Abstract
Giardia duodenalis has linear chromosomes capped with typical eukaryotic repeats [(TAGGG)n], subtelomeric rRNA genes, and telomere gene units. The absence of two closely associated NotI sites in the large-subunit rRNA gene was used as an indicator in hybridizations of one- and two-dimensional NotI-cleaved Giardia chromosome separations that some chromosomes carry only rearranged and, by deduction, nonfunctional rRNA genes.
We and others have shown that telomere repeats are associated with DNA encoding the rRNA gene unit or parts thereof (called herein the rRNA gene unit) on major chromosomes of the protozoan parasite Giardia duodenalis (1, 15). Adam et al. (1) proposed that rRNA gene units were juxtaposed with telomere repeats based on a sequence of recombinant DNA recovered from a library of Giardia DNA enriched for telomere sequences. Arkhipova and Morrison (2) have since published the finding that a transposable element in Giardia is juxtaposed to telomere repeats, and Burke et al. (3) have shown that close relatives of this element can be found associated with several chromosomes. We have mapped Giardia telomere regions and shown that, in addition to the rRNA gene unit, the telomere regions carry telomere gene units (TGU) (15). Two TGU have been fully sequenced (15). The TGU (Fig. 1A) terminate in ankyrin genes which are linked to the rRNA gene unit by a spacer region which inserts into the rRNA gene unit at exactly the same position, i.e., position 2523, in the large-subunit (LS) rRNA gene (15, 16). The evidence now suggests that rRNA gene units are not uniformly adjacent to telomere repeats but that all Giardia telomere regions are similarly structured according to the XbaI restriction enzyme cleavage patterns of whole chromosomes (15).
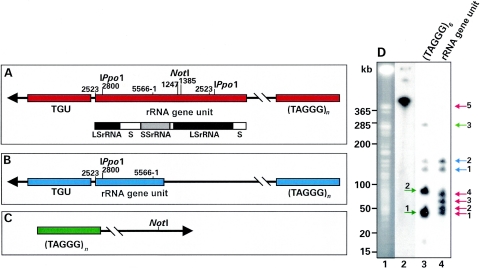
FIG. 1.
rRNA gene units in the subtelomeric regions of major Giardia chromosomes. The rRNA gene unit is inserted into the spacer separating it from the TGU at position 2523 in the LS rRNA (5, 10, 15). The rRNA gene unit repeat notation defines the last base of the unit (5566) within the SS rRNA gene. Numbering then recommences at 1 within the same gene. NotI sites are at positions 1247 and 1385, and an I-PpoI site is at position 2800 (5). The maximum size of subtelomeric regions of Giardia major chromosomes, including the rRNA gene unit and telomere repeats (TAGGG)6, is 35 kb. Excluding the TGU, the size is reduced to less than 30 kb. (A) Subtelomeric map with functional rRNA genes. NotI cleavage separates the telomere repeats from the rest of the chromosomal NotI segments. The functional rRNA gene transcription unit includes SS rRNA through the LS rRNA (black and grey bars). S, spacer region. (B) Map of rearranged subtelomeric region with NotI sites missing. NotI chromosome cleavage fails to separate telomere repeats from the rRNA gene unit. (C) Map of subtelomeric regions at chromosome ends devoid of rRNA gene units or TGU characterized by NotI segments larger than approximately 30 kb which hybridize only with (TAGGG)6 repeats. (D) One-dimensional separation of NotI-cleaved Giardia chromosomes (lane 1, ethidium bromide stained), hybridized with (TAGGG)6 (lane 3) or rRNA gene units (lane 4) (15). Bands larger than 30 kb hybridizing with rRNA gene units arise only from complete rRNA gene units (red arrows). Missing NotI sites in rearranged rRNA gene units result in bands which hybridize with both the rRNA gene unit and (TAGGG)6 (blue arrows). Bands greater than 30 kb hybridizing with (TAGGG)6 only are derived from telomeres at the distal chromosomal ends devoid of rRNA gene units or TGU (green arrows). Lane 2, uncleaved DNA hybridized with (TAGGG)6. Kilobase markers are derived from a 5-kb ladder, a lambda ladder, and yeast chromosomes (Bio-Rad). See the text for a description of the numbering scheme for the arrows.
