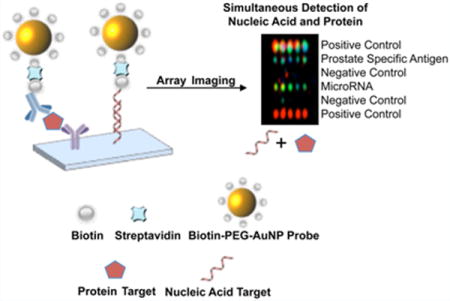
Abstract
Novel biotin—polyethylene glycol (biotin—PEG) gold nanoparticle probes have been synthesized and used as universal constructs for the detection of protein (prostate-specific antigen, PSA) and nucleic acid targets (microRNAs) from a single sample. Microarray assays based upon these probes enabled sensitive detection of biomarker targets (50 fM for nucleic acid targets and 1 pg/μL for the PSA target). Ways of detecting biomarkers, including nucleic acids and proteins, are necessary for the clinical diagnosis of many diseases, but currently available diagnostic platforms rely primarily on the independent detection of proteins or nucleic acids. In addition to the economic benefits associated with the use of a single platform to detect both classes of analytes, studies have shown that the simultaneous identification of multiple classes of biomarkers in the same sample could be useful for the detection and management of early stage diseases, especially when sample amounts are limited. Therefore, these new probes and the assays based upon them open the door for high-sensitivity combination-target assays for studying and tracking biological pathways and diseases.
Graphical abstract
INTRODUCTION
Molecular bioassays represent important diagnostic tools for the identification of disease-related biomarkers with significant research and clinical uses.1 Typical target biomolecules include nucleic acids (DNA,2 mRNA,3 and miRNA4) and proteins (antibodies and antigens)5 that are associated with states of disease, therapeutic treatment, or drug screening.6 Current diagnostic approaches such as the enzyme-linked immunosorbent assay (ELISA) enable the capture and detection of proteins through antibody—antigen binding interactions, and oligonucleotide microarray and sequencing technologies allow for the identification and discovery of specific nucleic acid targets, respectively. These methods have shown utility in early-stage disease detection.7–9
Despite the impressive capabilities of these conventional bioassays, there are limitations to current technologies. ELISA assays are typically only able to identify single types of molecules of interest.10 In addition, ELISA assays require larger volumes (mL) of reagents, and our assay uses much smaller sample volumes (μL). Nucleic acid detection by sequencing relies primarily on target amplification techniques that require multiple time-consuming processing steps along with challenges related to successful primer design and library generation.11,12 In addition, fluorophore-based nucleic acid detection probes may suffer from self-quenching,13 hydrolysis,14 and dye bleaching15 that lead to diminished assay sensitivity and misidentification of potential biomarkers.16 Beyond issues related to assay sensitivity, current techniques are typically only capable of detecting individual classes of biomarkers (i.e., nucleic acids or proteins)17 at a time. Both economic and medical arguments have been made for the need for multiplexed detection of nucleic acid and protein molecules in a single platform.18–24 To this end, a few advances have been made, including lateral flow devices,20 multifunctional biochips,21 microfluidic platforms,25 and combinatorial micro-arrays.18
In this work, a combinatorial assay that provides the ability to simultaneously detect multiple classes of biomolecules (nucleic acid and protein) has been developed. Gold nanoparticle (AuNP) conjugates have been used as alternatives to fluorophore-based probes,26 in part because they can be amplified through electroless metal deposition, which augments their ability to scatter light and enhances the sensitivity of the assay.16,27–39 The assay combines the high-throughput multiplexing capabilities of microarrays with biotin—PEG—linked gold nanoparticle probes to achieve a system that enables simultaneous detection of different classes of biomolecule targets (Scheme 1).
Scheme 1. Workflow for Simultaneous Capture and Detection of Nucleic Acid and Protein Target Molecules Using biotin—PEG AuNP Probesa.
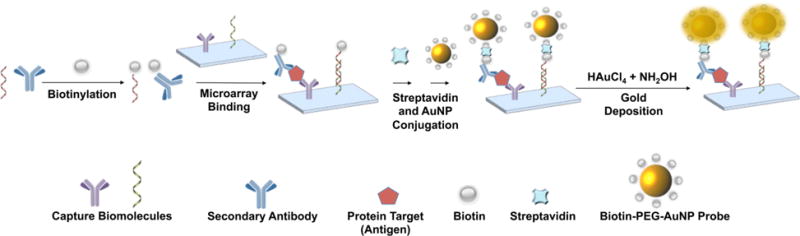
aBiomolecules are biotinylated and added to microarray slides containing capture molecules (complementary nucleic acid sequences or primary capture antibodies). Streptavidin and biotin—PEG—AuNPs are subsequently applied to the slide and then amplified with 1 mM HAuCl4 and 5 mM NH2OH to detect the captured targets.
In a typical assay, using a commercial kit, biomolecules from a sample of interest are conjugated to biotin and then added to array surfaces that are functionalized with capture antibodies (protein detection) and complementary nucleic acid sequences (nucleic acid detection) (Scheme 1). After slide washing, streptavidin and biotin—PEG—linked AuNP probes are subsequently added onto the microarray to detect the captured biomarker targets. To amplify the AuNP light-scattering signal intensity and modulate assay sensitivity, additional gold is further deposited onto the biotin—PEG—AuNPs using a solution of 1 mM HAuCl4 and 5 mM NH2OH. The array is finally imaged using a Tecan LS reloaded laser scanner at a wavelength of 633 nm to visualize the array surface and identify the captured biomarker targets.
