Abstract
Purpose
Epidermal growth factor receptor (EGFR) mutations confer sensitivity to EGFR tyrosine kinase inhibitors in patients with advanced non–small-cell lung cancer (NSCLC). There are limited and conflicting reports on the frequency of EGFR mutations in Latinos.
Patients and Methods
Samples from 642 patients with NSCLC from seven institutions in the United States and Latin America were assessed for EGFR mutations (exons 18 to 21) at Clinical Laboratory Improvement Amendments-certified central laboratories.
Results
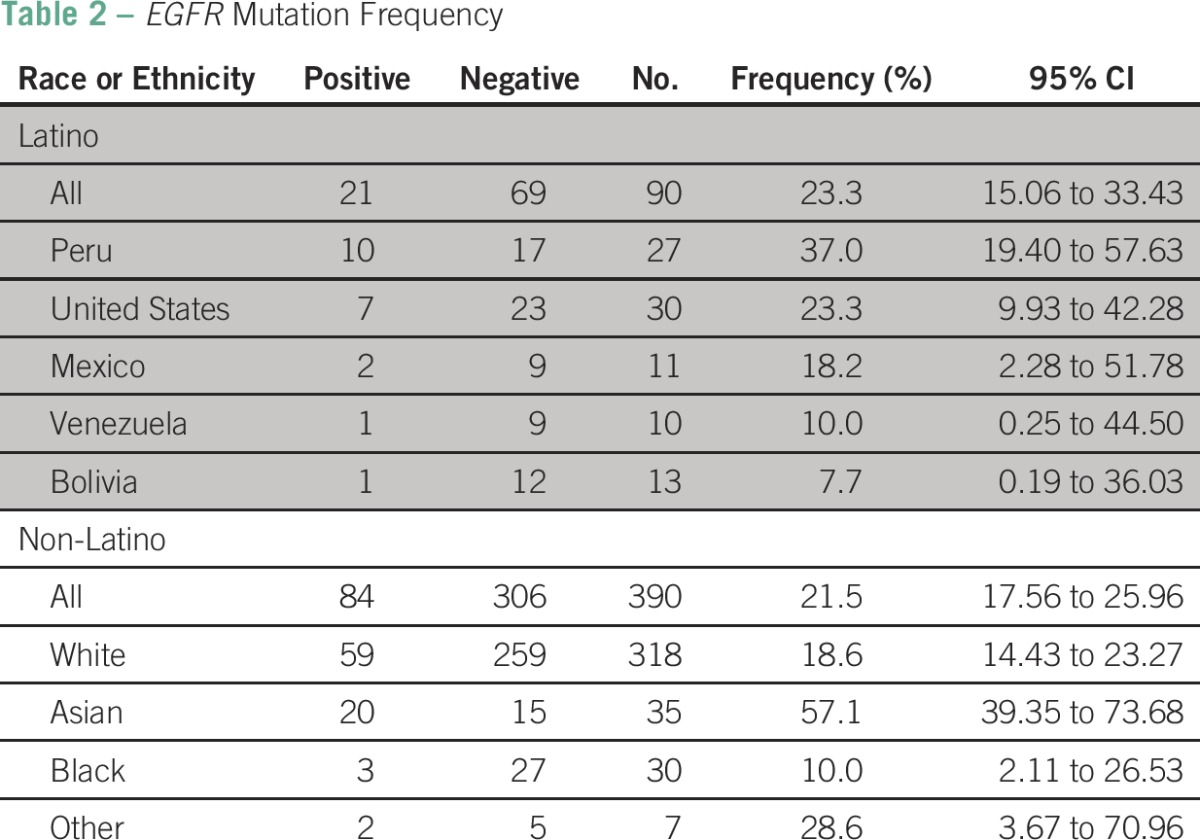
EGFR mutation analysis was successfully performed in 480 (75%) of 642 patients; 90 (19%) were Latinos, 318 (66%) were non-Latino whites, 35 (7%) were non-Latino Asians, 30 (6%) were non-Latino blacks, and seven (2%) were of other races or ethnicities. EGFR mutations were found in 21 (23%) of 90 Latinos with varying frequencies according to the country of origin; Latinos from Peru (37%), followed by the United States (23%), Mexico (18%), Venezuela (10%), and Bolivia (8%). In never-smoker Latinos and Latinos with adenocarcinoma histology, EGFR mutation frequencies were 38% and 30%, respectively. There was a significant difference in the frequency of EGFR mutations among the different racial and ethnic subgroups analyzed (P < .001), with non-Latino Asians having the highest frequency (57%) followed by Latinos (23%), non-Latino whites (19%), and non-Latino blacks (10%). There was no difference between Latinos (23%) and non-Latinos (22%; P = .78) and Latinos and non-Latino whites (P = .37). Patients from Peru had an overall higher frequency of mutations (37%) than all other Latinos (17%), but this difference only exhibited a trend toward significance (P = .058).
