Abstract
Dominant mutations in the MORC2 gene have recently been shown to cause axonal Charcot-Marie-Tooth (CMT) disease, but the cellular function of MORC2 is poorly understood. Here, through a genome-wide CRISPR/Cas9-mediated forward genetic screen, we identify MORC2 as an essential gene required for epigenetic silencing by the HUSH complex. HUSH recruits MORC2 to target sites in heterochromatin. We exploit a new method – Differential Viral Accessibility (DIVA) – to show that loss of MORC2 results in chromatin decompaction at these target loci, which is concomitant with a loss of H3K9me3 deposition and transcriptional derepression. The ATPase activity of MORC2 is critical for HUSH-mediated silencing, and the most common mutation affecting the ATPase domain found in CMT patients (R252W) hyper-activates HUSH-mediated repression in neuronal cells. These data define a critical role for MORC2 in epigenetic silencing by the HUSH complex and provide a mechanistic basis underpinning the role of MORC2 mutations in CMT disease.
Keywords: CRISPR/Cas9 forward genetic screen, heterochromatin, H3K9me3, gene silencing, HUSH complex, MORC2, DIVA, chromatin compaction, Charcot-Marie-Tooth disease
Introduction
Chromatin has historically been considered as existing in one of two distinct states: euchromatin or heterochromatin. Euchromatin is generally considered to be gene-rich, transcriptionally-active and associated with an open and accessible conformation, whereas heterochromatin is comparatively gene-poor, highly condensed and refractory to the transcription machinery1. Heterochromatin has traditionally been considered as occurring in two distinct flavors: H3K27me3-marked heterochromatin, which is dynamic during development2, and H3K9me3-marked heterochromatin, which is typically found at repeat-rich regions3.
A remarkable property of H3K9me3-marked heterochromatin is that it can ‘spread’ along the arm of a chromosome, which can result in the epigenetic repression of an active gene when it is placed in the vicinity of a heterochromatic domain4. This phenomenon gives rise to chromosomal position-effects, a term which refers to the difference in expression when an identical gene is positioned at different sites in the genome5,6. We previously exploited the near-haploid KBM7 cell line7,8 to identify genes required for position-effect variegation in human cells9. By isolating a population of cells that displayed epigenetic repression of an integrated green fluorescent protein (GFP) reporter construct and screening for mutants in which this repression was alleviated, we identified four genes that were essential for transgene repression: the histone lysine methyltransferase SETDB110, plus the Human Silencing Hub (HUSH) complex subunits TASOR, MPP8 and Periphilin9. Through the chromodomain of MPP811, HUSH preferentially localizes to genomic loci rich in the repressive histone modification H3K9me3. HUSH-mediated recruitment of the methyltransferase SETDB1 to these sites results in the ‘writing’ of additional H3K9me3, thereby mediating the spreading of heterochromatin across incoming transgenes9. The HUSH complex also plays a critical role in endogenous heterochromatin maintenance, as deletion of HUSH subunits results in decreased H3K9me3 at hundreds of genomic loci9 and early embryonic lethality in the mouse12,13.
HUSH-mediated silencing is achieved through the reading and writing of H3K9me3. However, in order to understand how the deposition of this modification ultimately results in epigenetic repression, knowledge of the full complement of HUSH components is essential. Although our original haploid screen identified the core HUSH complex members and SETDB1, our recent examination of the efficacy of the haploid gene-trap approach suggested that such screens are unlikely to achieve saturation14. HUSH-mediated silencing may therefore require additional factors not identified by the original gene-trap screen, and, in support of this idea, we recently identified ATF7IP as an additional factor required for HUSH-mediated repression using a proteomic approach15.
With the aim of identifying additional genes required for transgene silencing by the HUSH complex, we carried out a genome-wide forward genetic screen using CRISPR/Cas9 technology16,17. The CRISPR screen highlighted one additional gene as critical for HUSH-mediated repression: Microchidia CW-type zinc finger 2 (MORC2). We show that HUSH recruits MORC2 to heterochromatic sites, where its ATPase activity is required to mediate transgene silencing. Loss of MORC2 results in chromatin decompaction at HUSH target loci, concomitant with a loss of H3K9me3 and transcriptional derepression. Mutations in the ATPase domain of MORC2 have recently been implicated in Charcot-Marie-Tooth (CMT) disease, one of the most frequently inherited neurological disorders. We show that the most common of these mutations (R252W; p.Arg252Trp) results in hyper-activation of HUSH-mediated silencing in neuronal cells. Overall, this study identifies a critical role for MORC2 in HUSH-mediated repression and provides further insight into the mechanisms underlying HUSH complex function in health and disease.
Results
A genome-wide CRISPR screen identifies an essential role for MORC2 in HUSH-mediated silencing
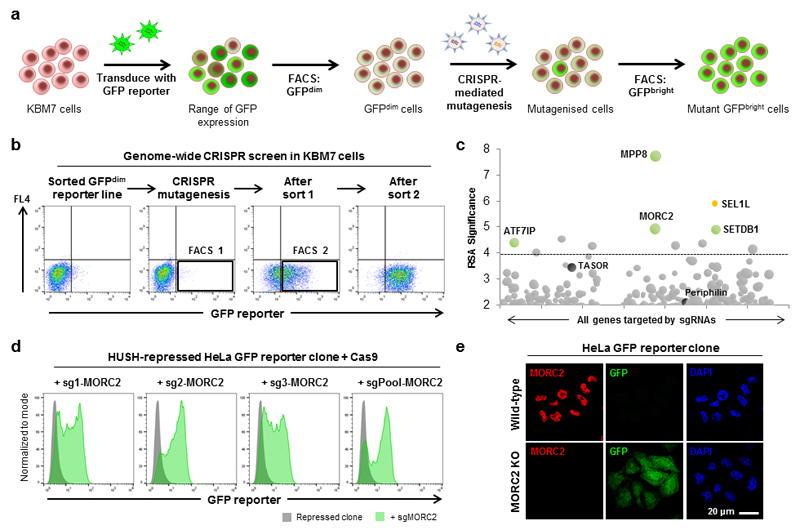
The rapid emergence of CRISPR/Cas9-mediated genome editing technologies has now allowed us to reevaluate the set of genes required for HUSH-mediated transgene silencing (Fig. 1a). The Cas9 nuclease was expressed in the same population of GFPdim KBM7 cells harboring epigenetically-repressed transgenes that we used previously9, and genome-wide CRISPR-mediated mutagenesis was performed using the GeCKO v2 sgRNA library18. Two rounds of fluorescence-activated cell sorting (FACS) were used to enrich for GFPbright cells in which epigenetic repression of the GFP reporter transgene was relieved (Fig. 1b), and sgRNA abundance in the selected GFPbright cells versus the unselected library of mutagenized cells was quantified by Illumina sequencing (Fig. 1c and Supplementary Fig. 1a).
Figure 1. A genome-wide CRISPR/Cas9-mediated forward genetic screen identifies an essential role for MORC2 in transgene silencing by the HUSH complex.
(a,b) A genome-wide CRISPR screen to identify genes required for transgene silencing. Cas9 was expressed in a population of GFPdim KBM7 cells harboring epigenetically repressed transgenes, and genome-wide mutagenesis carried out using the GeCKO v2 sgRNA library (a). Mutant GFPbright cells containing gene disruption events that prevented reporter repression were isolated through two sequential rounds of FACS (b). Black boxes indicate approximate sorting gates. (c) Bubble plot illustrating the hits from the screen. All genes targeted by sgRNAs are arranged alphabetically by gene name on the x-axis, with their statistical significance as determined by the RSA algorithm plotted on the y-axis. Bubble size is proportional to the number of active sgRNAs for each gene. Colored bubbles represent validated hits; the HUSH complex subunits TASOR and Periphilin (black) did not reach statistical significance, while SEL1L (orange) is involved in the degradation of the GFP fusion protein14. A fully annotated plot is provided in Fig. S1. (d,e) MORC2 is required for transgene silencing by the HUSH complex in HeLa cells. CRISPR/Cas9-mediated disruption of MORC2 results in derepression of a HUSH-repressed reporter as measured by flow cytometry (d) or immunofluorescence microscopy (e).
Five genes are known to be critical for HUSH-mediated transgene repression: the three HUSH complex subunits TASOR, MPP8 and Periphilin9, plus the histone methyltransferase SETDB19 and its accessory factor ATF7IP15. Three of these positive controls, MPP8, SETDB1 and ATF7IP, were successfully identified by the CRISPR screen; TASOR and Periphilin were not significantly enriched, however, suggesting that the screen failed to reach saturation (Fig. 1c and Supplementary Fig. 1a). Through individual CRISPR/Cas9-mediated gene disruption experiments, we assessed a potential role in transgene silencing for the other most significantly enriched genes (Supplementary Fig. 1b). This revealed an essential role for one additional gene, MORC2, a finding which we corroborated in an independent HeLa cell clone harboring a HUSH-repressed GFP reporter construct9 (Fig. 1d,e). With the exception of SEL1L, which mediates the degradation of the GFP reporter at the protein level14, we were unable to validate any of the additional candidate genes (Supplementary Fig. 1b). Therefore, the genome-wide CRISPR screen identified a single novel gene, MORC2, as a bona fide factor required for HUSH-mediated transgene repression.
