Abstract
Telomere maintenance is required for chromosome stability, and telomeres are typically replicated by the telomerase reverse transcriptase. In both tumor and yeast cells that lack telomerase, telomeres are maintained by an alternative recombination mechanism. By using an in vivo inducible Cre-loxP system to generate and trace the fate of marked telomeric DNA-containing rings, the efficiency of telomere-telomere recombination can be determined quantitatively. We show that the telomeric loci are the primary sites at which a marked telomeric ring-containing DNA is observed among wild-type and surviving cells lacking telomerase. Marked telomeric DNAs can be transferred to telomeres and form tandem arrays through Rad52-, Rad50-, and polymerase δ-mediated recombination. Moreover, increases of extrachromosomal telomeric and Y′ rings were observed in telomerase-deficient cells. These results imply that telomeres can use looped-out telomeric rings to promote telomere-telomere recombination in telomerase-deficient Saccharomyces cerevisiae.
Telomeres are dynamic DNA-protein complexes that protect the ends of linear chromosomes, prevent detrimental chromosome rearrangements, and defend against genomic instability and the associated risk of cancer (26, 35, 49). Telomeric DNA is synthesized by the enzyme telomerase (30, 36, 46). In certain human cells, telomerase activity is absent, and telomeres are gradually shortened with successive cell divisions due to incomplete replication, which eventually causes replicative senescence. Once telomeres become sufficiently short, they are thought to lose the ability to protect the ends of the chromosomes from being recognized as broken ends and being subjected to nuclease digestion and active recombinational repair. Continuous telomere shortening in human fibroblasts leads to chromosome fusions, crisis, and apoptosis (1). Very few human cells can bypass the crisis either through telomerase reactivation or through an alternative recombination pathway for lengthening of telomeres (ALT) (3, 8, 33).
Telomeric DNA in the yeast Saccharomyces cerevisiae consists of ∼350 ± 75 bp of TG1-3/C1-3A DNA repeats. Internal to the TG1-3/C1-3A tracts are middle repetitive DNA elements called X and Y′ (26, 49). The telomeric TG1-3/C1-3A DNA forms a complex nonnucleosomal chromatin structure. The major component in this complex is a double-stranded, sequence-specific DNA-binding protein-Rap1p complex which includes Rif1p and Rif2p. The copy number of the Rap1p complex negatively regulates telomere length (24, 47).
Even in organisms that normally rely on telomerase, telomerase-independent mechanisms of telomere maintenance exist. Although most cells in S. cerevisiae (22, 37), Kluyveromyces lactis (25), and Schizosaccharomyces pombe (27) that lack the gene for a telomerase component eventually enter cell cycle arrest, survivors arise relatively frequently. In both S. cerevisiae and K. lactis, the generation of survivors requires RAD52-dependent homologous recombination. In S. cerevisiae, the majority of cells that survive in the absence of telomerase activity have multiple tandem copies of the subtelomeric Y′ element and very short terminal tracts of TG1-3/C1-3A DNA (22, 42) (type I survivors). In a minor fraction (∼10%) of the survivors (type II), the lengths of the telomere sequence are increased variably from several hundred base pairs to 10 kb or longer (42). The generation of type II survivors depends on the presence of Rad50p, Rad59p, and Sgs1p (4, 5, 14, 17, 40), whereas the frequencies of type I and type II formation depend on a number of factors such as strain background (14), cell type, and various genes involved in nonhomologous end joining (21). The structure of type II telomeres in Saccharomyces resembles that of 10 to 15% of human cell lines and tumors that maintain telomeric DNA by the ALT pathway (3, 8, 33).
We previously showed that, prior to the appearance of type II survivors, the average telomere length in a telomerase-deficient culture is very short (40). However, type II lengthening is an abrupt recombinational process instead of a gradual incremental lengthening process (40) (see Fig. 6B). The sudden and dramatic increase in the length of telomere that accompanies the transition to the type II pattern of telomeric DNA is difficult to explain by conventional gene conversion events, because there is no telomere with sufficient length to act as a template for several kilobases of TG1-3 amplification prior to the appearance of type II survivors. Since the type II survivors with the characteristic of long telomeres arise suddenly in a population of telomerase-deficient cells, we speculated that the initial substrates for the type II recombination in S. cerevisiae may be the extrachromosomal circles of TG1-3 DNA (40). Recent results by Natarajan and McEachern suggested that transformed circles can lead to the production of extended stretches of telomeric DNA by serving as templates for rolling-circle synthesis in K. lactis (29). We show here in vivo that episomal telomeric DNA-containing rings could be generated spontaneously from telomerase-deficient S. cerevisiae and that these rings could be the substrates for telomere-telomere recombination. This telomeric circle-derived recombination pathway is mediated by Rad52 as well as Rad50 and is DNA polymerase δ (Polδ) dependent, displaying the same genetic requirement as the type II telomere-telomere recombination.
FIG. 6.
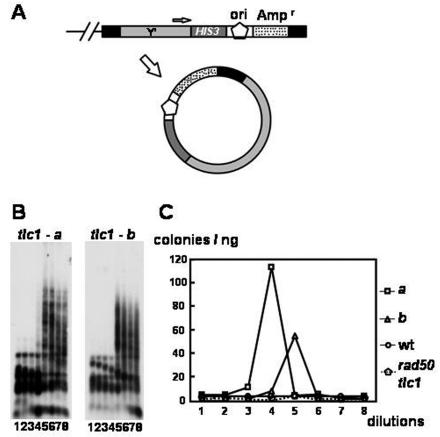
Increase of extrachromosomal Y′ rings in telomerase-deficient S. cerevisiae. (A) A marker-based, ring-releasing yeast strain. A HIS3-ori-Ampr gene marker was tagged at a telomere. We speculate that during telomerase-deficient yeast cell senescence, the whole genome may become more unstable and telomeres may loop out more easily. Black boxes indicate telomeric repeats, dark gray boxes indicate the HIS3 gene, light gray boxes indicate the Y′ sequence, dotted boxes indicate the Ampr gene, and pentagons indicate the E. coli replication origin. (B) Southern analysis of marked tlc1 strains. Two independent tlc1 spores (tlc1-a and tlc1-b) were subjected to a liquid serial dilution assay (see Materials and Methods). Genomic DNAs from each dilution were digested with a combination of AluI, HaeIII, HinfI, and MspI; fractionated through 1% agarose; and transferred to a nylon filter. The filter was hybridized to the C1-3A probe. (C) Rescue of circular DNA released from telomerase-deficient yeast telomeres. The frequencies of looped-out Y′ rings in genomic DNAs were determined by the colony numbers from E. coli transformation. wt, wild type.
