Abstract
Despite decades of work by structural biologists, there are still ~5200 protein families with unknown structure outside the range of comparative modeling. We show that Rosetta structure prediction guided by residue-residue contacts inferred from evolutionary information can accurately model proteins that belong to large families, and that metagenome sequence data more than triples the number of protein families with sufficient sequences for accurate modeling. We then integrate metagenome data, contact based structure matching and Rosetta structure calculations to generate models for 614 protein families with currently unknown structures; 206 are membrane proteins and 137 have folds not represented in the PDB. This approach provides the representative models for large protein families originally envisioned as the goal of the protein structure initiative at a fraction of the cost.
There are 14849 protein families in the PFAM (1) database with 50 or more residues, of which 4752 have at least one member with experimentally determined x-ray crystal or NMR structure, and an additional 3984 for which reliable comparative models can be built based on homologues of known structure detected using the powerful HHsearch fold recognition program (2; there are an additional 902 for which less confident comparative models can be built). There is no structural information available for 5211 of the remaining 6113 families (HHsearch E-value ≥ 1). Until recently, computational methods could not generate accurate models for these 5211 families as they lack homologues of known structure for comparative modeling, and the very large number of conformations accessible to a polypeptide chain made the sampling problem in de novo protein structure prediction intractable for all but the smallest proteins. The original goal of the protein structure initiative was to determine structures for at least one representative of such families, but this proved to be extremely challenging and the focus of the initiative shifted to targets of immediate biological interest (3).
The increase in the number of known amino acid sequences has enabled the accurate prediction of residue-residue contacts using evolutionary data (4 – 10) -- substitutions at positions close in space in the three dimensional structure covary. Such contact predictions have been used for a wide range of protein modeling efforts (11 – 22). Accurate contact prediction requires large numbers of aligned sequences so that residue-residue covariance is clearly distinguished from lineage effects. While coevolution based structure modeling has been used to generate models for individual proteins with fold-level accuracy (TMscore (23) > 0.5; 5, 7 – 8, 10 – 11, 14 – 18, 21, 22), it has not been clear whether such data combined with structure prediction methodology can generate accurate models on a larger scale.
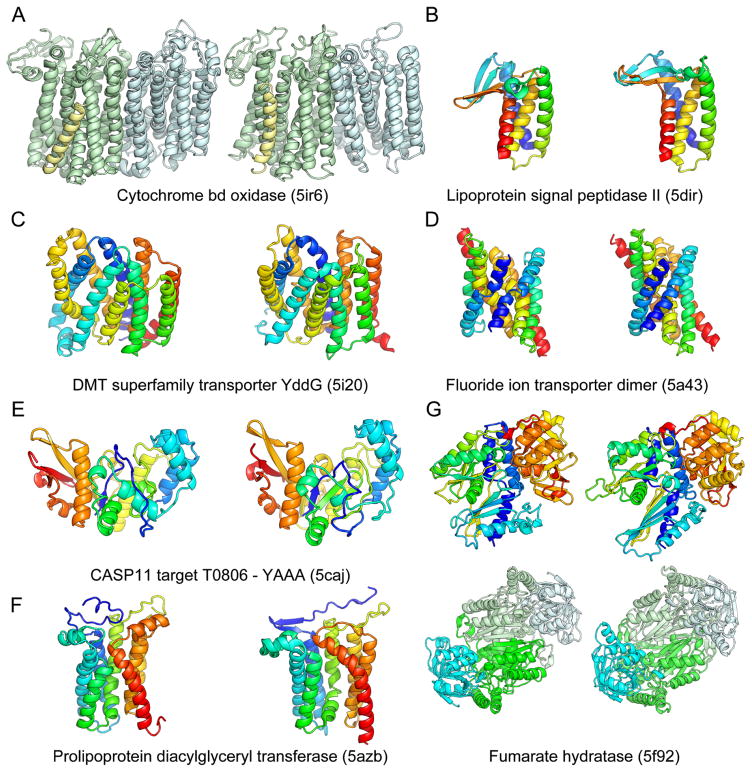
Rosetta de novo structure prediction calculations guided by evolutionary information were recently used to generate models for 58 large protein families (21). The structures of proteins in six of these families have since been published, providing an opportunity to assess this medium scale prediction effort. Recently solved structures of the Lipoprotein signal peptidase II (24), Prolipoprotein diacylglyceryl transferase (25), the fluoride ion transporter (26), cytochrome bd oxidase (27), DMT superfamily transporter YddG (28), and fumerate hydratase (29) a re all very close to computational models published and publicly released well before the structures were solved (Fig. 1). In the case of the three subunit cytochrome bd oxidase, the computational model of the 788 residue complex generated using both inter and intra subunit contact information was used together with experimental phase information obtained from the 3 heme irons and a single methionine to solve the structure. Because the phase information was weak, it was only possible to place the