The ATPase, CW and coiled-coil domains of MORC2 are essential for HUSH function
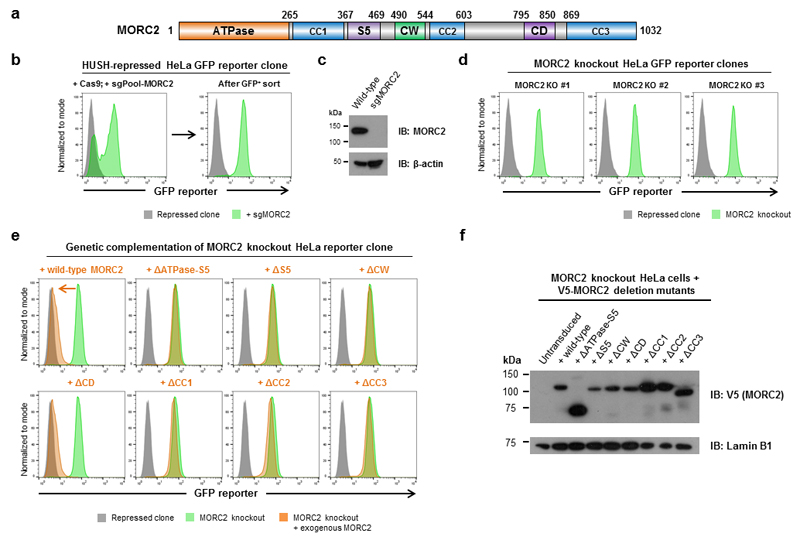
MORC2 is one of four MORC family proteins in humans (MORC1-4)19. Although little is known about their functional role in human cells, MORC family proteins have been implicated in epigenetic silencing in plants, worms, and mice20,21. MORC2 encodes a protein containing a GHKL-type ATPase, comprised of the N-terminal ATPase domain plus an associated ribosomal protein S5-like domain, followed by two putative histone binding modules: a CW-type zinc finger and a chromo-like domain (Fig. 2a). To assess which of these domains were required for the function of MORC2 in HUSH-mediated repression, we performed a series of genetic complementation experiments in a MORC2 knockout HeLa clone (Fig. 2b-d and Supplementary Table 1). While exogenous expression of full-length MORC2 or a MORC2 mutant lacking the chromo-like domain resulted in re-repression of the GFP reporter, deletion of both ATPase domains, the S5-like domain alone or the CW-type zinc finger abolished MORC2 activity (Fig. 2e,f). Sequence analysis also predicted the presence of three coiled-coil domains (denoted CC1, CC2 and CC3) (Fig. 2a); deletion of any one of these also abrogated MORC2 function (Fig. 2e,f). Thus, the MORC2 ATPase domain, CW-type zinc finger and coiled-coil domains, but not the chromo-like domain, appeared to be critical for HUSH-mediated epigenetic repression, with the caveat that although these mutant proteins are expressed in cells (Fig. 2f), we are unable to prove that they are correctly folded.
Figure 2. The ATPase, CW and coiled-coil domains of MORC2 are required for HUSH complex function.
(a) Schematic representation of the domain structure of MORC2. (CC, predicted coiled-coil; S5, ribosomal S5-like domain; CW, CW-type zinc finger; CD, chromo-like domain) (b-d) Generation and validation of MORC2 knockout HeLa cells. A HeLa reporter clone harboring a HUSH-repressed GFP reporter was transfected with a pool of plasmids to express Cas9 and three sgRNAs targeting MORC2. Cells in which the MORC2 gene was disrupted became GFP+ owing to derepression the GFP reporter and were isolated using FACS (b). The sorted GFP+ population no longer expressed MORC2 protein as assessed by immunoblot (c). MORC2 knockout single cell clones were then isolated from the GFP+ sorted population (d). (e,f) Assessing the domains required for MORC2 function through the genetic complementation of MORC2 knockout cells. Expression of wild-type MORC2 or a MORC2 mutant lacking the chromodomain (ΔCD) in MORC2 knockout cells resulted in re-repression of the GFP reporter transgene. In contrast, MORC2 variants lacking the ATPase, S5, CW or coiled-coil domains were non-functional and did not rescue HUSH-mediated repression (e). Immunoblot validation of expression of the MORC2 deletion mutants (f). All MORC2 variants were expressed with an N-terminal V5 epitope tag followed by an exogenous SV40 nuclear localization signal; detailed sequence information on the composition of the mutants is provided in Supplementary Table 1.
MORC2 interacts with the HUSH complex
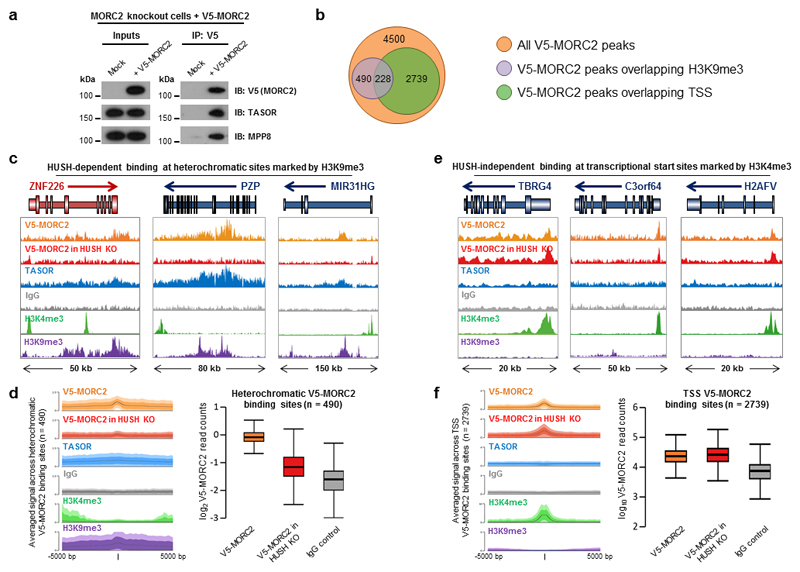
The genetic association between MORC2 and the HUSH complex suggested that MORC2 might physically interact with HUSH subunits. We were readily able to detect an interaction between a V5 epitope-tagged MORC2 immunoprecipitated from HeLa nuclei and the HUSH subunits TASOR and MPP8 (Fig. 3a). Repeating this assay with a series of V5-tagged N- and C-terminal truncation mutants of MORC2 (Supplementary Table 1) suggested that the second coiled-coil domain (CC2) of MORC2 (residues 548-603) was likely to be essential for this interaction (Supplementary Fig. 2a-c). The interaction between MORC2 and HUSH subunits also occurred in SETDB1 knockout cells (Supplementary Fig. 2d) and deletion of MORC2 did not prevent the interaction between SETDB1 and HUSH (Supplementary Fig. 2e), suggesting independent recruitment of these two potential effector proteins by the HUSH complex. However, like SETDB1, MORC2 did not appear to be a constitutive member of the HUSH complex, as, unlike loss of TASOR, MPP8 or Periphilin9, loss of MORC2 did not affect the protein levels of HUSH subunits (Supplementary Fig. 2f) nor their localization to chromatin (Supplementary Fig. 2g).
Figure 3. The HUSH complex recruits MORC2 to heterochromatic target sites.
(a) MORC2 interacts with the HUSH complex. TASOR and MPP8 coimmunoprecipitate with V5-tagged MORC2 expressed in HeLa cells. (b) Summary of MORC2 genome occupancy as measured by ChIP-seq. In total 4500 peaks of V5-MORC2 occupancy were detected, the majority of which fell into two categories: binding sites in heterochromatin marked by H3K9me3 (purple; left) and binding sites at transcriptional start sites (green; right). (c,d) The HUSH complex recruits MORC2 to heterochromatic sites marked by H3K9me3. TASOR and MORC2 occupancy was observed at a range of sites marked by H3K9me3; MORC2 recruitment at these sites was HUSH-dependent, as V5-MORC2 occupancy was lost in cells lacking all three HUSH subunits9 (HUSH KO). Three example loci are shown in (c), with summary data across all loci shown in (d). Full ChIP-seq traces including input DNA controls are shown in Supplementary Fig. 3a. (e,f) MORC2 recruitment to transcriptional start sites is independent of the HUSH complex. MORC2 occupancy was observed at a large number of transcriptional start sites (TSSs) marked by the H3K4me3 histone modification. This was independent of the HUSH complex, as TASOR occupancy was not observed at these sites and MORC2 occupancy was maintained in cells lacking HUSH subunits. Three example loci are shown in (e), with summary data across all loci shown in (f). Full ChIP-seq traces including input DNA controls are shown in Supplementary Fig. 3b.