Conclusion
There was no significant difference between the frequency of EGFR mutations in NSCLC in Latinos and non-Latinos.
INTRODUCTION
Racial and ethnic disparities in the incidence, stage at diagnosis, treatment, and survival of patients with lung cancer have been described; however, the reasons for these disparities are not completely understood.1 It is estimated that the ethnic and racial composition of the United States will change dramatically in the next few decades.2 Given this, it is paramount that racial and ethnic health care disparities be studied and steps taken to have a positive impact on health outcomes. To effectively target and eliminate lung cancer–related health care disparities, a better understanding of the molecular characteristics of the disease and their relationship with race and ethnicity is needed.
Activating mutations in the epidermal growth factor receptor (EGFR) gene confer sensitivity to EGFR tyrosine kinase inhibitors in patients with advanced non–small-cell lung cancer (NSCLC).3-6 Racial, ethnic, and demographic differences in the distribution of such mutations have been previously described. For example, activating EGFR mutations are significantly more common in East Asians, women, and never-smokers.7 The frequency of EGFR mutations varies from approximately 10% of lung adenocarcinomas in North America and Europe to as high as 50% to 60% in Asia.8,9 However, there are limited and conflicting reports of the frequency of EGFR mutations among Latinos (also referred to as Hispanics). Published reports have been limited to retrospective cohorts that are constrained, among other factors, by patient selection for mutation testing based on clinical characteristics.10,11
According to the US Census Bureau, Latinos currently comprise 17.4% of the US population and are projected to grow to 29% of the population by 2060—more than one quarter of the total population.2 Lung cancer is the third most commonly diagnosed cancer among Latino men and women; it is the leading cause of cancer death among Latino men and the second-leading cause among Latino women.12 The incidence, clinical course, and outcomes of lung cancer among Latinos are distinct from non-Latino whites, who currently constitute the largest racial and ethnic group in the United States. Lung cancer incidence rates are lower among Latinos13; however, despite the lower socioeconomic status, more limited access to care, and diagnoses at advanced stages of disease that would predict otherwise, lung cancer mortality rates are 50% lower among Latinos than non-Latino whites.13,14 Although this so-called Latino paradox is still a matter of debate, given the limited reports to date,10,11 there is a clear need for a more comprehensive molecular characterization of NSCLC in this ethnic group to better understand outcome disparities in the United States. In particular, patients with EGFR mutations are known to have better survival than patients with wild-type EGFR.15
In this prospective study, using central testing, we sought to characterize the types and frequencies of EGFR-activating mutations among Latino patients with NSCLC living in the United States and Latin America and compare them with US non-Latinos.
PATIENTS AND METHODS
Participants
This study was conducted at the following institutions: National Cancer Institute (NCI; Bethesda, MD; coordinating center), Oregon Health and Science University (OHSU; Portland, OR), Instituto Nacional de Enfermedades Neoplasicas (Lima, Peru), Instituto Nacional de Cancerologia (Mexico City, Mexico), Hospital Oncologico Luis Razetti (Caracas, Venezuela), Denver Health Medical Center (Denver, CO), and Universidad Mayor de San Simon (Cochabamba, Bolivia).
Samples from patients with histologically confirmed NSCLC were collected from two protocols that were registered with clinicaltrials.gov: NCT01306045 (CUSTOM; Molecular Profiling and Targeted Therapies in Advanced Thoracic Malignancies Trial) and NCT01255150 (Frequency of EGFR Mutations in Latinos With Non-Small Cell Lung Cancer). Patients eligible for NCT01306045 had histologically confirmed advanced NSCLC for whom surgical resection or multimodality therapy with curative intent was not feasible. Patients had to have biopsiable disease and be willing to undergo biopsy for molecular profiling or had to have enough and adequate archival material from a previous biopsy to perform molecular profiling analyses. For protocol NCT01255150, all samples were collected from patients with histologically confirmed NSCLC who were dead at the time of tissue collection. For the purposes of both studies, the terms Hispanic or Latino included individuals who self-identified as Hispanic or Latino or were born in any Latin American country.
EGFR mutation testing was performed centrally at Clinical Laboratory Improvement Amendments (CLIA) –certified central laboratories of the NCI or OHSU. EGFR mutation analysis of exons 18 through 21 was performed with either pyrosequencing at the NCI or Sequenom MassArray (San Diego, CA) at OHSU. The institutional review boards of the participating centers approved the research protocol, and all living participants provided written informed consent. Tumor samples from dead individuals for whom basic clinical information was available were included in this analysis.