transmembrane helices and a subset of the side-chains based on the density, but the loops, connectivity, location of the CydX subunit, and registration of the amino acid sequence on many of the helices were unclear. Our E. coli protein model closely overlapped with the traced helices, and Phenix-Rosetta refinement (30) of a model built for the Geobacillus thermodenitrificans protein resolved the above ambiguities enabling rapid completion of structure determination. The final deposited structure is very similar to our previously published model of the E. coli protein (Fig. 1A; TMalign score (23) of 0.8). The power of Rosetta structure prediction calculations coupled with coevolution data for soluble proteins is illustrated by an extremely accurate blind de novo prediction for a quite complex protein structure in the CASP11 structure prediction experiment (31) (Fig. 1E). In all of the cases shown in Fig. 1, standard threading or fold recognition methods fail to identify the correct fold. Taken together, these data show that Rosetta modeling guided by coevolutionary constraints generates quite accurate models (in all 6 cases, the TMalign score is greater than 0.7; the models also illustrate some of the limitations of the approach, including the lack of explicit modeling of ligands, cofactors, and lipids, see supplemental text).
Fig. 1.
Comparison of Rosetta models (left) to subsequently published crystal structures (right). The models accurately recapitulate the structural details of A) the Cytochrome bd oxidase (TMalign score 0.88) B) the Lipoprotein signal peptidase II (TMalign score 0.70) C) the DMT superfamily transporter YddG (TMalign score 0.70) D) the Fluoride ion transporter dimer (TMalign score 0.69) E) the CASP11 target T0806 F) Prolipoprotein diacylglyceryl transferase (TMalign score 0.69) and G) Fumarate hydratase (TMalign score 0.80 for monomer (top) and 0.76 for dimer (bottom)).
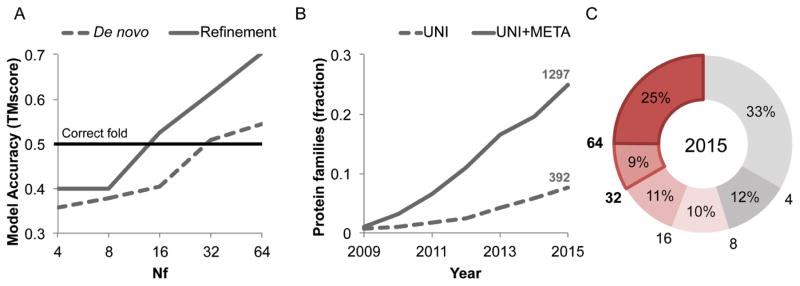
Structure models with the accuracy of those in Fig. 1 would have broad utility for framing biological hypotheses about function and interpreting mutational data, as well as guiding experimental structure determination. To determine the number of aligned sequences required for contact prediction accuracy sufficient to guide generation of accurate 3D models we carried out Rosetta structure prediction calculations for a benchmark set of 27 large protein families (Table S1) with known structure. We used both the full sequence alignments as well as alignments of subsets of the sequences for contact prediction. We also performed structure prediction calculations using Rosetta to hybridize and refine (32) partial structural matches identified by matching predicted contacts with the contact patterns of known protein structures. To do this, we developed an algorithm (map_align; see Supplementary info) that employs iterative double dynamic programming (33). The two approaches are complementary: de novo structure prediction (using only sequence information) (34) can succeed where there are no related structures in the PDB (Protein Data Bank), while making use of matches to known structures can help for large complex proteins that otherwise present a convergence challenge for de novo structure prediction (structural matches can occur in the absence of detectable sequence similarity since structural similarity is retained over larger evolutionary distances). For large sequence families, combining de novo structure prediction models and map_align structure matches using the Rosetta iterative hybridization protocol improved accuracy in 14 cases and decreased accuracy in only one (Fig. 2A solid line; Fig. S1; see Supplementary info). Contact prediction accuracy and hence predicted structure accuracy depends on the number of sequences in the family, the diversity of these sequences, and the length of the protein. A measure that incorporates all three factors (Nf, the number of sequence clusters at an 80% sequence identity clustering threshold divided by the square root of the protein length (21)) correlates well with contact prediction accuracy (21) and model accuracy (Fig. 2A, Fig. S1) over a broad range of families.