RESULTS
Thiol—PEG—Biotin Linker Synthesis
Novel biotin—PEG linkers with deliberately varied numbers of anchors (monothiol, cyclic disulfide, and trithiol) were synthesized for subsequent conjugation to spheril gold nanoparticles through gold—thiol adsorption chemistry40–43(Figure 1). The use of molecular linkers containing multiple anchoring groups has been shown to increase AuNP stability in biological environments through resistance to linker displacement by free thiols44,45 (Figure S1). As such, we chose to incorporate multianchor linkers in the context of AuNP probes for the dual target class biomarker detection assay. Monothiol biotin—PEG linkers (MW: 5000 Da) were purchased from Nanocs Inc. (Figure 1a) and used without further modification to synthesize monothiol—biotin—AuNP probes, and cyclic disulfide—PEG—biotin (Figure 1b) and trithiol—PEG—biotin linkers (Figure 1c) were synthesized in-house from commercially available reagents. Detailed linker synthesis protocols are included in the Materials and Methods section below.
Figure 1.
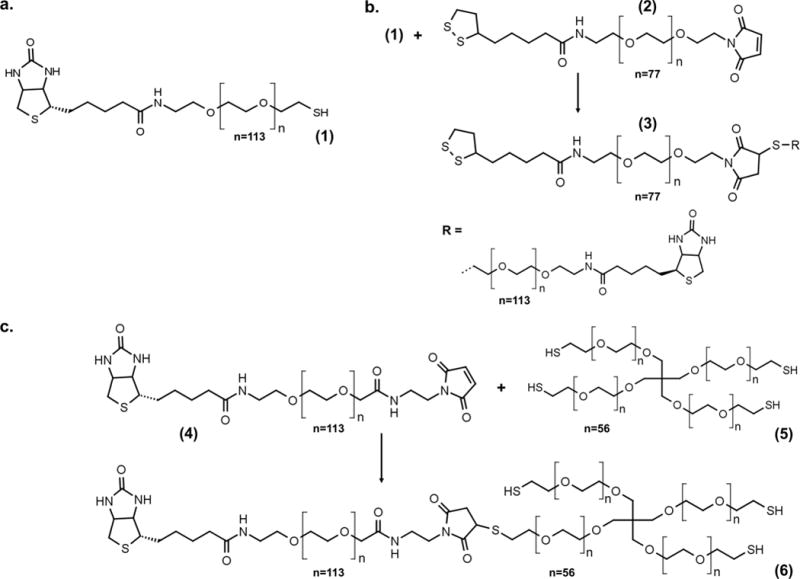
Synthesis of biotin—PEG linkers for AuNP probe conjugation. (a) Monothiol linker. (b) One-step synthesis of cyclic disulfide linker. (c) One-step synthesis of trithiol linker.
MicroRNA Detection Using Biotin-AuNP Probes
Biotin—PEG—AuNP probes with different thiol functionalities (monothiol, dithiol, and trithiol) were synthesized by adsorbing the linkers to the gold nanoparticle surface (Figure 2a–c). The functionality of the bioassay was initially characterized using synthetic miRNA target sequences before moving forward to simultaneous nucleic acid and protein detection experiments (oligonucleotide sequence information is included in Table S1). A target miRNA sequence (denoted as miRNA-1) was synthesized with a 3′ biotinylation modification for subsequent binding and detection using streptavidin and biotin—PEG—AuNP probes, respectively. Monothiol, dithiol, and trithiol biotin —PEG—AuNP probes were first used to investigate the ability of these probes to specifically detect miRNA-1.
Figure 2.
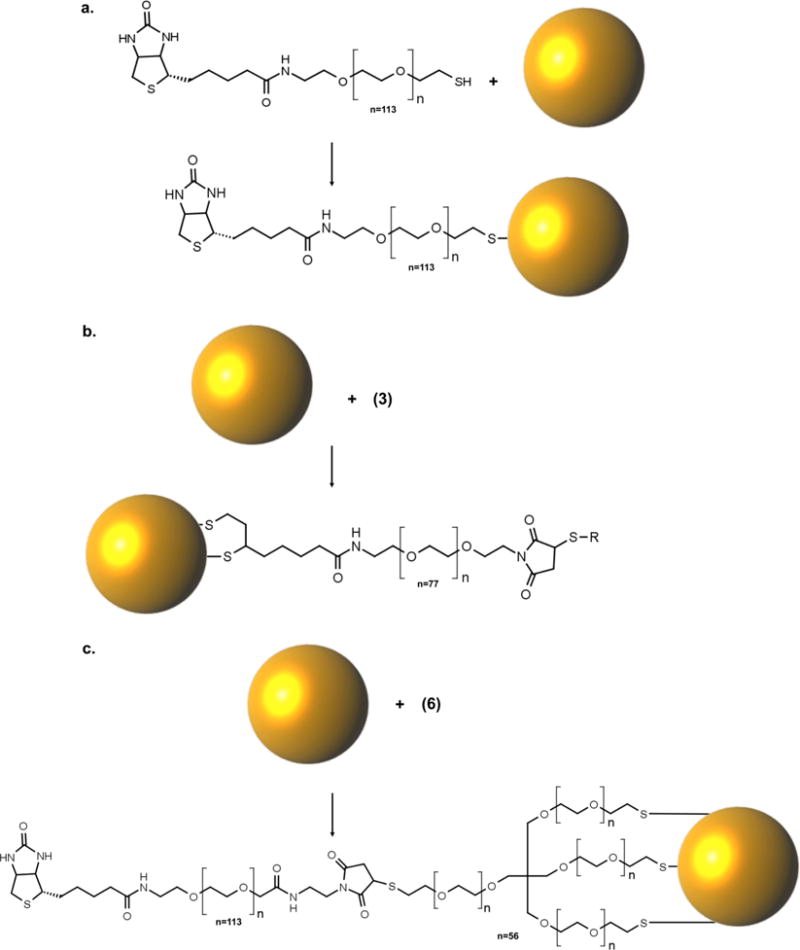
Biotin—PEG gold nanoparticle probe functionalization. (a) Conjugation of monothiol biotin—PEG linker to AuNP surface. (b) Conjugation of disulfide biotin—PEG linker to the AuNP surface. (c) Conjugation of trithiol biotin—PEG linker to AuNP surface.