The HUSH complex recruits MORC2 to heterochromatic loci
To ascertain the physiological relevance of the association between MORC2 and the HUSH complex, we performed chromatin immunoprecipitation followed by deep sequencing (ChIP-seq). Immunoprecipitation of a V5-tagged MORC2 construct expressed in MORC2 knockout cells revealed two modes of MORC2 binding to chromatin (Fig. 3b): a broad distribution across the bodies of genes located in heterochromatin marked by H3K9me3 (Fig. 3c,d and Supplementary Fig. 3a) and a series of discrete peaks at transcriptional start sites (TSSs) (Fig. 3e,f and Supplementary Fig. 3b). Comparing the occupancy of MORC2 with that of endogenous TASOR revealed significant overlap between the heterochromatic regions bound by MORC2 and those occupied by the HUSH complex (Fig. 3c,d). Indeed, MORC2 localization to these heterochromatic sites was dependent on the HUSH complex, as MORC2 occupancy was lost in cells lacking all three HUSH subunits (Fig. 3c,d). Thus, these datasets revealed HUSH-dependent recruitment of MORC2 to heterochromatic loci marked by H3K9me3. The functional relevance of MORC2 recruitment to TSSs remains unclear, but this was not dependent on the HUSH complex (Fig. 3e,f) and did not involve H3K4me3 recognition by the CW domain of MORC222 (Supplementary Fig. 4), as has been demonstrated for the CW domains of MORC3 and MORC423,24.
In further support of an association between HUSH and MORC2, we also found that the HUSH complex regulated the expression of MORC2. Knockout of HUSH subunits or SETDB1 resulted in an increase in MORC2 expression as measured by quantitative reverse transcription PCR (qRT-PCR) (Supplementary Fig. 2h). In support of the idea that this represented a direct feedback loop, our ChIP-seq data demonstrated both TASOR occupancy and HUSH-dependent recruitment of MORC2 to an H3K9me3-rich site in the MORC2 promoter (Supplementary Fig. 2i).
MORC2 loss results in chromatin decompaction at HUSH loci
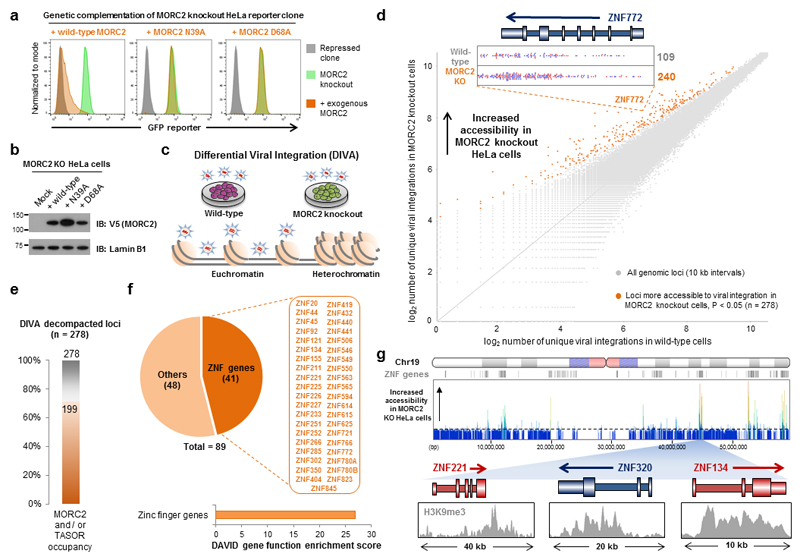
The catalytic activity of the MORC2 ATPase domain allows MORC2 to remodel nucleosomal templates in vitro25. Therefore, we hypothesized that ATP hydrolysis by MORC2 might be required to alter chromatin architecture at HUSH target sites in vivo. To directly test the functional requirement for MORC2 ATPase activity in HUSH-mediated silencing, we carried out further genetic complementation assays in MORC2 knockout HeLa cells (Fig. 4a). While exogenous expression of wild-type MORC2 restored HUSH function and resulted in the re-repression of the GFP reporter construct, MORC2 variants harboring point mutations in the critical residues of the ATPase domain required for ATP binding (N39A) or hydrolysis (D68A)25 were non-functional (Fig. 4a,b).
Figure 4. Loss of MORC2 results in chromatin decompaction at HUSH target sites.
(a,b) ATP hydrolysis by MORC2 is critical for HUSH-mediated transgene repression. Exogenous expression of MORC2 mutants unable to bind (N39A) or hydrolyze (D68A) ATP failed to rescue reporter repression in MORC2 knockout cells (a). Immunoblot validation of expression of the MORC2 point mutants (b). (c) Overview of the DIVA methodology, which utilizes large-scale mapping of lentiviral integration sites to probe accessible chromatin. See Supplementary Figure 5c,d for more details. (d) Loss of MORC2 results in chromatin decompaction. Scatter plot highlighting the 289 genomic loci (orange) which exhibit increased accessibility in MORC2 knockout cells. Unique lentiviral integration sites mapped to the ZNF772 locus in wild-type and MORC2 knockout cells are shown as an example. DIVA scores for all genomic loci are detailed in Supplementary Table 2. (e) The majority of loci exhibiting decompaction upon MORC2 knockout are direct targets of MORC2 and the HUSH complex. Of the 278 decompacted loci, MORC2 and/or TASOR occupancy can be detected at 199 loci (71%). (f,g) Loci exhibiting decompaction upon MORC2 knockout are highly enriched for ZNF genes (f), and the decompacted regions across chromosome 19 correspond to sites of ZNF gene clusters (g).
Should the ATPase activity of MORC2 be required to mediate chromatin compaction during HUSH-mediated repression in vivo, disruption of MORC2 should result in a more open chromatin structure at sites of HUSH activity. ATAC-seq is currently a leading method to assay changes in chromatin accessibility in vivo26, but our attempts to exploit this technology to examine potential chromatin decompaction in MORC2 knockout HeLa cells were hampered by a large proportion of contaminating mitochondrial reads27 and insufficient coverage of the heterochromatic portions of the genome at which HUSH functions (Supplementary Fig. 5a,b). We therefore developed a new approach, Differential Viral Accessibility (DIVA), to examine changes in chromatin accessibility in vivo. DIVA is conceptually similar to ATAC-seq, but, rather than identifying transposon integration sites to probe accessible chromatin, DIVA maps the integration sites of exogenous lentiviruses (Fig. 4c and Supplementary Fig. 5c). In contrast to the Tn5 transposase which preferentially integrates near transcriptional start sites, HIV-1-derived lentiviral vectors preferentially target gene bodies28.
We used DIVA to compare chromatin accessibility in wild-type and MORC2-null HeLa cells. Approximately 20 million independent viral integration sites were mapped in each cell type, and the two datasets were then compared to identify loci exhibiting significantly greater numbers of integrations in the MORC2 knockout cells compared to the parental wild-type cells (Fig. 4c and Supplementary Fig. 5d-g). Considering the genome as a series of 10 kb windows, we identified 278 loci exhibiting chromatin decompaction upon knockout of MORC2 (Fig. 4d and Supplementary Table 2). A large proportion of these loci were heterochromatic sites at which TASOR or MORC2 occupancy was observed by ChIP-seq (Fig. 4e). Notably, many of these loci contained zinc finger (ZNF) genes (Fig. 4f), and, on the ZNF-rich chromosome 19, for example, a striking concordance was observed between the positions of ZNF gene clusters and the loci exhibiting decompaction in MORC2 knockout cells (Fig. 4g). Employing an orthogonal technique based on micrococcal nuclease sensitivity, we validated increased chromatin accessibility in MORC2 knockout cells at four example ZNF loci (Supplementary Fig. 6).
MORC2 knockout results in chromatin decompaction, decreased H3K9me3 and transcriptional derepression
We also wanted to assess the relationship between MORC2-dependent changes in chromatin accessibility and H3K9me3 deposition. We found that loss of MORC2 in HeLa cells resulted in a decrease in H3K9me3 at the vast majority of loci exhibiting chromatin decompaction upon MORC2 knockout (Fig. 5a,b and Supplementary Fig. 7a), supporting the idea that chromatin decompaction also results in a concomitant decrease in H3K9me3 levels. Consistent with the functional association between MORC2 and the HUSH complex, 906 of the 918 loci (99%) that we previously found to have lost H3K9me3 upon deletion of either of the three HUSH subunits9 also showed decreased H3K9me3 levels in MORC2 knockout cells (Fig. 5c). At the vast majority of these sites, H3K9me3 deposition was dependent on the HUSH-associated methyltransferase SETDB1 (Supplementary Fig. 7a,b).