MATERIALS AND METHODS
Yeast strain and plasmid constructions.
All yeast operations were performed by standard methods. Yeast strains used in this study were derivatives of YPH501 (MATa/MATα ura3-52/ura3-52 lys2-801 amber/lys2-801 amber ade2-101 ochre/ade2-101 ochre trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 leu2-Δ1/leu2-Δ1). The yeast strains carrying tlc1, rad52, tlc1 rad50, tlc1 rad51, and tlc1 polδ were described previously (40, 42, 44). These heterozygous diploids were transformed with pSH47, a URA3 marker containing Cre recombinase expression plasmid, sporulated, and screened for segregants that were unable to grow at 37°C or for the URA3, HIS3, and LEU2 markers by replica plating to selective medium. Spore cells were serially diluted into or restreaked onto yeast extract-peptone-dextrose (YEPD) medium as described previously (44) for making type I and type II survivors. The fragment for tagging the 3′ untranslated region of the Y′ elements with the HIS3-ori-Ampr gene region was amplified by PCR with 50-bp Y′ sequences that spanned the stop codon of the Y′ open reading frame 2 at the ends of the primers. pRS303 was used as a template. The resulting Y′-HIS3-ori-Ampr gene-Y′ PCR-amplified fragment was transformed into the YPH501 tlc1::LEU2 rad50::HIS3/TLC1 strain. The Y′-HIS3-ori-Ampr gene-tagged strains were selected on medium lacking histidine. Tagging of individual telomeres by the HIS3-ori-Ampr gene region was confirmed by Southern blot analysis with a HIS3 probe as described below. These heterozygous diploids were then sporulated and screened for the HIS3 and LEU2 markers by replica plating them onto selective medium. To generate pUG6TG1-3, the BamHI- and EcoRV-digested 550-bp TG1-3 fragment in pCRTG1-3550 (42) was cloned into the BstEII-digested Klenow fragment-treated and BglII-digested 4-kb pUG6 plasmid (11), a vector having the loxP-kan-loxP marker.
In vivo telomeric ring-targeting system.
The pSH47-containing haploid strains were cultured overnight in 50 ml of synthetic complete medium minus uracil (Sc−Ura) to an optical density at 600 nm (OD600) of 1 to 2. The overnight cultures were diluted to 300 ml of yeast extract-peptone-adenine galactose to an OD600 of 0.25 and grown in an incubator at 200 rpm with shaking to an OD600 of 0.6. The cultures were transformed with the pUG6TG1-3 plasmid as described previously (10). The transformants were then plated on Sc−Ura galactose medium and incubated at 30°C overnight to allow telomeric circle-mediated telomere-telomere recombination to occur. The cells grown overnight were replica plated onto YEPD plates plus 200 μg of G418/ml for selection and for stopping the CRE expression. Since the telomeric arrays generated by the excised circles can themselves be the targets of Cre, presumably, during the period of Cre induction in galactose medium, there would be both array formation and destruction. Because the cleavage of the arrays would regenerate circles, circles should still be present after Cre expression stops. The tlc1 polδ strain was also cultured at 30°C, its semipermissive temperature. Data shown in Table 1 are the average of three independent transformations for each yeast strain.
TABLE 1.
pUG6TG1-3 transformation in different genetic backgrounds of yeast strains
| Strain | Frequency of G418r eventsa | % of circle-mediated telomeric targetingb in G418r events (no. of resistant colonies/total no.) | Frequency of circle-mediated telomeric targetingd |
|---|---|---|---|
| Wild type | 7.90 ± 2.65 | 100 (32/32) | 7.90 ± 2.65 (0.44) |
| tlc1 mutant | 18.6 ± 4.34 | 96.1 (73/76) | 17.9 ± 4.17 (1.00) |
| tlc1 rad50 mutant | 1.10 ± 0.39 | 80.3 (41/51) | 0.88 ± 0.31 (0.05) |
| tlc1 rad51 mutant | 17.0 ± 2.41 | 85.7 (36/42) | 14.6 ± 2.07 (0.82) |
| tlc1 type I | 22.1 ± 2.2 | 100 (39/39) | 22.1 ± 2.2 (1.23) |
| tlc1 type II | 23.8 ± 9.41 | 91.3 (42/46) | 21.7 ± 8.59 (1.21) |
| rad52 mutant | 0.20 ± 0.12 | 85.7 (6/7) | 0.17 ± 0.10 (0.01) |
| tlc1 polδ mutant | 0.00 ± 0.00 | NAc | NA |
Results demonstrated as the number of G418-resistant colonies from transformation per microgram of pUG6TG1-3 DNA. Experiments using the control plasmid pRS314 showed that all strains exhibited similar efficiencies for plasmid transformation. The calculation is based on three independent measurements. A standard deviation is given.
The telomeric targeting was determined by Southern analysis using the Kanr gene fragment as a probe, as described for Fig. 2B and C.
Not available (NA) because no G418-resistant colony was recovered from this strain.
The frequency of circle-mediated telomeric targeting was derived by multiplying the values obtained in the previous two columns. Relative frequencies (in parentheses) were calculated by dividing each frequency by that of the tlc1 strain.
Culture condition, DNA preparation, enzyme digestion, gel electrophoresis, and Southern blot analysis.
Genomic DNA preparation and Southern blot analysis were performed as previously described (44). To examine the DNA from an individual colony, the colony was expanded in 2 ml of liquid medium to obtain DNA for Southern analysis. The DNA was digested with KpnI, with SacI, or with a mixture of AluI, HaeIII, HinfI, and MspI. When the DNA was examined by two-dimensional electrophoresis, the wild-type culture and tlc1 culture that was at the dilution point for type II development (40) were expanded in 500 ml of liquid medium to obtain enough DNA for Southern analysis (16). For the Bal 31 exonuclease digestion experiment, 70 μg of genomic DNA from G418 survivors was digested with 3 U of Bal 31 (New England BioLabs) in a 200-μl final volume. A 50-μl volume of digested DNAs was removed every 10 min, subjected to phenol-chloroform extraction and ethanol precipitation, and digested with KpnI and SacI. For pulsed-field gel electrophoresis (PFGE), yeast chromosomal DNA blocks were prepared by mixing equal volumes of yeast cells from stationary-phase cultures with 1% low-melting-point agarose (FMC BioProducts) as described previously (42). PFGE was performed with the dynamically regulated contour-clamped homogeneous electric field CHEF-DR III system (Bio-Rad). Chromosomes were separated on a 1% agarose gel in 0.5× Tris-borate-EDTA (TBE) buffer at 14°C for 30 h at 6.0 V/cm (200 V) with a 120° included angle and a 60- to 120-s linear switch time ramp.