Fig. 2.
Metagenome data greatly increased fraction of structures which can be accurately modeled. A) Dependence of coevolution guided Rosetta structure prediction accuracy on the effective number of sequences Nf (a function of both sequence number and diversity; see Methods definition) in the protein family. For each of 27 proteins of known structure, the multiple sequence alignment was subsampled and residue-residue contacts predicted using GREMLIN. Rosetta structure prediction calculations were then used to generate ~20,000 models, and a single model was selected based on the Rosetta energy and the fit to the coevolution constraints; the average TMscore of these selected models over all 27 cases is shown on the y axis (dashed line). Hybridization based refinement of the top 20 models together with the top 10 map_align based models for each case increases the average accuracy (solid line); models with fold-level accuracy (TMscore > 0.5) are obtained for Nf ≥ 16, and models with accuracy typical of comparative modeling, for Nf of 64. B) Fraction of protein families of unknown structure with at least 64 Nf. Dashed line: including only sequences in UniRef100 database; solid line: including sequences in UniRef100 database together with metagenome sequence data from JGI (37). C) Distribution of Nf values for 5211 PFAM families with currently unknown structure, after the addition of metagenomic sequences; 25% of the protein-families have Nf > 64, 34% have Nf > 32 and 45% have Nf > 16.
How many protein families with currently unknown structure have Nf values in the range where accurate models can be built? The models in Fig. 1 were all generated for families with Nf > 64; accuracy falls off for lower values of Nf (Fig. 2A). As shown in Fig. 2B, less than 8% of families have Nf values of 64 or better. Modeling the remaining 92% of families of unknown structure at reasonable accuracy is not currently possible using the sequence information in the UniRef100 database (35).
This limitation in structure modeling can be largely overcome by taking advantage of progress in a completely different research area. Metagenome sequencing projects, in which complex biological samples are shotgun sequenced, have provided insights into biological communities and provide a treasure trove of new sequence data (36, 37). The number of protein sequences determined in metagenome sequence projects is growing considerably faster than the UniRef100 database (Fig. 2B, solid versus dashed line). With the inclusion of metagenome sequence data, the number of sequences increases by as much as 100 fold for some families (Table S2), and the fraction of families with unknown structure that can be accurately modeled using coevolution guided structure prediction methods increases dramatically. At Nf ≥ 64, the fraction increases from 0.08 to 0.25, and at Nf ≥ 32 (where fold level accuracy can be achieved (Fig. 2A)), the fraction increases from 0.16 to 0.33. To assess structure prediction and model evaluation accuracy using metagenome data, we carried out a second benchmark of 81 PFAMs with recently solved structures and Nf ≥ 64 (Fig. S1E–F, Table S5). Structure prediction accuracy was correlated with the extent of convergence of the lowest energy models and the fraction of predicted contacts present in these models (Fig. S1F and S2). For 42 families, the predictions converged with most of the predicted contacts satisfied (see Supplementary information for convergence criteria) and of these, 25 had a TMscore > 0.7 and 13 a TMscore > 0.6 (in 3 of the 4 remaining cases, NMR structures of small transmembrane proteins, our models fit the predicted contacts much better, and in the last case, an intertwined dimer, our monomer model contained all the correct contacts (Fig. S13)).