The majority of tandemly arrayed rRNA gene units are found on accessory chromosomes in strain WB (16); Le Blancq (8) and Hou et al. (6) described specific chromosomes which undergo frequent rearrangements resulting in size variation due mostly to changes in the rRNA gene unit repeat numbers. In order to answer the question of whether rRNA genes on the major and/or accessory chromosomes are transcribed, it is important to establish whether complete rRNA gene units (consisting of at least one contiguous segment encoding small-subunit [SS] rRNA, intervening sequences, 5.8S rRNA, and LS rRNA) (Fig. 1A) are present on the major chromosomes, since cotranscription of all rRNA genes is regarded as the most likely scenario (7).
An intact rRNA gene unit extends more than 8,000 bp from the rRNA gene unit insertion site at position 2523 of the fragmented LS rRNA gene (Fig. 1A). Close to this site is the I-PpoI site within the fragment at position 2800 of the rRNA gene unit sequence (5) (Fig. 1A). Following the fragmented LS rRNA gene shown in Fig. 1A is a spacer region (S), and following this are (i) an intact rRNA gene unit encoding the SS rRNA gene, (ii) transcribed spacer regions, (iii) 5.8S rRNA, and (iv) LS rRNA genes (12), all of which are required for transcription (Fig. 1A) (reference 7 and references therein). Well into the complete LS rRNA gene are two NotI sites at positions 1247 and 1385 of the rRNA gene unit (Fig. 1A). These NotI sites are not present in the fragmented LS rRNA sequence joined to the TGU. In general, NotI sites in the Giardia genome are infrequent (11). We already know that the maximum size of the TGU-rRNA gene unit-telomere repeat region in Giardia is approximately 35 kb and significantly smaller than this when the TGU are not included (TGU1, 6.8 kb; TGU2, 8.4 kb [15]). If we cleave chromosomal DNA with I-PpoI, all of the detectable rRNA gene units are removed from the chromosomes (data not shown). If, however, the chromosomes carrying complete rRNA gene units are cleaved with NotI, more than 4,000 bp of the rRNA gene units will remain associated with the TGU and other chromosomal DNA, while the telomere repeats will be found on chromosome segments of less than 30 kb. The rRNA gene units from accessory chromosomes carrying tandemly arrayed rRNA genes will mostly be reduced to individual rRNA gene units of 5,566 bp (5).
If the rRNA gene units on the major chromosomes are rearranged or if there is only a remnant of the rRNA gene unit at a particular site such that the two NotI sites are missing, then large NotI chromosome segments (most NotI Giardia chromosome segments fall in the 50- to 650-kb range [see reference 11] [Fig. 1D, lane 1]) carrying rRNA gene units will also hybridize with telomere repeats (Fig. 1B). Telomere repeats associated with chromosome ends not carrying TGU or rRNA gene units are likely to be associated with a range of NotI-cleaved chromosome segments, not with rRNA gene units (Fig. 1C).
Successive hybridization of NotI-cleaved chromosomes (separated in a single dimension as previously described [11]) with telomere repeats [(TAGGG)6] and complete rRNA gene units (15) showed (i) bands greater than 30 kb that hybridized with rRNA gene units only, consistent with intact rRNA gene units (Fig. 1D, red arrows 1 to 5); (ii) bands that hybridized with rRNA gene units and telomere repeats, indicating rearranged rRNA gene units (NotI sites missing) (Fig. 1D, blue arrows 1 and 2); and (iii) telomere-hybridizing bands only indicating telomeres located on the rRNA gene unit distal end of the chromosome (Fig. 1D, green arrows 1 to 3). The bands of approximately 45 to 50 kb (Fig. 1D, lanes 3 and 4, green arrow 1 and red arrow 1), while apparently of the same size, are not convincingly similar spots, suggesting that they are different bands. To identify which chromosomes carried rearranged rRNA gene units and which carried intact NotI sites (and were therefore likely to carry complete rRNA gene units), we separated WB-1B chromosomes, cleaved the separated chromosomes with NotI, and separated the NotI segments in the second dimension (under cleavage and separation conditions similar to those described for one-dimensional separations [11]). The gel was blotted and hybridized as described above. The logic applied is the same as that described above for a one-dimensional separation of NotI segments. Identical spots in Fig. 2B and C which align with chromosomes shown in Fig. 2A indicate missing NotI sites in the rRNA gene units. Unique spots in Fig. 2C suggest intact rRNA gene units associated with a specific chromosome.