The following probes were used for Southern hybridization: a 690-bp HindIII-PstI fragment of the Kanr gene (Fig. 2B and 3), a 2.4-kb PvuII-SpeI fragment of pUG6 (Fig. 2C), a 377-bp PCR fragment from the 3′ end of Y′ (Fig. 2D), a 1-kb 5′ EcoRI fragment of PIF1 (Fig. 3B), an EcoRI-digested yeast genomic DNA (Fig. 4B), the 4.5-kb pRS303 plasmid (Fig. 4B), the 3-kb pBSKSII plasmid (Fig. 4B), a 270-bp C1-3A fragment (Fig. 5A and 6B), a 341-bp XhoI-KpnI fragment from the 3′ end of Y′ (Fig. 5A), a 2.1-kb EcoRI fragment of rRNA (Fig. 5A), the pRS423 containing the plasmid 2μm and an autonomously replicating sequence (ARS) (Fig. 5A), and a 586-bp NdeI-NsiI fragment of HIS3 (Fig. 6A). Probes were randomly labeled with the random prime labeling system (Invitrogene). Data shown here are representatives of two or more experiments from independent spores.
FIG. 2.
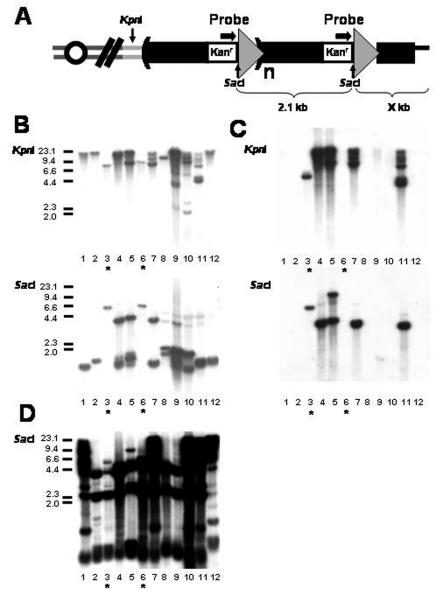
TG1-3-kan-loxP markers targeted to chromosomes. (A) Predicted structure of the TG1-3-kan-loxP markers on chromosomes if telomeric circle-mediated targeting occurs. The formation of one long telomere is postulated to occur via the gene conversion copying a 2.1-kb (TG1-3-kan-loxP) circle. The arrows above markers indicate the probe used in panel B. Subtelomeric KpnI sites are located 0.5 kb from the telomeric ends in untransformed yeast cells. Positions of KpnI and SacI sites are indicated. Tandem arrays of TG1-3-kan-loxP formed at telomeres after targeting are shown here. (B through D) Southern analysis of G418-resistant colonies from the telomeric ring-targeting assay. Equal amounts of genomic DNAs from 12 independent G418-resistant survivors were digested with KpnI and SacI and fractionated in a 0.7% agarose gel. After the survivors were transferred to a nylon filter, the filters were hybridized with a kan probe (B), a pUG6 vector probe (C), and a Y′ 3′-end probe (D), sequentially. Asterisks mark two events (lanes 3 and 6) with only one copy of the marker targeted to the internal regions of chromosomes. Size markers (in kilobases) are shown on the left.
FIG. 3.
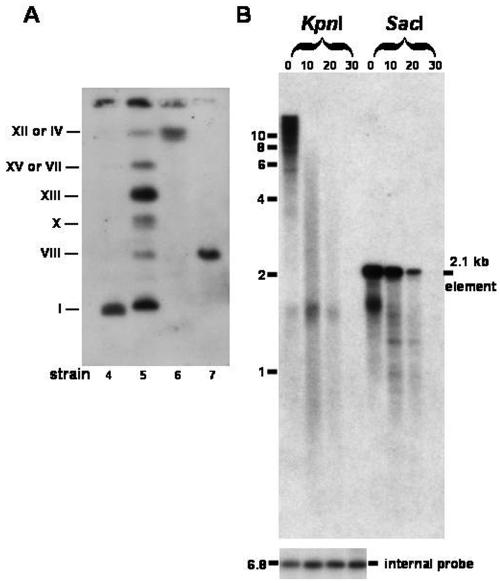
kan sequences targeted to the physical ends of chromosomes. (A) PFGE analysis of G418-resistant survivors from telomerase-deficient S. cerevisiae. Chromosomes from four G418-resistant survivors (survivors 4 through 7 in Fig. 2) were separated by PFGE. After the survivors were transferred to a nylon filter, the filter was hybridized with a kan probe. Chromosome numbers are shown on the left. (B) The kan marker targets telomeres of G418-resistant survivors. Genomic DNA from a G418-resistant tlc1 survivor was digested with Bal 31 exonuclease for increasing lengths of time. Samples were removed at 10-min intervals, subjected to phenol-chloroform extraction and ethanol precipitation, and digested with KpnI and SacI. Digested DNAs were fractionated in a 0.7% agarose gel and analyzed by Southern hybridization sequentially with a kan probe (top) and an internal probe (the PIF1 gene) (bottom).
FIG. 4.
