Abstract
Through exome sequencing we identified six individuals with biallelic loss-of-function mutations in TRIP13. All six developed Wilms tumor. Constitutional mosaic aneuploidies, microcephaly, developmental delay and seizures, which are features of mosaic variegated aneuploidy (MVA) syndrome1,2, were more variably present. Through functional studies we show that TRIP13-mutant patient cells have no detectable TRIP13 and have substantial impairment of the spindle assembly checkpoint (SAC) leading to a high rate of chromosome missegregation. Accurate segregations as well as SAC proficiency are rescued by restoring TRIP13 function. Individuals with biallelic TRIP13 or BUB1B mutations have a high risk of embryonal tumors3, and here we show that their cells display severe SAC impairment. MVA due to biallelic CEP57 mutations4, or of unknown cause, is not associated with embryonal tumors and their cells show minimal SAC deficiency. These data provide insights into the complex relationships between aneuploidy and carcinogenesis.
Accurate chromosome segregation during cell division is required to maintain the correct number of chromosomes in cells. Errors of chromosome segregation can lead to aneuploidy, a term that describes cells with loss or gain of one or more chromosomes. Aneuploidy is an important cause of human disease, implicated in diverse pathologies, including recurrent miscarriage, infertility, developmental disorders and cancer5–7. Many biological processes, including spindle assembly, chromatid-spindle attachment, attachment error-correction, and the spindle assembly checkpoint (SAC) are involved in ensuring chromosome segregation proceeds flawlessly and that aneuploidy is prevented6,8.
Rare individuals with constitutional mosaic aneuploidies involving varying chromosomes are well documented1,2. Affected individuals often have other clinical features such as microcephaly, developmental delay and various congenital abnormalities, and the term ‘mosaic variegated aneuploidy (MVA)’ is used to describe this condition1–3. Cancer predisposition is one of the most important associations of MVA, with substantial increased risk of childhood malignancies, particularly Wilms tumor and rhabdomyosarcoma3,9,10.
We have been studying this rare condition for over a decade. We previously reported biallelic mutations in the spindle assembly checkpoint gene BUB1B as a cause of MVA and childhood cancer3. To date we have identified 14 individuals with biallelic BUB1B mutations. More recently we identified biallelic mutations in CEP57, which encodes a centrosomal protein involved in kinetochore attachment, in four individuals with MVA, none of whom have developed cancer4. Together these two genes only account for a proportion of MVA cases.
To identify additional MVA genes we undertook exome sequencing in 43 individuals from 20 families, including 21 probands with MVA (Supplementary Table 1). We generated exome data using Illumina exome capture assays and called variants using the OpEx pipeline as previously described11,12. We performed two analyses to prioritize variants for further evaluation. We undertook an individual proband analysis to identify genes with two rare variants, as MVA is a recessively inherited condition. We also identified genes with protein truncating variants (PTVs) present in more than one proband, using the PTV prioritization method4,12.
We identified the homozygous stop-gain mutation TRIP13 c.1060C>T_p.Arg354X in three probands (ID_0319, ID_0644, ID_7054) (Table 1, Supplementary Fig. 1). The mutation leads to nonsense-mediated mRNA decay (Supplementary Fig. 2) and no detectable TRIP13 protein (Supplementary Fig. 3). Protein expressed from exogenous cDNA is present at a significantly lower level compared to the expression of wild-type TRIP13 (Supplementary Fig. 4). This shows that in addition to mRNA stability, the mutation also directly affects the TRIP13 protein. Therefore, the TRIP13 c.1060C>T_p.Arg354X allele is highly unlikely to produce any functional TRIP13 protein.
Table 1. Summary of molecular and clinical findings in individuals with biallelic TRIP13 mutations.
| ID | TRIP13 mutations | Aneupl oidy | Premature chromatid separation | Wilms tumor age at diagnosis | Current status | Other clinical features |
|---|---|---|---|---|---|---|
| ID_0319 | c.1060C>T_p.Arg354X c.1060C>T_p.Arg354X |
yes | uk | 2 yrs | alive, 6 yrs | microcephaly, developmental delay, growth retardation arthrogryposis, |
| ID_0644 | c.1060C>T_p.Arg354X c.1060C>T_p.Arg354X |
yes | yes | 4 yrs, relapse 5 yrs | alive, 43 yrs | growth retardation |
| ID_7054 | c.1060C>T_p.Arg354X c.1060C>T_p.Arg354X |
yes | yes | 2 yrs | alive, 5 yrs | café au lait patches |
| ID_0649 | c.1060C>T_p.Arg354X c.1060C>T_p.Arg354X |
no | yes | 2 yrs relapse 10 yrs | died, 10 yrs | microcephaly, growth retardation, seizures, skin pigmentation |
| ID_6112 | c.1060C>T_p.Arg354X c.1060C>T_p.Arg354X |
uk | uk | 5 yrs | alive, 6 yrs | |
| ID_7679 | c.673-1G>C c.693-1G>C |
no | uk | 1.3 yrs | alive, 1.5 yrs | seizures, developmental de |
Fuller details are provided in Supplementary Fig 1.
The three individuals had been independently recruited and there was no known relationship between them, but they were from families of Asian origin. Interestingly, all three had Wilms tumor. To further explore the association of TRIP13 and Wilms tumor we performed exome sequencing in 11 UK individuals of reported Asian origin with Wilms tumor. Two, ID_0649 and ID_6112, were also homozygous for TRIP13 c.1060C>T_p.Arg354X (Table 1 and Supplementary Fig. 1). ID_0649 had been noted to have premature chromatid separation, but no mosaic aneuploidies in lymphocytes. No constitutional karyotype in ID_6112 has been performed, but the tumor karyotype was reported to be normal. Of note, the sister of ID_6112 died at four years of age after developing a pelvic Sertoli-Leydig cell tumor and acute myeloid leukemia (AML). No sample is available for mutation testing, but this observation suggests biallelic TRIP13 mutations may also predispose to cancers other than Wilms tumor.
