Abstract
The promoter selectivity of Escherichia coli RNA polymerase (RNAP) is determined by the sigma subunit. The model prokaryote Escherichia coli K-12 contains seven species of the sigma subunit, each recognizing a specific set of promoters. For identification of the “constitutive promoters” that are recognized by each RNAP holoenzyme alone in the absence of other supporting factors, we have performed the genomic SELEX screening in vitro for their binding sites along the E. coli K-12 W3110 genome using each of the reconstituted RNAP holoenzymes and a collection of genome DNA segments of E. coli K-12. The whole set of constitutive promoters for each RNAP holoenzyme was then estimated based on the location of RNAP-binding sites. The first successful screening of the constitutive promoters was achieved for RpoD (σ70), the principal sigma for transcription of growth-related genes. As an extension, we performed in this study the screening of constitutive promoters for four minor sigma subunits, stationary-phase specific RpoS (σ38), heat-shock specific RpoH (σ32), flagellar-chemotaxis specific RpoF (σ28) and extra-cytoplasmic stress-response RpoE (σ24). The total number of constitutive promoters were: 129~179 for RpoS; 101~142 for RpoH; 34~41 for RpoF; and 77~106 for RpoE. The list of constitutive promoters were compared with that of known promoters identified in vivo under various conditions and using varieties of E. coli strains, altogether allowing the estimation of “inducible promoters” in the presence of additional supporting factors.
Introduction
The genome of Escherichia coli K-12, the most well-characterized model prokaryote, contains a total of more than 4,500 genes, which are transcribed by a single species of the RNA polymerase (RNAP). The intracellular concentration of RNAP is, however, approximately 2,000 molecules per genome, which is less than the total number of genes or operons [1–3]. The pattern of genome expression is therefore determined by the selective distribution of a limited number of RNAP within the genome [4,5]. For adaptation to stressful environments, the pattern of genome transcription is, however, altered by modulating the promoter selectivity of RNAP through two-step interaction with two groups of the regulatory factor, i.e., 7 species of the sigma factor with promoter recognition activity at the first step [5,6] and then approximately 300 species of the transcription factor (TF) including both protein and nucleotide factors at the second step [4,5,7,8]. For understanding the genome regulation at molecular level, therefore, three kinds of the basic knowledge are absolutely needed for both all the sigma and TF factors [8,9]: (1) the whole set of regulatory target promoters, genes or operons under the control of each regulatory factor; (2) the binding affinity of the test regulatory protein to target DNA; and (3) the intracellular concentrations of the functional forms of each regulatory protein [note that the activity of TF is often controlled by effector ligands or protein modification such as phosphorylation]. Once we get these three lines of knowledge, we will be able to predict the pattern of genome transcription.
After the complete genome sequencing of E. coli K-12, its transcription pattern or transcriptome in vivo has been analyzed for various E. coli wild-type and mutant strains growing under various stress conditions, including niches within host animals, using modern technologies such as the microarray system [10,11]. The localization of RNAP and TFs on the genome was also analyzed by using ChIP-chip system [12–14]. More recently microarray was replaced by direct sequencing of RNAs [15–17] or mapping of transcription start sites [18,19]. These data are assembled in the databases such as RegulonDB [20,21] and EcoCyc [22,23]. The huge accumulation of background knowledge is absolutely needed for understanding the regulation mechanism of genome transcription as a whole in a single organism, and thus at this stage, E. coli is reassessed as the model organism. The binding sites of RNAP and TF identified in vivo using these modern techniques, however, do not represent the whole set of their binding sites because: i) their binding to regulatory sites is often interfered by other DNA-binding proteins, thereby masking their binding target sequences by antagonistic inhibitory proteins [8,9,24]; and ii) in the case of activator-dependent transcription, their binding to targets depends on the simultaneous presence of supporting factors [8,9,25]. Under the in vivo situations, therefore, it is in principle impossible to obtain the whole set of binding sites for both RNAP and TFs. In addition, the transcription-related data listed in the databases include different levels of accuracy. For instance, a number of TF-binding sites are estimated in silico relying on the consensus sequences that often include the inaccurate prediction. Another serious problem is originated from the use of various E. coli strains with different genetic background and of different culture conditions used in each experiment (for details see Discussion).
In order to avoid the problems associated with these in vivo experiments, we then decided to employ the in vitro approaches. For identification of the binding sites of RNAP and TFs, we developed the Genomic SELEX system [26] and successfully employed for search of regulatory targets for a number of TFs [8,9]. We also employed the Genomic SELEX for mapping of promoters. As described in the previous report [27], we identified a total of 2,071 sites on the E. coli K-12 genome of binding of RNAP holoenzyme containing RpoD (σ70), the major sigma for transcription of most of the growth-related genes, and mapped the location of “constitutive promoters” that are recognized by RpoD holoenzyme alone in the absence of other DNA-binding proteins [Note that the “constitutive promoter” is defined as the promoter that is recognized by RNAP alone in the absence of supporting factors while the promoters that are detected only in vivo are defined as the “inducible promoters”, supposedly under the support of accessory regulatory factors].
Besides this major house-keeping RpoD sigma (σ70), E. coli K-12 contains six alternative minor sigma factors, i.e., nitrogen-regulated gene-specific RpoN (σ54), stationary-phase nutrient-starvation specific RpoS (σ38), heat-shock response-specific RpoH (σ32), flagellar-chemotaxis specific RpoF (σ28), extra-cytoplasmic stress-response RpoE (σ24), and iron-starvation specific FecI (σ28) [4–6]. In this study, we identified the list of constitutive promoters for four minor sigma factors, RpoS, RpoH, RpoF and RpoE. Since RpoN sigma requires an additional TF such as NtrC for promoter binding, the set of promoters recognized by RpoN sigma differs depending on the species of collaborative TF. The list of promoters recognized by RpoN will be described elsewhere. On the other hand, FecI sigma is rather a unique sigma that recognizes only a specific target of the gene for fecA encoding transport of ferric citrate [28]. Thus, these two sigma factors, RpoN and FecI, are not included in this report. The list of constitutive promoters herein described provides the fundamental catalogs for the promoters recognized by the four minor sigma factors alone. The data described in this report will be deposited into TEC (Transcription Profile of Escherchia coli) database (https://shigen.nig.ac.jp/ecoli/tec/) [9]. The data of each minor sigma will be shown by ordering the sigma name (RpoS, RpoH, RpoF or RpoE) [https://shigen.nig.ac.jp/ecoli/tec/tfmap]. For details follow the instruction in TEC [9].
Results
Genomic SELEX screening for the constitutive promoters
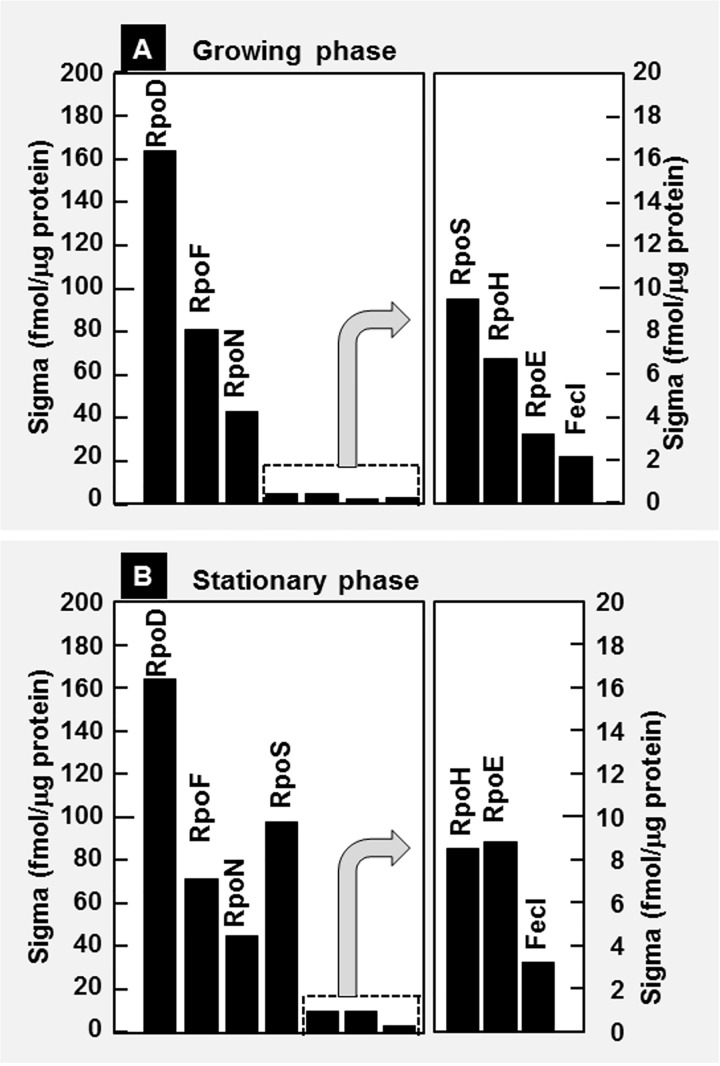
The constitutive promoters are transcribed in vitro by the RNA polymerase holoenzyme alone in the absence of supporting factors. In order to identify the whole set of constitutive promoters on the entire genome of E. coli K-12 W3110, we performed a mass-screening in vitro of the whole set of sequences that are recognized by the reconstituted holoenzymes, each containing only one specific minor sigma factor. The sigma-free core enzyme was prepared by passing the purified RNA polymerase three times through phosphocellulose column chromatography in the presence of 5% glycerol, the stabilizer of holoenzyme complexes in the storage buffer [29]. The level of remaining sigma subunits was less than 0.1%, if any, as detected by both protein staining and immuno-staining against each of all seven species of sigma factors (RpoD, RpoN, RpoS, RpoH, RpoF, RpoE and FecI). The stoichiometry between core enzyme subunits was also checked by immune-staining with antibodies against the core subunits, RpoA, RpoB, RpoC and RpoZ. The holoenzymes fully saturated with each sigma subunit were reconstituted by mixing this sigma-free core enzyme and 4-fold molar excess of purified sigma factors, RpoS, RpoH, RpoF and RpoE. Since these sigma subunits alone are unable to bind to promoter DNA, the presence of excess sigma does not interfere with the function of RNAP holoenzymes. For the identification of DNA sequences that are recognized by each holoenzyme, we employed the Genomic SELEX screening system [26], in which a library of E. coli genome DNA fragments of 200–300 bp in length was used instead of synthetic oligonucleotides with all possible sequences used in the original SELEX method [30–32].