We generated coevolution based contact predictions using GREMLIN (4, 12) for the 1297 protein families with Nf ≥ 64, and built models for the 921 protein families (1024 domains) with many contacts between positions separated by more than five residues along the linear sequence (number of long range contacts > half the number of residues in protein). The structure prediction calculations converged on models with predicted TM scores greater than 0.65 for 614 of the 1024 domains according to the benchmarks. A list of the PFAM families covered by these models is in Table S3; the models are available at <https://gremlin2.bakerlab.org/meta.php>, along with an interactive 3D interface powered by 3Dmol.js (38) and D3.js (39) for visualization of coevolution contacts on the models. These structures provide close templates for comparative modeling of 487,306 UniRef100 and 3,868,268 IMG metagenomic unique (less than 80% pairwise identity) sequences.
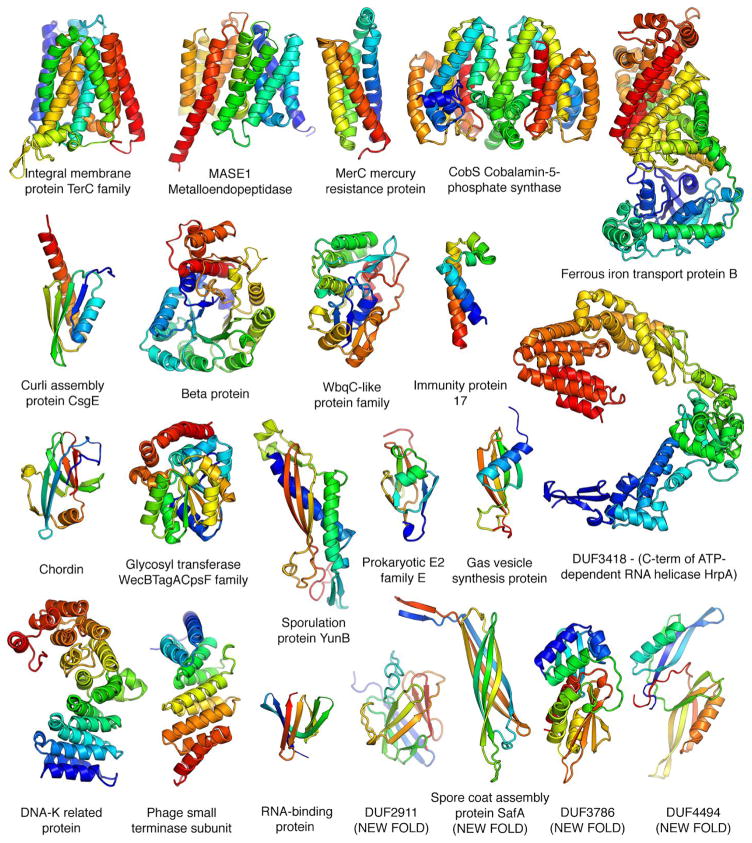
The converged models for the 614 PFAM families (Table S3) provide a view of the hitherto unseen protein universe. To determine if the models belong to known protein folds, we carried out structure-structure comparisons against the SCOP (40) domain database. For 477 of the families, the models matched a protein of known structure over nearly the entire length and hence can be assigned to SCOP folds (52 distinct all alpha, 29 alpha/beta, 51 alpha+beta, and 28 all beta folds). In a number of cases, the SCOP classifications are consistent with previous functional information, for example the restriction endonuclease Xho1 is assigned to the restriction enzyme fold, and a family of prokaryotic putative ubiquitin like proteins is assigned the beta-grasp fold (to which ubiquitin belongs). For 137 of the domains, there were no significant structure matches of the models to the PDB (TMalign score < 0.5) and hence these have new folds. Space limitations preclude showing here even a small number of the 614 models; instead we show a small selection of the 3D structures in Fig. 3. They include the key developmental regulator Chordin, CobS a key enzyme in cobalbumin synthesis, a metalloendopeptidase, and mercury and iron transporters; six are transmembrane proteins, four have new folds and several have quite complex topologies. These and the remaining 590 structure models not shown in Fig. 3 should provide a basis for understanding molecular function, mechanism and guide experimental structure determination (such efforts should be informed of the limitations of the modeling approach described in the Supplementary text). While this manuscript was in preparation, crystal structures of members of five of the 614 families were published and are very similar to the corresponding models (TMalign score ≥ 0.7; See Fig. S3 and Table S4).