The four available parental samples were all heterozygous for the mutation, consistent with recessive inheritance. The mutation is not present in the Exome Aggregation Consortium (ExAC) or ICR1000 series13,14, nor is it present in 11,677 other exomes we have analyzed with the same pipeline. Multidimensional scaling analysis strongly suggests the families originate from Pakistan (Supplementary Fig. 5). Exploration of the available family history suggests the families come from the Azad Kashmir region of Pakistan. Many Azad Kashmir families were given work permits for the UK in the 1960s due to the construction of the Mangla Dam, which led to large-scale local displacement. The incidence of Wilms tumor in this region of Pakistan is very high (M Rashid, personal communication). This may be related to the TRIP13 mutation we have identified. Evaluation of the contribution of the TRIP13 mutation to Wilms tumor in this population would therefore be of interest.
We subsequently became aware of a Norwegian girl, ID_7679, who developed Wilms tumor at 15 months, who is homozygous for a different truncating TRIP13 mutation. The mutation, c.697-1G>C, is predicted to disrupt the canonical 3’ splice-site in intron 7 of TRIP13 and a new splice-site 2bp upstream is predicted to be used, resulting in a 2bp frameshift and premature protein truncation. No constitutional mosaic aneuploidies were observed in her lymphocytes.
These data provide compelling genetic evidence that TRIP13 is a cancer predisposition gene. Biallelic loss-of-function TRIP13 mutations confer a high risk of Wilms tumor and also predispose to chromosome segregation dysfunction which can manifest as mosaic aneuploidies and/or premature chromatid separation. There were no consistent phenotypic features amongst the six probands, though developmental delay, microcephaly, seizures, growth retardation and skin pigmentation were each noted in more than one individual (Table 1 and Supplementary Fig. 1).
TRIP13 encodes a highly conserved AAA+ATPase that contributes to homologue pairing, synapsis and recombination during meiosis15. In mitosis, TRIP13 remodels the crucial SAC effector MAD2 from a ‘closed’ (active) into an ‘open’ (inactive) form16,17. This has dual impact on SAC function: in prometaphase, MAD2 remodeling is thought to enable continuous replenishment of MAD2 pools that can be activated by unattached kinetochores for the generation of the initial SAC inhibitor. TRIP13 thus ensures robust and long-lasting SAC signaling. In metaphase however, when no new active MAD2 is generated by kinetochores, MAD2 inactivation by TRIP13 promotes disassembly of the inhibitor, causes SAC silencing and mitotic exit18–20.
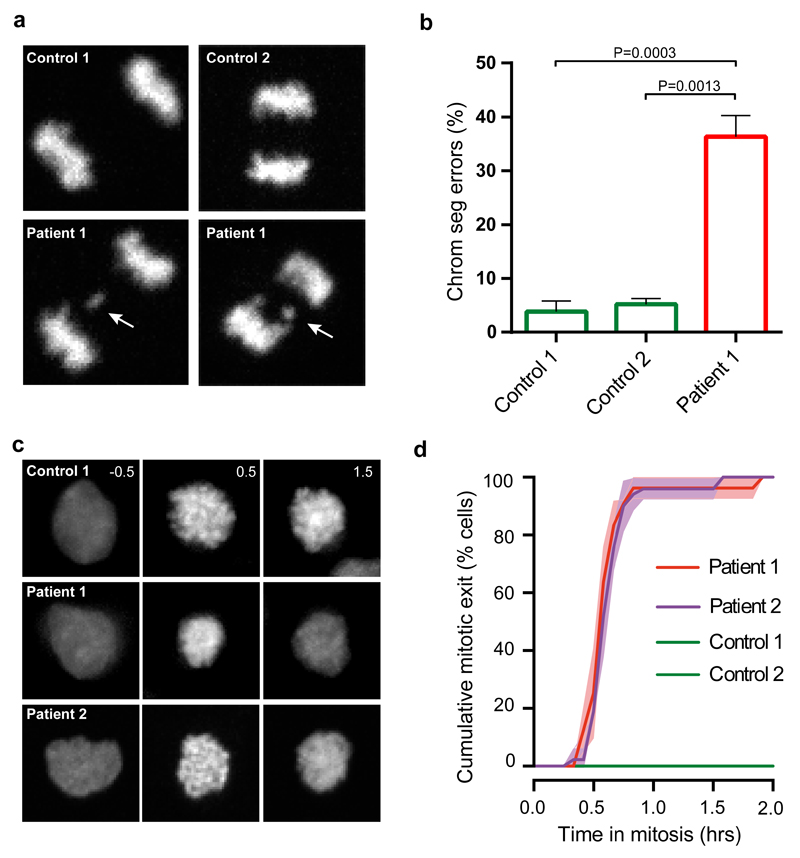
We sought to examine which defective molecular processes underlie aneuploidy and chromosome missegregation in TRIP13-mutant patients. We first infected immortalized TRIP13-mutant patient lymphoblasts with virus carrying H2B-mNeon to visualize chromatin. Live cell imaging showed that patient lymphoblasts displayed high levels of chromosome segregation errors such as lagging chromosomes and chromosome bridges (Fig. 1a,b). To understand how TRIP13 mutations cause chromosomal instability (CIN) in patient cells, we examined the fidelity of the SAC, the main chromosome segregation surveillance mechanism in which TRIP13 has been implicated. To this end, we analyzed cells for their ability to maintain mitotic arrest after exposure to the spindle poison nocodazole (Fig. 1c and Supplementary Fig. 6). Control cells maintained the arrest for >2 hours, whereas all cells from two different TRIP13-mutant patients escaped the arrest within one hour (Fig. 1c,d). Mitotic exit despite unattached chromosomes is indicative of a compromised SAC.
Figure 1. TRIP13 loss-of-function mutations cause chromosome segregation errors and SAC deficiency.
(a) Representative anaphases of immortalized control (upper) and TRIP13-mutant patient (lower) lymphoblasts expressing H2B-mNeon, showing a lagging chromosome in the lower panels indicated with a white arrow.
