectal cancer in families with Lynch syndrome. Gastroenterology. 2010;138:2300–2306. doi: 10.1053/j.gastro.2010.02.053. [DOI] [PubMed] [Google Scholar]
- 12.Lindor NM, Petersen GM, Hadley DW, et al. Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: A systematic review. JAMA. 2006;296:1507–1517. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
- 13.Vasen HF, Blanco I, Aktan-Collan K, et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): Recommendations by a group of European experts. Gut. 2013;62:812–823. doi: 10.1136/gutjnl-2012-304356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ait Ouakrim D, Dashti SG, Chau R, et al. Aspirin, ibuprofen, and the risk of colorectal cancer in Lynch syndrome. J Natl Cancer Inst. 2015;107:djv170. doi: 10.1093/jnci/djv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kastrinos F, Steyerberg EW, Mercado R, et al. The PREMM1,2,6 model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology. 2011;140:73–81. doi: 10.1053/j.gastro.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Comprehensive Cancer Network Guidelines for detection, prevention, and risk reduction: Genetic/Familial High-Risk Assessment: Colorectal (Version1.2016) https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf
- 17.Barnetson RA, Tenesa A, Farrington SM, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006;354:2751–2763. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Wang W, Lee S, et al. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA. 2006;296:1479–1487. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balmaña J, Stockwell DH, Steyerberg EW, et al. Prediction of MLH1 and MSH2 mutations in Lynch syndrome. JAMA. 2006;296:1469–1478. doi: 10.1001/jama.296.12.1469. [DOI] [PubMed] [Google Scholar]
- 20.Steyerberg EW, Balmaña J, Stockwell DH, et al. Data reduction for prediction: A case study on robust coding of age and family history for the risk of having a genetic mutation. Stat Med. 2007;26:5545–5556. doi: 10.1002/sim.3119. [DOI] [PubMed] [Google Scholar]
- 21.Marshall A, Altman DG, Holder RL, et al. Combining estimates of interest in prognostic modelling studies after multiple imputation: Current practice and guidelines. BMC Med Res Methodol. 2009;9:57–64. doi: 10.1186/1471-2288-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ligtenberg MJ, Kuiper RP, Geurts van Kessel A, et al. EPCAM deletion carriers constitute a unique subgroup of Lynch syndrome patients. Fam Cancer. 2013;12:169–174. doi: 10.1007/s10689-012-9591-x. [DOI] [PubMed] [Google Scholar]
- 23. Steyerberg EW: Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. New York, NY, Springer, 2009. doi: 10.1007/978-0-387-77244-8. [Google Scholar]
- 24.Steyerberg EW, Vergouwe Y. Towards better clinical prediction models: Seven steps for development and an ABCD for validation. Eur Heart J. 2014;35:1925–1931. doi: 10.1093/eurheartj/ehu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickers AJ, Elkin EB. Decision curve analysis: A novel method for evaluating prediction models. Med Decis Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vickers AJ, Cronin AM. Traditional statistical methods for evaluating prediction models are uninformative as to clinical value: Towards a decision analytic framework. Semin Oncol. 2010;37:31–38. doi: 10.1053/j.seminoncol.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Localio AR, Goodman S. Beyond the usual prediction accuracy metrics: Reporting results for clinical decision making. Ann Intern Med. 2012;157:294–295. doi: 10.7326/0003-4819-157-4-201208210-00014. [DOI] [PubMed] [Google Scholar]
- 29.Austin PC, Steyerberg EW. Graphical assessment of internal and external calibration of logistic regression models by using loess smoothers. Stat Med. 2014;33:517–535. doi: 10.1002/sim.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dana-Farber Cancer Institute: PREMM5 Model: Lynch syndrome prediction model for MLH1, MSH2, MSH6, PMS2 and EPCAM gene mutations. http://premm.dfci.harvard.edu/
- 31.Balaguer F, Balmaña J, Castellví-Bel S, et al. Validation and extension of the PREMM1,2 model in a population-based cohort of colorectal cancer patients. Gastroenterology. 2008;134:39–46. doi: 10.1053/j.gastro.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercado RC, Hampel H, Kastrinos F, et al. Performance of PREMM(1,2,6), MMRpredict, and MMRpro in detecting Lynch syndrome among endometrial cancer cases. Genet Med. 2012;14:670–680. doi: 10.1038/gim.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan O, Blanco A, Conrad P, et al. Performance of Lynch syndrome predictive models in a multi-center US referral population. Am J Gastroenterol. 2011;106:1822–1827, quiz 1828. doi: 10.1038/ajg.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kastrinos F, Ojha RP, Leenen C, et al. Comparison of prediction models for Lynch syndrome among individuals with colorectal cancer. J Natl Cancer Inst. 2015;108:djv308. doi: 10.1093/jnci/djv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Win AK, Macinnis RJ, Dowty JG, et al. Criteria and prediction models for mismatch repair gene mutations: A review. J Med Genet. 2013;50:785–793. doi: 10.1136/jmedgenet-2013-101803. [DOI] [PubMed] [Google Scholar]
- 36. doi: 10.1158/1055-9965.EPI-16-0693. Win AK, Jenkins MA, Dowty JG, et al: Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol Biomarkers Prev doi: 10.1158/1055-9965.EPI-16-0693 [epub ahead of print on October 31, 2016] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senter L, Clendenning M, Sotamaa K, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suerink M, van der Klift HM, Ten Broeke SW, et al. The effect of genotypes and parent of origin on cancer risk and age of cancer development in PMS2 mutation carriers. Genet Med. 2016;18:405–409. doi: 10.1038/gim.2015.83. [DOI] [PubMed] [Google Scholar]
- 39.Yurgelun MB, Allen B, Kaldate RR, et al. Identification of a variety of mutations in cancer predisposition genes in patients with suspected Lynch syndrome. Gastroenterology. 2015;149:604–13.e20. doi: 10.1053/j.gastro.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]