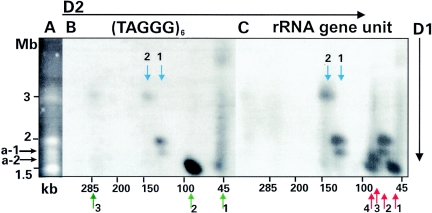
FIG. 2.
Chromosome localization of rearranged rRNA gene units on major and minor chromosomes of Giardia. Chromosomes of Giardia strain WB were separated in the first dimension (D1) as indicated by a vertical arrow (A) and after NotI cleavage in the second dimension (D2) as indicated by a horizontal arrow (15). A blot of the gel was hybridized sequentially with (TAGGG)6 (B) and the rRNA gene unit (C). Blue arrows indicate similar spots in panels B and C which arose from chromosomes with rearranged or fragmented LS rRNA genes. Red arrows indicate spots which arose from chromosomes with intact rRNA gene unit NotI sites consistent with functional rRNA genes. Kilobase markers are derived from a 5-kb ladder, a lambda ladder, and yeast chromosomes (Bio-Rad). Chromosome sizes (in megabases) are derived from our published mapping study (4), yeast markers (Bio-Rad), and a study which revealed that the 3-Mb chromosome was cleaved once internally by I-PpoI into 0.9- and 2.1-Mb segments (data not shown). See the text for a description of accessory chromosomes a-1 and a-2 and of the numbering scheme for the arrows.
The obvious conclusions to be drawn from Fig. 2 are that the 3-Mb major chromosome carries incomplete rRNA genes (blue arrow 2) and that the 2-Mb chromosome, which is a single linkage group (4), carries both incomplete rRNA genes (blue arrow 1) and rRNA gene units with intact NotI sites (red arrow 2). The non-rRNA gene unit telomere end of the 3-Mb chromosome is also evident (Fig. 2B, green arrow 3).
The chromosome band of 1.5 Mb carries two chromosomes (13), and NotI segments derived from these two chromosomes hybridize with two rRNA gene unit bands (Fig. 2C, red arrows 1 and 4) and the telomere probe (Fig. 2B, green arrow 2). The approximately 90-kb bands (Fig. 2, red arrow 4 and green arrow 2), while appearing similar in Fig. 2, are not identical in Fig. 1D, lanes 3 and 4 (green arrow 2 and red arrow 4, respectively). The 45- to 50-kb bands (Fig. 1D, green arrow 1 and red arrow 1) can now be viewed as two-dimensional spots, one of which aligns with the 1.5-Mb chromosomes (Fig. 2C, red arrow 1), but there are no identical spots in Fig. 2B. This finding is consistent with the presence of intact rRNA genes on the 1.5-Mb chromosomes (Fig. 2C).
The approximately 50-kb intense band (Fig. 1D, lane 3, green arrow 1) is visible as a smear at approximately 50 kb (Fig. 2B, green arrow 1). We interpret this smear to be the non-rRNA gene unit-carrying telomere ends of accessory chromosomes which separate as smears in the first dimension due to the stickiness of the rRNA gene unit arrays (15, 16).
In addition to the major chromosomes, we see two prominent accessory chromosomes, a-1 and a-2 (Fig. 2A), which carry intact rRNA genes (Fig. 2C, red arrows 2 and 3). Chromosome a-1 also carries rearranged rRNA gene units associated with telomere repeats (spot of approximately 130 kb) (Fig. 2B and C, blue arrow 1).
We were not able to make any conclusions regarding chromosomes larger than 3 Mb, due, we believe, to a generalized loss of DNA during successive electrophoretic runs that was particularly evident with large chromosomes. Thus, the rRNA gene unit-hybridizing band indicated in Fig. 1D (lane 4, red arrow 5) is likely to be associated with the largest chromosome. Conversely, smaller spots tend to be more intense (see especially the band indicated by green arrow 2, which may also carry many telomere repeats).