(b) Quantification of chromosome segregation errors of lymphoblasts as visualized in (a). Each bar depicts the mean of 3-4 experiments ±SEM, >60 cells in total. P-values ≤ 0.05, from unpaired Student’s t-tests, are shown. Patient 1 cells show increased levels of chromosome segregation errors.
(c) Representative images of H2B-mNeon expressing control (upper) and patient (lower) lymphoblasts going through mitosis (time in hours with mitotic entry at t=0.0) in the presence of nocodazole. Unlike control cell lines both patient cell lines have exited from mitosis after 1.5 hours (third column of second and third row panels).
(d) Analysis of mitotic delay of cells as visualized in (c), indicating the cumulative percentage of cells that exited from mitosis as a function of time (mean of three experiments ±SEM, 10-30 cells per experiment). Both patient cell lines escaped mitotic arrest within one hour.
Key: patient 1, ID_0644; patient 2, ID_7054; SEM, standard error of the mean
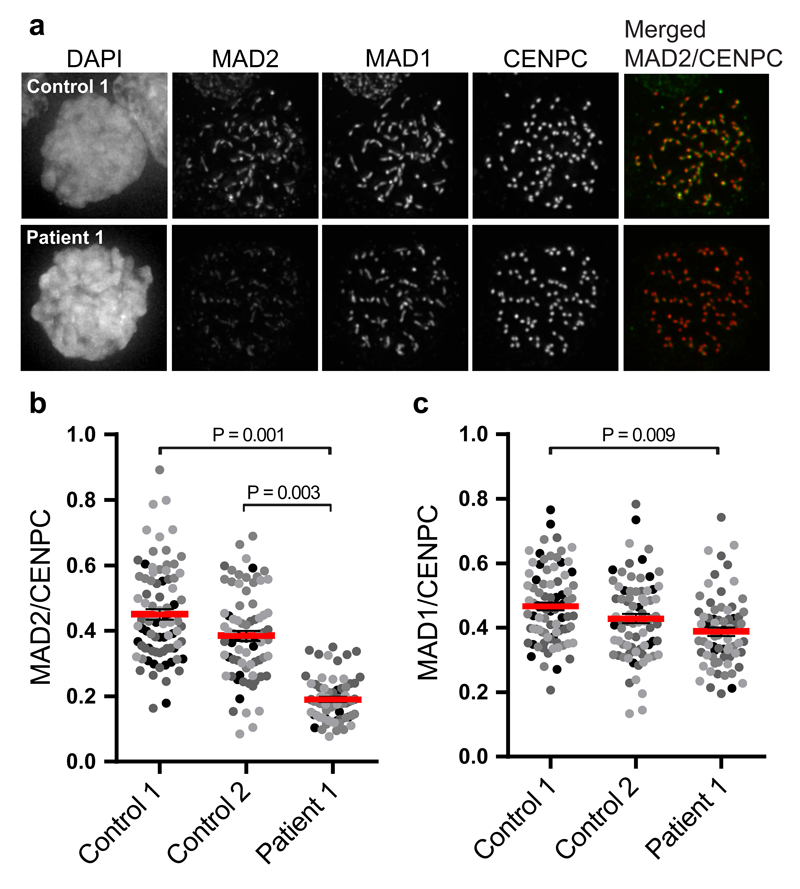
To gain insight into the molecular defect causing SAC impairment, we analyzed SAC protein expression and subcellular localization. Immunofluorescence imaging of the SAC effector MAD2 in nocodazole-treated cells showed that TRIP13-mutant patient cells recruited ~50% fewer molecules of MAD2 to their unattached kinetochores (Fig. 2a,b). Kinetochore levels of the MAD2 receptor MAD1 were unaffected (Fig. 2c). Absence of TRIP13 caused increased overall p31comet expression in one patient and reduced overall MAD2 expression (Supplementary Fig. 7) in both patients, consistent with data reported in TRIP13 knockout HeLa cells20.
Figure 2. TRIP13 loss-of-function mutations cause reduced levels of MAD2 on unattached kinetochores.
(a-c) Immunofluorescence labelling (a) and quantification (b-c) of indicated proteins in nocodazole-arrested control or patient lymphoblasts. Each dot represents one cell, with dots from separate experiments in different shades of grey. The red bar depicts the mean of four experiments ±SEM, >70 cells in total. P-values ≤ 0.05, from unpaired Student’s t-tests, are shown. Patient 1 cells have reduced kinetochore levels of MAD2, but not MAD1, compared to the controls.
Key: patient 1, ID_0644; SEM, standard error of the mean
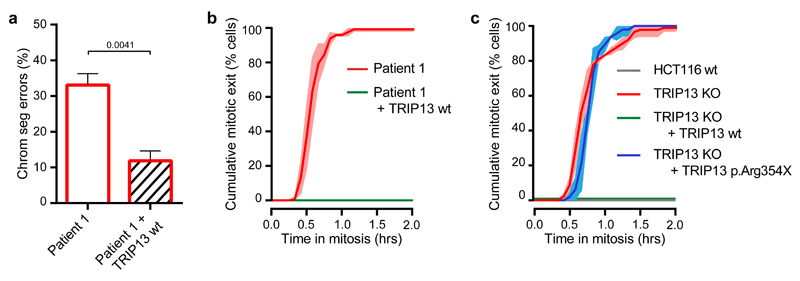
We next restored TRIP13 function by expressing GFP-TRIP13 in patient lymphoblasts using lentiviral delivery. Importantly, GFP-TRIP13 expression rescued the CIN as well as the SAC defect (Fig. 3a,b). Moreover, GFP-tagged TRIP13 p.Arg354X was unable to rescue an impaired SAC caused by CRISPR/Cas9-mediated knock out of the TRIP13 gene in HCT116 cells (Fig. 3c and Supplementary Fig. 8,9). These observations provide an explanation for the chromosome segregation defects observed in individuals with biallelic TRIP13 loss-of-function mutations. They also show that the SAC defects and resulting CIN are directly due to the loss of TRIP13 function caused by the homozygous TRIP13 c.1060C>T_p.Arg354X mutation.