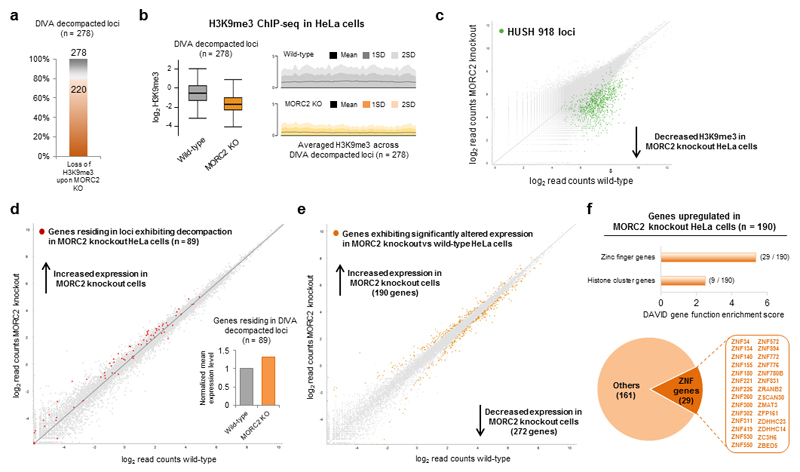
Figure 5. Chromatin decompaction in MORC2 knockout cells is accompanied by a loss of H3K9me3 and transcriptional depression.
(a,b) H3K9me3 is lost across the majority of loci exhibiting decompaction upon MORC2 knockout. Of the 278 decompacted loci, a loss of H3K9me3 upon MORC2 knockout in HeLa cells was observed at 220 loci (79%), as measured by ChIP-seq (a). Summary data across all 278 decompacted loci is shown in (b). (c) Knockout of MORC2 results in loss of H3K9me3 at the same sites which lose H3K9me3 upon knockout of HUSH subunits. Previously we identified 918 genomic loci in HeLa cells that lose H3K9me3 upon knockout of either TASOR, MPP8 or Periphilin (green dots)9; these same loci also exhibited decreased H3K9me3 levels upon knockout of MORC2. (d,e) The effect of knockout of MORC2 on the transcriptome. RNA-seq analysis was performed to compare the transcriptome of wild-type and MORC2 knockout HeLa cells. In (d), genes residing in loci exhibiting decompaction in MORC2 knockout cells (n = 89) are highlighted in red; loci exhibiting decompaction also exhibit increased expression as measured by RNA-seq (inset). In (e), all genes exhibiting significantly altered expression between wild-type and MORC2 knockout cells (n = 462; DEseq P < 0.05) are highlighted in orange. (f) Functional classification of the genes upregulated in MORC2 knockout cells using the DAVID functional annotation tool. Genes upregulated upon MORC2 knockout are highly enriched for ZNF genes.
Finally we considered the effect of chromatin decompaction at these sites on gene expression. In addition to a decrease in H3K9me3, we found that decompaction following knockout of MORC2 resulted in an increase in the mean expression level of the 89 genes residing within these loci (Fig. 5d), with 42 genes (47%) displaying >1.2-fold increase in expression. Furthermore, of all 190 genes significantly upregulated upon MORC2 knockout (Fig. 5e), ZNF genes represented the most significantly enriched functional group (Fig. 5f). Consistent with a repressive role for MORC2, the majority of the upregulated genes were direct targets of TASOR and/or MORC2 as assessed by ChIP-seq (Supplementary Fig. 7c,d), and many also exhibited a loss of H3K9me3 and decompaction upon MORC2 knockout (Supplementary Fig. 7e). In contrast, ablation of MORC2 did not result in a global transcriptional change across genes exhibiting HUSH-independent MORC2 occupancy at their TSSs (Supplementary Fig. 7f). Altogether, these data support a model whereby loss of MORC2 results in chromatin decompaction, a loss of SETDB1-mediated H3K9me3 deposition and transcriptional derepression at HUSH target sites.
The R252W CMT mutation in MORC2 hyper-activates HUSH silencing
Five independent studies have recently reported that dominant mutations in the ATPase domain of MORC2 can cause axonal Charcot-Marie-Tooth (CMT) disease (Fig. 6a)29–33. We focused on understanding the functional impact of the most prevalent mutation, an arginine to tryptophan substitution at residue 252 (R252W or p.Arg252Trp in Uniprot: Q9Y6X9-1), which results in a severe axonal form of CMT230. This is the identical mutation to the reported R190W variant of MORC229,31,32, which refers to a putative alternative isoform of the protein (Uniprot: Q9Y6X9-2) that lacks 62 amino acids at the N-terminus.
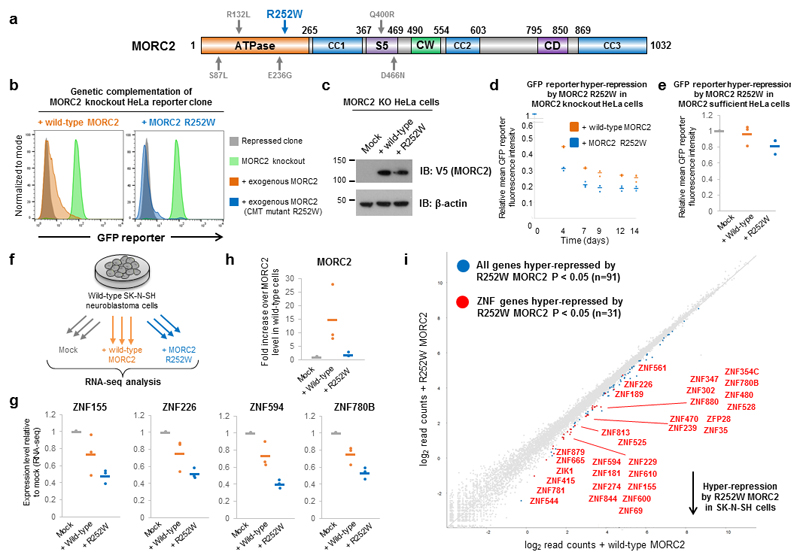
Figure 6. The R252W mutation in MORC2 associated with Charcot-Marie-Tooth disease hyper-activates HUSH-mediated epigenetic repression.
(a) Schematic representation of the mutations associated with CMT affecting the ATPase domain of MORC2. (b,c) Assessing the effect of the R252W CMT mutation on MORC2 function through genetic complementation of MORC2 knockout cells. The R252W CMT mutant MORC2 is functional, restoring HUSH-mediated transgene silencing when expressed in MORC2 knockout cells (b). Immunoblot validation of expression of wild-type or R252W mutant MORC2 (c). (d,e) The R252W CMT mutation in MORC2 hyper-activates HUSH-mediated transgene silencing in HeLa cells. Time-course of transgene re-repression in MORC2 knockout HeLa cells (d): the R252W MORC2 mutant increases both the rate and overall extent of transgene re-repression. Hyper-repression of a GFPdim HUSH reporter in MORC2-sufficient cells by over-expression of R252W MORC2 (e). (f) Schematic representation of the RNA-seq experiment in wild-type SK-N-SH neuroblastoma cells. (g – i) Expression of either wild-type MORC2 or the R252W mutant results in hyper-repression of HUSH target genes. RNA-seq in SK-N-SH cells overexpressing either wild-type or R252W MORC2 reveals hyper-repression of example HUSH target genes (g); despite R252W MORC2 being expressed at a much lower level than the wild-type protein (h), this effect was more pronounced with the R252W mutant. In total, 91 genes were hyper-repressed by the R252W mutant (edgeR P < 0.05), of which 31 were ZNF genes (i).
Consistent with the genetic data which suggests a gain-of-function mechanism underlying the pathogenicity of the mutations, MORC2 harboring the R252W mutation did encode a functional protein capable of restoring HUSH-mediated transgene repression in MORC2 knockout HeLa cells (Fig. 6b,c). Moreover, the R252W mutant consistently outperformed the wild-type protein, resulting in enhanced re-repression of the GFP reporter. Indeed, despite being expressed at a lower level that the wild-type protein, the R252W mutation both accelerated the rate of reporter re-repression and enhanced the overall degree of reporter repression observed (Fig. 6d and Supplementary Fig. 8). To verify that this effect was also observed at endogenous genes targeted by HUSH and MORC2, we performed RNA-seq analysis to compare the transcriptome of MORC2 knockout HeLa cells reconstituted with wild-type MORC2 versus the R252W mutant (Supplementary Fig. 9). In agreement with the results from the reporter assays, we found that the R252W mutation resulted in the hyper-repression of MORC2 target genes (Supplementary Fig. 9).