We conclude that while all major and minor chromosomes carry rRNA gene units, two major WB-1B chromosomes carry incomplete, rearranged, or fragmented subtelomeric rRNA genes. The two NotI sites are present even in the most diverse G. duodenalis isolates yet described (14). Therefore, it can be deduced that such rearrangements in the highly conserved G. duodenalis LS rRNA genes (14) will result in nonfunctional LS rRNA genes and rRNA gene units (7). The precise function of the rRNA gene unit fragments embedded in the telomere regions remains unknown, but the telomere regions of Giardia chromosomes are highly plastic (9, 16), and they appear to be indicative of the plasticity and rearrangements typical of this region of eukaryotic chromosomes.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia and by the Australian Research Council.
REFERENCES
- 1.Adam, R. D., T. E. Nash, and T. E. Wellems. 1991. Telomeric location of Giardia rDNA genes. Mol. Cell. Biol. 11:3326-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arkhipova, I. R., and H. G. Morrison. 2001. Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead. Proc. Natl. Acad. Sci. USA 98:14497-14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke, W. D., H. S. Malik, S. M. Rich, and T. H. Eickbush. 2002. Ancient lineages of non-LTR retrotransposons in the primitive eukaryote, Giardia lamblia. Mol. Biol. Evol. 19:619-630. [DOI] [PubMed] [Google Scholar]
- 4.Chen, N., J. A. Upcroft, and P. Upcroft. 1994. Physical map of a 2 Mb chromosome of the intestinal parasite Giardia duodenalis. Chromosome Res. 2:307-313. [DOI] [PubMed] [Google Scholar]
- 5.Healey, A., R. Mitchell, J. A. Upcroft, P. F. L. Boreham, and P. Upcroft. 1990. Complete nucleotide sequence of the ribosomal RNA tandem repeat unit from Giardia intestinalis. Nucleic Acids Res. 18:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou, G., S. M. Le Blancq, Y. E., H. Zhu, and M. G.-S. Lee. 1995. Structure of a frequently rearranged rRNA-encoding chromosome in Giardia lamblia. Nucleic Acids Res. 23:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacob, S. T. 1995. Regulation of ribosomal gene transcription. Biochem. J. 306:617-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Blancq, S. M. 1994. Chromosome rearrangements in Giardia lamblia. Parasitol. Today 10:177-179. [DOI] [PubMed] [Google Scholar]
- 9.Le Blancq, S. M., and R. D. Adam. 1998. Structural basis of karyotype heterogeneity in Giardia lamblia. Mol. Biochem. Parasitol. 97:199-208. [DOI] [PubMed] [Google Scholar]
- 10.Le Blancq, S. M., R. S. Kase, and L. H. T. Van der Ploeg. 1991. Analysis of a Giardia lamblia rRNA encoding telomere with [TAGGG]n as the telomere repeat. Nucleic Acids Res. 19:5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Upcroft, J. A., N. Chen, and P. Upcroft. 1996. Mapping variation in chromosome homologues of different Giardia strains. Mol. Biochem. Parasitol. 76:135-143. [DOI] [PubMed] [Google Scholar]
- 12.Upcroft, J. A., A. Healey, R. Mitchell, P. F. Boreham, and P. Upcroft. 1990. Antigen expression from the ribosomal DNA repeat unit of Giardia intestinalis. Nucleic Acids Res. 18:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Upcroft, J. A., A. Healey, and P. Upcroft. 1993. Chromosomal duplication in Giardia duodenalis. Int. J. Parasitol. 23:609-616. [DOI] [PubMed] [Google Scholar]
- 14.Upcroft, J. A., A. Healey, and P. Upcroft. 1994. A new rDNA repeat unit in human Giardia. J. Eukaryot. Microbiol. 41:639-642. [DOI] [PubMed] [Google Scholar]
- 15.Upcroft, P., N. Chen, and J. A. Upcroft. 1997. Telomeric organization of a variable and inducible toxin gene family in the ancient eukaryote Giardia duodenalis. Genome Res. 7:37-46. [DOI] [PubMed] [Google Scholar]
- 16.Upcroft, P., and J. A. Upcroft. 1999. Organization and structure of the Giardia genome. Protist 150:17-23. [DOI] [PubMed] [Google Scholar]