The multi-copy plasmid library of 200–300 bp-long random DNA fragments was constructed from the E. coli K-12 W3110 genome [26]. The library used in this study contained 6.5-fold molar excess of the entire genome, and thus a single and the same sequence might be included in 6 different overlapping segments on average, thereby increasing the resolution of mapping of SELEX fragments. In each experiment of Genomic SELEX screening, the mixture of genome DNA fragments, which was regenerated by PCR from the genome DNA library, was mixed with 2-fold molar excess of the reconstituted each RNAP holoenzyme, and subjected to Genomic SELEX screening. DNA-holoenzyme complexes formed were recovered using the anti-RpoC antibody, which gave the highest level of RNAP recovery among all the anti-core subunit antibodies. RNA polymerase-associated DNA was isolated from the antibody precipitates, amplified by PCR, and subjected to next cycles of SELEX. After repeated SELEX screening, the final products of holoenzyme-bound DNA fragments were subjected to mapping on the genome using a DNA tilling microarray (Oxford Gene Technology, Oxford, UK) [14]. The binding intensity was measured as the ratio of holoenzyme-bound DNA labeled by Cy3 against original library DNA labeled by Cy5 on an array and plotted along E. coli genome about each holoenzyme. On the DNA tilling array used, the 60 b-long probes are aligned along the E. coli genome at 105 bp-intervals, and therefore approximately 300 bp-long SELEX fragments should bind to two or more consecutive probes. This criterion was employed to avoid the background noise of non-specific binding of holoenzyme-bound DNA fragments to the tilling array [note that peaks showing hybridization to only a single probe was judged as a false-positive noise].
The binding sites were classified into two groups, one ‘within spacers’ and another ‘inside genes’. The binding sites on ‘within spacers’ were further classified into 3 types; type-A spacer located between bidirectional transcription units, type-B spacer located upstream of one transcription unit but downstream of another transcription unit, and type-C spacer located downstream of both transcription units. Based on the transcription direction of flanking genes, the total number of the constitutive promoters was predicted to range between the minimum [number of type-A spacer plus number of type-B spacer] and the maximum [number of type-A spacer x 2 plus number of type-B spacer]. The intragenic binding site was referred to type-D site. The height of binding intensity identified by SELEX-chip system is generally in good agreement with the number of clones identified by SELEX-clos (cloning-sequencing) system, indicating that these two parameters correlate with the binding affinity of test TF to DNA [4,5,8,9].
The whole set of constitutive promoters for the stationary-phase sigma RpoS
In laboratory culture of E. coli, cell growth enters into the stationary phase mainly due to the limited availability of nutrients. Upon entry into the stationary phase, the pattern of genome expression is markedly altered by turning down the growth-related genes and instead up-regulation of the stress-response genes. In switching the transcription pattern, the stationary-phase specific minor sigma RrpoS is involved [33,34]. The rpoS gene is not essential for growth under non-stress conditions, but strains carrying mutations affecting rpoS activity are extremely sensitive to environmental stresses. As in the case of other sigma factors, RpoS interacts with RNAP core enzyme and modulates its promoter recognition specificity so as to recognize a specific but large set of genes.
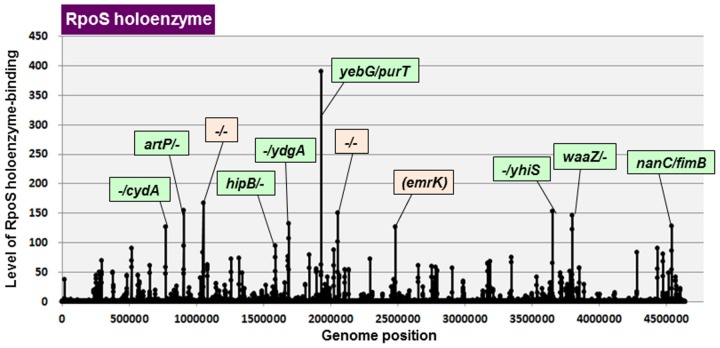
As noted above, the set of genes identified in vivo include a number of genes under the indirect control of RpoS. On the other hand, some target promoters of RpoS are masked in vivo due to competitive interference by other DNA-binding proteins. In order to identify the constitutive promoters directly recognized by RpoS in the absence of other DNA-binding proteins, the Genomic SELEX screening in vitro was performed using the reconstituted RNAP RpoS holoenzyme. The sequences with binding affinity to the RpoS holoenzyme formed a number of peaks along the entire E. coli genome (Fig 1). Location of peaks was aligned along the map of E. coli K-12 genome (Table 1). By setting the cut-off level of 3.0 fold-higher intensity over the background of original library DNA, a total of 218 peaks were identified, of which 125 (67%) are located within intergenic spacers and 73 (33%) are inside of open reading frames (Table 2). Since the majority of hitherto identified promoters are located within spacers and generally upstream of open reading frames, detailed search for the constitutive promoters was focused on these 125 spacer peaks. These spacers can be classified into three types: 50 peaks are located within type-A spacer between bidirectional transcription units; 79 peaks are located within type-B spacers located upstream of one transcription unit but downstream of another transcription unit; and 16 peaks are located within type-C located spacers downstream of both transcription units. Based on the transcription direction of flanking genes, the total number of RpoS constitutive promoters was predicted to range between minimum 129 (50 type-A plus 79 type-B) and maximum 179 (50x2 type-A plus 79 type-B) (Table 2). Type-A spacers should contain two promoters for bidirectional transcription, at least one of which should be RpoS-dependent promoter. The RpoS holoenzyme-binding sites identified in a total of 50 type-A spacers should represent promoters for one or both of bidirectional transcription.
Fig 1. SELEX-chip search for RNAP RpoS holoenzyme-binding sequences on the E. coli K-12 genome.
The y-axis represents the relative number of RpoS holoenzyme-bound DNA fragments whereas x-axis represents the position on the E. coli K-12 genome, in base pair. The adjacent gene on E. coli K-12 genome of peak position was indicated for high intensity peaks. The peaks located within spacer regions are shown in green color, while peaks located within open reading frames are shown in orange color. The list of RpoS holoenzyme-binding sites is described in Table 1.