The mutant MORC2 proteins exert a dominant effect in CMT patients, driving the disease phenotype when co-expressed with a wild-type MORC2 allele. Therefore, we considered whether the R252W mutant MORC2 could hyper-repress HUSH target genes when expressed alongside the wild-type protein in MORC2-sufficient cells. First, we found that overexpression of R252W MORC2, but not wild-type, resulted in hyper-repression of a HUSH-responsive “GFPdim” HeLa reporter clone that exhibited weak expression at steady state (Fig. 6e). We then considered whether this effect would also occur at endogenous genes in neuronal cells. Transcriptome analysis of SK-N-SH neuroblastoma cells by RNA-seq (Fig. 6f) showed that overexpression of either wild-type or R252W MORC2 enhanced repression at example HUSH target sites (Fig. 6g); however, this effect was substantially greater with the R252W mutant, despite it being expressed at ~7.5-fold lower levels than the wild-type protein (Fig. 6h). Indeed, of the 91 genes significantly over-repressed by R252W compared to the wild-type protein, 31 (34%) were ZNF genes marked by H3K9me3 (Fig. 6i). Overall these data suggested that the R252W mutation in MORC2 can hyper-activate HUSH-mediated epigenetic repression, resulting in over-repression of HUSH target genes in neuronal cells.
Discussion
The recently-identified HUSH complex is an important regulator of mammalian heterochromatin that is critical for silencing of newly-integrated retroviruses9 and for normal embryonic development12,13. Here, through a genome-wide CRISPR/Cas9-mediated forward genetic screen, we have identified MORC2 as an additional accessory member of the HUSH complex. MORC2 is recruited by the HUSH complex to heterochromatic loci, where its ATPase activity is essential for HUSH-mediated silencing. Exploiting a new method, Differential Viral Accessibility (DIVA), we found that loss of MORC2 resulted in chromatin decompaction at these sites, accompanied by a decrease in H3K9me3 levels and transcriptional derepression. Furthermore, we show that the most commonly identified Charcot-Marie-Tooth disease mutation in the MORC2 ATPase domain, R252W, results in hyper-activation of HUSH-mediated repression at heterochromatic loci.
Previously we identified the HUSH complex through a gene-trap mutagenesis screen in the near-haploid KBM7 human cell line9. The rapid emergence of CRISPR/Cas9-mediated genome editing technologies has now allowed us to reexamine the complement of genes required for transgene repression, using a genome-wide library of sgRNAs to create the starting pool of mutant cells. The results of the two screens proved to be complementary, as, although the CRISPR screen failed to identify two of the core HUSH complex subunits detected by the original haploid gene-trap screen, it did reveal an essential role for one additional gene, MORC2 (Fig. 1). The CRISPR screening technique therefore represents a powerful alternative to the haploid gene-trap approach14, and, with considerable interest in the design of more efficacious guide RNA libraries34–36, it is likely that the accuracy of such genome-wide CRISPR screens will continue to improve.
In humans, the MORC protein family comprises four proteins (MORC1-4) plus the more divergent SMCHD119. MORC1 represses transposable elements in the male mouse germ line21 and MORC2 has been implicated in the response to DNA damage25, but the molecular function of the mammalian MORC family members remains largely uncharacterized. All MORC proteins share a similar domain architecture, comprising an N-terminal GHKL-type ATPase domain and a CW-type zinc finger, but their functions are likely to be distinct. For example, the inability of the CW-type zinc fingers of MORC1 and MORC2 to recognize methylated H3K4 suggests a different mode of recruitment to chromatin compared to MORC3 and MORC422, whilst it is only mutations in MORC2 that are associated with CMT disease29–33. A key challenge, therefore, is to elucidate how MORC proteins are recruited to chromatin and to determine their effect on nucleosomal architecture at target sites. Here we demonstrate that HUSH regulates the localization of MORC2 to target sites on chromatin, and that the ATPase activity of MORC2 plays a critical role in altering chromatin architecture at these sites during HUSH-mediated silencing.
The ATPase activity of MORC2 is required to drive nucleosome remodeling in vitro25, and was essential for transgene silencing by the HUSH complex in vivo (Fig. 4a,b). Therefore, it followed that the remodeling activity of MORC2 might be required to alter chromatin architecture at these sites to promote gene silencing. Although ATAC-seq has been widely used to study the degree of chromatin “opening” at transcriptional start sites, we found it unsuitable to probe decompaction of heterochromatic regions due to the low proportion of transposon integrations into such sites. Instead of mapping transposon integration sites, DIVA examines the integration preferences of exogenous lentiviruses to probe accessible chromatin. Lentiviruses preferentially target the bodies of genes28, and indeed our data show that this approach affords far greater coverage of the heterochromatic sites at which the HUSH complex functions. We therefore anticipate that DIVA may prove a useful approach to interrogate changes in heterochromatin structure in vivo in other experimental systems. However DIVA may not be suitable for comparisons between non-isogenic cell lines, where it may be impossible to control for differences in the abundance or activity of other factors influencing viral integration preferences. Furthermore DIVA relies on high-efficiency transduction with lentiviral vectors, which may not be achievable in all cell types, and, compared to the simplicity of ATAC-seq, requires a more involved protocol for the preparation of sequencing libraries to map virus-genome junctions.
Our application of DIVA to compare wild-type and MORC2-null cells revealed that loss of MORC2 resulted in chromatin decompaction at HUSH target sites. The repetitive ZNF gene family represented the most enriched functional group. Many of the ZNF genes are located in clusters covered by high levels of H3K9me3, which also coats other repetitive regions of the genome, including centromeres and tandem repeats. While we did not observe decompaction of these additional repetitive regions upon MORC2 disruption, we do not know how well DIVA will capture these other sites of repressive heterochromatin. Further experiments will be needed to address the specific physiological role of MORC2 with HUSH and SETDB1 in maintaining H3K9me3 across the body of ZNF genes.
The DIVA results suggest that HUSH target loci normally adopt a compact chromatin state which needs to be reversed to alleviate repression. Although repressed genomic regions have canonically been associated with dense chromatin domains that are refractory to the transcription machinery, there may be distinct mechanisms through which such nucleosome compaction is achieved. In particular, the specific requirement for ATP hydrolysis to drive compaction during HUSH-mediated silencing seems to be distinct from other key repressive complexes. For example, ATP hydrolysis has not been implicated in heterochromatin formation through the SUV39H1-HP1 axis, another major route to H3K9me3-mediated silencing. Compact heterochromatic structures have also been associated with Polycomb-driven repression37, but again this compaction function does not seem to require an ATP motor and has instead been attributed to a disordered region rich in basic amino acids in PRC138. However, ATP-dependent chromatin remodelers have previously been shown to play an indirect role in transcriptional repression by establishing the appropriate nucleosome spacing for subsequent silencing39. For example, the Nucleosome Remodeling Deacetylase (NuRD) complex utilizes the chromatin remodeling function of its CHD subunits to space adjacent nucleosomes, thereby making the histone tails accessible for deacetylation40. Therefore, one possibility is that MORC2 is recruited by HUSH to position nucleosomes in a manner that renders the chromatin permissive to transcriptional repression.
The mechanism through which dominant mutations in MORC2 cause axonal CMT disease29–33 and severe spinal muscular atrophy (SMA)-like disease41 is unknown. Given that these patients retain one functional wild-type MORC2 allele, the disease phenotype is unlikely to result from loss-of-function of the mutant protein. Our data suggest that a gain-of-function effect underlies the phenotype of the MORC2 R252W CMT mutation, which results in hyper-activation of HUSH complex function at both exogenous transgenes and endogenous genomic loci. Indeed, the closely-related MORC1 protein – which represses transposable elements in the male germ line21 – contains a tryptophan residue at the equivalent position.
Further work will be required to elucidate the biochemical basis underlying the hyper-activation phenotype resulting from the MORC2 R252W mutation. Structural insight into the organization of the MORC2 ATPase domain would be invaluable in assessing the impact of the R252W mutation, and may also guide the rational design of small molecule inhibitors. Inhibition of Hsp90, the prototypical member of the GHKL-family of ATPases, has been successfully achieved using the small molecules geldanamycin and radicicol42,43, and the latter is also effective against the distantly-related GHKL-family member SMCHD144. Overall, these data suggest that inhibition of the ATPase domain of MORC2 provides an attractive target for the therapeutic modulation of HUSH complex function.
Online Methods
Cell culture
HeLa cells were obtained from ECACC and were grown in RPMI 1640 plus 10% fetal calf serum (FCS) and penicillin/streptomycin (100 U/ml); approximate doubling time was 22-23 hours for both wild-type cells all derivative cell lines. KBM7 cells, obtained from Dr. Brent Cochran7, and HEK 293ET cells, a gift from Dr. Felix Randow, and were cultured in IMDM plus 10% FCS and penicillin/streptomycin (100 U/ml). SK-N-SH cells were a gift from Prof. David Rubinsztein and were grown in DMEM plus 10% FCS and penicillin/streptomycin (100 U/ml); approximate doubling time was 55-60 hours for both wild-type cells and all derivative cell lines. All cell lines were routinely tested for mycoplasma contamination (ATCC Universal Mycoplasma Detection Kit).