Array slide surfaces were prepared using 3′ amine terminal DNA capture oligonucleotides complementary to the miRNA target. These DNA sequences were synthesized in-house, purified by reverse-phase high-performance liquid chromatography (RP-HPLC), and adsorbed onto CodeLink activated NHS-modified glass slides (Surmodics). Next, the biotinylated miRNA target was hybridized onto the capture oligonucleotides. After stringency washes at 54°C to remove unbound miRNA, streptavidin and biotin—PEG—AuNP probes were subsequently added onto the microarray to label the captured miRNA targets. To amplify the light-scattering signal intensity, additional gold was deposited onto the biotin—PEG—AuNP using HAuCl4 and NH2OH.16,27 The microarray slide was finally imaged using a Tecan LS reloaded laser scanner at a wavelength of 633 nm, and the images were transferred to GenePix software for analysis. A control well was also included that contained the glass slide-bound capture DNA oligonucleotide and biotin—PEG—AuNP probe in buffer solution (no miRNA target was added to this well).
Each biotin—PEG—AuNP probe (monothiol, dithiol, and trithiol) was able to detect the miRNA target. Robust light scattering signal from the bound biotin—PEG—AuNP probe was present for each nanoparticle probe type, as shown in Figure 3a–d. Importantly, nonspecific binding between the biotin—PEG— AuNP probe, DNA capture sequence, and glass surface was undetectable (Figure 3a). Quantitative analysis of the array slide showed that the light scattering signal intensity increased with increasing numbers of nanoparticle surface-bound anchors (monothiol, dithiol, and trithiol) (Figure 3e). The biotin—PEG linkers used for each AuNP probe vary in molecular weight (monothiol: 5 kDa, cyclic disulfide: 8.4 kDa, trithiol: 15 kDa), and we hypothesize that this difference in signal intensity is due to decreased steric hindrance (lower biotin—PE— surface loading) with increasing linker anchors46 that ultimately enhances nanoparticle-target binding. In addition, biotin—PEG—AuNP probes were also synthesized using different sizes of gold nanoparticles (15 to 80 nm in diameter), and all of the probes demonstrated functionality in detecting biotinylated miRNA (Figure S2).
Figure 3.
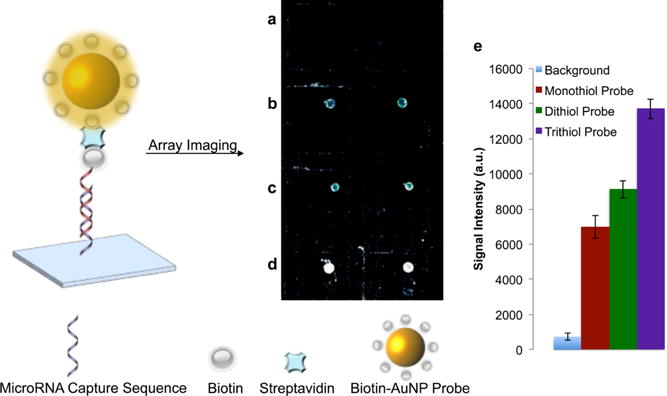
MicroRNA detection using biotin—PEG—AuNP probes. (a) Control well containing biotin—PEG—AuNP probe and complementary capture oligonucleotide (without miRNA target). (b) Monothiol—PEG—AuNP probe. (c) Dithiol—PEG—AuNP probe. (d) Trithiol—PEG—AuNP probe. (e) Signal intensity values generated from biotin—PEG—AuNP probes.
An additional miRNA detection study was conducted to characterize the ability of the assay to detect multiple miRNA sequences simultaneously. In addition, we investigated the sensitivity of the assay toward miRNA targets. Monothiol biotin—PEG—AuNP probes were used for the remaining studies due to their ease of synthesis and satisfactory assay performance. A total of four distinct miRNA sequences (sequence information provided in Table S1) were synthesized and functionalized with 3′ biotin modifications for subsequent detection using strep-tavidin and biotin—PEG—AuNP probes. After RP-HPLC purification, the biotinylated miRNA strands were hybridized onto a 7th generation miRCURY locked nucleic acid (LNA) microarray (Exiqon) that contains LNA and DNA capture sequences for the four biotinylated miRNA targets of interest. After stringency washes to remove unbound miRNA, streptavidin and biotin—PEG—AuNP probes were added onto the microarray to label the captured miRNA targets. Gold deposition and microarray imaging were performed as previously described.
The bioassay enabled simultaneous capture of all four miRNA targets at concentrations of 1 μM(Figure 4a), 10 pM (Figure 4b), and 1 pM (Figure 4c). Light-scattering signals from the biotin—PEG—AuNP probes after a single gold deposition were visualized at each concentration. Next, the sensitivity of the assay was investigated using the four different miRNA targets at various concentrations (5 pM, 1 pM, 200 fM, and 50 fM), as shown in panels d and e of Figure 4. In addition, quantification of the light-scattering signal generated by the biotin—PEG—AuNP probes is included in Figure 4f. Importantly, with this assay, one can detect and differentiate target concentrations from 50 fM to 5 pM. The large range of detection (fM—pM) along with a sensitive assay limit of detection (50 fM) enabled by these biotin—PEG—AuNP probes demonstrates the versatility of the assay and prompted us to investigate detection of nucleic acid and protein targets, simultaneously.