Figure 3. SAC deficiency and CIN caused by TRIP13 loss-of-function is rescued with wild type but not mutant TRIP13.
(a) Quantification of chromosome segregation errors of TRIP13-mutated patient lymphoblasts expressing H2B-mNeon or co-expressing GFP-TRIP13 wt. Each bar depicts the mean of 3-4 experiments ±SEM, >85 cells in total. P-values ≤ 0.05 from an unpaired Student’s t-test are shown. Addition of GFP-TRIP13 wt to patient 1 cells reduced the rate of chromosome missegregation.
(b) Analysis of mitotic delay as in (Fig 1d) of nocodazole-treated patient lymphoblasts expressing H2B-mNeon or co-expressing GFP-TRIP13 wt. Mean of three experiments ±SEM, >35 cells in total. Patient 1 cells expressing GFP-TRIP13 now maintain mitotic arrest.
(c) Analysis of mitotic delay as in (Fig 1d) and (b) of nocodazole-treated HCT116 wt or HCT116 TRIP13 KO cells expressing H2B-mNeon, co-expressing GFP-TRIP13 wt, or co-expressing TRIP13 p.Arg354X. Mean of three experiments ± SEM, >45 cells in total. HCT116 wt and TRIP13 KO + TRIP13 wt cells both maintain mitotic arrest unlike TRIP13 KO and TRIP13 KO + TRIP13 p.Arg354X cells.
Key: patient 1, ID_0644; wt, wild-type; KO, knockout; CIN, chromosomal instability; SAC, spindle assembly checkpoint; SEM, standard error of the mean
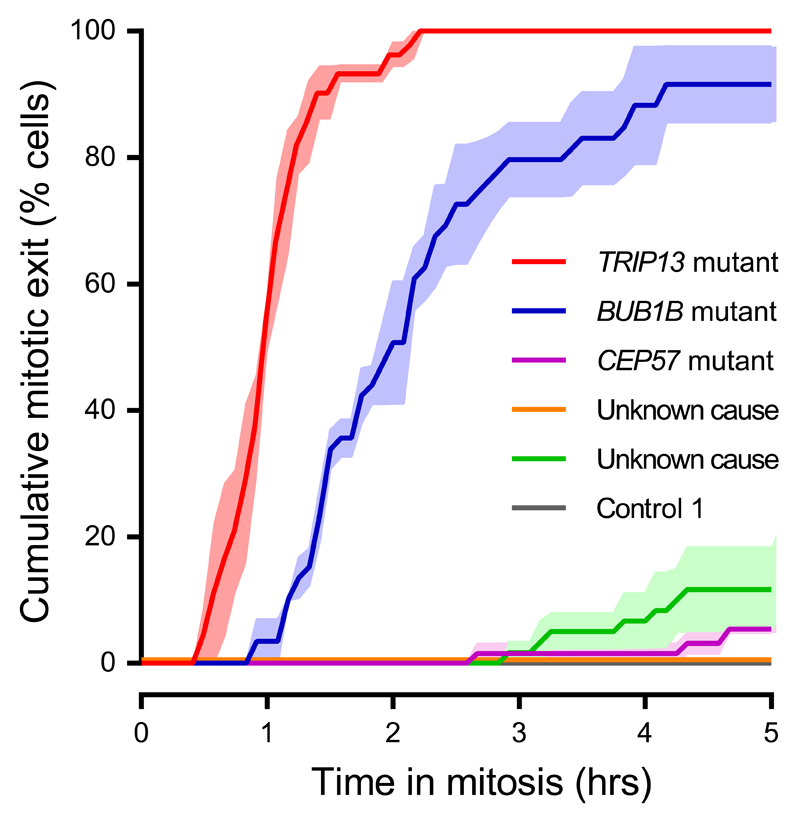
TRIP13 and BUBR1 (the protein encoded by BUB1B) are close functional partners in the spindle assembly checkpoint15. BUBR1 is part of the anaphase inhibitory complex MCC which also includes MAD2, the main target of TRIP13’s remodeling activity in mitosis21. Notably, severe SAC impairment was observed in TRIP13-mutant and BUB1B-mutant patient cells but not in cells from patients with CEP57 mutations or in whom the cause of MVA remains unclear after exome sequencing (Fig. 4).
Figure 4. Patient cells with TRIP13 or BUB1B mutations have severely compromised SAC.
Analysis of mitotic delay as in Fig 1d and Fig 3b of nocodazole-treated MVA patient lymphoblasts treated with far-red DNA dye to visualize the DNA, showing the cumulative percentage of cells that exited from mitosis as a function of time (mean of three experiments ±SEM, 20 cells per experiment). Only TRIP13-mutant and BUB1B-mutant patient cells rapidly escape from mitotic arrest.
Key: SAC, spindle assembly checkpoint; SEM, standard error of the mean
All six children with biallelic TRIP13 mutations developed Wilms tumor in childhood, five of whom were successfully treated (Table 1). Limited information is available, but there were no obvious distinctive histopathological features in the tumors. Individuals with biallelic BUB1B mutations are also at high risk of childhood embryonal tumors. In fact, all MVA cases with childhood solid tumors in our series have either BUB1B or TRIP13 mutations. By contrast, only one MVA individual in our series without mutations in BUB1B or TRIP13 has developed cancer, an ALL at 3 years in a child in whom the cause of MVA remains unknown10. It is therefore tempting to speculate that the high cancer risk may be causally related to severe impairment of the SAC. Irrespective of the mechanism, our findings have clinical utility indicating that BUB1B and TRIP13 mutation-positive individuals are at high risk of cancer and require close surveillance, whereas other individuals with MVA may be at lower cancer risk.
The data also suggest that the mechanism generating aneuploidy in affected individuals determines the cancer risk, not the aneuploidy per se. If confirmed, this is an important distinction as there has long been a debate regarding whether aneuploidy is a cause or consequence of cancer5. Further studies into the biological sequelae of these rare human mutations may therefore provide insights into the complex relationships between aneuploidy and carcinogenesis.