Pyrosequencing
DNA was extracted from paraffin-embedded tissue sections using the Qiagen QIAamp DNA FFPE Tissue Kit (Hilden, Germany), according to manufacturer’s instructions. For identification of point mutations, coamplifications at lower denaturation temperature–polymerase chain reaction (PCR; cold PCR) were performed either individually or in a single 96-well microtiter plate (complete gene panel) in an Applied Biosystems (ABI) 9700 thermocycler (Foster City, CA). After PCR, the products were subjected to pyrosequencing on a Qiagen PyroMark Q24 system. For identification of deletions and insertions, independent PCR reactions were performed with fluorescein-labeled primers in the ABI 9700, and the products were analyzed by capillary electrophoresis on an ABI 3130xl Genetic Analyzer. Five PCR reactions were designed to interrogate the most commonly occurring mutations, including deletion mutations in exon 19, point mutations (codons 858, 861, and 863) in exon 21, insertions and point mutations in exon 20 (codon 790), and mutations at codon 719 in exon 18. The products of the exon 20, exon 21, and exon 18 reactions were analyzed by pyrosequencing, and the products of deletion mutations in exon 19 and insertions in exon 20 were analyzed by capillary electrophoresis, as described.16
Sequenom MassArray
Initial PCR reactions were set up with an EpMotion 5075 liquid handler (Eppendorf, Hamburg, Germany) and used 10-ng DNA per multiplex in a total volume of 5 μl, with 100 nmol/L of primers, 2 mmol/L of magnesium chloride, 500 μmol/L of deoxynucleotide, and 0.1 units of Taq polymerase. Amplification included one cycle at 94°C for 4 minutes, followed by 45 cycles at 94°C for 20 seconds, 56°C for 30 seconds, and 72°C for 1 minute and one final cycle at 72°C for 3 minutes. Unincorporated nucleotides were inactivated by addition of 0.3 units of shrimp alkaline phosphatase and incubation at 37°C for 40 minutes, followed by heat inactivation of shrimp alkaline phosphatase at 85°C for 5 minutes. Single-base primer extension reactions were performed with 0.625 to 1.25 μmol/L of extension primer and 1.35 units of TypePLEX thermosequenase DNA polymerase (Sequenom). Extension cycling included one cycle at 94°C for 30 seconds, and 40 cycles at 94°C for 5 seconds, five cycles at 52°C for 5 seconds and 80°C for 5 seconds, followed by one cycle at 72°C for 3 minutes. Extension products were purified by incubation for 30 minutes with an ion exchange resin (SpectroCLEAN; Sequenom), and approximately 10 nl of purified product was spotted onto SpectroChip II matrices (Sequenom). A Bruker matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MassARRAY Compact; Sequenom) was used to resolve extension products. MassARRAY Typer Analyzer software (Sequenom) was used for automated data analysis, accompanied by visual inspection of extension products.
Statistical Analyses
Analyses were performed using Fisher’s exact test or Mehta’s modification to Fisher’s exact test to determine relationships among parameters, whether for all patients or by adenocarcinoma or not, smoking or not, and center and country effects. All P values are two tailed and reported without adjustment for multiple comparisons.
RESULTS
Demographic Characteristics
We collected samples from 642 patients with NSCLC from seven institutions in the United States and Latin America. EGFR mutation analysis was successfully performed in 480 (75%) of 642. Among patients who had a successful mutation analysis, 90 (19%) were Latinos, 318 (66%) were non-Latino whites, 35 (7%) were non-Latino Asians, 30 (6%) were non-Latino blacks, and seven (2%) were of other races or ethnicities. Women accounted for 53% (255 of 480) of patients and men for 46% (220 of 480). Most patients were either age 40 to 64 years (259 [54%] of 480) or age 65 years or older (203 [42%] of 480; Table 1). Adenocarcinoma was the most frequent histology (370 [77%] of 480), and never-smokers accounted for 34% (165 of 480) of patients. The majority of patients were from the United States (420 [88%] of 480), whereas patients from Peru, Bolivia, Venezuela, and Mexico accounted for 6%, 3%, 2%, and 2%, respectively. Most of the patients who enrolled were current or former smokers (306 [64%] of 480) and had advanced disease (stage IIIb to IV; 447 [93%] of 480).
Table 1.