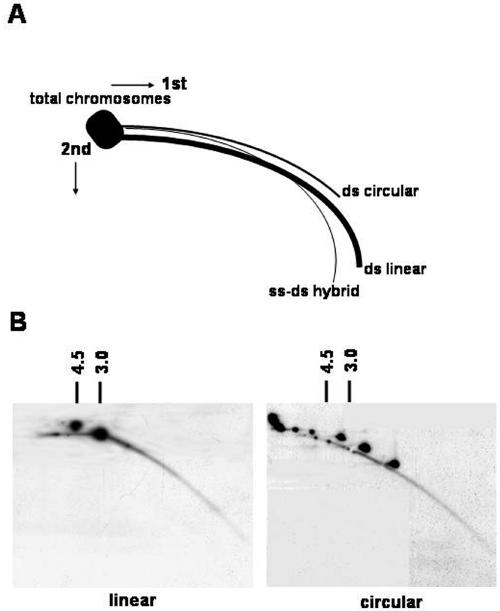
Detection of extrachromosomal circular molecules by two-dimensional gel analysis. (A) Diagram of two-dimensional gel electrophoretic patterns of genomic DNA generated by populations of linear and circular molecules heterogeneous in size (adapted from previous studies [7]). Each arc consists of molecules sharing the same structure but having different masses. (B) Two-dimensional gel analysis of DNA from plasmid DNAs and yeast genomic DNAs. The HinfI-digested linear yeast genomic DNA samples, mixed with either SalI-digested pRS303 (4.5-kb) and pBSKS (3.0-kb) plasmids (left panel) or undigested plasmid circles (right panel), were separated on a two-dimensional gel. The blot was hybridized with combined probes containing yeast genomic DNA, pRS303, and pBSKS. The multiple spots of circles reflect the different topologies as well as sizes of these plasmids.
FIG. 5.
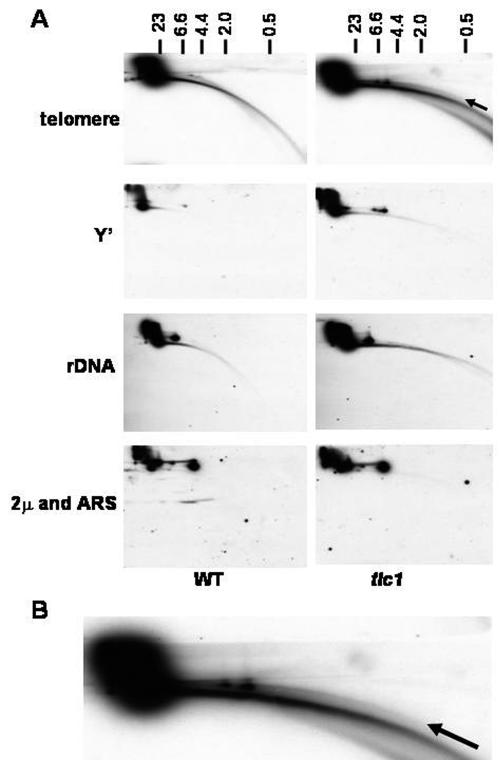
Detection of extrachromosomal circular telomeres in telomerase-deficient S. cerevisiae. (A) Two-dimensional gel analysis of DNA from cells lacking telomerase. DNA was isolated from wild-type (WT) and tlc1 cells at the stage when survival were being generated. Similar amounts of DNA were separated on a two-dimensional gel. The blot was first hybridized with a C1-3A probe to detect telomeric rings and was hybridized sequentially with Y′, rRNA genes (labeled rDNA), or ARS and 2μm probes with exposure times of equal lengths. An arrow indicates extrachromosomal rings of telomeres. Size markers (in kilobases) in the first dimension are shown on the top. (B) The enlarged telomere signal of tlc1 cells.
Neutral-neutral two-dimensional agarose gel electrophoresis.
Two-dimensional gel electrophoresis was performed according to the method of Brewer and Fangman (2). In the first dimension, samples were separated on 0.7% agarose gels in 1× TBE with 0.5 μg of ethidium bromide/ml at 0.5 to 0.6 V/cm for 20 h. Subsequently, the entire lane was excised and embedded in a 1.3% agarose gel. Separation in the second dimension was at 5 V/cm in 1× TBE containing 0.5 μg of ethidium bromide/ml. After electrophoresis, the DNA was transferred onto nylon membranes and hybridized with the corresponding DNA probes as described above. Imaging and quantification of the data were performed with an Alpha Innotech photodocumentation system. The amount of 2μm in each sample was used as a normalization factor for determining the severalfold enhancement of the Y′ amount.
The marker-based Y′ ring-releasing assay.
The wild-type, tlc1, and tlc1 rad50 spores tagged with Y′-HIS3-ori-Ampr gene were used to inoculate cultures. Cultures were inoculated directly from the tetrad master plate into 10 ml of liquid YEPD medium and allowed to grow to stationary phase on a 30°C roller drum. The cultures were diluted repeatedly 1:10,000 into fresh medium for 48 or 72 h. Genomic DNA from each culture was extracted, and 2.5 ng of DNA was electroporated into ElecoMax DH10B cells (Invitrogen). Data shown in the results are averages of three independent Escherichia coli transformations.
RESULTS
In vivo telomeric ring-targeting assay demonstrates that the markers target in tandem the wild-type and tlc1 yeast genomes.
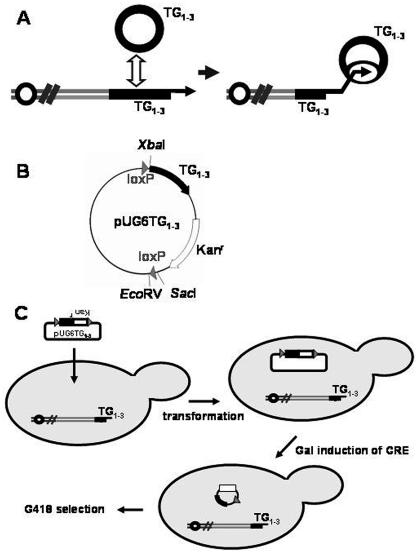
Due to the repetitive nature of telomeric DNA, telomeric circles could be generated by intramolecular recombination (19). We speculated that these extrachromosomal circles of TG1-3 DNA could be the substrate for type II recombination (Fig. 1A). To test this possibility, we generated an in vivo telomeric ring-targeting assay. A marker plasmid, pUG6TG1-3, that contains a 550-bp telomeric repeat and a kanamycin-resistant gene, both flanked by loxP sequences, was constructed (Fig. 1B). This plasmid did not contain an ARS element, and therefore it could not replicate extrachromosomally. Additionally, in this plasmid, there is no sequence other than the TG1-3 fragment that is found naturally in the S. cerevisiae chromosome. By this design, we reduced the possibility that the plasmid would integrate at sites other than the telomere. This plasmid was then transformed into cultures from Cre recombinase-expressing freshly dissected spores. We expected that a 2.1-kb telomeric DNA-containing marker would be generated in vivo and recombined to telomeres in these yeast cells (Fig. 1C).