Table 1. RpoS holoenzyme-binding sites on the E. coli K-12 genome.
| No | Type | Map | Gene Function | Left | D | RpoS | D | Right | Gene Function | Intensity |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D | 17732 | sokC | > | nhaA | > | nhaR | transcriptional activator | 5.3 | |
| 2 | A | 41946 | predicted transporter | caiT | < | > | fixA | electron transfer flavoprotein | 3.6 | |
| 3 | B | 63256 | RNAP-associated helicase | hepA | < | < | polB | 3.8 | ||
| 4 | D | 238050 | aspV | > | yafT | < | ykfM | 19.0 | ||
| 5 | B | 251970 | dinB | > | > | yafN | YafO-YafN toxin-antitoxin system | 45.1 | ||
| 6 | B | 289870 | ornithine carbamoyltransferase 2 | argF | < | < | insB | 25.0 | ||
| 7 | D | 296234 | CP4-6 prophage protein | yagN | < | intF | > | ptwF | Xaa tRNA | 70.4 |
| 8 | A | 296336 | CP4-6 prophage integrase | intF | < | > | ptwF | Xaa tRNA | 45.5 | |
| 9 | A | 328672 | transcriptional repressor | betI | < | > | betT | choline transporter | 3.8 | |
| 10 | B | 379186 | frmRAB operon regulator | frmR | < | < | yaiO | 50.8 | ||
| 11 | A | 383868 | conserved protein | yaiS | < | > | tauA | taurine transporter | 24.1 | |
| 12 | D | 451740 | cytochrome o ubiquinol oxidase | cyoA | < | ampG | < | yajG | 5.9 | |
| 13 | B | 455746 | tig | > | > | clpP | ATP-dependent serine protease | 7.4 | ||
| 14 | B | 467530 | cof | > | > | ybaO | DNA-binding transcriptional regulator | 11.5 | ||
| 15 | D | 479868 | modulator of gene expression with H-NS | hha | < | tomB | < | acrB | 45.1 | |
| 16 | B | 480454 | predicted protein | tomB | < | < | acrB | 22.6 | ||
| 17 | A | 515034 | membrane anchored protease | qmcA | < | > | ybbL | ABC superfamily transporter | 70.3 | |
| 18 | D | 532860 | allA | > | allR | > | gcl | glyoxylate carboligase | 3.5 | |
| 19 | D | 562966 | sfmH | > | sfmF | < | fimZ | 45.3 | ||
| 20 | D | 569366 | emrE | > | ybcK | > | ybcL | DLP12 prophage kinase inhibitor | 6.8 | |
| 21 | B | 570042 | ybcK | > | > | ybcL | DLP12 prophage kinase inhibitor | 19.7 | ||
| 22 | A | 576032 | IS5 transposase and trans-activator | insH | < | > | essD | DLP12 prophage lysis protein | 34.0 | |
| 23 | B | 582672 | nohB | > | > | *appY | DLP12 prophage transcriptional activator | 12.9 | ||
| 24 | D | 583166 | nohB | > | appY | < | ompT | 11.4 | ||
| 25 | D | 606570 | predicted inner membrane protein | ybdJ | < | ybdK | > | hokE | toxic polypeptide | 17.3 |
| 26 | A | 651466 | citrate lyase synthetase | citC | < | > | dpiB | CitBA TCS sensory histidine kinase | 8.3 | |
| 27 | D | 654932 | dpiA | > | dcuC | > | pagP | palmitoyl transferase for Lipid A | 60.7 | |
| 28 | D | 659134 | tatE | > | lipA | < | ybeF | 5.9 | ||
| 29 | B | 692668 | nucleoside triphosphate hydrolase | ybeZ | < | < | miaB | 19.9 | ||
| 30 | C | 720060 | ybfK | > | < | kdpE | 3.6 | |||
| 31 | B | 770146 | mngB | > | > | cydA | cytochrome d terminal oxidase, subunit I | 127.5 | ||
| 32 | D | 799356 | pgl | > | ybhD | > | ybhH | conserved protein | 4.8 | |
| 33 | A | 812472 | hypothetical protein | ybhU | < | > | uvrB | nucleotide excision repair nuclease | 12.9 | |
| 34 | B | 837732 | conserved protein | *ybiI | < | < | ybiX | 19.6 | ||
| 35 | A | 849568 | threonine and homoserine efflux system | rhtA | < | > | ompX | outer membrane protein | 5.4 | |
| 36 | A | 852256 | ncRNA | rybA | < | > | mntR | DNA-binding transcriptional regulator | 26.0 | |
| 37 | A | 877270 | SAM-dependent methyltransferase | yliG | < | > | bssR | conserved protein | 5.3 | |
| 38 | B | 891170 | nfsA | > | > | rimK | ribosomal protein S6 modification protein | 4.1 | ||
| 39 | B | 903170 | arginine transporter | *artP | < | < | ybjP | 154.2 | ||
| 40 | A | 931668 | thioredoxin reductase | trxB | < | > | lrp | DNA-binding transcriptional dual regulator | 5.9 | |
| 41 | D | 943972 | dmsB | > | dmsC | < | ycaC | 24.3 | ||
| 42 | B | 953940 | formate transporter | focA | < | < | ycaO | 3.4 | ||
| 43 | B | 959450 | aroA | > | > | ycaL | peptidase with chaperone function | 6.6 | ||
| 44 | B | 962934 | rpsA | > | > | *ihfB | integration host factor (IHF) | 3.6 | ||
| 45 | D | 1028042 | hemimethhylated DNA-binding protein | hspQ | < | yccW | > | yccX | predicted acylphosphatase | 40.5 |
| 46 | C | 1049830 | insB | > | < | cspH | 167.2 | |||
| 47 | B | 1084156 | efeB | > | > | phoH | conserved protein with NTPase domain | 62.7 | ||
| 48 | B | 1120372 | predicted protein | bssS | < | < | dinI | 6.6 | ||
| 49 | B | 1120772 | DNA damage-inducible protein I | dinI | < | < | pyrC | 3.7 | ||
| 50 | D | 1144848 | yceQ | > | rluC | < | yceF | 23.7 | ||
| 51 | D | 1147330 | rpmF | > | plsX | > | fabH | 3-oxoacyl-[acyl-carrier-protein] synthase III | 31.2 | |
| 52 | A | 1168238 | DNA-binding transcriptional regulator | ycfQ | < | > | bhsA | predicted protein | 17.6 | |
| 53 | D | 1187848 | conserved protein | ycfD | < | phoQ | < | phoP | 5.5 | |
| 54 | D | 1211030 | 5-methyl-C-specific restriction nuclease | mcrA | > | elbA | < | ycgX | 4.7 | |
| 55 | D | 1212068 | predicted protein | elbA | < | ycgX | < | ycgE | 27.8 | |
| 56 | A | 1214962 | predicted FAD-binding phosphodiesterase | ycgF | < | > | *ycgZ | predicted protein | 14.2 | |
| 57 | B | 1215830 | ariR | > | > | ymgC | predicted protein | 6.7 | ||
| 58 | C | 1218960 | ymgF | > | < | ymgD | 15.1 | |||
| 59 | A | 1250254 | dihydroxyacetone kinase | *dhaK | < | > | *dhaR | DNA-binding transcriptional regulator | 15.7 | |
| 60 | A | 1257834 | peptidyl-tRNA hydrolase | pth | < | > | ychH | predicted inner membrane protein | 72.5 | |
| 61 | D | 1318064 | indole-3-glycerol-P synthetase | trpC | < | trpD | < | trpE | 73.6 | |
| 62 | B | 1341438 | lipoprotein | *osmB | < | < | yciT | 3.4 | ||
| 63 | D | 1342872 | hypothetical protein | yciZ | < | gmr | < | rnb | 48.2 | |
| 64 | A | 1359040 | gamma-Glu-putrescine synthase | *puuA | < | > | *puuD | gamma-Glu-GABA hydrolase | 21.8 | |
| 65 | D | 1393856 | mppA | > | ynaI | > | insH | IS5 transposase and trans-activator | 4.0 | |
| 66 | B | 1432638 | DNA-binding transcriptional regulator | ynaE | < | < | uspF | 10.2 | ||
| 67 | A | 1438872 | pyruvate-flavodoxin oxidoreductase | ydbK | < | > | ydbJ | predicted protein | 5.1 | |
| 68 | D | 1447730 | feaB | > | tynA | < | maoC | 5.0 | ||
| 69 | C | 1463278 | paaY | > | < | insD | 6.2 | |||
| 70 | A | 1489640 | ncRNA | rydC | < | > | ydcA | predicted protein | 14.9 | |
| 71 | B | 1500460 | tehB | > | > | ydcL | predicted lipoprotein | 7.4 | ||
| 72 | A | 1515332 | hypothetical protein | yncL | < | > | ydcX | predicted inner membrane protein | 28.5 | |
| 73 | B | 1518146 | yncB | > | > | mcbR | DNA-binding transcriptional regulator | 4.4 | ||
| 74 | D | 1528272 | yncH | > | ydcD | > | ydcC | conserved protein | 9.2 | |