Patient Demographic and Clinical Characteristics
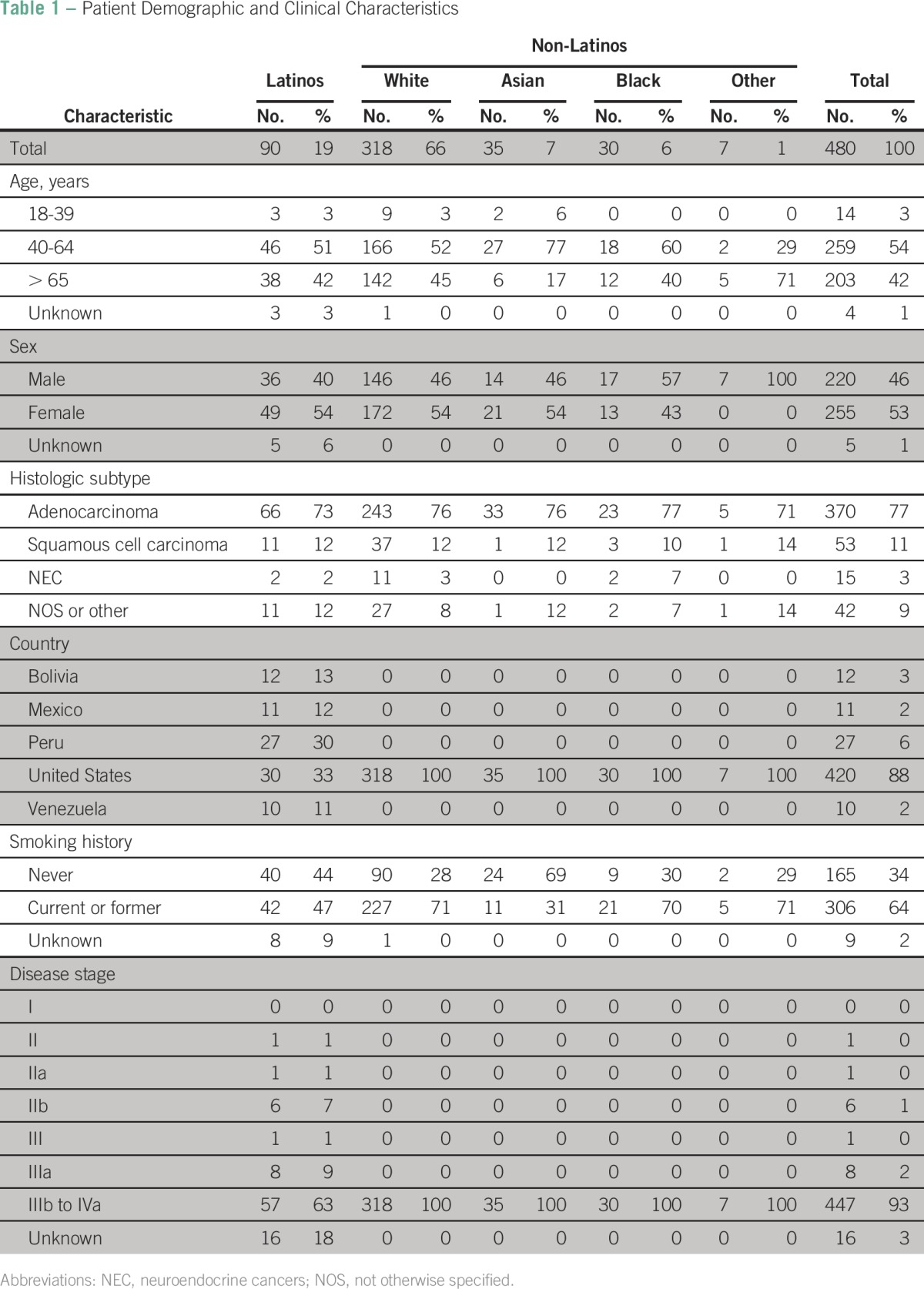
Among Latinos who had a successful EGFR mutation analysis, those from the United States (30 [33%] of 90) and Peru (27 [30%] of 90) accounted for the majority. In this population, 42% (38 of 90) were age older than 65 years, 54% (49 of 90) were women, 73% (66 of 90) had adenocarcinoma histology, 47% (42 of 90) were current or former smokers, and 63% (57 of 90) had advanced disease.
EGFR Mutations in Latinos
EGFR mutations were detected in 22% (105 of 480) of patients overall (Table 2). Among Latinos, EGFR mutations were found in 23.3% (21 of 90), and the frequency varied based on the country of origin. The highest frequency was observed in Latinos from Peru (37%) followed by the United States (23%), Mexico (18%), Venezuela (10%), and Bolivia (8%). Among non-Latinos, the overall frequency of EGFR mutations was 21.5% (84 of 390), and the frequency varied based on race. The highest frequency was observed in Asians (57.1%) followed by whites (18.6%) and blacks (10%; 95% CI, 2.11 to 26.53).