Figure 4.
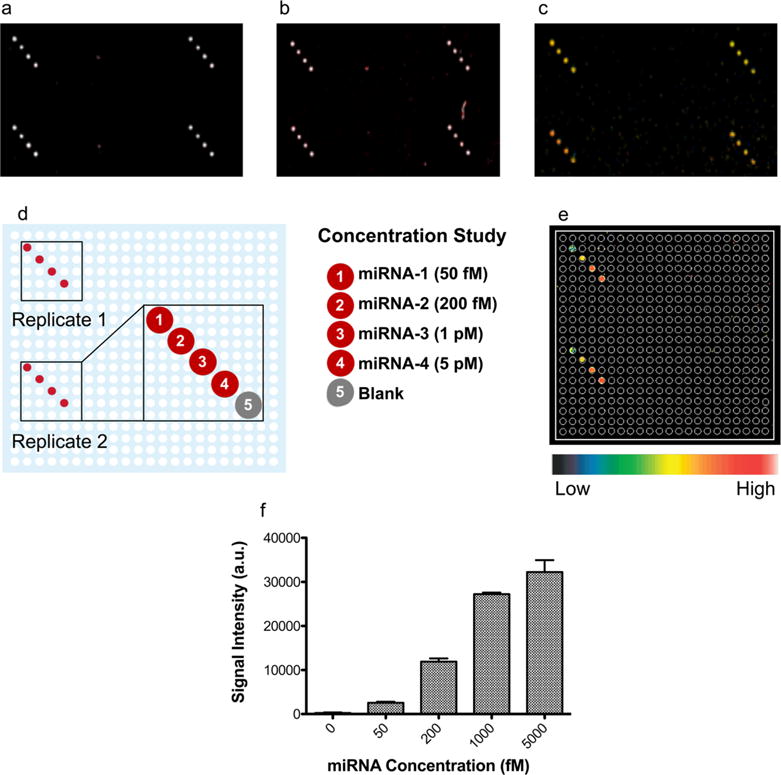
Analysis of bioassay performance and sensitivity for miRNA detection. Simultaneous visualization of four distinct miRNA sequences at (a) 1 μM, (b) 10 pM, and (c) 1 pM concentrations. (d) Layout of microarray subgrid showing location of miRNA capture sites. (e) Concentration-dependent detection of four distinct miRNA targets. (f) Light scattering signal quantification of biotin—PEG—AuNPs bound to miRNA targets from (e).
Simultaneous and Combinatorial miRNA and Protein Detection
We used our previously studied miRNA target sequence (miRNA-1) and prostate-specific antigen (PSA, Bios-Pacific) as target molecules for this proof-of-concept experiment. PSA was chosen as a well-studied and extensively characterized protein marker that has primary and secondary monoclonal antibodies that are commercially available (Bios-Pacific). Detection of miRNA and protein antigen was performed using slides obtained from Full Moon BioSystems that were printed with arrays of amine-modified capture DNA oligonucleotides (10 nmol) and PSA-specific monoclonal antibody (100 ng/100 μL, BiosPacific) along with positive-control biomolecules (biotinylated bovine serum albumin (BSA) and biotinylated PEG) and negative-control biomolecules (BSA and a DNA sequence that is not complementary to miRNA-1). All of the capture molecules were printed on the slide in the order shown in Figure 5.
Figure 5.
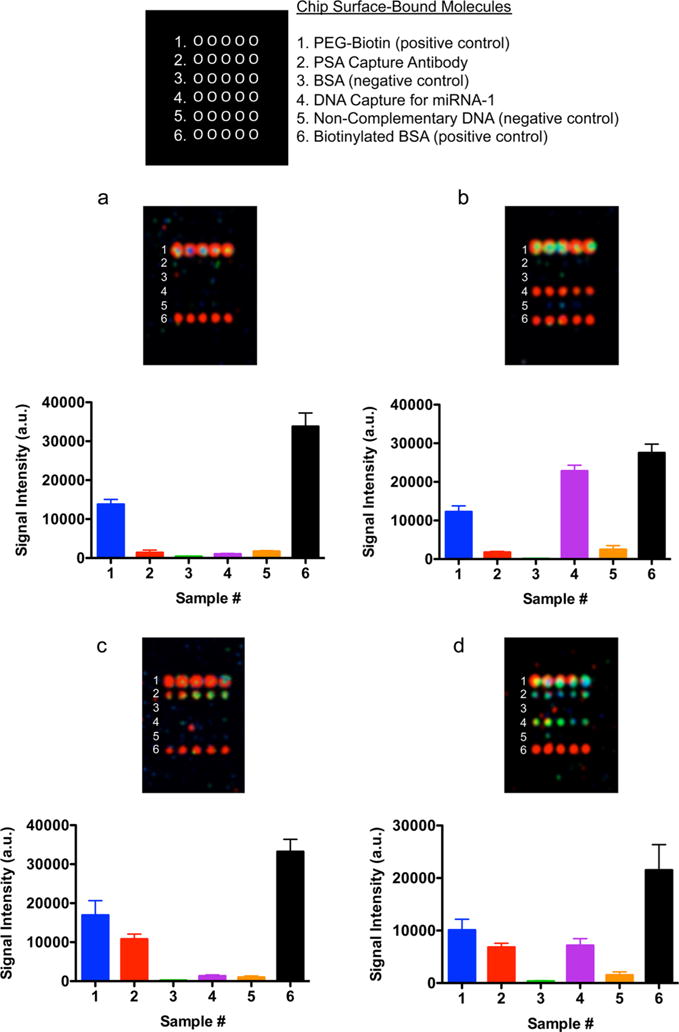
Simultaneous detection of nucleic acid and protein targets using biotin—PEG—AuNP probes. (a) Detection of positive control molecules (PEG—biotin and biotinylated BSA); 3σ threshold = 315 au. (b) Detection of miRNA and positive control molecules (PEG—biotin and biotinylated BSA);, 3σ threshold = 216 au. (c) Detection of PSA protein and positive control molecules (PEG—biotin and biotinylated BSA); 3σ threshold = 105 au. (d) Simultaneous detection of miRNA, PSA, and positive control molecules (PEG—biotin and biotinylated BSA); 3σ threshold = 339 au.