FIG. 1.
(A) Schematic of the hypothesis of the initial step of telomere-telomere recombination. The yeast telomere locus consists of tandem telomeric repeats from which circles may be liberated by recombination. The arrows indicate the 3′-end hydroxyl group at telomeres. The liberated telomeric rings may be used as templates for telomeric circle-mediated telomere-telomere recombination. (B) Map of the 4.6-kb pUG6TG1-3 plasmid. Black boxes indicate the telomeric repeats, white boxes indicate the Kanr gene, and gray triangles indicate the loxP sequences. (C) In vivo telomeric ring targeting system. A Cre expression telomeric, kan-marked, ring-releasing yeast strain was generated for the experiments. The TG1-3-kan-loxP ring used for telomeric circle-mediated targeting was released from pUG6TG1-3 under the galactose induction of Cre. The recombinants can be selected on G418 plates.
To determine if this telomeric circle-mediated telomere-telomere recombination exists, we conducted the in vivo telomeric ring-targeting assay with the wild-type and freshly dissected tlc1 cells that existed prior to activation of the alternative recombination pathway. Three independent experiments were conducted, and the frequencies were averaged. As expected, for each microgram of pUG6TG1-3 transformation, 18.6 colonies of G418-resistant tlc1 survivors were recovered. Interestingly, G418-resistant colonies were also recovered from the wild-type strain. Relative to the respective frequency in the tlc1 strains, the frequency of the G418-resistant colonies in the wild-type strains was reduced to 42% (7.90/18.6) (Table 1), suggesting that a more efficient marker targeting occurs in the telomerase-deficient strain. As a control, when a CEN plasmid, pRS314, was used to transform both strains, no difference was observed in the transformation efficiency (data not shown). We checked 76 of the G418-resistant tlc1 survivors by Southern analysis. The genomic DNA was digested with KpnI, which does not cleave within the 2.1-kb TG1-3-kan-loxP marker sequence but cuts at the 3′-most end of the Y′ element (Fig. 2A). The digested DNA was subjected to electrophoresis and analyzed by hybridization to a kan probe (12 samples are shown in Fig. 2B). As expected, heterogeneous signals, a sign of the size of telomeres with major signals being over 2.1 kb, were detected in most survivors (Fig. 2B). This result suggested that kan markers were inserted into the chromosomes of these G418-resistant tlc1 survivors. If circular markers would serve as templates for recombination, tandem copies of the markers might be generated at telomeres before the DNA polymerases fall off. A 1.6- to 2.1-kb band should be detected when a similar Southern analysis is conducted using SacI, a restriction enzyme which cleaves once in the 2.1-kb circle (Fig. 2A). The exact size depends on the size of the telomeric tract, ranging from 50 to 550 bp after recombination, because this 550-bp TG1-3 tract may be highly unstable in E. coli and in yeast (28, 42). A band at 1.6 to 2.1 kb was observed in 73 of 76 survivors (Fig. 2B and data not shown). The remaining three events showed fixed bands at different sizes (Fig. 2B, lanes 3 and 6, and data not shown). These results implied that the TG1-3-kan-loxP markers in tandem may target the yeast chromosome in tlc1 cells. The actual copy number of segments arrayed in tandem was determined to range from 2 to more than 10 copies (Fig. 2B). As shown in Fig. 2C, four lanes (lanes 4, 5, 7, and 11) in Fig. 2B contained bands at around 4 kb which were generated from ring targeting of pUG6TG1-3 without the Cre-mediated cleavage. To determine if sequences from circles associated with telomeres, the same blot was hybridized with a Y′ probe (Fig. 2D). This probe showed a shifting of some subtelomeric signal to the large bands hybridizing to the kan probe with the KpnI digest. To further confirm that the formation of tandem copies of markers in chromosomes depends on the circular element, we tested the ability of formation of tandem copies of markers in chromosomes by using a 2.1-kb TG1-3-kan-loxP linear fragment generated from the XbaI-EcoRV digestion of pUG6TG1-3. This fragment contains only one copy of loxP that is incapable of generating a CRE-mediated ring (Fig. 1B). For each microgram of XbaI-EcoRV-digested pUG6TG1-3 transformation, 0.74 of the colonies of G418-resistant tlc1 survivors were recovered. As expected, none of them (0 of 18) showed any sign of tandem targeting of markers based on the SacI Southern analysis (data not shown), suggesting that a circular structure is required for the formation of tandem copies of telomeric repeats in our assay. KpnI- and SacI-digested Southern blot analysis also demonstrated that the kan markers in all G418-resistant wild-type clones that we examined (32 of 32) were integrated in tandem at the chromosomes (data not shown). This result suggests that telomeric circle-mediated telomere-telomere recombination also occurs in wild-type cells.
TG1-3-kan-loxP repeats target the physical ends of chromosomes.
To obtain a better understanding of the kinds of changes that can occur during the formation of G418-resistant strains, we used PFGE to determine if the kan markers were located exactly on chromosomes in four independent G418-resistant tlc1 colonies (Fig. 2, survivors 4, 5, 6, and 7). PFGE can separate full-length chromosomes from each other, although certain chromosomes comigrate (XII and IV; VII and XV) and cannot be distinguished. As shown in Fig. 3A, strains 4, 6, and 7 contained one tandem array of G418-resistant markers on chromosomes I, XII or IV, and VIII, respectively. In contrast, strain 5 contains multiple G418-resistant markers on six chromosomes. These multiple arrays could be generated from additional interchromosomal and/or ring-targeting events. All these results support the observation that the TG1-3-kan-loxP markers indeed target the yeast chromosomes.
Recombination may also occur in places other than the telomere in chromosomes. If the marker were at the physical end of a chromosome, it would be sensitive to digestion by the exonuclease Bal 31. To address this possibility, genomic DNA was prepared from a G418-resistant event (Fig. 2, survivor 2). This DNA was digested with Bal 31, and the samples were removed at 10-min intervals. The DNA was then digested with KpnI and SacI, subjected to electrophoresis, and analyzed by hybridization to a kan probe (Fig. 3B). Since the KpnI-digested kan hybridizing sequences were found to shorten and the SacI-digested Kan hybridizing signals were found to decrease as time increased, kan sequences should have been near a free end in the chromosomes of the survivor. An internal probe demonstrated that the nontelomeric sequences were not Bal 31 sensitive. A 1.6-kb kan-containing band which corresponds to the SacI-digested kan fragment from the most distal copy of the array was also observed in the strain. Some of the other clones that we examined also had one smaller kan fragment that rapidly disappeared, such as the 1.6-kb band shown in Fig. 3B (data not shown). Similar Bal 31 digestion results were obtained with five other G418-resistant survivors and wild-type strains (data not shown). These data showed that these telomere-containing rings target telomeres.