Table 2.
EGFR Mutation Frequency
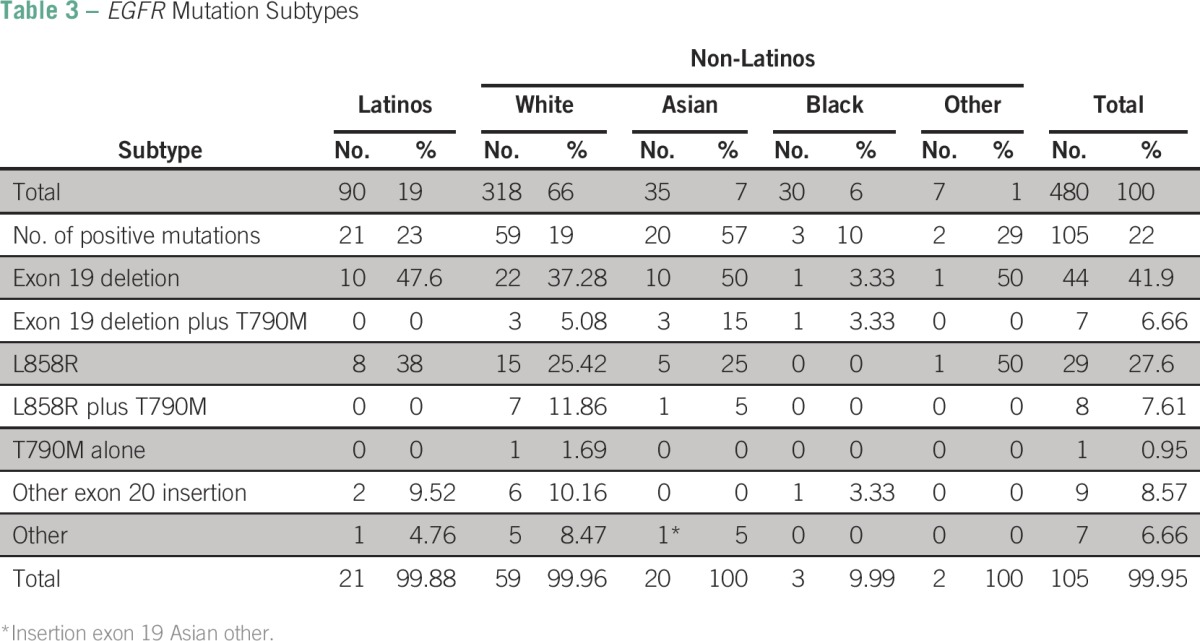
EGFR exon 19 deletions were the most frequent type of mutation among Latinos (10 [47.6%] of 21; Table 3) followed by EGFR L858R and exon 20 insertions, which were found in 38% (eight of 21) and 9.5% (two of 21) of patients, respectively. EGFR exon 19 deletions were also the most frequent subtype of mutations among whites, Asians, and blacks, with frequencies ranging from 10% (three of 30) in blacks to 57% (20 of 35) in Asians. The frequency of EGFR L858R mutations was 25% each in Asians (five of 20) and whites (15 of 59); additionally, one (5%) of 20 Asians and seven (11.8%) of 59 whites had concurrent EGFR L858R and T790M mutations.
Table 3.