The assay enabled the detection of miRNA, protein antigen, and miRNA and antigen in combination simultaneously on one microarray slide. An assay control well without the addition of miRNA or PSA (Figure 5a) displayed light-scattering signals only from the positive control spots (biotinylated BSA and biotinylated PEG). Wells that contained either miRNA or PSA targets (panels b and c of Figure 5, respectively) gave nanoparticle light-scattering signals in the correct array rows. In the final well (Figure 5d), both miRNA and PSA were detected simultaneously, along with the positive control biomolecules. In addition, we have calculated a 3σ threshold for the BSA (negative control) wells for each experiment to ascertain the statistical significance of each measurement (n = 5). The 3σ values are included in the Figure 5 caption, and the results show that in each case the signal intensities of the target spots are above this threshold and as such represent statistically significant signals. From these experiments, we conclude that this assay is capable of detecting both antigen and RNA individually or in a combinatorial fashion on the same slide using biotinylated gold nanoparticle probes.
An important observation from this experiment is that in all cases the signal intensity from the negative control positions (BSA and noncomplementary DNA) was low and distinguishable from the target positions of interest (Figure 5). We also prove the utility of using this assay for direct detection of biotinylated surface-bound proteins (biotinylated BSA) and polymers (biotinylated PEG), the two positive-control biomolecules. The use of biotin—PEG—AuNP probes enabled simultaneous biomolecule detection with low background signal.
DISCUSSION
Cancer progression is characterized by the presence of multiple biomarkers whose expression changes throughout various stages of the disease.47 As such, the ability to simultaneously detect and identify multiple classes of biomarkers (nucleic acids and proteins) in one sample is important for disease diagnosis and management,48 especially when only small amounts of patient samples are available.49 A variety of strategies related to DNA and protein detection on the same platform are currently in development.25,50,51 Many of these platforms are based on either fluorescence52 or electrochemical53,54 detection techniques that are experimentally complicated, laborious, and expensive (with respect to reagents or experimental setups). We have developed a high-throughput, array-based assay using biotin—PEG—linked gold nanoparticle probes that exhibits high sensitivity and has the capability to detect nucleic acid and protein targets in a multiplexed manner. Importantly, this platform includes inexpensive detection probes that are easy to synthesize from commercially available reagents. Furthermore, these biotin-linked gold nanoparticle probes are chemically stable for at least six months without a loss of function.
In this work, three unique biotin—PEG—AuNP probes were synthesized for investigating probe stability toward free-thiol molecules in solution. Dithiothreitol (DTT) studies indicated that dithiol and trithiol-linked AuNP probes showed greater colloidal stability toward free thiols in comparison with the monothiol AuNP probes (Figure S1). It is also important to note that the sensitivity and dynamic range of biotin—PEG—AuNP assay can be tuned through additional gold reductions that result in enhanced light scattering for low concentration targets.16 The ability to modulate the nanoparticle probe size, biotin—PEG linker chemistry (number of terminal anchors), and light-scattering signal amplification enables the synthesis of detection probes with high tailorability for different assay conditions. This tunability is important when assaying multiple types of biomarkers that may be present at varying concentrations in a single sample. In addition, we have shown that these probes are able to interface with conventional microarray platforms (Exiqon 7th generation miRCURY LNA microRNA microarrays and microarrays from Full Moon Biosystems) without the generation of false-positive signals.
The methodology and unique nanoscale detection probes developed in this work have several important advantages over existing technologies. The use of biotin—streptavidin interactions in a sandwich format results in specific target detection and allows for the identification of essentially any biotinylated molecule. In addition, the incorporation of PEG linkers onto the gold nanoparticle surface reduces the tendency for nonspecific probe adsorption and increases the specificity of the assay for a wide variety of molecules55 (proteins, nucleic acids, and polymers). This assay is ideally suited for sensitive, selective, high-throughput screening of any biotinylated biomarker, and future studies will investigate the utility of using this assay for identification of unique disease states through simultaneous profiling of diverse biomarkers. Finally, because DNA-modified gold nanoparticle probes have been used to detect a wide class of targets beyond nucleic acid and proteins, including small molecules56 and metal ions,57 one can also envision future applications of the biotin—PEG—AuNP probes developed in this work for detection of these classes of analytes.
EXPERIMENTAL PROCEDURES
Synthesis of Biotin—PEG Linkers
Cyclic disulfide—PEG— biotin linkers were synthesized by addition of a 100 μM solution of lipoic acid—PEG—maleimide (Nanocs Inc.; MW: 3400 Da) to a 100 μM solution of thiol—PEG—biotin (N anocs Inc.; MW: 5000 Da). The mixture was placed on a shaker at 25°C for 17 h. After incubation, the mixture was filtered using a 7000 Da cutoff filter (Thermo Fisher Scientific).
Trithiol—PEG—biotin linkers were synthesized by addition of a 100 μM solution of 4-arm—PEG—thiol (Laysan Bio, MW: 10 000 Da) to a 100 μM solution of biotin—PEG—maleimide (Laysan Bio, MW: 5000 Da). The mixture was placed on a shaker at 25°C for 17 h. After incubation, the mixture was filtered using a 10 000 Da cutoff filter (Amicon, EMD Millipore).