Fig. 3.
Representative structure models for selected PFAM families. Membrane proteins are on the top row; new folds on the bottom right. The multidomain models of the iron transporter and RNA helicase and the dimeric model of CobS, an enzyme in vitamin B synthesis, are guided by both intra- and inter-chain coevolution restraints.
The models presented in this paper fill in about 12% of the structural information missing for known protein families. That this could be accomplished using computational modeling methods was not at all apparent five years ago. This progress required integration of advances in quite disparate research areas: metagenome sequencing, coevolutionary analysis, and de novo protein structure prediction methodology. This combined approach has a bright future: extrapolating from the data in Fig. 2B suggests that in several years the majority of families will have sufficient number of sequences for accurate structure modeling. A current limitation is that most sequence data is for prokaryotes, but as fungal and other simple eukaryote genome sequencing projects ramp up the approach should become applicable to eukaryote specific protein families.
Supplementary Material
Acknowledgments
We would like to thank Pietro Di Lena, Noel Malod-Dognin and Rumen Andonov for providing the source code for their software (Al-eigen and a_purva) and for their discussion and advice on contact map alignment. 3-D structures of 614 PFAMs modeled in the study is available at https://gremlin2.bakerlab.org/meta.php. We also thank Rosetta@home and Charity engine participants for donating their computer time. The work performed by NV, GAP and NCK, was supported by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, under Contract No. DE-AC02-05CH11231. Research reported in this publication was supported by NIGMS of the National Institutes of Health under award number R01GM092802. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Materials and Methods
References (41–63)
References
- 1.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–85. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Söding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 3.Montelione GT. The Protein Structure Initiative: achievements and visions for the future. F1000 Biol Rep. 2012;4:7. doi: 10.3410/B4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci U S A. 2013;110:15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marks DS, et al. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morcos F, et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci U S A. 2011;108:E1293–1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopf TA, et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 2012;149:1607–1621. doi: 10.1016/j.cell.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nugent T, Jones DT. Accurate de novo structure prediction of large transmembrane protein domains using fragment-assembly and correlated mutation analysis. Proc Natl Acad Sci U S A. 2012;109:E1540–1547. doi: 10.1073/pnas.1120036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones DT, Buchan DW, Cozzetto D, Pontil M. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28:184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- 10.Marks DS, Hopf TA, Sander C. Protein structure prediction from sequence variation. Nat Biotechnol. 2012;30:1072–1080. doi: 10.1038/nbt.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sułkowska JI, Morcos F, Weigt M, Hwa T, Onuchic JN. Genomics-aided structure prediction. Proc Natl Acad Sci U S A. 2012;109:10340–10345. doi: 10.1073/pnas.1207864109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balakrishnan S, Kamisetty H, Carbonell JG, Lee SI, Langmead CJ. Learning generative models for protein fold families. Proteins. 2011;79:1061–1078. doi: 10.1002/prot.22934. [DOI] [PubMed] [Google Scholar]
- 13.Ekeberg M, Lövkvist C, Lan Y, Weigt M, Aurell E. Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87:012707. doi: 10.1103/PhysRevE.87.012707. [DOI] [PubMed] [Google Scholar]