EGFR Mutation Subtypes
EGFR Mutation Frequency Among Racial or Ethnic Subgroups
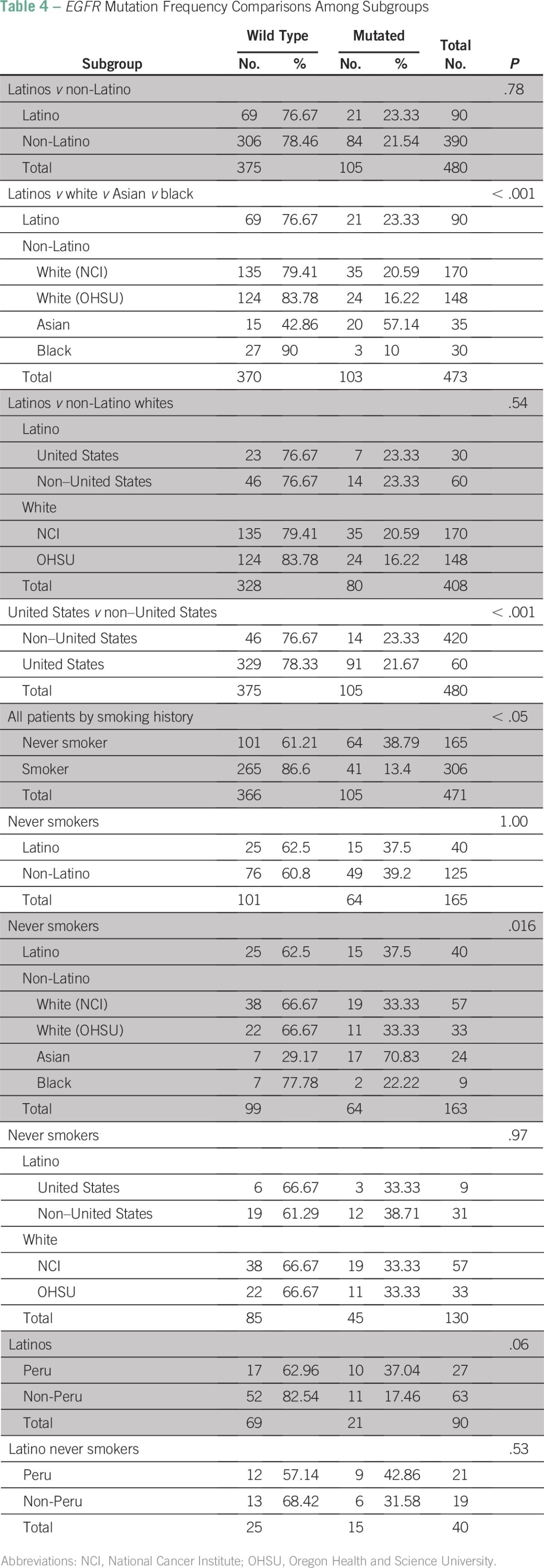
There was a significant difference in the frequency of EGFR mutations among the different racial and ethnic subgroups analyzed in the study (P < .05), which was primarily driven by the high frequency of EGFR mutations seen in non-Latino Asians and the low frequency observed in non-Latino blacks. However, there was no significant difference in EGFR mutation frequency overall between Latinos (23%) and non-Latinos (22%; P = .78; Table 4). In addition, there was no significant difference in the frequency of EGFR mutations between Latinos and non-Latino whites (overall, 21 of 90 v 59 of 318; P = .37 v .54 accounting for location) and between patients enrolled in the United States and those enrolled in Latin America (non-US patients; P = .74). Although the EGFR mutation frequency seemed to be higher in patients from Peru compared with patients of other races or ethnicities, this difference only exhibited a trend toward being statistically significant (P = .058), and the association was further diminished greatly when smoking and adenocarcinoma histology subgroups were analyzed independently (data not shown).
Table 4.
EGFR Mutation Frequency Comparisons Among Subgroups
DISCUSSION
In this study, which is the largest to our knowledge to prospectively evaluate the frequency of EGFR mutations in CLIA-certified laboratories in Latino patients with NSCLC from the United States and Latin America, there was no difference in the frequency of EGFR mutations between Latinos and non-Latinos. Although we observed a significant difference in the frequency of EGFR mutations among the different racial and ethnic subgroups studied, this difference was not significant when non-Latino Asians and blacks were removed from the analysis. This is consistent with previous reports that have observed a significantly higher frequency of EGFR mutations in Asians. Furthermore, the samples analyzed were part of a multiethnic and international cohort of patients with NSCLC, which allowed us to make comparisons among racial and ethnic groups rather than relying on historical controls. Finally, an additional strength of this study was that molecular testing was performed in two CLIA-certified central laboratories, avoiding quality control issues and potential variability in EGFR mutation frequency resulting from differences in testing methods used in different countries.
However, this study has limitations that underscore important aspects of cancer molecular characterization studies in Latinos and other under-represented minority populations in the United States. First, enrollment of Latino patients in the United States was low (30 [7%] of 420), despite the fact that one of the protocols in this study (NCT01255150) was specifically geared toward enrollment of such patients. Furthermore, despite the establishment of collaborations with several large cancer centers in Latin America to address the challenges described in this article, the number of Latinos overall was low. This problem highlights the significant difficulties of research collaborations with developing countries in which resource constraints, logistic, and legal challenges may significantly affect enrollment. This is in line with previous experience in other molecular characterization studies and clinical trials. Furthermore, it partially explains the paucity of tumor-specific molecular characterization data in Latinos and other minority populations in the United States. In this particular case, although the first reports of the frequency of EGFR mutations in NSCLC in the United States date back to 2004,17 the initial report for Latinos in the United States came almost a decade later,11 and the total number of reports and patients tested by race or ethnicity differs significantly between majority and minority populations. Of utmost importance is that in the event the cancer molecular profile of a particular minority population would differ significantly from the majority, such a disparity in molecular data availability could lead to the worsening of the already existing outcome disparities among patients of different races or ethnicities in the United States.