Antibodies
The following primary antibodies were used: rabbit α-MORC2 (Bethyl Laboratories, A300-149A used for immunoblot), rabbit α-MORC2 (Santa Cruz Biotechnology, sc-366271, used for immunofluorescence), rabbit α-TASOR (Atlas Antibodies, HPA006735), rabbit α-MPP8 (Proteintech, 16796-1-AP), rabbit α-SETDB1 (Proteintech, 11231-1-AP), mouse α-GFP (Life Technologies, A11120), mouse α-V5 (Abcam, ab27671), rabbit α-V5 (Abcam, ab15828), goat α-Lamin B1 (Santa Cruz, sc-6217), rabbit α-H3K9me3 (Abcam, ab8898), rabbit α-Histone H3 (Biolegend, 601902), rabbit IgG (Cell Signaling, #2729), mouse α-calnexin (AF8, a kind gift from M. Brenner, Harvard Medical School) and mouse α-β-actin (Sigma-Aldrich, A5316). Alexa Fluor 488- and Alexa Fluor 546-conjugated secondary antibodies for immunofluorescence were obtained from Molecular Probes. HRP-conjugated secondary antibodies for immunoblot were obtained from Jackson ImmunoResearch.
CRISPR/Cas9-mediated forward genetic screen
Previously we generated a polyclonal population of GFPdim KBM7 cells harboring epigenetically-repressed transgenes through lentiviral transduction with a vector expressing a GFP-fusion protein from the SFFV LTR promoter (pHRSIN-PSFFV-GFP-HLA-A2). The Cas9 nuclease was stably expressed in this population through lentiviral transduction followed by hygromycin selection (500 μg/ml). One hundred million Cas9-expressing reporter KBM7 cells were transduced with the GeCKO v2 sgRNA library at a multiplicity of infection of ~0.1 and untransduced cells removed from the population through puromycin selection (0.75 μg/ml) commencing 48 hours post-transduction. GFPbright cells resulting from the mutagenesis were enriched by two rounds of FACS, with the first sort taking place 7 days post-mutagenesis and the second sort taking place a further 7 days later. Genomic DNA was extracted (Gentra Puregene Kit) from the selected GFPbright cells after the second sort together with a representative sample of the unsorted mutagenized library that had been grown in parallel for the equivalent amount of time.
The abundance of sgRNAs in each sample was quantified by Illumina sequencing. The variable region of the sgRNAs was amplified in a nested PCR reaction (Q5 High-Fidelity Polymerase, NEB) with forward primers binding the U6 promoter region and reverse primers binding the constant region of the sgRNA. For PCR1, 32 x 100 μl reactions each using 4 µg of DNA were performed for the unselected library sample, and 32 x 100 μl reactions each using 4 µg of DNA were performed for selected cells sample. The products of these reactions were pooled, one-tenth was purified using Agencourt AMPure XP beads (Beckman Coulter), and one-tenth of resulting DNA was used as a template for 12 cycles of PCR2 with Illumina P5- and P7- adapted primers. Following a further AMPure XP bead purification, PCR products were quantified, pooled and sequenced on an Illumina HiSeq 2500 instrument using 50 bp single-end reads from a custom primer binding immediately upstream of the sgRNA sequence. All primer sequences are detailed in Supplementary Table 3.
The resulting sequence reads were trimmed of the constant portion of the sgRNA using the fastx toolkit, and then aligned to an index of the GeCKO v2 library using Bowtie 2. Uniquely aligning reads were used to generate sgRNA count tables for each sample, which were analyzed further using the RSA algorithm to identify genes significantly enriched for gene disruption events in the selected cells versus the unselected library.
Individual CRISPR/Cas9-mediated gene disruption
Oligonucleotides (Sigma-Aldrich) for top and bottom strands of the sgRNA were phosphorylated with T4 PNK (NEB), annealed by heating to 95ºC followed by slow cooling to room temperature, and then cloned into either the dual Cas9/sgRNA expression vector pSpCas9(BB)-2A-Puro (Addgene #48139, kindly deposited by Dr. Feng Zhang45) or the lentiviral sgRNA expression vector pKLV-U6gRNA(BbsI)-PGKpuro2ABFP (Addgene #50946, kindly deposited by Dr. Kosuke Yusa46).
Lentiviral expression
Exogenous gene expression was achieved using the expression vectors pHRSIN-PSFFV-GFP-PPGK-Hygro, pHRSIN-PSFFV-GFP-IRES-mCherry-PPGK-Hygro and pHRSIN-PSFFV-GFP-PPGK-Blasto, with the gene of interest inserted in place of GFP. For lentiviral expression of shRNA constructs, the pHR-SIREN vector was used with hairpins cloned in as BamHI-EcoRI fragments. In all cases, lentivirus was generated through the triple transfection of HEK 293ET cells with the lentiviral transfer vector plus the two packaging plasmids pCMVΔR8.91 and pMD.G using TransIT-293 transfection reagent (Mirus) according to the manufacturer’s recommendations. Viral supernatant was typically harvested 48 h post-transfection, cell debris removed using a 0.45 µm filter, and target cells transduced by spin infection at 800 x g for 60 min. Transduced HeLa cells were selected with the following drug concentrations: puromycin, 2 μg/ml; hygromycin 50 μg/ml; blasticidin, 3 μg/ml. Transduced SK-N-SH cells were selected using 50 μg/ml hygromycin.
Flow cytometry
Cells were fixed in 1% PFA and analyzed on a FACSCalibur (BD) or a FACSFortessa (BD) instrument. Data was analyzed using FlowJo software. For cell sorting, cells were resuspended in PBS + 2% FCS and FACS was carried on an Influx cell sorter (BD).
Immunofluorescence
HeLa cells were grown overnight on glass coverslips, fixed with the addition of 4% PFA for 10 minutes, permeabilized with 0.5% Triton X-100 for 5 minutes, and then blocked for at least 30 min using 4% BSA dissolved in PBS + 0.1% Tween-20 (PBS-T). The primary antibody was diluted in 40 µl of blocking solution and was applied for 1 h, and, following five washes in PBS-T, fluorophore-conjugated secondary antibody (Molecular Probes) was applied for 45 min in the same way. Following a further five washes in PBS-T and a final rinse in distilled water, coverslips were mounted in 4 µl ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific) and imaged using a Nikon LSM880 laser scanning confocal microscope (Zeiss). Images were processed using Adobe Photoshop (Adobe, CA) or GIMP 2.
Co-immunoprecipitation
Cells were washed once in PBS and then lysed in cell lysis buffer (0.5% IGEPAL, 85 mM KCl and 10 mM HEPES in distilled water). Nuclei were harvested by centrifugation at 800 x g for 5 minutes at 4ºC, and then lysed in nuclear lysis buffer (1% IGEPAL plus 1:100 Benzonase in TBS). Insoluble nuclear material was removed by centrifugation (10,000 x g for 5 minutes at 4ºC) and then the supernatant was pre-cleared with protein G magnetic beads (Thermo Fisher Scientific) for 1 h at 4°C. Immunoprecipitation was performed by adding 1 µg of antibody and protein G magnetic beads for 2 h at 4°C, and, following three washes of the beads in lysis buffer, samples were eluted in SDS sample buffer.
Immunoblotting
Cells were lysed in 1% SDS plus 1:100 Benzonase (Sigma-Aldrich) for 20 minutes at room temperature, and then heated to 70°C in SDS sample buffer for 10 min. Following separation by SDS-PAGE, proteins were transferred to a PVDF membrane (Millipore) which was then blocked in 5% milk in PBS + 0.2% Tween-20. Membranes were probed overnight with the indicated antibodies, washed three times PBS + 0.2% Tween-20, and then incubated with HRP-conjugated secondary antibodies (Jackson ImmunoResearch) for 45 minutes at room temperature. Reactive bands were visualized using SuperSignal West Pico or West Dura (Thermo Fisher Scientific). All blots presented in the figures have been cropped; the original uncropped images can be found in Supplementary Figure 10.
Subcellular fractionation
One million cells were washed in PBS and then again in Buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and an EDTA-free protease inhibitor cocktail tablet (Roche)). The cells were then lysed upon resuspension in Buffer A plus 0.1% (v/v) IGEPAL on ice for 10 min. Nuclei were pelleted by centrifugation (1400 x g for 4 min at 4°C) and the supernatant, which contains the cytosolic fraction, was collected. The nuclear pellet was resuspended in Buffer B (20 mM HEPES, 1.5 mM MgCl2, 300 mM NaCl, 0.5 mM DTT, 25% v/v glycerol, 0.2 mM EDTA and an EDTA-free protease inhibitor cocktail tablet) for 10 min on ice, and, following centrifugation at 1700 x g for 4 min at 4°C, the supernatant (containing the nucleosolic fraction) was removed. The insoluble pellet, constituting the chromatin fraction, was subsequently solubilized in 1% SDS plus 1:100 Benzonase for 20 min at room temperature.