Online Methods
Samples
The study was approved by the London Multicentre Research Ethics Committee (05/MRE02/17). For mutational analyses DNA was obtained through our ongoing research of cancer predisposition syndromes. Appropriate consent was obtained from patients and/or parents as applicable. DNA was extracted from whole blood or an EBV-transformed lymphoblastoid cell line (ID_0644, ECACC ID:96061307) using standard protocols.
For the functional experiments the following patient LCLs were used: ID_0644 (patient 1, biallelic TRIP13 mutations, ECACC ID:96061307), ID_7054 (patient 2, biallelic TRIP13 mutations), ID_0675 (patient with biallelic BUB1B mutations, Coriell ID:GM22006), ID_0663 (patient with biallelic CEP57 mutations, Coriell ID:GM21654), ID_0639 (Coriell ID:GM09703) and ID_5728_01 (ECACC ID:FACT5728DLB).
Two control LCLs were also used: ID_5728_03, an unaffected sibling of ID_5728_01 (control 1, ECACC ID:FACT5728KC) and C0106 (control 2, ECACC ID:91071212). Cells were cultured in RPMI supplemented with 15% fetal bovine serum (FBS), 100 μg/ml penicillin/streptomycin and 2 mM alanyl-glutamine. Cells expressing H2B-mNeon were created by lentiviral transduction, using standard procedures.
Exome sequencing
We performed exome sequencing using the TruSeq Exome Enrichment Kit (Illumina) or the Nextera Rapid Capture Exome Kit (Illumina). For TruSeq exome sequencing we prepared libraries from 1.5 μg genomic DNA using the TruSeq Paired-End DNA Sample Preparation Kit (Illumina). DNA was fragmented using Covaris technology and libraries were prepared without gel size selection. For Nextera exome sequencing we prepared libraries from 50 ng genomic DNA using the Nextera DNA Sample Preparation Kit (Illumina). Captured DNA libraries were PCR amplified using the supplied paired-end PCR primers. Paired-end sequencing was performed on an Illumina HiSeq 2000 or HiSeq 2500 (high output mode) using v3 chemistry.
Alignment and variant calling
We used OpEx v1.0 pipeline to perform variant calling (www.icr.ac.uk/opex)11. Raw data was converted to FASTQs using CASAVA version 1.8.2 with default settings. The OpEx v1.0 pipeline uses Stampy22 to map to the human reference genome, Picard (http://picard.sourceforge.net) to flag duplicates, Platypus23 to call variants, and CAVA24 to provide consistent annotation of variants with the HGVS-compliant CSN (Clinical Sequencing Notation) standard v1.0 24. Only high quality calls, as defined by OpEx, were used in the analyses.
Reference datasets
We used the following datasets as control samples: the Exome Aggregation Consortium (ExAC) data13 version 3 accessed on 13/11/2015 (excluding the TCGA samples) (http://exac.broadinstitute.org), the 1000 Genomes Project data25 (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20110521/) and the ICR1000 UK exome series14 (www.icr.ac.uk/icr1000exomes). The ICR1000 UK exome series was generated using the same sequencing and analysis pipelines as the cases.
PTV prioritization method
The protein truncating variant (PTV) prioritization method is a gene-based strategy that aims to prioritize potential disease-associated genes for follow-up by leveraging two properties of protein truncating variants: (1) the strong association of rare truncating variants with disease, and (2) collapsibility; different PTVs within a gene typically result in the same functional effect and can be combined equally. We output all the predicted protein truncating variants: stop gains, coding frameshifts and essential splice site variants (-2, -1, +1, +2). For this experiment we defined ‘rare’ as PTVs that had an alternate allele frequency <1% in the control datasets. We next stratified the genes according to the number of different rare PTVs called in each affected individual.
Recessive analysis
We prioritized variants for evaluation in each affected individual assuming a recessive inheritance model. We removed all intronic, 3’/5’ UTR, and non-essential splice site variants and any variant that had an allele frequency >1% in control datasets. We also removed any heterozygous variant combination pair or homozygous alternate variant in which both alleles were from a single parent.
Sanger sequencing
We used Sanger sequencing to confirm the TRIP13 mutations identified in patients through exome sequencing and to confirm the presence of mutation following CRISPR-Cas9-mediated knockout of TRIP13 in HCT116 cells. For patient samples, primers were designed using Exon-Primer from the UCSC genome browser (http://genome.ucsc.edu/). PCRs were prepared using the QIAGEN Multiplex PCR Kit (QIAGEN) according to the manufacturer’s instructions. Amplicons were bidirectionally sequenced using the BigDyeTerminator Cycle sequencing kit and an ABI 3730 automated sequencer (Life Technologies). We analyzed sequencing traces using Mutation Surveyor software (SoftGenetics) and by visual inspection. For samples derived from HCT116 cells, primers were designed manually and PCRs were prepared using the Phusion® High-Fidelity DNA Polymerase PCR protocol (New England BioLabs) according to the manufacturer’s instructions. Amplicons were sent to Macrogen for sequencing and sequencing traces were analyzed using ApE version V2.0.49.10 software and by visual inspection. All primer sequences are available on request.
Multidimensional scaling analysis
We used the multidimensional scaling (MDS) analysis tool provided by PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/) to identify which population in the 1000 Genomes Project was most similar to the individuals with the TRIP13 c.1060C>T_p.Arg354X mutation. To identify the set of variants to use in the MDS analysis, we took the union of all coding and splice-site base substitutions in the 45 individuals with exome sequencing data and the 1000 Genomes Project individuals. We then removed variants for which 20 or more individuals had less than 15x coverage. We selected a subset of the individuals to use in the MDS analysis (Supplementary Fig. 5). We then used PLINK to remove variants with a minor allele frequency <0.01 or a Hardy-Weinberg equilibrium p-value <1x10-10. We removed non-independent variants using default settings for the --indep-pairwise method in PLINK (--indep-pairwise 50 5 0.5). We then used the MDS algorithm (--mds-plot) to extract the first four dimensions from the variant data.