| 75 | B | 1543272 | predicted protein | yddJ | < | < | yddG | 3.5 | ||
| 76 | D | 1553260 | dehydrogenase/acetaldehyde reductase | adhP | < | maeA | < | sra | 4.9 | |
| 77 | D | 1566238 | predicted diguanylate cyclase | yddV | < | yddW | < | gadC | 14.5 | |
| 78 | B | 1570272 | glutamate decarboxylase B | *gadB | < | < | pqqL | 14.9 | ||
| 79 | B | 1580646 | conserved protein | ydeN | < | < | ydeO | 8.0 | ||
| 80 | B | 1590548 | DNA-binding transcriptional regulator | hipB | < | < | ydeU | 94.4 | ||
| 81 | B | 1596530 | predicted lipoprotein | ydeK | < | < | lsrK | 12.9 | ||
| 82 | D | 1613844 | yneJ | > | yneK | > | ydeA | predicted arabinose transporter | 4.3 | |
| 83 | B | 1618030 | marA | > | > | marB | predicted protein | 3.9 | ||
| 84 | B | 1627238 | ydfH | > | > | ydfZ | conserved protein | 5.5 | ||
| 85 | A | 1630740 | predicted mannonate dehydrogenase | ydfI | < | > | ydfK | Qin prophage transcriptional regulator | 10.8 | |
| 86 | A | 1655452 | predicted protein | ynfC | < | > | ynfD | predicted protein | 32.3 | |
| 87 | B | 1682244 | rstB | > | > | tus | inhibitor of replication at Ter | 77.2 | ||
| 88 | B | 1687868 | manA | > | > | ydgA | conserved protein | 132.4 | ||
| 89 | B | 1745130 | ydhR | > | > | ydhS | protein with FAD/NAD(P)-binding domain | 3.8 | ||
| 90 | A | 1753168 | predicted protein | ydhZ | < | > | *pykF | pyruvate kinase I | 4.2 | |
| 91 | D | 1769372 | conserved protein | ydiL | > | ydiM | > | ydiN | predicted transporter | 3.2 |
| 92 | C | 1793764 | integration host factor (IHF) | ihfA | < | pheT | < | pheS | 8.1 | |
| 93 | D | 1811050 | ydjN | > | ydjO | < | cedA | 29.2 | ||
| 94 | C | 1841754 | gdhA | > | < | ynjI | 79.5 | |||
| 95 | B | 1894766 | nudL | > | > | sdaA | L-serine deaminase I | 55.4 | ||
| 96 | B | 1905768 | predicted protein | yobF | < | < | yebO | 11.2 | ||
| 97 | D | 1921260 | ncRNA | ryeA | > | ryeB | < | yebY | 11.5 | |
| 98 | B | 1927030 | protease II | ptrB | < | < | yebE | 6.1 | ||
| 99 | A | 1928846 | conserved protein regulated by LexA | yebG | < | > | purT | P-ribosylglycinamide formyltransferase 2 | 63.2 | |
| 100 | D | 1928972 | conserved protein regulated by LexA | yebG | < | purT | < | eda | 390.6 | |
| 101 | A | 1944202 | RuvABC resolvasome | ruvA | < | > | yebB | predicted protein | 11.3 | |
| 102 | B | 1956162 | TMAO reductase III (TorYZ) | torY | < | < | cutC | 17.4 | ||
| 103 | C | 1966932 | CheAB TCS chemotaxis regulator | cheB | < | cheR | < | tap | 8.7 | |
| 104 | B | 1994970 | DNA-binding transcriptional activator | sdiA | < | < | yecC | 21.7 | ||
| 105 | A | 2023030 | predicted protein | *dsrB | < | > | yodD | predicted protein | 88.4 | |
| 106 | B | 2031954 | rseX | > | > | *hchA | Hsp31 molecular chaperone | 7.7 | ||
| 107 | D | 2049968 | asnT | > | yeeJ | > | shiA | shikimate transporter | 150.8 | |
| 108 | D | 2057732 | Asn tRNA | asnW | < | yeeO | > | asnU | Asn tRNA | 3.2 |
| 109 | B | 2061434 | L,D-transpeptidase linking Lpp to murein | erfK | < | < | cobT | 44.9 | ||
| 110 | D | 2103732 | predicted acyl transferase | wbbJ | < | wbbI | < | rfc | 4.3 | |
| 111 | D | 2104956 | conserved protein | wbbI | < | rfc | < | glf | 14.6 | |
| 112 | D | 2106934 | UDP-galactopyranose mutase | glf | < | rfbX | < | rfbC | 54.2 | |
| 113 | B | 2134130 | protein-tyrosine phosphatase | wzb | < | < | wza | 53.9 | ||
| 114 | B | 2226932 | NAD(P)-binding oxidoreductase | yohF | < | < | dusC | 3.3 | ||
| 115 | A | 2247636 | DNA-binding transcriptional regulator | yeiE | < | > | yeiH | conserved inner membrane protein | 12.7 | |
| 116 | D | 2255672 | predicted nucleoside transporter | psuT | < | psuG | < | psuK | pseudouridine kinase | 7.7 |
| 117 | B | 2284170 | yejM | > | > | proL | Pro tRNA | 9.5 | ||
| 118 | A | 2311066 | outer membrane porin protein C | ompC | < | > | micF | ncRNA | 16.1 | |
| 119 | B | 2311354 | micF | > | > | rcsD | RcsBC TCS phosphotransfer protein | 13.8 | ||
| 120 | D | 2389234 | yfbP | > | nuoN | < | nuoM | 13.0 | ||
| 121 | B | 2454170 | predicted fimbrial-like adhesin protein | yfcV | < | < | sixA | 25.4 | ||
| 122 | D | 2467360 | yfdH | > | yfdI | < | yfdK | 38.3 | ||
| 123 | C | 2468764 | yfdI | > | < | yfdK | 26.5 | |||
| 124 | D | 2480972 | predicted multidrug efflux system | emrY | < | emrK | > | *evgA | EvgAS TCS response regulator | 126.6 |
| 125 | A | 2510860 | manganese/divalent cation transporter | mntH | < | > | nupC | nucleoside (except guanosine) transporter | 13.5 | |
| 126 | D | 2532356 | ptsH | > | ptsI | > | *crr | glucose-specific PTS enzyme IIA | 9.2 | |
| 127 | B | 2574144 | carboxysome structural protein | eutS | < | < | maeB | 11.3 | ||
| 128 | D | 2587966 | narQ | > | acrD | < | ypfM | 18.4 | ||
| 129 | B | 2651536 | sseA | > | > | ryfA | ncRNA | 40.6 | ||
| 130 | B | 2689548 | ncRNA | glmY | < | < | purL | 24.9 | ||
| 131 | B | 2753630 | smpB | > | > | ssrA | tmRNA | 60.7 | ||
| 132 | B | 2763338 | CP4-57 prophage; predicted protein | yfjL | < | < | yfjM | 13.8 | ||
| 133 | B | 2765760 | yfjO | > | > | yfjP | CP4-57 prophage GTP-binding protein | 5.2 | ||
| 134 | D | 2771468 | yfjT | > | yfjW | > | yfjX | CP4-57 prophage antirestriction protein | 32.6 | |
| 135 | D | 2772262 | yfjT | > | yfjW | > | yfjX | CP4-57 prophage antirestriction protein | 26.4 | |
| 136 | B | 2773166 | yfjW | > | > | yfjX | CP4-57 prophage antirestriction protein | 41.0 | ||
| 137 | D | 2779640 | psaA | > | ypjA | < | ileY | 28.8 | ||
| 138 | B | 2783272 | adhesin-like autotransporter | ypjA | < | < | ileY | 58.9 | ||
| 139 | A | 2784466 | Ile tRNA | ileY | < | > | *csiD | predicted protein | 3.7 | |
| 140 | A | 2786358 | Ile tRNA | ileY | < | > | *csiD | predicted protein | 31.8 | |
| 141 | A | 2795168 | predicted membrane protein | yqaE | < | > | ygaV | DNA-binding transcriptional regulator | 53.2 | |
| 142 | B | 2882250 | predicted protein | ygcL | < | < | ygcB | 6.3 | ||
| 143 | A | 2903434 | conserved protein | ygcF | < | > | ygcG | predicted protein | 57.0 | |
| 144 | B | 2985568 | yqeG | > | > | yqeH | protein with bipartite regulator domain | 32.6 | ||
| 145 | C | 2987942 | yqeJ | > | < | yqeK | 9.1 | |||
| 146 | D | 2990738 | ygeG | > | ygeH | > | ygeI | predicted protein | 6.8 | |
| 147 | C | 2992070 | ygeI | > | < | insD | 6.4 | |||
| 148 | C | 2992950 | ygeI | > | < | insD | 6.9 | |||
| 149 | C | 2993358 | ygeI | > | < | insD | 13.2 | |||
| 150 | B | 3134436 | phosphate transporter | pitB | < | < | gsp | 12.6 | ||
| 151 | A | 3145934 | predicted protein | yghW | < | > | yghZ | aldo-keto reductase | 3.2 | |
| 152 | B | 3166762 | predicted cyanide hydratase | mqsR | < | < | ygiV | 65.1 | ||
| 153 | C | 3181642 | zupT | > | < | ribB | 10.0 | |||