ChIP-seq
Cells were washed once in PBS, resuspended in growth medium, and then cross-linked in 1% formaldehyde for 10 min. The reaction was quenched by adding glycine to a final concentration of 0.125 M for 5 min before the cells were lysed in cell lysis solution (1 mM HEPES, 85 mM KCl, 0.5% IGEPAL). Nuclei were pelleted by centrifugation, and then resuspended in nuclear lysis solution (5 mM Tris, 10 mM EDTA, 1% SDS) for 10 min. The chromatin was sheared using a Bioruptor (Diagenode; high power, 20 cycles of 30 s on, 30 s off) to obtain a mean fragment size of ~300 bp. Insoluble material was removed by centrifugation. The chromatin solution was pre-cleared with protein A sepharose (Sigma-Aldrich) and then immunoprecipitated overnight using 5 µg primary antibody and protein A sepharose, or, for V5-MORC2, anti-V5 agarose affinity gel (Sigma-Aldrich, A7345). The next day the beads were washed a total of five times, and then bound protein-DNA complexes eluted in 150 mM NaHCO3 and 1% SDS. Cross-links were reversed by overnight incubation at 67ºC with 0.3 M NaCl and RNaseA. Proteinase K was then added and the samples incubated for 2 h at 45ºC, and then the DNA was purified using a spin column (Qiagen PCR purification kit). Illumina sequencing libraries were produced from this material using the TruSeq ChIP sample prep kit (Illumina), and 50 bp single-end reads generated on a HiSeq 2500 instrument. Reads were aligned to the human genome (GRCh37) using Bowtie 2, and reads with a MAPQ score >10 were imported into SeqMonk and EaSeq47 for further analysis. V5-MORC2 binding sites were identified using the implementation of the MACS peak caller in SeqMonk (windows size = 300 bp; p-value < 0.0001) and the resulting 4500 peaks were centered and plotted using the Average Signal Tracks tool in EaSeq. H3K4me3 ChIP-seq data generated by the Bernstein laboratory was downloaded from the ENCODE project (ENCSR000AOF).
qRT-PCR
RNA was prepared using the RNeasy Plus kit (Qiagen) and converted into cDNA using Super RT reverse transcriptase (HT Biotechnology) and a poly(dT) primer. Quantification by qPCR was performed on an ABI 7500 Real Time PCR System (Applied Biosystems) using SYBR green PCR mastermix (Life Technologies) in a final reaction volume of 25 μl. Thermal cycling parameters were: 50°C for 2 min; 95°C for 5 min; 40 cycles of 95°C for 15 s and 58°C for 1 min. Error bars represent the standard deviation of at least three technical replicates. Primer sequences are detailed in Supplementary Table 3.
Expression and purification of the CW domain of MORC2
A synthetic, E. coli codon-optimized DNA construct (IDT) encoding the MORC2 CW domain (residues 490-546) plus a C-terminal FLAG tag was cloned into the expression vector pET-28a-N-His-SUMO between the BamHI and NotI restriction sites for production of the N-terminally His-SUMO-tagged protein product. Protein was produced in E. coli BL21(DE3) cells through overnight incubation at 18 ºC with shaking at 220 rpm following induction with 200 µM IPTG. Cell pellets were resuspended in 30 ml lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 10 mM imidazole, 0.1 mM ZnSO4, 1 mM DTT, 1x Complete EDTA-free protease inhibitors, pH 8.0), flash frozen in liquid nitrogen and stored at -80 ºC.
Pellets were thawed and further lysed by sonication on ice, before clarification by centrifugation (45 min, 40,000 x g, 4ºC). The resulting His-SUMO-CW-FLAG-containing supernatant was subjected to immobilized Ni-NTA affinity chromatography with wash (50 mM Tris.HCl, 500 mM NaCl, 10 mM imidazole, 0.1 mM ZnSO4, 1 mM DTT, pH 8.0) and elution (50 mM Tris.HCl, 500 mM NaCl, 300 mM imidazole, 0.1 mM ZnSO4, 1 mM DTT, pH 8.0) buffers, and then desalted using a Bio-Rad Econo-Pac 10DG column into cleavage buffer (20 mM HEPES, 150 mM NaCl, 0.1 mM ZnSO4, 1 mM DTT, pH 7.4). The His-SUMO tag was cleaved from the protein by the addition of His-tagged SENP1 protease (1:10 w/w) for 18 h on ice. Subtractive immobilized Ni-NTA affinity chromatography was then used to deplete the tagged protease and the free His-SUMO tag, and the protein was further purified by size exclusion chromatography on a Superdex 75 (10/300) column in a buffer containing 20 mM HEPES, 200 mM NaCl, 0.1 mM ZnSO4, 1 mM TCEP, pH 7.3. The mass of the intact, pure proteins were confirmed using a Waters Xevo G2 Q-ToF mass spectrometer.
Differential Viral Accessibility (DIVA)
Six million wild-type and MORC2 knockout HeLa cells were seeded at a density of 1x106 cells per well of a 6-well plate, and transduced on successive days with the lentiviral vector pHRSIN-PSFFV-mCherry-WPRE (encoding the mCherry fluorescent protein driven by the spleen focus-forming virus LTR promoter) at high multiplicity of infection such that ~100% of the cells were mCherry+. One day after the second transduction genomic DNA was extracted (Gentra Puregene Kit) and digested overnight with NlaIII (NEB). The distribution of NlaIII sites in the lentiviral vector is such that DNA fragments smaller than ~1.8 kb cannot contain viral-genome junctions; large fragments were therefore enriched through a size-selection step using a 0.5x Agencourt AMPure XP bead clean-up (Beckman Coulter). An annealed dsDNA adaptor with an NlaIII-compatible overhang was then ligated onto the ends of the genomic DNA fragments using T4 DNA Ligase (NEB) overnight at 16°C, and the ligated products purified through a further AMPure bead clean-up. Virus-genome junctions were then amplified in 200 cycles of a linear PCR reaction using Accuprime Taq (Thermo Fisher Scientific) using a biotinylated primer binding the just upstream of the 5’ LTR in the integrated provirus, and the resulting products were annealed to streptavidin-coated M-280 dynabeads (Thermo Fisher Scientific). Following 5 x 10 minute washes with PBS + 0.1% Tween-20, virus-genome junctions were amplified through 12 cycles of on-bead exponential PCR reaction with an Illumina P5-adapted primer binding the proviral 5’LTR and a P7-adapted reverse primer binding the splinkerette adaptor. After a final AMPure XP clean-up, PCR products were quantified, pooled and sequenced on an Illumina HiSeq2500 instrument generating 50 bp single-end reads from a custom primer annealing to the extreme end of the proviral 5’ LTR. All oligonucleotide sequences are detailed in Supplementary Table 3.
The resulting sequence reads were trimmed of adaptor sequences using the fastx toolkit and aligned to the human genome (hg19) using Bowtie 2. Reads with a MAPQ score > 10 were imported into SeqMonk and converted into a set of unique points of integration. The Intensity Difference Filter in Seqmonk was used to identify 10 kb genomic windows (overlapping by 5 kb) significantly enriched for lentiviral integration sites in MORC2 knockout cells versus the parental wild-type cells; individual loci were only considered to exhibit decompaction if at least two adjacent windows reached statistical significance. Full data for all genomic loci is detailed in Supplementary Table 2.
Micrococcal nuclease accessibility assay
Wild-type and MORC2 knockout HeLa cells were washed once in PBS, resuspended in growth medium, and then cross-linked by the addition of 1% formaldehyde for 10 min. The reaction was quenched by addition of glycine to a final concentration of 0.125 M for 5 min. The cells were then lysed in cell lysis solution (1 mM HEPES, 85 mM KCl, 0.5% IGEPAL) and nuclei pelleted by centrifugation (800 x g, 5 min, 4°C). The nuclei (5 x 105 per reaction) were then resuspended in micrococcal nuclease buffer supplemented with BSA (NEB), and chromatin digested by the addition of 2 units of micrococcal nuclease (NEB) for 30 min at 37°C. EDTA was added to a final concentration of 45 mM to stop the reaction. Nuclei were then lysed in 1% SDS, RNA digested with RNaseA (37°C for 30 min) and cross-links reversed overnight at 65°C. Proteins were then digested with proteinase K (37°C for 2 h) and DNA isolated by phenol-chloroform extraction followed by ethanol precipitation. Size-selection of the digested DNA was achieved using SPRIselect beads (Beckman Coulter) using 0.6 volumes of beads. The bound fraction contained large fragments (>~1000bp); the supernatant containing unbound fragments (<~1000bp) was removed and subsequently purified by the addition of an extra 1 volume of beads. The amount of DNA from target loci in the accessible and inaccessible fractions versus the pre-size-selected starting material was measured by qPCR performed on a CFX96 Touch Real-Time PCR machine (Bio-Rad) using iTaq Universal SYBR Green Supermix (Bio-Rad) in a final reaction volume of 25 μl. Thermal cycling parameters were: 50°C for 2 min; 95°C for 5 min; 40 cycles of 95°C for 15 s and 58°C for 1 min. Error bars represent the standard deviation of at least three technical replicates. Primer sequences are detailed in Supplementary Table 3.