Another potential limitation of our study is that we did not account for the significant racial and genetic differences that exist within the Latino population. This population is significantly heterogeneous and in our study was defined based on either a geographic definition (ie, Latin America) or self-identification18 and included individuals of different races (eg, Latino whites, Latino American Indians, and Latino blacks). In this study, we did not collect information on race in the Latino population, nor did we perform genetic ancestry analyses or germline ancestry informative markers that could characterize the genetic origin within admixed populations.19 It is possible that we may have primarily sampled a subset of Latino patients with NSCLC, such as the Latino white population. Because this population is similar, in terms of genetic ancestry, to the non-Latino white population in the United States, this may have obscured a potential difference in EGFR mutation frequency between the two groups. However, Latinos with American Indian ancestry are of special interest, given that they represent the majority population in Mexico, Central America, and parts of South America (eg, Peru), and Latinos from such geographic areas comprise the largest subgroup of Latinos in the United States. Thus, we may not have captured a significant portion of the population of interest (ie, Latino American Indians), resulting in the negative results presented here. Conversely, the observation of a high EGFR mutation frequency in patients from Peru, which is known to have a large percentage of Latinos with high American Indian ancestry, could be an indication that there may in fact be a higher EGFR mutation frequency in a subset of Latinos defined by high American Indian ancestry. Alternatively, this could be related to sampling of a Peruvian population of Chinese and Japanese descent, which is also among the largest in Latin America.20 This underscores the need to account for race and genetic ancestry analysis in future studies.
Finally, it is important to note that the EGFR mutation frequency in the non-Latino white population of our cohort was considerably higher than what has been described in historical controls. This is likely the result of an unintended patient selection bias related to the fact that the populations of patients seeking care at large research institutions in the United States (eg, NCI and OHSU) tend to have clinical characteristics that have been associated with a higher frequency of EGFR mutations (eg, never smokers and adenocarcinoma histology).21 This is of utmost importance, because our conclusions are based on a comparator arm for which EGFR mutation frequency may have been overestimated as a result of selection bias. Thus, this comparison may not be appropriate, and our conclusions may not be accurate. This may explain the contrasting conclusions reached by this and other studies, particularly that conducted by Arrieta et al,10 in which the EGFR mutation frequency of a large (N = 5,738) retrospective cohort of NSCLC samples from different countries in Latin America was analyzed. In that study, a frequency of 26% was found, which was compared with a historical control of 15% in non-Latino patients from the United States; thus, the authors concluded that the EGFR mutation frequency in Latinos was higher. In contrast, Zhang et al11 evaluated EGFR mutations in lung adenocarcinomas from US Latino patients and found an EGFR mutation frequency of 15% (six of 40), which was similar to that observed in non-Latino white patients (eight [18.6%] of 43).
Our study provides what is to our knowledge the first prospective assessment of EGFR mutation frequency in CLIA-certified laboratories among Latinos from the United States and Latin America and allows comparison with frequencies among US non-Latino patients of different races. Although we did not observe a difference in the frequency of EGFR mutations between Latinos and non-Latinos, our results should be interpreted with caution, given the significant limitations of the study. Furthermore, our observations are inconclusive, and more research is needed to better understand the molecular characteristics of NSCLC in Latinos, with a particular focus on the role of race and genetic admixture and its molecular implications and the determination of its impact on the treatment and outcome of all patients with NSCLC regardless of race or ethnicity.
Footnotes
Supported by the National Cancer Institute (NCI) Intramural Research Program, the Knight Cancer Institute at Oregon Health and Science University, and the NCI Cancer Therapy Evaluation Program.
Presented in part at the 51st Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, May 29-June 2, 2015.
Authors’ disclosures of potential conflicts of interest and contributions are found at the end of this article.