Rad52p, Rad50p, and replicative Polδ are involved in telomeric circle-mediated targeting and telomere lengthening.
To better understand the effect of proteins involved in telomeric circle-mediated targeting, we began to examine the factors which might be involved in this pathway. We conducted the in vivo telomeric ring-targeting assay with yeast cells of several mutation backgrounds. As a control experiment, when a CEN plasmid, pRS314, was used to transform these strains, no difference in transformation efficiency was observed (data not shown). To determine if Rad52, Rad50, and Rad51 proteins may specially govern the formation of ring targeting, we tested their effects in their mutant strains. Since tlc1 rad52 mutants senesced extremely fast, this experiment was limited to a rad52 strain. As shown in Table 1, the telomeric circle-mediated G418-resistant colonies were observed but were almost abolished in the rad52 and tlc1 rad50 strains. Thus, Rad52 and Rad50 functions are involved to generate telomeric circle-mediated targeting. In contrast, tlc1 rad51 strains showed only a slight decrease in telomeric circle-mediated targeting, suggesting that telomeric circle-mediated telomere-telomere recombination does not rely on the strand transfer ability of Rad51p. We previously showed that DNA Polδ is required to generate elongated telomeres and/or subtelomeres during telomere-telomere recombination (44). In the partial absence of function of Polδ at its semipermissive temperature, the 2.1-kb telomeric ring should be replicated only incompletely. As expected, no G418 colony was recovered from the tlc1 polδ strain (Table 1), suggesting that Polδ is required for telomeric circle-mediated targeting. In addition, established type I and type II tlc1 survivors both displayed a 20% higher frequency in telomeric circle-mediated targeting than the tlc1 cells right from sporulation and prior to the survivor formation (Table 1), suggesting that telomeric circle-mediated targeting occurs more frequently in established type I and type II survivors.
Neutral-neutral two-dimensional agarose gel electrophoresis revealed the presence of telomeric rings in telomerase-deficient cells.
The technique of two-dimensional electrophoresis has been useful in a variety of versions to analyze replicative intermediates of prokaryotic and eukaryotic organisms (9). To analyze ring structures, ethidium bromide was added to the running buffer. Ethidium bromide intercalates into double-stranded DNA (dsDNA), reducing its mobility in comparison to that of single-stranded DNA and adding the positive superhelical turns into negatively supercoiled circular DNA. The two-dimensional system was relatively insensitive to nicking during the preparation, in comparison to that with alkaline gels. The hybrid of single-stranded DNA-dsDNA, forming more straight lines, was separated from dsDNA, which formed arcs. Circular DNA was easily recognized by the separation of topoisomers or the resulting higher arcs appearance in molecules of lower mobility (7) (Fig. 4A). The assignment of these linear and circular species was confirmed by a comparison of blots with linear and circular controls of plasmid DNAs (Fig. 4B).
To determine if telomeric rings were increased in telomerase-deficient cells, total DNAs were prepared from the wild type and the tlc1 strain that was at the point when the type II survivors were being generated (see Materials and Methods). The samples were then separated by two-dimensional gel electrophoresis. Southern hybridization analysis with a 32P-labeled C1-3A fragment showed a discrete arc of dsDNA linear fragments migrating differently from the main population of chromosomes near the well in wild-type cells (Fig. 5A). In the tlc1 cells, additional continuous signals corresponding to dsDNA rings and ssDNA-dsDNA hybrids were observed as the arcs above and below the main arc of dsDNA linear fragments, respectively (Fig. 5A and shown enlarged in Fig. 5B). Experimental results after stripping the nylon membrane and reprobing with a Y′ probe showed that there were Y′ rings popping out in both wild-type and tlc1 cells in the upper arc. But the tlc1 cells displayed relatively stronger signals of the Y′ rings (Fig. 5A). The total Y′ copy number was elevated 1.57-fold, and the extrachromosomal Y′ rings were elevated 5.60-fold in tlc cells. Other repetitive sequences in chromosomes (rRNA genes) and episomal plasmids (2μm) did not display any major differences with wild-type and tlc1 cells, suggesting that the increase of ring structures is specific to the telomeric regions in telomerase-deficient cells.
Marker-based ring-releasing assay showed that telomeric Y′ rings are released from chromosomes during the formation of type II survivors.
To test the possibility that episomal telomeric-Y′ rings are induced during the survivor formation in telomerase-deficient cells, a more sensitive assay to quantitatively detect telomeric-Y′ rings was developed. We first tagged the 3′ end of Y′ elements with a HIS3-ori-Ampr gene marker (Fig. 6A). Two independent Y′-HIS3-ori-Ampr gene-tagged tlc1 spores were used to inoculate liquid cultures. In the liquid culture assay (40), when cultures starting from freshly dissected spores were repeatedly diluted 1:10,000 at 48-h intervals, type II-like dramatic telomere lengthening could be observed after several dilutions. The genomic DNA from each culture was extracted and subjected to Southern analysis. In the Southern blot analysis, the genomic DNA was digested with a mixture of four restriction enzymes to very small fragments. The telomere sequences, which were not cut by these enzymes, remained relatively large. As shown in Fig. 6B, two independent Y′-HIS3-ori-Ampr gene-tagged tlc1 spores, a and b, revealed a type II pattern developing at the fourth and the fifth dilutions, respectively. This observation was not unexpected because individual telomeres of telomerase-deficient cells are not static and the turning point for the formation of survivors is dynamic. The genomic DNA at each time point was then electroporated into E. coli. As shown in Fig. 6C, dramatic increases in the transformation efficiency were observed at the fourth and the fifth dilutions of spores a and b, respectively. In contrast, DNAs from the wild-type and tlc1 rad50-tagged spores showed no increase in transformation efficiency throughout eight dilutions. Additionally, plasmid DNAs from these E. coli transformants were recovered and confirmed to be the Y′ HIS3-ori-Ampr gene rings by restriction digestion and Southern analysis (data not shown). These results demonstrated that telomeric Y′ rings released from chromosomes are increased during or immediately after the formation of type II survivors.