RNA-seq
Total RNA was extracted using the miRNEasy kit (Qiagen) as per the manufacturer’s instructions and multiplexed Illumina sequencing libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit (Illumina). For the comparison between wild-type and MORC2 knockout HeLa cells, ribosomal RNA was depleted using the Ribo-Zero Gold rRNA Removal Kit (Epicentre) and 150 bp paired-end reads were generated on an Illumina HiSeq2500 instrument. For all comparisons between wild-type and R252W MORC2, polyA+ RNA was sequenced on an Illumina HiSeq2500 instrument using 50 bp single-end reads. Sequence reads were aligned to the human genome (GRCh37) using HISAT2. Mapped reads with a MAPQ score >40 were imported into SeqMonk and further analyzed using the RNA-seq quantitation pipeline followed by DEseq2 or edgeR analysis.
Statistics
No statistical methods were used to predetermine sample size. Experiments were not randomized and the experimenters were not blinded to the outcome. Genes significantly enriched for inactivating mutations in the CRISPR screen were evaluated using the RSA algorithm comparing the frequency of sgRNAs in the GFPbright selected population versus the unselected mutangenized pool. All qRT-PCR data is represented as the mean +/- standard deviation of at least three technical replicates, and the data are representative of at least two independent experiments. For the RNA-seq data presented in Fig. 5e, three independent MORC2 knockout clones were used; genes exhibiting significantly altered expression in MORC2 knockout cells were identified using the implementation of DEseq2 in Seqmonk (P < 0.05). For the RNA-seq data presented in Fig. 6g-i, cells were transduced in triplicate with lentiviral vectors to express either wild-type or R252W mutant MORC2; genes exhibiting significantly reduced expression upon expression of the R252W mutant as compared to wild-type MORC2 were identified using the implementation of edgeR in Seqmonk (P < 0.05).
Data availability
The sequence data from the CRISPR/Cas9 forward genetic screen, ChIP-seq, DIVA and RNA-seq experiments that support the findings of this study have been deposited in GEO with the primary accession code GSE95480.
Supplementary Material
Acknowledgements
We are indebted to CIMR core facilities: R. Schulte and his team for FACS, and M. Gratian and M. Bowen for microscopy. We thank S. Andrews for assistance with data analysis using SeqMonk, and S. Kundu and M. Tolstorukov for helpful discussions. This work was supported by the Wellcome Trust, through a Principal Research Fellowship to P.J.L. (101835/Z/13/Z), a Senior Research Fellowship to Y.M. (101908/Z/13/Z), a Sir Henry Wellcome Postdoctoral Fellowship to R.T.T. (201387/Z/16/Z) and a Ph.D studentship to I.A.T, and by the BBSRC, through a Future Leader Fellowship to C.H.D.. I.A.T. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2277-16). The CIMR is in receipt of a Wellcome Trust strategic award.
Footnotes
Author contributions
I.A.T., R.T.T. and P.J.L. conceived the study. Except for the in vitro expression and purification experiments which were carried out by C.H.D., I.A.T. and R.T. performed all of the experiments, and, together with Y.M. and P.J.L., analysed the data and wrote the manuscript. G.D., R.C.R. and R.E.K. contributed essential reagents.
Competing Financial Interests
The authors declare that no competing financial interests exist.
References
- 1.Wang J, Lawry ST, Cohen AL, Jia S. Chromosome boundary elements and regulation of heterochromatin spreading. Cell Mol Life Sci. 2014;71:4841–4852. doi: 10.1007/s00018-014-1725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker JS, Nicetto D, Zaret KS. H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends Genet. 2015;32:29–41. doi: 10.1016/j.tig.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 5.Schotta G, Ebert A, Dorn R, Reuter G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol. 2003;14:67–75. doi: 10.1016/s1084-9521(02)00138-6. [DOI] [PubMed] [Google Scholar]
- 6.Akhtar W, et al. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–27. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Kotecki M, Reddy PS, Cochran BH. Isolation and characterization of a near-haploid human cell line. Exp Cell Res. 1999;252:273–80. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- 8.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–5. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 9.Tchasovnikarova IA, et al. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348:1481–5. doi: 10.1126/science.aaa7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–32. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kokura K, Sun L, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010;29:3673–87. doi: 10.1038/emboj.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harten SK, et al. The first mouse mutants of D14Abb1e (Fam208a) show that it is critical for early development. Mamm Genome. 2014;25:293–303. doi: 10.1007/s00335-014-9516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soehn AS, et al. Periphilin is strongly expressed in the murine nervous system and is indispensable for murine development. Genesis. 2009;47:697–707. doi: 10.1002/dvg.20553. [DOI] [PubMed] [Google Scholar]
- 14.Timms RT, et al. Genetic dissection of mammalian ERAD through comparative haploid and CRISPR forward genetic screens. Nat Commun. 2016;7:11786. doi: 10.1038/ncomms11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timms RT, et al. ATF7IP-Mediated Stabilization of the Histone Methyltransferase SETDB1 Is Essential for Heterochromatin Formation by the HUSH Complex. Cell Rep. 2016;17:653–659. doi: 10.1016/j.celrep.2016.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–4. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D-Q, Nair SS, Kumar R. The MORC family: new epigenetic regulators of transcription and DNA damage response. Epigenetics. 2013;8:685–93. doi: 10.4161/epi.24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moissiard G, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–51. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pastor WA, et al. MORC1 represses transposable elements in the mouse male germline. Nat Commun. 2014;5:5795. doi: 10.1038/ncomms6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, et al. Family-wide characterization of histone binding abilities of human CW domain containing proteins. J Biol Chem. 2016;291:9000–13. doi: 10.1074/jbc.M116.718973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, et al. Structure and function of CW domain containing proteins. Curr Protein Pept Sci. 2016;17:497–506. doi: 10.2174/1389203717666160125115130. [DOI] [PubMed] [Google Scholar]
- 24.Li S, et al. Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin. Proc Natl Acad Sci U S A. 2016;113:E5108–16. doi: 10.1073/pnas.1609709113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D-Q, et al. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2012;2:1657–69. doi: 10.1016/j.celrep.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–7. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 28.Kvaratskhelia M, Sharma A, Larue RC, Serrao E, Engelman A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014;42:10209–25. doi: 10.1093/nar/gku769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sevilla T, et al. Mutations in the MORC2 gene cause axonal Charcot-Marie-Tooth disease. Brain. 2016;139:62–72. doi: 10.1093/brain/awv311. [DOI] [PubMed] [Google Scholar]
- 30.Albulym OM, et al. MORC2 mutations cause axonal Charcot-Marie-Tooth disease with pyramidal signs. Ann Neurol. 2015;79:419–27. doi: 10.1002/ana.24575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laššuthová P, et al. Severe axonal Charcot-Marie-Tooth disease with proximal weakness caused by de novo mutation in the MORC2 gene. Brain. 2016;139:e26. doi: 10.1093/brain/awv411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyun YS, Hong Y Bin, Choi B-O, Chung KW. Clinico-genetics in Korean Charcot-Marie-Tooth disease type 2Z with MORC2 mutations. Brain. 2016;139:e26. doi: 10.1093/brain/aww082. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, et al. MORC2 mutations in a cohort of Chinese patients with Charcot-Marie-Tooth disease type 2. Brain. 2016;139:e56. doi: 10.1093/brain/aww156. [DOI] [PubMed] [Google Scholar]
- 34.Wang T, et al. Identification and characterization of essential genes in the human genome. Science (80-. ) 2015;350:1096–101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horlbeck MA, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife. 2016;5 doi: 10.7554/eLife.19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–7. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 38.Grau DJ, et al. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–21. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 41.Schottmann G, Wagner C, Seifert F, Stenzel W, Schuelke M. MORC2 mutation causes severe spinal muscular atrophy-phenotype, cerebellar atrophy, and diaphragmatic paralysis. Brain. 2016 doi: 10.1093/brain/aww252. aww252. [DOI] [PubMed] [Google Scholar]
- 42.Stebbins CE, et al. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–50. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 43.Sharma SV, Agatsuma T, Nakano H. Targeting of the protein chaperone, HSP90, by the transformation suppressing agent, radicicol. Oncogene. 1998;16:2639–2645. doi: 10.1038/sj.onc.1201790. [DOI] [PubMed] [Google Scholar]
- 44.Chen K, et al. Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc Natl Acad Sci U S A. 2015;112:E3535–44. doi: 10.1073/pnas.1504232112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–73. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 47.Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23:349–57. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data from the CRISPR/Cas9 forward genetic screen, ChIP-seq, DIVA and RNA-seq experiments that support the findings of this study have been deposited in GEO with the primary accession code GSE95480.