AUTHOR CONTRIBUTIONS
Conception and design: Ariel Lopez-Chavez, Seth M. Steinberg, Giuseppe Giaccone
Collection and assembly of data: Ariel Lopez-Chavez, Anish Thomas, Moses O. Evbuomwan, Liqiang Xi, Guinevere Chun, Tatiana Vidaurre, Oscar Arrieta, George Oblitas III, Ana Belen Oton, Arun Rajan, Mark Raffeld, Lorena Arze-Aimaretti, Giuseppe Giaccone
Data analysis and interpretation: Ariel Lopez-Chavez, Anish Thomas, Liqiang Xi, Alejandro R. Calvo, Seth M. Steinberg, Lorena Arze-Aimaretti, Giuseppe Giaccone
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Ariel Lopez-Chavez
Employment: Genentech
Stock or Other Ownership: Genentech
Anish Thomas
No relationship to disclose
Moses O. Evbuomwan
No relationship to disclose
Liqiang Xi
No relationship to disclose
Guinevere Chun
No relationship to disclose
Tatiana Vidaurre
No relationship to disclose
Oscar Arrieta
No relationship to disclose
George Oblitas III
No relationship to disclose
Ana Belen Oton
Employment: Eli Lilly
Honoraria: Pfizer
Consulting or Advisory Role: Pfizer
Travel, Accommodations, Expenses: Pfizer, Eli Lilly
Alejandro R. Calvo
Speakers’ Bureau: Genentech, Boehringer Ingelheim
Arun Rajan
No relationship to disclose
Mark Raffeld
Stock or Other Ownership: Pfizer, GlaxoSmithKline
Seth M. Steinberg
No relationship to disclose
Lorena Arze-Aimaretti
No relationship to disclose
Giuseppe Giaccone
Consulting or Advisory Role: Astex Pharmaceuticals, Boehringer Ingelheim, Clovis Oncology, Celgene
Research Funding: Karyopharm Therapeutics, AstraZeneca
REFERENCES
- 1.Clegg LX, Li FP, Hankey BF, et al. Cancer survival among US whites and minorities: A SEER (Surveillance, Epidemiology, and End Results) program population-based study. Arch Intern Med. 2002;162:1985–1993. doi: 10.1001/archinte.162.17.1985. [DOI] [PubMed] [Google Scholar]
- 2.Colby SL, Ortman JM. Projections of the Size and Composition of the U.S. Population: 2014 to 2060—Population Estimates and Projections. https://www.census.gov/content/dam/Census/library/publications/2015/demo/p25-1143.pdf.
- 3.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 4.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 6.Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213–222. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 7.Jänne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non–small-cell lung cancer: Implications for treatment and tumor biology. J Clin Oncol. 2005;23:3227–3234. doi: 10.1200/JCO.2005.09.985. [DOI] [PubMed] [Google Scholar]
- 8.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrieta O, Cardona AF, Martín C, et al. Updated frequency of EGFR and KRAS mutations in nonsmall-cell lung cancer in Latin America: The Latin-American Consortium for the Investigation of Lung Cancer (CLICaP) J Thorac Oncol. 2015;10:838–843. doi: 10.1097/JTO.0000000000000481. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, McQuitty EB, Olsen R, et al. EGFR mutations in US Hispanic versus non-Hispanic white patients with lung adenocarcinoma. Arch Pathol Lab Med. 2014;138:543–545. doi: 10.5858/arpa.2013-0311-OA. [DOI] [PubMed] [Google Scholar]
- 12.American Cancer Society . Cancer Facts and Figures for Hispanics/Latinos 2012-2014. http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-034778.pdf. [Google Scholar]
- 13.National Cancer Institute SEER Stat Fact Sheets: Lung and Bronchus Cancer. http://seer.cancer.gov/statfacts/html/lungb.html.
- 14.Saeed AM, Toonkel R, Glassberg MK, et al. The influence of Hispanic ethnicity on nonsmall cell lung cancer histology and patient survival: An analysis of the Survival, Epidemiology, and End Results database. Cancer. 2012;118:4495–4501. doi: 10.1002/cncr.26686. [DOI] [PubMed] [Google Scholar]
- 15.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non–small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhuri SR, Xi L, Pham TH, et al. EGFR and KRAS mutation analysis in cytologic samples of lung adenocarcinoma enabled by laser capture microdissection. Mod Pathol. 2012;25:548–555. doi: 10.1038/modpathol.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 18.Yaeger R, Avila-Bront A, Abdul K, et al. Comparing genetic ancestry and self-described race in African Americans born in the United States and in Africa. Cancer Epidemiol Biomarkers Prev. 2008;17:1329–1338. doi: 10.1158/1055-9965.EPI-07-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Araujo LH, Timmers C, Bell EH, et al. Genomic characterization of non–small-cell lung cancer in African Americans by targeted massively parallel sequencing. J Clin Oncol. 2015;33:1966–1973. doi: 10.1200/JCO.2014.59.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Central Intelligence Agency The World Factbook Peru. https://www.cia.gov/library/publications/the-world-factbook/geos/pe.html.
- 21.Brewin CR, Bradley C. Patient preferences and randomised clinical trials. BMJ. 1989;299:313–315. doi: 10.1136/bmj.299.6694.313. [DOI] [PMC free article] [PubMed] [Google Scholar]