DISCUSSION
In this paper, we tested the possibility of telomeric circles being the template for telomere-telomere recombination in S. cerevisiae. An in vivo Cre-loxP system was used to generate telomeric rings in yeast cells. Qualitative and quantitative results presented in Fig. 2 and 3 and Table 1 suggest a rolling-circle replication model. Although our observations could also be explained by multiple exchanges with an extrachromosomal element, the fact that type II lengthening is abrupt rather than gradual (40) argues against this possibility. We provide two lines of evidence suggesting that telomeric circles exist in S. cerevisiae, especially at the point of occurrence of survivors. First, the increase in the telomeric TG1-3 rings and Y′ rings in tlc1 cells is detected by two-dimensional gels. Second, these Y′ rings can be quantified and confirmed in E. coli by a marker-based system. However, our assays cannot distinguish whether these telomeric (Fig. 5) and Y′ (Fig. 6) rings were generated during or immediately after the formation of survivors. Interestingly, this pathway is favored but does not occur exclusively in telomerase-deficient cells (Table 1). Moreover, established type I and type II survivors displayed higher frequency for telomeric circle-mediated targeting than tlc1 cells prior to the occurrence of survivors (Table 1), suggesting that telomeric circle-mediated targeting occurs more easily in established survivors. These data also indicate that there is no difference in ring targeting between type I and type II survivors once the alternative recombination pathway is activated and established.
The formation of variable lengths of extrachromosomal circular DNA of telomeric repeats has been shown in human ALT cells (31, 34, 43, 48) and during the development of Xenopus laevis (6). The mechanism by which minicircles of telomeres arise may be similar to that involved in the generation of extrachromosomal rRNA gene circles from the cluster of 100 to 200 rRNA gene units repeated in tandem on chromosome XII of S. cerevisiae. Horowitz and Haber demonstrated the existence of circles of subtelomeric tandem Y′ elements in the wild-type S. cerevisiae. Presumably, they arose from either Y′-Y′ or tract-tract recombination (13). Taken together, the most reasonable interpretation of our results is that, in telomerase-deficient cells, especially at the time for type II survivor development, critically short telomeres increase instability that causes telomeric and subtelomeric fragments to loop out by telomere rapid deletion (TRD)-mediated intrachromosomal recombination (19). Telomere-telomere recombination promoted by telomeric rings was first demonstrated in telomerase-deficient K. lactis (29). It was recently reported that DNA circles as short as 18 nucleotides in vitro (20) and 100 nucleotides in vivo (28, 29) can act as catalytic templates for efficient synthesis of long telomeres by DNA polymerase. Additionally, as shown in Fig. 4 and 5, Y′ rings might contribute to type I recombination. However, utilization of Y′ telomere circles to form telomeric tandem arrays might be RAD50 independent due to the high level of homology provided by the Y′ element.
We demonstrated previously that type I survivors arise by a RAD51-dependent process, whereas type II formation is RAD50 dependent (40). Rad51p, on which type I recombination depends, is a RecA-like, strand transfer protein (39). Our in vivo assay quantitatively showed that telomeric circle-mediated recombination is Rad50p dependent and Rad51 independent. Collectively, creation of this transforming activity in our telomeric ring-targeting assay is recombination dependent and it exhibits the same genetic requirement as the type II recombination pathway. The DNA ends are processed to generate 3′ overhangs which can invade homologous duplex DNA to initiate gene conversion. Genetic and biochemical evidences indicate that Rad51p acts in concert with Rad54p, Rad55p, and Rad57p. Furthermore, Rad54p is proposed to remodel chromatin to allow Rad55p/Rad57p-enhanced and Rad51p-mediated strand exchange (38). These activities are critical for gene conversion events of regular chromatin structure. Based on our results, the Rad51p-independent extrachromosomal telomeric circle-mediated telomere-telomere recombination indicates that the substrate for these events may not have a regularly packed chromatin structure and might be more like a recombinosome-accessible, Rap1p-protected telomere structure. The Rad50p protein, on which type II survivors and extrachromosomal telomeric circle-mediated telomere-telomere recombination depend, functions in a complex with Xrs2p and Mre11p (18, 45). A potential role for the Rad50/Mre11/Xrs2 complex to process double-strand breaks to single-strand ends (32) could be a critical step for telomeric circle-mediated telomere-telomere recombination. Moreover, RAD51-independent recombination requires much less homology for strand invasion than does RAD51-dependent repair (15). This may explain why Rad50p-dependent recombination machinery is better suited for finding short, sufficiently homologous regions within TG1-3 sequences in TG1-3-containing rings to permit the initiation of recombination.
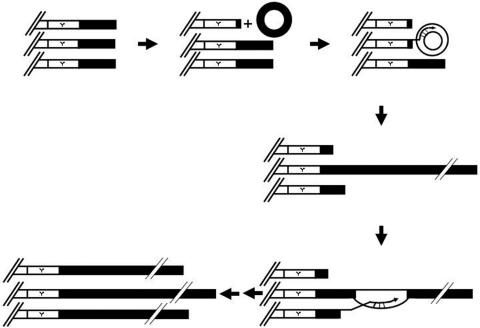
We previously demonstrated that the type II pathway may be initiated from a single telomere (41). When telomeres become critically short, a single telomere may use a telomeric ring as a template and the 3′ hydroxyl group as a primer for telomere-telomere recombination (Fig. 7). We speculate that the initial template for type II recombination is the extrachromosomal circles of TG1-3 DNAs. Due to the repetitive nature of telomeric DNA, telomeric circles could be generated by intramolecular recombination. Additionally, telomeric circles could be generated by intramolecular t-loop formation (23). If a telomere complex invades a TG1-3 circle, a potential rolling-circle replication pathway would allow an abrupt, dramatic, and variable increase in telomere length. Initiation of the first recombinant telomere presumably would be the rate-limiting step in telomere-telomere recombination. This long telomere would preferentially be the template of gene conversion for other critically short telomeres. This result is consistent with the recent finding in K. lactis by the McEachern group (29) suggesting that telomeric rings can promote Rad52-, Rad50-, and Polδ-dependent telomere-telomere recombination in both K. lactis and S. cerevisiae. This mechanism might also be relevant to the ALT pathway as reviewed in reference 12. Whether the formation of extrachromosomal telomeric rings in some ALT-prone tumors would be easier remains to be elucidated.