Biotin—PEG—AuNP Probe Synthesis
A 5 nM solution of gold nanoparticles was mixed with a 4 μM solution of PEG—biotin linker (monothiol—PEG—biotin, cyclic disulfide— PEG—biotin, or trithiol—PEG—biotin) and shaken at 25°C for 24 h. After incubation, buffer solution containing 1.5 M NaCl, 100 mM phosphate (pH 7.2), 1% BSA, and 0.2% Tween was added to bring the final buffer concentration to 0.15 M NaCl, 10 mM phosphate, 0.1% BSA, and 0.02% Tween. After an additional 1 h, the salt concentration was raised to 0.3 M NaCl using 5 M NaCl, and the mixture was kept shaking at 25°C for 1 h. The biotin—PEG—AuNP probes were aliquoted into low retention tubes (Eppendorf) and centrifuged at 12000g for 25 min at 25°C to remove excess reagents. After removal of the supernatant and two additional washes with nanopure water, the probes were resuspended in buffer (1× PBS, 0.1% BSA, 0.02% Tween) and stored at 4°C until further use.
Oligonucleotide Preparation
MicroRNA and DNA oligonucleotide sequences were synthesized on a MerMade 12 system (Bioautomation) and MM48 oligonucleotide synthesizer (BioAutomation), respectively, using standard solid-phase synthesis protocols and reagents (Glen Research). Oligonucleotide sequences were purified by RP-HPLC using a Microsorb C18 column (Varian) with 0.1 M triethylammonium acetate (TEAA) at pH 7 with a linear gradient of 100% CH3CN. Elution of the oligonucleotides was monitored at a wavelength of 254 nm.
Buffer Solution Preparation
Assay buffer solution containing 1× PBS (Sigma-Aldrich, pH 7), 1% BSA (Sigma-Aldrich), and 0.02% Tween (Sigma-Aldrich) was used for sample dilution (AuNP probe and biomolecule targets) and all slide-washing steps.
MicroRNA Biotinylation
The 3′ terminal biotinylation of microRNA target molecules was accomplished using a commercial kit (RNA3′ End Biotinylation Kit, Pierce) according to the manufacturer’s specifications.
Antibody Biotinylation
The biotinylation of secondary PSA antibody (BiosPacific) was accomplished using a commercial kit (Antibody Biotinylation Kit for IP, Pierce) according to the manufacturer’s specifications.
Simultaneous Nucleic Acid and Protein Detection
MicroRNA and prostate-specific antigen (BiosPacific) targets were diluted in assay buffer and captured onto a custom array from Full Moon BioSystems at 54°C for 2 h. The location of the surface-bound capture molecules that were adsorbed onto each well of the array is given in Figure 5. The slide was then washed with prewarmed (54°C) assay buffer. Biotinylated secondary antibody (10 μg/mL, BiosPacific) was added to the wells that contained PSA target for 30 min at 25°C and then washed with buffer. Next, the microarray was treated with streptavidin (10 μg/mL, Thermo Fisher Scientific) at 25°C for 20 min, washed with 1× PBS buffer, and then dried. This binding step was performed at 25°C to avoid denaturation of the streptavidin protein. The arrays were subsequently treated with 100 pM of 30 nm monothiol biotin—PEG—AuNP probe for 20 min at 25 °C, washed with 1× PBS, and dried. The microarray was treated with HAuCl4 (1 mM) and NH2OH (5 mM) for 45 s, thoroughly washed with deionized water, and dried. The microarrays were finally imaged using a LS Reloaded laser scanner (Tecan) using 633 nm laser wavelength, and the images were transferred to GenePix software for analysis.
Supplementary Material
Acknowledgments
The research is based on work supported by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via the Federal Bureau of Investigation contract no. DJF-15-1200-K-0001730. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the ODNI, IARPA, or the U.S. Government. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes, notwithstanding any copyright annotation thereon. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award nos. U54CA151880 and U54CA199091. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. A.W.S. acknowledges a Terminal Year Fellowship from the Northwestern University Biomedical Engineering Department.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.6b00529.
A table showing oligonucleotide sequences. Figures showing biotin—PEG—AuNP probe stability study and miRNA detection using different sized biotin—PEG—AuNP probes. (PDF)
Special Issue: Interfacing Inorganic Nanoparticles with Biology
Notes
The authors declare no competing financial interest.