HCT116 cell culture and TRIP13 knockout
HCT116 cells were cultured in McCoy’s-5A medium supplemented with 10% Tet-approved FBS, 100 μg/ml penicillin/streptomycin and 2 mM alanyl-glutamine. HCT116 TRIP13 knockout cells were generated by transient expression of pSpCas9(BB)-2A-GFP (Addgene 48138) with sgRNA against exon 11 (5’-CACCgCAATCATCTCTAGCTCTCGG-3’), followed by single-cell FACS sorting of GFP+ cells. Knockouts were verified by immunoblotting for protein expression and Sanger sequencing (Supplementary Figs. 8 and 9).
HeLa cell culture and stable Trex Flp-In cell line creation
HeLa Trex Flp-In cells were grown in DMEM high glucose supplemented with 10% Tet-approved FBS, 100 μg/ml penicillin/streptomycin, 2 mM alanyl-glutamine, 4 μg/ml blasticidin and 200 μg/ml hygromycin. To generate stably integrated HeLa Flp-In cell lines, pCDNA5-constructs were co-transfected with pOG44 recombinase in a 1:9 ratio using Fugene HD (Roche) according to the manufacturer's instructions. Constructs were expressed by addition of 1 μg/ml doxycycline for 24 hours.
Plasmids, cloning and virus production
For the HeLa Trex Flp-In cell lines stably expressing TRIP13, pcDNA5-LAP-TRIP13-WT was created by cloning TRIP13 cDNA derived from HeLa cells into empty pCDNA5-LAP using the following PCR primers: 5’-GCGCGGGATCCgacgaggccgtgggcgacctg-3’ and 5’-gcgcgactagtttaaatgtaagctgcaagcttc-3’ and digestion with BamHI and SpeI. pcDNA5-LAP-TRIP13-p.Arg354X was created by PCR mutagenesis of pcDNA5-LAP-TRIP13-WT using the Gibson Assembly strategy26 with two fragments generated using the following primer pairs: 5’-ctcgccagcagctgctgaccctctaagagctagagatgattggcttc-3’ + 5’-GGCAGCACTGCATAATTCT-3’ and 5’-gaagccaatcatctctagctcttagagggtcagcagctgctggcgag-3’ + 5’-AGAATTATGCAGTGCTGCC-3’.
Lentiviral TRIP13 constructs were derived from a lentiviral construct encoding fluorescently tagged histone 2B (H2B) and a puromycin-resistance cassette (pLV-H2B-mNeon-ires-Puro)27. The fluorescently tagged H2B was substituted by LAP-TRIP13-WT derived from pcDNA5-LAP-TRIP13-WT (see above) with PCR primers 5’-atatggcgcgccagcttatggtgagcaagggc-3’ and 5’-atatgctagcttaaatgtaagctgcaagcttctttc-3’ and digestion with AscI and NheI. For the subsequent steps the Gibson Assembly strategy was used26. The puromycin resistance cassette was substituted by fluorescently tagged H2B (pLV primers: 5’-acctggtgcatgacccgcaa-3’ and 5’-ggcgaccgttgtggccatat-3’; H2B primers: 5’-atatgggataatatggccacaacggtcgccatgccagagccagcgaagtc-3’ and 5’-ggcaccgggcttgcgggtcatgcaccaggtcgaccggttagctagccttgtac-3’). The CMV promoter was substituted by the ubiquitin promoter from pUB6/V5-His vector (Invitrogen) (pLV primers: 5’-cgtttagtgaccgtcagatcgc-3' and 5’-cggaactcccaagcttatcga-3'; pUB6 primers: 5’-aaaattttatcgataagcttgggagttccgCGAAAAGTGCCACCTGACGTC-3' and 5’-gtctccaggcgatctgacggtcactaaacgAAGCTTCGTCTAACAAAAAAGCCA-3'). Finally, the TRIP13 c.1060C>T_p.Arg354X mutation was inserted by assembling two PCR fragments generated with the following primer pairs: 5’-ctcgccagcagctgctgaccctctaagagctagagatgattggcttc-3’ + 5’-GGCAGCACTGCATAATTCT-3’ and 5’-gaagccaatcatctctagctcttagagggtcagcagctgctggcgag-3’ + 5’-AGAATTATGCAGTGCTGCC-3’.
Virions were generated by transient transfection of HEK 293T cells with the transfer vector and separate plasmids that express Gag-Pol, Rev, Tat and VSV-G. Supernatants were clarified by filtration.
Live cell imaging analysis of chromosome segregation errors
Lymphoblasts stably expressing H2B-mNeon were arrested in early S-phase for 24 hours by addition of 2 mM thymidine. Cells were then released from thymidine for 4 hours and mitotic progression was assayed. Cells were plated in 8-well Ibidi μ-slides in a thin layer of 0.15% UltraPure agarose with a layer of 0.3% agar on top. Cells were imaged in a heated chamber (37°C and 5% CO2). Images were acquired every 3 minutes at 1 x 1 binning in 15 x 1.5 μmz layers and projected to a single layer by maximum intensity projection using NIS-Elements software 4.45.
Imaging was performed with a Nikon Ti-Eclipse microscope equipped with CSUW spinning disk (yokogawa), Borealis, Andor iXon Ultra 888 EMCCD camera, 40x water objective NA 1.15 WD 0.6 mm, and 488 nm laser. Analysis of these experiments was carried out with ImageJ software. Chromosome segregation errors include one or more lagging chromosomes, misaligned chromosomes, chromosome bridges, and multipolar mitoses. Percentage of chromosome segregation errors is the amount of divisions with one or more mitotic errors divided by the total amount of divisions. Three or four replicate experiments were performed. Quantifications were subjected to unpaired Student’s t-tests; P<0.05.
Live cell imaging analysis of mitotic delay
Lymphoblasts or HCT116 cells were synchronized and plated as described above (HCT116 cells were plated in 24-well plates without agarose). After release from thymidine, mitotic progression was assayed in the presence of 0.83 μM nocodazole. For experiments in Fig. 4, lymphoblasts were incubated with 100 nM SiR-DNA dye (Spirochrome) for 4 hours prior to imaging, to facilitate visualization of mitotic arrest. For experiments in Fig. 3, cells were infected with a lentivirus expressing LAP-TRIP13 (wt or p.Arg354X)-IRES-H2B-mNeon, ensuring that H2B-mNeon-positive cells co-express LAP-TRIP13.