FIG. 7.
A model for telomeric circle-mediated telomere-telomere recombination. See text for details.
Acknowledgments
We thank M. Gartenberg for providing the pCRE and pUG6 plasmids, as well as M. McEachern, V. Zakian, T.-K. Li, M.-K. Chern, and our anonymous reviewers for their critical comments on the manuscript.
This work was begun in V. Zakian's lab and supported by grants from the NIH GM26938 (V. Zakian), a postdoctoral fellowship from the U.S. Army Breast Cancer Program, and grants from the National Science Council and National Health Research Institute of Taiwan (S.-C.T.).
REFERENCES
- 1.Artandi, S. E., and R. A. DePinho. 2000. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10:39-46. [DOI] [PubMed] [Google Scholar]
- 2.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 3.Bryan, T. M., A. Englezou, L. Dalla-Pozza, M. A. Dunham, and R. R. Reddel. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271-1274. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, H., and D. A. Sinclair. 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. USA 98:3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, S., and M. Mechali. 2002. Formation of extrachromosomal circles from telomeric DNA in Xenopus laevis. EMBO Rep. 3:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, S., S. Menut, and M. Mechali. 1999. Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol. Cell. Biol. 19:6682-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447-450. [DOI] [PubMed] [Google Scholar]
- 9.Friedman, K. L., and B. J. Brewer. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262:613-627. [DOI] [PubMed] [Google Scholar]
- 10.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henson, J. D., A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21:598-610. [DOI] [PubMed] [Google Scholar]
- 13.Horowitz, H., and J. E. Haber. 1985. Identification of autonomously replicating circular subtelomeric Y′ elements in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:2369-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, P., F. E. Pryde, D. Lester, R. L. Maddison, R. H. Borts, I. D. Hickson, and E. J. Louis. 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11:125-129. [DOI] [PubMed] [Google Scholar]
- 15.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivessa, A. S., J. Q. Zhou, and V. A. Zakian. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100:479-489. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, F. B., R. A. Marciniak, M. McVey, S. A. Stewart, W. C. Hahn, and L. Guarente. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20:905-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johzuka, K., and H. Ogawa. 1995. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139:1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, B., and A. J. Lustig. 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10:1310-1326. [DOI] [PubMed] [Google Scholar]
- 20.Lindstrom, U. M., R. A. Chandrasekaran, L. Orbai, S. A. Helquist, G. P. Miller, E. Oroudjev, H. G. Hansma, and E. T. Kool. 2002. Artificial human telomeres from DNA nanocircle templates. Proc. Natl. Acad. Sci. USA 99:15953-15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liti, G., and E. J. Louis. 2003. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol. Cell 11:1373-1378. [DOI] [PubMed] [Google Scholar]
- 22.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 23.Lustig, A. J. 2003. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 4:916-923. [DOI] [PubMed] [Google Scholar]
- 24.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 25.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822-1834. [DOI] [PubMed] [Google Scholar]
- 26.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 28.Natarajan, S., C. Groff-Vindman, and M. J. McEachern. 2003. Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot. Cell. 2:1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Natarajan, S., and M. J. McEachern. 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22:4512-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nugent, C. I., and V. Lundblad. 1998. The telomerase reverse transcriptase: components and regulation. Genes Dev. 12:1073-1085. [DOI] [PubMed] [Google Scholar]
- 31.Ogino, H., K. Nakabayashi, M. Suzuki, E. Takahashi, M. Fujii, T. Suzuki, and D. Ayusawa. 1998. Release of telomeric DNA from chromosomes in immortal human cells lacking telomerase activity. Biochem. Biophys. Res. Commun. 248:223-227. [DOI] [PubMed] [Google Scholar]
- 32.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddel, R. R., T. M. Bryan, L. M. Colgin, K. T. Perrem, and T. R. Yeager. 2001. Alternative lengthening of telomeres in human cells. Radiat. Res. 155:194-200. [DOI] [PubMed] [Google Scholar]
- 34.Regev, A., S. Cohen, E. Cohen, I. Bar-Am, and S. Lavi. 1998. Telomeric repeats on small polydisperse circular DNA (spcDNA) and genomic instability. Oncogene 17:3455-3461. [DOI] [PubMed] [Google Scholar]
- 35.Shay, J. W., Y. Zou, E. Hiyama, and W. E. Wright. 2001. Telomerase and cancer. Hum. Mol. Genet. 10:677-685. [DOI] [PubMed] [Google Scholar]
- 36.Shore, D. 2001. Telomeric chromatin: replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11:189-198. [DOI] [PubMed] [Google Scholar]
- 37.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 38.Sugawara, N., E. L. Ivanov, J. Fishman-Lobell, B. L. Ray, X. Wu, and J. E. Haber. 1995. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 373:84-86. [DOI] [PubMed] [Google Scholar]
- 39.Sung, P. 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265:1241-1243. [DOI] [PubMed] [Google Scholar]
- 40.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 41.Teng, S. C., C. Epstein, Y. L. Tsai, H. W. Cheng, H. L. Chen, and J. J. Lin. 2002. Induction of global stress response in Saccharomyces cerevisiae cells lacking telomerase. Biochem. Biophys. Res. Commun. 291:714-721. [DOI] [PubMed] [Google Scholar]
- 42.Teng, S.-C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tokutake, Y., T. Matsumoto, T. Watanabe, S. Maeda, H. Tahara, S. Sakamoto, H. Niida, M. Sugimoto, T. Ide, and Y. Furuichi. 1998. Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem. Biophys. Res. Commun. 247:765-772. [DOI] [PubMed] [Google Scholar]
- 44.Tsai, Y.-L., S.-F. Tseng, S.-H. Chang, C.-C. Lin, and S.-C. Teng. 2002. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 22:5679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 46.Weilbaecher, R. G., and V. Lundblad. 1999. Assembly and regulation of telomerase. Curr. Opin. Chem. Biol. 3:573-577. [DOI] [PubMed] [Google Scholar]
- 47.Wotton, D., and D. Shore. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748-760. [DOI] [PubMed] [Google Scholar]
- 48.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175-4179. [PubMed] [Google Scholar]
- 49.Zakian, V. A. 1996. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30:141-172. [DOI] [PubMed] [Google Scholar]