| 154 | B | 3183246 | yqiC | > | > | ygiL | predicted fimbrial-like adhesin protein | 3.7 | ||
| 155 | D | 3189250 | yqiH | > | yqiI | < | glgS | 68.5 | ||
| 156 | D | 3210472 | rpsU | > | dnaG | > | rpoD | RNA polymerase sigma 70 | 21.3 | |
| 157 | D | 3259830 | L-PSP (mRNA) endoribonuclease | tdcF | < | tdcE | < | tdcD | 3.6 | |
| 158 | D | 3266232 | tdcR | > | yhaB | > | yhaC | predicted protein | 9.6 | |
| 159 | C | 3326238 | yhbY | > | < | greA | 3.6 | |||
| 160 | B | 3335948 | ABC-type organic solvent transporter | yrbC | < | < | yrbD | 14.0 | ||
| 161 | D | 3345766 | ptsN | > | yhbJ | > | npr | N-regulated PTS system (Npr) | 75.4 | |
| 162 | D | 3348770 | ncRNA | ryhA | > | arcB | < | yhcC | 5.9 | |
| 163 | D | 3395840 | ribonuclease G | rng | < | yhdE | < | mreD | 4.3 | |
| 164 | D | 3453834 | general secretory pathway component | gspA | < | gspC | > | gspD | general secretory pathway component | 9.3 |
| 165 | B | 3467930 | periplasmic endochitinase | chiA | < | < | tufA | 5.0 | ||
| 166 | B | 3497370 | cysG | > | > | yhfL | conserved secreted peptide | 13.0 | ||
| 167 | D | 3533142 | pck | > | envZ | < | ompR | 41.3 | ||
| 168 | D | 3542562 | yhgA | > | bioH | > | gntX | gluconate periplasmic binding protein | 15.9 | |
| 169 | B | 3576742 | DNA-binding transcriptional repressor | gntR | < | < | yhhW | 12.2 | ||
| 170 | B | 3582734 | yrhD | > | > | yrhB | predicted protein | 4.3 | ||
| 171 | A | 3584864 | gamma-glutamyltranspeptidase | ggt | < | > | yhhA | conserved protein | 12.9 | |
| 172 | D | 3604756 | yhhN | > | zntA | < | sirA | 8.8 | ||
| 173 | B | 3621930 | yhhH | > | > | yhhI | predicted transposase | 3.3 | ||
| 174 | A | 3632156 | predicted protein | yhiJ | < | > | *yhiM | conserved inner membrane protein | 32.2 | |
| 175 | A | 3632742 | predicted protein | yhiJ | < | > | *yhiM | conserved inner membrane protein | 13.3 | |
| 176 | A | 3637868 | universal stress protein B | *uspB | < | > | uspA | universal stress global response regulator | 22.7 | |
| 177 | D | 3647644 | arsR | > | arsB | > | arsC | arsenate reductase | 31.8 | |
| 178 | C | 3648872 | arsC | > | < | insH | 153.6 | |||
| 179 | B | 3656130 | hdeD | > | > | gadE | DNA-binding transcriptional activator | 9.1 | ||
| 180 | B | 3708672 | predicted metal dependent hydrolase | eptB | < | < | yhjX | 48.9 | ||
| 181 | B | 3717944 | yiaG | > | > | cspA | major cold shock protein | 6.1 | ||
| 182 | B | 3720058 | insK | > | > | sokA | ncRNA | 31.7 | ||
| 183 | A | 3749938 | predicted protein | yiaT | < | > | yiaU | DNA-binding transcriptional regulator | 22.4 | |
| 184 | B | 3794944 | rfaC | > | > | rfaL | O-antigen ligase | 3.8 | ||
| 185 | D | 3796942 | rfaL | > | waaU | < | rfaZ | 46.2 | ||
| 186 | D | 3798472 | lipopolysaccharide core synthesis protein | rfaZ | < | rfaY | < | rfaJ | 145.8 | |
| 187 | D | 3800958 | UDP-D-glucose:LPS glucosyltransferase | rfaJ | < | rfaI | < | rfaB | 15.8 | |
| 188 | D | 3802448 | UDP-galactose:LPS galactosyltransferase | rfaB | < | rfaS | < | rfaP | 21.5 | |
| 189 | B | 3834856 | Sec tRNA | selC | > | > | setC | predicted sugar efflux system | 22.6 | |
| 190 | A | 3851272 | ncRNA | istR | < | > | tisB | lexA-regulated toxic peptide | 57.3 | |
| 191 | B | 3886640 | tnaC | > | > | tnaA | tryptophanase/L-cysteine desulfhydrase | 29.5 | ||
| 192 | D | 4001054 | conserved inner membrane protein | yigF | < | yigG | < | rarD | 5.5 | |
| 193 | D | 4002164 | predicted inner membrane protein | yigG | < | rarD | < | yigI | 13.0 | |
| 194 | B | 4076572 | yiiD | > | > | yiiE | predicted transcriptional regulator | 4.5 | ||
| 195 | D | 4110740 | conserved protein | yiiQ | < | yiiR | > | yiiS | conserved protein | 10.8 |
| 196 | A | 4116232 | glycerol facilitator | glpF | < | > | yiiU | conserved protein | 15.6 | |
| 197 | B | 4120330 | HslUV protease | hslV | < | < | ftsN | 3.6 | ||
| 198 | D | 4220334 | aceK | > | arpA | < | iclR | 5.4 | ||
| 199 | B | 4281230 | conserved protein | yjcF | < | < | actP | 83.6 | ||
| 200 | A | 4380530 | fumarate reductase | frdA | < | > | poxA | predicted lysyl-tRNA synthetase | 4.7 | |
| 201 | A | 4417636 | L-ascorbate 6-phosphate lactonase | ulaG | < | > | ulaA | L-ascorbate-specific PTS enzyme IIC | 4.9 | |
| 202 | A | 4432132 | NAD(P)H:quinone oxidoreductase | ytfG | < | > | ytfH | predicted transcriptional regulator | 90.2 | |
| 203 | A | 4434562 | 2':3'-cyclic-nucleotide 2'-phosphodiesterase | cpdB | < | > | cysQ | PAPS 3'(2'),5'-bisphosphate nucleotidase | 4.3 | |
| 204 | B | 4472740 | yjgJ | > | > | yjgK | conserved protein | 15.2 | ||
| 205 | C | 4475268 | yjgL | > | < | argI | 80.9 | |||
| 206 | A | 4477770 | predicted acetyltransferase | yjgM | < | > | yjgN | conserved inner membrane protein | 4.1 | |
| 207 | A | 4492648 | L-idonate 5-dehydrogenase, NAD-binding | idnD | < | > | idnK | D-gluconate kinase, thermosensitive | 6.0 | |
| 208 | B | 4504448 | yjhC | > | > | ythA | expressed protein | 8.4 | ||
| 209 | C | 4505142 | ythA | > | < | insI | 5.3 | |||
| 210 | A | 4538166 | N-acetylnuraminic acid OM channel protein | nanC | < | > | fimB | Tyr recombinase/fimA inversion regulator | 5.5 | |
| 211 | A | 4538758 | N-acetylnuraminic acid OM channel protein | nanC | < | > | fimB | Tyr recombinase/fimA inversion regulator | 127.7 | |
| 212 | D | 4543668 | fimC | > | fimD | > | fimF | minor component of type 1 fimbriae | 5.0 | |
| 213 | C | 4570642 | yjiS | > | < | mcrC | 41.4 | |||
| 214 | D | 4575670 | yjiS | > | mcrC | < | mcrB | 12.1 | ||
| 215 | D | 4576468 | 5-methylcytosine-specific restriction enzyme | mcrC | < | mcrB | < | symE | 26.3 | |
| 216 | D | 4588370 | conserved protein | yjiX | < | *yjiY | > | tsr | methyl-accepting chemotaxis protein I | 10.5 |
| 217 | A | 4601330 | predicted inner membrane protein | yjjP | < | > | yjjQ | DNA-binding transcriptional regulator | 22.9 | |
| 218 | B | 4609336 | prfC | > | > | osmY | periplasmic protein | 3.7 |
Genomic SELEX was performed for search of the binding sites of RNAP RpoS holoenzyme. By setting the cut-off level of 3.0, a total of 218 binding sites were identified (see Fig 1 for SELEX pattern), which are aligned along the map of E. coli K12 genome. A total of 125 sites are located within intergenic spacers: 50 within type-A spacers (shown under orange background); and 79 within type-B spacers (shown under green background). The constitutive promoters of RpoS were predicted based on the adjacent genes [note that only the genes next to the RpoS holoenzyme-binding sites are shown] and the gene orientation (shown by arrows in the column of transcription direction). A total of 73 RpoS holoenzyme-binding sites are located inside open reading frames as indicated by the gene symbols shown in RpoS column.