Images (far-red for Fig. 4; mNeon for Fig. 1d, Fig. 3b,c) were acquired every 5 minutes on a Nikon Ti-Eclipse widefield microscope equipped with an Andor Zyla 4.2 sCMOS camera, 40x oil objective NA 1.3 WD 0.2 mm, and Lumencor SpectraX light engine. Analysis of these experiments was carried out with ImageJ software. Time in mitosis was defined as the time between chromosome condensation and anaphase onset. Three replicate experiments were performed. Quantifications were subjected to unpaired Student’s t-tests; P<0.05.
Immunofluorescence imaging
Cells were plated on round 12 mm coverslips (coated with poly-L-lysine (Santa Cruz) according to manufacturer's instructions for LCLs) and treated for 2.5 hours with 3.3 μM nocodazole and 5 μM MG132 before being pre-extracted with 0.1% Triton X-100 in PEM (100 mM PIPES pH 6.8, 1 mM MgCl2 and 5 mM EGTA) for ±60 seconds before fixation with zinc formalin fixative, 1:1 in PEM with 0.1% Triton X-100 for 5-10 minutes. Coverslips were washed twice with cold PBS and blocked with 3% BSA in PBS for 16 hours at 4°C, incubated with primary antibodies for 16 hours at 4°C, washed 4 times with PBS containing 0.1% Triton X-100, and incubated with secondary antibodies for 1 hour at room temperature. Coverslips were then washed 4 times with PBS/0.1% Triton X-100 and mounted using ProLong Gold Antifade with DAPI (Molecular Probes). All images were acquired on a deconvolution system (DeltaVision Elite; Applied Precision/GE Healthcare) equipped with a 100x/1.40 NA UPlanSApo objective (Olympus) using SoftWorx 6.0 software (Applied Precision/GE Healthcare). Images are maximum intensity projections of deconvolved stacks. A CellProfiler40 28 pipeline was used to threshold and select all kinetochores and all chromosome areas (excluding kinetochores) using the DAPI and CENP-C. This was used to calculate the relative average kinetochore intensity of various proteins ((kinetochores– chromosome arm intensity (kinetochore localized protein of interest))/(kinetochores– chromosome arm intensity (CENP-C))). Quantifications were subjected to unpaired Student’s t-tests; P<0.05. Primary antibodies used were guinea pig anti-CENP-C (MBL International, PD030), rabbit anti-MAD2 (custom raised against full-length 6 x His-tagged MAD2 as antigen) and mouse anti-MAD1 (Millipore, MABE867). Secondary antibodies used were goat anti-guinea pig Alexa Fluor 647 (A21450), goat anti–rabbit Alexa Fluor 488 (A11034) and anti–mouse Alexa Fluor 568 (A11031), all obtained from Invitrogen Molecular Probes.
Quantitative real-time PCR
Lymphoblasts were seeded at 500,000 cells/ml in 20 ml medium and split in two the following day. 50% of cells were treated with 100 μg/ml cycloheximide (Sigma) for 4.5 hours, and the remaining cells were left untreated. Total mRNA was extracted using the RNeasy Mini Kit (Qiagen). Equal amounts of RNA (0.5 μg) were reverse-transcribed using the iScript cDNA Synthesis Kit (Bio-Rad) with an optimum blend of oligo(dT) and random hexamers. Real-time quantification was performed with IQ SYBR green Supermix (Bio-Rad) on a CFX connect Real-Time system (Bio-Rad) using probes generated against TRIP13 and GAPDH (TRIP13 FW: 5’-CGTGCTGATTGATGAGGTGG-3’, TRIP13 RV: 5’-ACGTCGATCTTCTCGGTGAT-3’, GAPDH FW: 5‘-AATCCCATCACCATCTTCCA-3’, GAPDH RV: 5’-TGGACTCCACGACGTACTCA-3’). The data were normalized to GAPDH expression and patient sample data were compared to the average expression of two control samples to quantify relative expression levels of TRIP13, using the 2-ΔΔCt method. Three independent experiments were performed.
Immunoblotting analysis of TRIP13 expression
For the data in Supplementary Figs. 3, 4 and 7) cells were treated as indicated and lysed in Laemmli lysis buffer (4% SDS, 120 mM Tris pH 6.8 and 20% glycerol). Lysates were processed for SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Immunoblotting was performed using standard protocols. Visualization of signals was performed on an Amersham Imager 600 scanner using enhanced chemiluminescence. For quantification of immunostainings, all images of similarly stained experiments were acquired with identical illumination settings. Primary antibodies used were mouse anti-Tubulin (Sigma, T5168), mouse anti-GFP (Roche, 11-814-460-001), rabbit anti-Histone H3 (Abcam, ab1791) and rabbit anti-TRIP13 (Abcam, ab128171). Secondary antibodies used and obtained from Bio-Rad were goat anti-mouse HRP (170-6516) and goat anti-rabbit HRP (170-6515), both obtained from Bio-Rad.
Immunoblotting analysis of TRIP13 p31comet and MAD2 expression
For the data in Supplementary Fig. 7 cell pellets were resuspended in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP-40 supplemented with 1 tablet per 10 ml of protease inhibitor cocktail (Roche) and 10 μM microcystin), vortexed and incubated for 20 minutes on ice. They were clarified by centrifugation for 10 minutes at 4ºC at 14000 rpm. Protein concentration was measured by Bradford assay using Coomassie Plus Proten Assay reagent (Thermo Scientific). Absorbance at 595 nm was measured on a spectrophotometer (Amersham). Primary antibodies used were Mad2 (Bethyl, A300-301A), p31comet (Clone E29.19.14; a kind gift of Andrea Musacchio), beta tubulin (Sigma, T4026) and TRIP13 (Bethyl lab, A303-605A). Secondary antibodies used were IRDye 800CW donkey anti-rabbit (926-32213) and IRDye 680CW donkey anti-mouse (926-68072). The molecular marker used was Odyssey® One-Color Protein Molecular Weight Marker (928-40000). Quantitative immunoblotting was performed using an Odyssey infrared imager (LICOR Biosciences) running Odyssey v1.2 software. Images were opened in ImageStudio Lite and the mean intensity values were extracted by drawing rectangles around the band of interest. The background was measured close to each individual band and automatically subtracted by the software. Protein levels were normalised to tubulin as a loading control.