References
- 1.Giljohann DA, Mirkin CA. Drivers of biodiagnostic development. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debouck C, Goodfellow PN. DNA microarrays in drug discovery and development. Nat Genet. 1999;21:48–50. doi: 10.1038/4475. [DOI] [PubMed] [Google Scholar]
- 3.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Kingsmore SF. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat Rev Drug Discovery. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nat Rev Drug Discovery. 2003;2:566–580. doi: 10.1038/nrd1130. [DOI] [PubMed] [Google Scholar]
- 7.Jia HL, Ye QH, Qin LX, Budhu A, Forgues M, Chen YD, Liu YK, Sun HC, Wang L, Lu HZ, et al. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res. 2007;13:1133–1139. doi: 10.1158/1078-0432.CCR-06-1025. [DOI] [PubMed] [Google Scholar]
- 8.Visintin I, Feng Z, Longton G, Ward DC, Alvero AB, Lai YL, Tenthorey J, Leiser A, Flores-Saaib R, Yu H, et al. Diagnostic markers for early detection of ovarian cancer. Clin Cancer Res. 2008;14:1065–1072. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Ba Y, Ma LJ, Cai X, Yin Y, Wang KH, Guo JG, Zhang YJ, Chen JN, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 10.Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 11.Baker M. MicroRNA profiling: separating signal from noise. Nat Methods. 2010;7:687–692. doi: 10.1038/nmeth0910-687. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkins Stevens P, Hall JG, Lyamichev V, Neri BP, Lu M, Wang L, Smith LM, Kelso DM. Analysis of single nucleotide polymorphisms with solid phase invasive cleavage reactions. Nucleic Acids Res. 2001;29:E77. doi: 10.1093/nar/29.16.e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hrdlicka PJ, Babu BR, Sorensen MD, Harrit N, Wengel J. Multilabeled pyrene-functionalized 2′-amino-LNA probes for nucleic acid detection in homogeneous fluorescence assays. J Am Chem Soc. 2005;127:13293–13299. doi: 10.1021/ja052887a. [DOI] [PubMed] [Google Scholar]
- 15.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–24. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 16.Alhasan AH, Kim DY, Daniel WL, Watson E, Meeks JJ, Thaxton CS, Mirkin CA. Scanometric MicroRNA array profiling of prostate cancer markers using spherical nucleic acid-gold nanoparticle conjugates. Anal Chem. 2012;84:4153–4160. doi: 10.1021/ac3004055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ullal AV, Peterson V, Agasti SS, Tuang S, Juric D, Castro CM, Weissleder R. Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci Transl Med. 2014;6:219ra9. doi: 10.1126/scitranslmed.3007361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrin A, Duracher D, Perret M, Cleuziat P, Mandrand B. A combined oligonucleotide and protein microarray for the codetection of nucleic acids and antibodies associated with human immunodeficiency virus, hepatitis B virus, and hepatitis C virus infections. Anal Biochem. 2003;322:148–55. doi: 10.1016/j.ab.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Kochan J, Wawro M, Kasza A. Simultaneous detection of mRNA and protein in single cells using immunofluorescence-combined single-molecule RNA FISH. Bio Techniques. 2015;59:209–221. doi: 10.2144/000114340. [DOI] [PubMed] [Google Scholar]
- 20.Mao X, Gurung A, Xu H, Baloda M, He YQ, Liu GD. Simultaneous detection of nucleic acid and protein using gold nanoparticles and lateral flow device. Anal Sci. 2014;30:637–642. doi: 10.2116/analsci.30.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Askari MDF, Miller GH, Vo-Dinh T. Simultaneous detection of the tumor suppressor FHIT gene and protein using the multi-functional biochip. Cancer Detect Prev. 2002;26:331–342. doi: 10.1016/s0361-090x(02)00091-0. [DOI] [PubMed] [Google Scholar]
- 22.Shipp G. Ultrasensitive measurement of protein and nucleic acid biomarkers for earlier disease detection and more effective therapies. Biotechnol Healthc. 2006;3:35–40. [PMC free article] [PubMed] [Google Scholar]
- 23.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 24.Flamini E, Mercatali L, Nanni O, Calistri D, Nunziatini R, Zoli W, Rosetti P, Gardini N, Lattuneddu A, Verdecchia GM, Amadori D. Free DNA and carcinoembryonic antigen serum levels: An important combination for diagnosis of colorectal cancer. Clin Cancer Res. 2006;12:6985–6988. doi: 10.1158/1078-0432.CCR-06-1931. [DOI] [PubMed] [Google Scholar]
- 25.Falconnet D, She J, Tornay R, Leimgruber E, Bernasconi D, Lagopoulos L, Renaud P, Demierre N, van den Bogaard P. Rapid, sensitive and real-time multiplexing platform for the analysis of protein and nucleic-acid biomarkers. Anal Chem. 2015;87:1582–1589. doi: 10.1021/ac502741c. [DOI] [PubMed] [Google Scholar]
- 26.Bailey RC, Nam JM, Mirkin CA, Hupp JT. Real-time multicolor DNA detection with chemoresponsive diffraction gratings and nanoparticle probes. J Am Chem Soc. 2003;125:13541–13547. doi: 10.1021/ja035479k. [DOI] [PubMed] [Google Scholar]