*The genes listed in RegulonDB as the regulated targets of RpoS.
Table 2. Distribution of the binding sites of each RNAP holoenzyme.
| Sigma | Total no. holoenzyme binding sites | Within Spacers | Inside Genes | Constitutive promoters | ||||
|---|---|---|---|---|---|---|---|---|
| Type-A | Type-B | Type-C | Type-A spacer | Type-B spacer | Total | |||
| RpoD | 1320 | 177 | 317 | 49 | 777 (60%) | 177~354 | 317 | 494~671 |
| 543 (40%) | ||||||||
| RpoS | 218 | 50 | 79 | 16 | 73 (33%) | 50~100 | 79 | 129~179 |
| 125 (67%) | ||||||||
| RpoH | 133 | 41 | 60 | 6 | 26 (20%) | 41~82 | 60 | 101~142 |
| 107 (80%) | ||||||||
| RpoF | 105 | 7 | 27 | 3 | 68 (65%) | 7~14 | 27 | 34~41 |
| 37 (35%) | ||||||||
| RpoE | 126 | 29 | 48 | 7 | 42 (33%) | 29~58 | 48 | 77~106 |
| 84 (67%) | ||||||||
RNAP holoenzyme was reconstituted from the sigma-free core enzyme and 4-fold molar excess of each sigma subunit. The binding site of each holoenzyme on the genome of E. coli K-12 W3110 was determined in vitro using the improved Genomic SELEX screening system. Details of the experimental procedures are described previously [26]. The number of constitutive promoters were estimated based on the location of holoenzyme-binding sites. The number of constitutive promoters recognized by RpoD holooenzyme were described in the previous report [27].
Up to the present, two general approaches have been employed to define the RpoS regulon: the proteome analysis using two-dimensional gels of whole cell lysates [35]; and the transcriptome analysis using ChIP-chip or ChIP-Seq systems [37–39]. These studies altogether indicated that RpoS regulates, directly or indirectly, 10% (approximately 500 genes) of the E. coli genes, of which only about 140 genes were predicted to be under the direct control in vivo of RpoS [38]. The total number of RpoS promoters (or the transcription initiation sites) listed in the current RegulonDB database is as many as 164 [note that all these promoters were detected in vivo]. Of which 21 were identified by setting the cut-off level at 3.0 (Table 1, marked by asterisk), indicating that only these promoters represent the constitutive promoters and the majority of other known RpoS promoters represent the inducible promoters that are expressed only under the support of regulatory factors. Genomic SELEX analysis identified minimum 129 and maximum 179 RpoS constitutive promoters including 21 known RpoS-dependent promoters (Table 2). The highest peak (390-fold higher than the background of original library alone) was located at the 5’-proximal region of the purT gene (Fig 1), which encodes a bifunctional enzyme with both phosphoribosylglycinamide formyltransferase using formate (the third-step reaction of purine nucleotide synthesis) and acetate kinase for the synthesis of acetylphosphate (AcP). AcP might be utilized as the general phosphate donor for phosphorylation of most of the stress-response TCS (two-component system) response regulators under stressful conditions. A high-level peak (154-fold higher than the background of DNA library) was detected upstream of the artPIQM operon encoding L-arginine ABC transporter. This promoter was also identified in vivo to be RpoS dependent [36] (Table 1). High-level binding of the RpoS holoenzyme was also identified upstream of the cydAB operon encoding cytochrome oxidase for anaerobic respiration, and the hipBA operon encoding anti-toxin-toxin pair for control the persistence (Fig 1 and Table 1). RpoS-dependent constitutive promoter(s) also exists upstream of the nanCMS operon (N-acetylneuraminic acid transport and utilization) and/or the fimB gene (regulator for fimA encoding fimbrin, the major type-1 pili) (Fig 1 and Table 1). Noteworthy is that most of the RpoS-dependent promoters listed in the current databases might be those under the indirect control of RpoS [8,9,27]. Otherwise a set of RpoS-dependent promoters, designated as the inducible promoters, might be activated in the presence of additional supporting factors.
Using the newly constructed collection of E. coli promoters expressing two-fluorescent reporters, one attached to the test promoter and another to the reference promoter, we performed a systematic quantitative search in vivo for E. coli promoters that are activated in the stationary phase [39]. The activity of RpoS-dependent promoters was measured at various growth phases under various growth conditions. The results indicated that the constitutive promoters exhibited low but steady-state activity while the inducible promoters generally showed high activity during the transition from exponential growth to stationary phase.
The RpoS regulon is involved in not only cell survival in the stationary phase, but also in cross protection against various stresses, including nutrient starvation, osmotic stress, acid shock, cold shock, heat shock, and oxidative DNA damage [33,34]. Beyond entry into stationary phase, E. coli forms aggregates or biofilms that are morphologically and physiologically distinct from cells of planktonic growth. This requires coordinated production of an extracellular matrix of polysaccharide polymers and protein fibers that facilitate cell aggregation and adhesion to solid surface. The genes involved in biofilm formation and transformation into persister cells were included in the list of RpoS constitutive promoters [40,41].
The whole sets of constitutive promoters for heat-shock response sigma RpoH
When E. coli cells are exposed to higher temperature, a set of heat-shock proteins (HSPs) is markedly and transiently induced. Heat shock-induced proteins (HSPs) play major roles in controlling the structure and function of various proteins, including protein folding, assembly, transport, repair and degradation during normal growth as well as under stress conditions [42,43]. The heat-shock response is a cellular protective system for maintenance of protein homeostasis. The set of HSPs include the GroEL (HSP60) and DnaK (HSP70) chaperones and the Lon and the Clp proteases. RpoH is specifically required for expression of the genes encoding a set of HSPs as identified by proteome [44,45] and also by transcriptome analyses [46]. Genome-wide transcription profiling of the regulatory targets of RpoH was identified under the moderate induction of a plasmid-borne rpoH gene under defined, steady state growth conditions [47]. A total of 126 genes were influenced in the absence or in the over-expression of RpoH, which are organized in 85 operons. The set of genes identified in vivo by changing the level of RpoH include a large number of indirect targets, which are affected in response to the changes in the level of direct target [8,9,27]. The total number of RpoH promoters (or the transcription initiation sites) listed in the current RegulonDB database is as many as 322, but the majority of RpoH targets are predicted by the computational analysis using the consensus sequence that was predicted based on a few experimentally identified RpoH promoters.
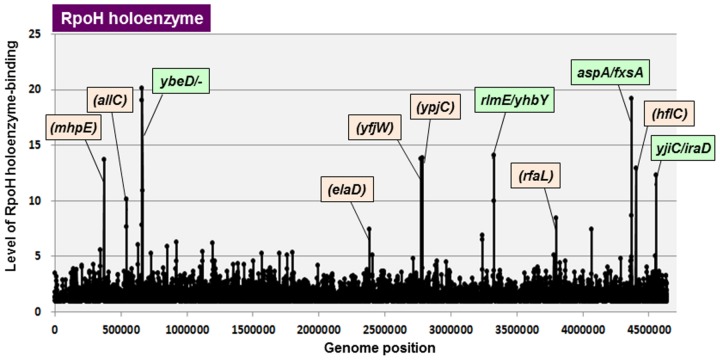
We isolated RpoH protein for the first time and confirmed its recognition in vitro of the known HSP gene promoters [48]. Since then no serious examination in vitro has been performed to identify the RpoH function and it regulatory targets. To get insights into the regulatory role of RpoH sigma, we then performed in this study the Genomic SELEX screening using the reconstituted RNAP RpoH holoenzyme. By setting the cut-off level of 3.0 fold higher than the background of original library DNA, a total of 133 RpoH holoenzyme-binding peaks were identified (Fig 2 and Table 3), of which 107 (80%) are located within intergenic spacers and 26 (20%) are inside of open reading frames (Table 2). Since the majority of hitherto identified promoters are located within spacers, detailed search for the constitutive promoters was focused on the total of 107 peaks within spacers. The spacers containing RpoH holoenzyme-binding sites were also classified into three types (Tables 2 and 3 for the whole list): 41 peaks are located within type-A spacer; 60 peaks are located within type-B spacers; and 6 peaks are located within type-C spacers. Based on the transcription direction of flanking genes, the total number of RpoH constitutive promoters was predicted to range between minimum 101 (41 type-A plus 60 type-B) and maximum 142 (41x2 type-A plus 60 type-B) (Table 2).