Supplementary Material
Acknowledgements
We thank the families for their participation and the researchers that recruited them including K. Asakura-Hay, S. Bernardo de Sousa, P. Callier, D. Chitayat, J. Clayton-Smith, S. Fernandes. D. FitzPatrick, L. Florentin, J. Hurst, B. Isidor, S. Jacquemont, R. Marin Iglesias, M. Micale, J. Tolmie. We thank H.J. Snippert for providing the Lentiviral H2B plasmid. We thank Anthony Renwick, Shazia Mahamdallie, Chey Loveday and members of the Kops lab for helpful discussions and Ann Strydom and Brittany Rex for assistance in preparing the manuscript. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre. This research was supported by the Wellcome Trust (100210/Z/12/Z), by the Netherlands Organisation for Scientific Research (NWO-ALW 823.02.004 to GJPLK) and by the Dutch Cancer Society (KWF Kankerbestrijding to RdV).
Footnotes
URLs
OpEx variant calling pipeline: www.icr.ac.uk/opex
ICR1000 UK exome series: www.icr.ac.uk/icr1000exomes
Exome Aggregation Consortium (ExAC): http://exac.broadinstitute.org.
1000 Genomes Project: ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20110521/
Author contributions
N.R. designed and oversaw the study. G.K designed and oversaw the functional experiments. E. Ra. undertook the exome sequencing. S.H., H.W. and S.Se. performed the molecular analyses. S.Y., M.C., and E.Ru. performed bioinformatic analyses. B.dW., E.U., R.dV., B.E. and C.M. undertook functional analyses under the supervision of G.K., and J.P. S.P., A.S., S.Sm. provided samples and data, coordinated by A.Z. and A.E. S.Y., S.H., B.dW., A.Z., G.K. and N.R. wrote the paper with input from the other authors.
Competing financial interests
The authors declare that they have no competing financial interests.
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information.
References
- 1.Garcia-Castillo H, Vasquez-Velasquez AI, Rivera H, Barros-Nunez P. Clinical and genetic heterogeneity in patients with mosaic variegated aneuploidy: delineation of clinical subtypes. Am J Med Genet A. 2008;146a:1687–95. doi: 10.1002/ajmg.a.32315. [DOI] [PubMed] [Google Scholar]
- 2.Warburton D, Anyane-Yeboa K, Taterka P, Yu CY, Olsen D. Mosaic variegated aneuploidy with microcephaly: a new human mitotic mutant? Ann Genet. 1991;34:287–92. [PubMed] [Google Scholar]
- 3.Hanks S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 4.Snape K, et al. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat Genet. 2011;43:527–9. doi: 10.1038/ng.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricke RM, van Deursen JM. Aneuploidy in health, disease, and aging. J Cell Biol. 2013;201:11–21. doi: 10.1083/jcb.201301061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–46. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–95. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajii T, et al. Cancer-prone syndrome of mosaic variegated aneuploidy and total premature chromatid separation: report of five infants. Am J Med Genet. 2001;104:57–64. doi: 10.1002/ajmg.1580. [DOI] [PubMed] [Google Scholar]
- 10.Jacquemont S, Boceno M, Rival JM, Mechinaud F, David A. High risk of malignancy in mosaic variegated aneuploidy syndrome. Am J Med Genet. 2002;109:17–21. doi: 10.1002/ajmg.10281. discussion 16. [DOI] [PubMed] [Google Scholar]
- 11.Ruark E, et al. OpEx - a validated, automated pipeline optimised for clinical exome sequence analysis. Sci Rep. 2016;6:31029. doi: 10.1038/srep31029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahamdallie SS, et al. Mutations in the transcriptional repressor REST predispose to Wilms tumor. Nat Genet. 2015:1471–1474. doi: 10.1038/ng.3440. [DOI] [PubMed] [Google Scholar]
- 13.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruark E, et al. The ICR1000 UK exome series: a resource of gene variation in an outbred population. F1000Res. 2015;4:883. doi: 10.12688/f1000research.7049.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vader G. Pch2(TRIP13): controlling cell division through regulation of HORMA domains. Chromosoma. 2015;124:333–9. doi: 10.1007/s00412-015-0516-y. [DOI] [PubMed] [Google Scholar]
- 16.Eytan E, et al. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31(comet) Proc Natl Acad Sci U S A. 2014;111:12019–24. doi: 10.1073/pnas.1412901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye Q, et al. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. Elife. 2015;4 doi: 10.7554/eLife.07367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, et al. Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J Biol Chem. 2014;289:23928–37. doi: 10.1074/jbc.M114.585315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson CR, Hwang T, Chen PH, Bhalla N. TRIP13PCH-2 promotes Mad2 localization to unattached kinetochores in the spindle checkpoint response. J Cell Biol. 2015;211:503–16. doi: 10.1083/jcb.201505114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma HT, Poon RY. TRIP13 Regulates Both the Activation and Inactivation of the Spindle-Assembly Checkpoint. Cell Rep. 2016;14:1086–99. doi: 10.1016/j.celrep.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Vleugel M, Hoogendoorn E, Snel B, Kops GJ. Evolution and function of the mitotic checkpoint. Dev Cell. 2012;23:239–50. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–9. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rimmer A, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46:912–8. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munz M, et al. CSN and CAVA: variant annotation tools for rapid, robust next-generation sequencing analysis in the clinical setting. Genome Med. 2015;7:76. doi: 10.1186/s13073-015-0195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 27.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–7. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 28.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.