- 14.Wickles S, et al. A structural model of the active ribosome-bound membrane protein insertase YidC. Elife. 2014;3:e03035. doi: 10.7554/eLife.03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian P, et al. Structure of a functional amyloid protein subunit computed using sequence variation. J Am Chem Soc. 2015;137:22–25. doi: 10.1021/ja5093634. [DOI] [PubMed] [Google Scholar]
- 16.Hayat S, Sander C, Elofsson A, Marks DS. Accurate prediction of transmembrane β-barrel proteins from sequences. 2014 doi: 10.1073/pnas.1419956112. bioRxiv, 006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopf TA, et al. Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat Commun. 2015;6:6077. doi: 10.1038/ncomms7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abriata LA. An homology-and coevolution-consistent structural model of bacterial copper-tolerance protein CopM supports function as a “metal sponge”and suggests regions for …. 2015 bioRxiv. [Google Scholar]
- 19.Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopf TA, et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife. 2014;3 doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovchnnikov S, et al. Large-scale determination of previously unsolved protein structures using evolutionary information. Elife. 2015;4:e09248. doi: 10.7554/eLife.09248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antala S, Ovchinnikov S, Kamisetty H, Baker D, Dempski RE. Computation and Functional Studies Provide a Model for the Structure of the Zinc Transporter hZIP4. J Biol Chem. 2015;290:17796–17805. doi: 10.1074/jbc.M114.617613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 24.Vogeley L, et al. Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science. 2016;351:876–880. doi: 10.1126/science.aad3747. [DOI] [PubMed] [Google Scholar]
- 25.Mao G, et al. Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat Commun. 2016;7:10198. doi: 10.1038/ncomms10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stockbridge RB, et al. Crystal structures of a double-barrelled fluoride ion channel. Nature. 2015;525:548–551. doi: 10.1038/nature14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Safarian S, et al. Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases. Science. 2016;352:583–586. doi: 10.1126/science.aaf2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuchiya H, et al. Structural basis for amino acid export by DMT superfamily transporter YddG. Nature. 2016;534:417–420. doi: 10.1038/nature17991. [DOI] [PubMed] [Google Scholar]
- 29.Feliciano PR, Drennan CL, Nonato MC. Crystal structure of an Fe-S cluster-containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold. Proc Natl Acad Sci U S A. 2016;113:9804–9809. doi: 10.1073/pnas.1605031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiMaio F, et al. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat Methods. 2013;10(11):1102–1104. doi: 10.1038/nmeth.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovchinnikov S, Kim DE, Wang R. Improved de novo structure prediction in CASP11 by incorporating Co-evolution information into rosetta. Proteins: Struct Funct Bioinf. 2015 doi: 10.1002/prot.24974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor WR. Protein structure comparison using iterated double dynamic programming. Protein Sci. 1999;8:654–665. doi: 10.1110/ps.8.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simons KT, et al. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 35.Suzek BE, et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P. A bioinformatician’s guide to metagenomics. Microbiol Mol Biol Rev. 2008;72(4):557–78. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markowitz VM, et al. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 2014;42:D568–73. doi: 10.1093/nar/gkt919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rego N, Koes D. 3Dmol.js: molecular visualization with WebGL. Bioinformatics. 2015;31:1322–1324. doi: 10.1093/bioinformatics/btu829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bostock M, Ogievetsky V, Heer J. D #x0B3; Data-Driven Documents. IEEE Trans Vis Comput Graph. 2011;17:2301–2309. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- 40.Andreeva A, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–25. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.