Fig 2. SELEX-chip search for RNAP RpoH holoenzyme-binding sequences on the E. coli K-12 genome.
The y-axis represents the relative number of RpoH holoenzyme-bound DNA fragments whereas x-axis represents the position on the E.coli K-12 genome, in base pair. The adjacent gene on E. coli K-12 genome of peak position was indicated for high intensity peaks. The peaks located within spacer regions are shown in green color, while peaks located within open reading frames are shown in orange color. The list of RpoH holoenzyme-binding sites is described in Table 3.
Table 3. RpoH holoenzyme-binding sites on the E. coli K-12 genome.
| No | Type | Map | Gene Function | Left | D | RpoH | D | Right | Gene Function | Intensity |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A | 12044 | predicted protein | yaaI | < | > | *dnaK | chaperone Hsp70 | 3.2 | |
| 2 | A | 142736 | carbonic anhydrase | *can | < | > | yadG | ABC superfamily transporter | 3.9 | |
| 3 | B | 155442 | outer membrane usher protein | htrE | < | < | ecpD | 3.0 | ||
| 4 | B | 164658 | hrpB | > | > | mrcB | glycosyl transferase and transpeptidase | 3.8 | ||
| 5 | B | 202068 | lpxD | > | > | fabZ | hydroxymyristol acyl carrier dehydratase | 4.0 | ||
| 6 | B | 229134 | aspU | > | > | dkgB | 2,5-diketo-D-gluconate reductase B | 3.0 | ||
| 7 | C | 262270 | thrW | > | < | ykfI | 3.6 | |||
| 8 | B | 292170 | CP4-6 prophage protein | yagK | < | < | yagL | 4.3 | ||
| 9 | B | 331456 | betT | > | > | yahA | DNA-binding transcriptional regulator | 3.5 | ||
| 10 | D | 343660 | yahK | > | yahL | > | yahM | predicted protein | 5.6 | |
| 11 | B | 379186 | frmRAB operon regulator | frmR | < | < | yaiO | 3.0 | ||
| 12 | B | 406536 | yaiA | > | > | aroM | conserved protein | 3.6 | ||
| 13 | B | 477848 | conserved inner membrane protein | ylaB | < | < | ylaC | 3.3 | ||
| 14 | D | 543470 | conserved protein | ylbA | < | allC | < | allD | 10.1 | |
| 15 | B | 557960 | sfmA | > | > | sfmC | pilin chaperone | 3.5 | ||
| 16 | D | 559456 | sfmC | > | sfmD | > | sfmH | fimbrial-like adhesin protein | 3.1 | |
| 17 | B | 581644 | nohB | > | > | appY | DLP12 transcriptional activator | 3.8 | ||
| 18 | B | 592452 | phage N4 receptor IM protein | nfrB | < | < | cusS | 3.2 | ||
| 19 | B | 629042 | ybdB | > | > | cstA | carbon starvation protein | 6.0 | ||
| 20 | D | 655854 | anaerobic C4-dicarboxylate transport | dcuC | < | pagP | > | cspE | DNA-binding transcriptional repressor | 3.3 |
| 21 | B | 661936 | conserved protein | *ybeD | < | < | dacA | 20.1 | ||
| 22 | D | 732870 | ybfA | > | *rhsC | > | ybfB | predicted inner membrane protein | 3.2 | |
| 23 | D | 747240 | predicted regulator | abrB | < | ybgO | < | ybgP | 3.5 | |
| 24 | B | 784068 | zinc efflux system | zitB | < | < | ybgS | 3.5 | ||
| 25 | A | 784656 | conserved protein | ybgS | < | > | aroG | D-arabino-heptulosonate-7P synthase | 3.0 | |
| 26 | B | 837732 | conserved protein | ybiI | < | < | ybiX | 3.1 | ||
| 27 | B | 913136 | anaerobic terminal reductases | hcp | < | < | ybjE | 3.8 | ||
| 28 | A | 918368 | conserved protein | ybjX | < | > | macA | macrolide transporter | 6.3 | |
| 29 | B | 959450 | aroA | > | > | ycaL | peptidase with chaperone function | 3.2 | ||
| 30 | B | 985134 | aspartate aminotransferase | aspC | < | < | ompF | 3.4 | ||
| 31 | A | 1050632 | CspA-family stress protein | *cspH | < | > | cspG | DNA-binding transcriptional regulator | 3.6 | |
| 32 | A | 1102554 | DNA-binding transcriptional activator | csgD | < | > | csgB | curlin nucleator protein | 4.0 | |
| 33 | B | 1120230 | predicted protein | *bssS | < | < | dinI | 5.4 | ||
| 34 | C | 1195868 | icd | > | < | ymfD | 6.2 | |||
| 35 | D | 1198640 | e14 prophage inner membrane protein | ymfE | < | lit | < | intE | 3.8 | |
| 36 | D | 1200062 | e14 prophage integrase | intE | < | xisE | > | ymfI | e14 prophage; predicted protein | 3.2 |
| 37 | B | 1218154 | ycgG | > | > | ymgF | predicted protein | 3.4 | ||
| 38 | C | 1219948 | ymgF | > | < | ymgD | 3.9 | |||
| 39 | A | 1233950 | sodium:proton antiporter | nhaB | < | > | fadR | DNA-binding transcriptional regulator | 3.1 | |
| 40 | B | 1255834 | predicted adhesin | ycgV | < | < | ychF | 3.2 | ||
| 41 | B | 1308330 | voltage-gated potassium channel | kch | < | < | yciI | 3.7 | ||
| 42 | B | 1349272 | enoyl-[acyl-carrier-protein] reductase | fabI | < | < | ycjD | 4.3 | ||
| 43 | A | 1389946 | predicted hydrolase | *ycjY | < | > | *ycjZ | DNA-binding transcriptional regulator | 4.4 | |
| 44 | B | 1432738 | Rac prophage transcriptional regulator | ynaE | < | < | uspF | 4.2 | ||
| 45 | B | 1486246 | ydcF | > | > | aldA | aldehyde dehydrogenase A | 3.7 | ||
| 46 | B | 1565470 | predicted diguanylate cyclase | yddV | < | < | yddW | 5.2 | ||
| 47 | B | 1568560 | glutamate:aminobutyric acid antiporter | gadC | < | < | gadB | 2.3 | ||
| 48 | B | 1580550 | conserved protein | ydeN | < | < | ydeO | 3.3 | ||
| 49 | B | 1585730 | fimbrial-like adhesin protein | ydeQ | < | < | ydeR | 3.7 | ||
| 50 | B | 1613766 | yneJ | > | > | *yneK | predicted protein | 3.0 | ||
| 51 | A | 1630638 | mannonate dehydrogenase | ydfI | < | > | ydfK | Qin prophage transcriptional regulator | 4.0 | |
| 52 | B | 1639072 | Qin prophage S lysis protein | essQ | < | < | cspB | 3.3 | ||
| 53 | A | 1639660 | Qin prophage cold shock protein | cspB | < | > | cspF | Qin prophage cold shock protein | 3.7 | |
| 54 | B | 1646444 | dicA | > | > | *ydfA | Qin prophage protein | 3.2 | ||
| 55 | B | 1669358 | ynfM | > | > | asr | acid-shock periplasmic protein | 3.7 | ||
| 56 | B | 1751846 | predicted oxidoreductase | ydhV | < | < | ydhY | 3.1 | ||
| 57 | B | 1762570 | Fe-S cluster assembly protein | sufA | < | < | rydB | 5.1 | ||
| 58 | A | 1801256 | threonyl-tRNA synthetase | thrS | < | > | yniD | predicted protein | 5.3 | |
| 59 | A | 1801758 | threonyl-tRNA synthetase | thrS | < | > | yniD | predicted protein | 3.4 | |
| 60 | B | 1990832 | PG phosphate synthase | pgsA | < | < | uvrC | 4.2 | ||
| 61 | D | 2055332 | amn | > | yeeN | < | asnW | 3.0 | ||