- 27.Kim D, Daniel WL, Mirkin CA. Microarray-based multiplexed scanometric immunoassay for protein cancer markers using gold nanoparticle probes. Anal Chem. 2009;81:9183–7. doi: 10.1021/ac9018389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nam JM, Stoeva SI, Mirkin CA. Bio-bar-code-based DNA detection with PCR-like sensitivity. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 29.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 30.Cao YWC, Jin RC, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 31.Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. Multiplexed detection of protein cancer markers with biobarcoded nanoparticle probes. J Am Chem Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 32.Thaxton CS, Elghanian R, Thomas AD, Stoeva SI, Lee JS, Smith ND, Schaeffer AJ, Klocker H, Horninger W, Bartsch G, et al. Nanoparticle-based bio-barcode assay redefines “undetectable” PSA and biochemical recurrence after radical prostatectomy. Proc Natl Acad Sci U S A. 2009;106:18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niemeyer CM, Ceyhan B. DNA-directed functionalization of colloidal gold with proteins. Angew Chem, Int Ed. 2001;40:3685–3688. doi: 10.1002/1521-3773(20011001)40:19<3685::aid-anie3685>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 34.Niemeyer CM. Nanoparticles, proteins, and nucleic acids: Biotechnology meets materials science. Angew Chem, Int Ed. 2001;40:4128–4158. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 35.Wang ZX, Lee J, Cossins AR, Brust M. Microarray-based detection of protein binding and functionality by gold nanoparticle probes. Anal Chem. 2005;77:5770–5774. doi: 10.1021/ac050679v. [DOI] [PubMed] [Google Scholar]
- 36.Goluch ED, Nam JM, Georganopoulou DG, Chiesl TN, Shaikh KA, Ryu KS, Barron AE, Mirkin CA, Liu C. A bio-barcode assay for on-chip attomolar-sensitivity protein detection. Lab Chip. 2006;6:1293–1299. doi: 10.1039/b606294f. [DOI] [PubMed] [Google Scholar]
- 37.Roco MC, Mirkin CA, Hersam MC. Nanotechnology Research Directions for Societal Needs in 2020: Retrospective and Outlook. Sci Pol Rep. 2011;1:1–690. [Google Scholar]
- 38.Thaxton CS, Hill HD, Georganopoulou DG, Stoeva SI, Mirkin CA. A bio-bar-code assay based upon dithiothreitol-induced oligonucleotide release. Anal Chem. 2005;77:8174–8178. doi: 10.1021/ac0514265. [DOI] [PubMed] [Google Scholar]
- 39.Shaikh KA, Ryu KS, Goluch ED, Nam JM, Liu JW, Thaxton S, Chiesl TN, Barron AE, Lu Y, Mirkin CA, et al. A modular microfluidic architecture for integrated biochemical analysis. Proc Natl Acad Sci U S A. 2005;102:9745–9750. doi: 10.1073/pnas.0504082102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim WJ, Choi SH, Rho YS, Yoo HJ. Bio-functionalized gold nanoparticles for surface-plasmon-absorption-based protein detection. Bull Korean Chem Soc. 2011;32:4171–4175. [Google Scholar]
- 41.Zijlstra P, Paulo PMR, Orrit M. Optical detection of single non-absorbing molecules using the surface plasmon resonance of a gold nanorod. Nat Nanotechnol. 2012;7:379–382. doi: 10.1038/nnano.2012.51. [DOI] [PubMed] [Google Scholar]
- 42.Kim NH, Baek TJ, Park HG, Seong GH. Highly sensitive biomolecule detection on a quartz crystal microbalance using gold nanoparticles as signal amplification probes. Anal Sci. 2007;23:177–181. doi: 10.2116/analsci.23.177. [DOI] [PubMed] [Google Scholar]
- 43.Caswell KK, Wilson JN, Bunz UHF, Murphy CJ. Preferential end-to-end assembly of gold nanorods by biotin-streptavidin connectors. J Am Chem Soc. 2003;125:13914–13915. doi: 10.1021/ja037969i. [DOI] [PubMed] [Google Scholar]
- 44.Li Z, Jin R, Mirkin CA, Letsinger RL. Multiple thiol-anchor capped DNA-gold nanoparticle conjugates. Nucleic Acids Res. 2002;30:1558–1562. doi: 10.1093/nar/30.7.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letsinger RL, Elghanian R, Viswanadham G, Mirkin CA. Use of a steroid cyclic disulfide anchor in constructing gold nanoparticle-oligonucleotide conjugates. Bioconjugate Chem. 2000;11:289–291. doi: 10.1021/bc990152n. [DOI] [PubMed] [Google Scholar]
- 46.Oh E, Susumu K, Makinen AJ, Deschamps JR, Huston AL, Medintz IL. Colloidal stability of gold nanoparticles coated with multithiol-Poly(ethylene glycol) ligands: importance of structural constraints of the sulfur anchoring groups. J Phys Chem C. 2013;117:18947–18956. [Google Scholar]
- 47.Mukundan H, Xie HZ, Anderson AS, Grace WK, Shively JE, Swanson BI. Optimizing a waveguide-based sandwich immunoassay for tumor biomarkers: evaluating fluorescent labels and functional surfaces. Bioconjugate Chem. 2009;20:222–230. doi: 10.1021/bc800283e. [DOI] [PubMed] [Google Scholar]
- 48.Das J, Cederquist KB, Zaragoza AA, Lee PE, Sargent EH, Kelley SO. An ultrasensitive universal detector based on neutralizer displacement. Nat Chem. 2012;4:642–648. doi: 10.1038/nchem.1367. [DOI] [PubMed] [Google Scholar]
- 49.Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, Wappett M, Sharpe A, Dymond M, Barrett JC, et al. Evaluating robustness and sensitivity of the NanoString Technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res. 2015;75:2587–2593. doi: 10.1158/0008-5472.CAN-15-0262. [DOI] [PubMed] [Google Scholar]
- 50.Polsky R, Gill R, Kaganovsky L, Willner I. Nucleic acid-functionalized Pt nanoparticles: catalytic labels for the amplified electrochemical detection of biomolecules. Anal Chem. 2006;78:2268–2271. doi: 10.1021/ac0519864. [DOI] [PubMed] [Google Scholar]
- 51.Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH. Advancing the speed, sensitivity and accuracy of biomolecular detection usingmulti-length-scale engineering. Nat Nanotechnol. 2014;9:969–980. doi: 10.1038/nnano.2014.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 54.Zhu CZ, Dong SJ. Energetic graphene-based electrochemical analytical devices in nucleic acid, protein and cancer diagnostics and detection. Electroanalysis. 2014;26:14–29. [Google Scholar]
- 55.Wang L, Lofton C, Popp M, Tan WH. Using luminescent nanoparticles as staining probes for affymetrix GeneChips. Bioconjugate Chem. 2007;18:610–613. doi: 10.1021/bc060365u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han MS, Lytton-Jean AKR, Mirkin CA. Agold nanoparticle based approach for screening triplex DNA binders. J Am Chem Soc. 2006;128:4954–4955. doi: 10.1021/ja0606475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee JS, Mirkin CA. Chip-based scanometric detection of mercuric ion using DNA-functionalized gold nanoparticles. Anal Chem. 2008;80:6805–6808. doi: 10.1021/ac801046a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