| 62 | A | 2060070 | DNA-binding transcriptionall regulator | nac | < | > | asnV | Asn tRNA | 3.2 | |
| 63 | B | 2096360 | LPS O-antigen length regulator | *cld | < | < | ugd | 3.2 | ||
| 64 | D | 2104956 | conserved protein | wbbI | < | *rfc | < | glf | 3.4 | |
| 65 | A | 2261530 | fructose-specific PTS enzyme IIA | fruB | < | > | setB | lactose/glucose efflux system | 3.3 | |
| 66 | B | 2276452 | predicted protein | yejG | < | < | bcr | 3.1 | ||
| 67 | A | 2342846 | hypothetical protein | ypaB | < | > | nrdA | ribonucleoside diphosphate reductase | 3.4 | |
| 68 | D | 2355640 | glpC | > | yfaD | > | ypaA | predicted protein | 3.4 | |
| 69 | D | 2363146 | nudI | > | ais | > | arnB | uridine aminotransferase | 3.8 | |
| 70 | B | 2403570 | NADH:ubiquinone oxidoreductase | nuoA | < | < | lrhA | 5.1 | ||
| 71 | A | 2405036 | DNA-binding transcriptional regulator | lrhA | < | > | yfbQ | predicted aminotransferase | 3.2 | |
| 72 | D | 2476546 | DNA-binding transcriptional regulator | dsdC | < | dsdX | > | dsdA | D-serine ammonia-lyase | 3.6 |
| 73 | D | 2489940 | predicted transporter | yfdV | < | oxc | < | frc | 3.6 | |
| 74 | D | 2490262 | predicted oxalyl-CoA decarboxylase | oxc | < | frc | < | yfdX | 3.6 | |
| 75 | A | 2492430 | predicted protein | yfdX | < | > | ypdI | lipoprotein for colanic acid biosynthesis | 3.0 | |
| 76 | A | 2493362 | predicted inner membrane protein | yfdY | < | > | lpxP | palmitoleoyl-ACP acyltransferase | 3.6 | |
| 77 | D | 2532356 | ptsH | > | ptsI | > | crr | glucose-specific PTS enzyme IIA | 3.2 | |
| 78 | A | 2597838 | dihydrodipicolinate synthase | dapA | < | > | gcvR | DNA-binding transcriptional repressor | 3.1 | |
| 79 | B | 2599140 | bcp | > | > | hyfA | hydrogenase 4, 4Fe-4S subunit | 3.1 | ||
| 80 | D | 2626032 | ppx | > | yfgF | > | yfgG | predicted protein | 3.6 | |
| 81 | A | 2696642 | conserved protein | yfhB | < | > | yfhH | DNA-binding transcriptional regulator | 3.3 | |
| 82 | A | 2714742 | pyruvate formate lyase subunit | yfiD | < | > | ung | uracil-DNA-glycosylase | 4.8 | |
| 83 | A | 2739338 | D-arabino-heptulosonate-7P synthase | aroF | < | > | yfiL | predicted protein | 3.5 | |
| 84 | A | 2823870 | lytic murein transglycosylase B | mltB | < | > | srlA | glucitol/sorbitol-specific PTS IIC | 3.1 | |
| 85 | A | 2837534 | DNA-binding transcriptional repressor | ascG | < | > | ascF | cellobiose/arbutin/salicin PTS IIB-IIC | 3.5 | |
| 86 | D | 2884946 | predicted protein | ygcL | < | ygcB | < | cysH | 3.4 | |
| 87 | A | 2898372 | deoxygluconate dehydrogenase | ygcW | < | > | yqcE | predicted transporter | 4.6 | |
| 88 | A | 2932264 | L-fuculose-1-phosphate aldolase | fucA | < | > | fucP | L-fucose transporter | 3.3 | |
| 89 | B | 2967056 | nucleotide hydrolase | rppH | < | < | ygdT | 4.5 | ||
| 90 | A | 2976930 | diaminopimelate decarboxylase | lysA | < | > | lysR | DNA-binding transcriptional regulator | 3.4 | |
| 91 | B | 2985164 | yqeG | > | > | yqeH | protein with bipartite regulator domain | 3.3 | ||
| 92 | D | 2985970 | yqeG | > | yqeH | > | yqeI | transcriptional regulator | 3.2 | |
| 93 | D | 2991152 | ygeG | > | ygeH | > | ygeI | predicted protein | 3.7 | |
| 94 | C | 2991858 | ygeI | > | < | insD | 3.5 | |||
| 95 | B | 3067930 | mechanosensitive channel | mscS | < | < | fbaA | 3.0 | ||
| 96 | B | 3166268 | DNA-binding transcriptional regulator | ygiT | < | < | mqsR | 3.0 | ||
| 97 | B | 3237654 | alx | > | > | sstT | sodium:serine/threonine symporter | 6.9 | ||
| 98 | D | 3266066 | tdcR | > | yhaB | > | yhaC | predicted protein | 3.0 | |
| 99 | A | 3276944 | DNA-binding transcriptional regulator | agaR | < | > | kbaZ | tagatose 6-phosphate aldolase 1 | 3.6 | |
| 100 | B | 3319952 | predicted hydrolase, inner membrane | yhbX | < | < | leuU | 3.2 | ||
| 101 | A | 3325832 | 23S rRNA methyltransferase | rrmJ | < | > | yhbY | predicted RNA-binding protein | 14.1 | |
| 102 | D | 3360232 | gltF | > | *yhcA | > | yhcD | predicted outer membrane protein | 3.2 | |
| 103 | B | 3375554 | stringent starvation protein A | sspA | < | < | rpsI | 3.7 | ||
| 104 | B | 3387142 | p-hydroxybenzoic acid efflux system | *aaeA | < | < | aaeX | 3.1 | ||
| 105 | A | 3559934 | thiosulfate:cyanide sulfurtransferase | glpE | < | > | glpD | sn-glycerol-3-phosphate dehydrogenase | 3.5 | |
| 106 | D | 3629568 | predicted HlyD family secretion protein | yhiI | < | yhiJ | > | yhiM | conserved inner membrane protein | 3.6 |
| 107 | C | 3634072 | yhiM | > | < | yhiN | 3.8 | |||
| 108 | B | 3655654 | hdeD | > | > | gadE | DNA-binding transcriptional activator | 4.1 | ||
| 109 | C | 3661646 | mdtF | > | < | gadW | 3.7 | |||
| 110 | B | 3681552 | C4-dicarboxylic acid citrate transporter | dctA | < | < | yhjK | 3.0 | ||
| 111 | A | 3717248 | conserved protein | yiaF | < | > | yiaG | transcriptional regulator | 3.0 | |
| 112 | B | 3717858 | yiaG | > | > | cspA | major cold shock protein | 3.0 | ||
| 113 | B | 3720058 | insK | > | > | sokA | ncRNA | 3.0 | ||
| 114 | A | 3826772 | glutamate transporter | gltS | < | > | yicE | predicted transporter | 4.4 | |
| 115 | B | 3834954 | selC | > | > | setC | predicted sugar efflux system | 3.4 | ||
| 116 | A | 3865668 | heat shock chaperone | *ibpA | < | > | yidQ | conserved outer membrane protein | 4.6 | |
| 117 | A | 3939432 | predicted transcriptional regulator | yieP | < | > | *rrsC | 16S ribosomal RNA of rrnC operon | 3.6 | |
| 118 | A | 4002730 | conserved protein | yigI | < | > | *pldA | outer membrane phospholipase A | 3.7 | |
| 119 | B | 4068432 | predicted aldose-1-epimerase | yihR | < | < | yihS | 7.5 | ||
| 120 | B | 4360430 | DNA-binding transcriptional activator | cadC | < | < | pheU | 4.0 | ||
| 121 | B | 4364770 | C4-dicarboxylate antiporter | dcuA | < | < | aspA | 4.6 | ||
| 122 | A | 4366568 | aspartate ammonia-lyase | aspA | < | > | *fxsA | inner membrane protein | 19.2 | |
| 123 | D | 4371850 | yjeI | > | *yjeJ | < | yjeK | 4.9 | ||
| 124 | D | 4486746 | yjgQ | > | yjgR | < | idnR | 4.1 | ||
| 125 | B | 4494638 | leuX | > | > | insC | KpLE2 phage IS2 element repressor | 3.1 | ||
| 126 | B | 4518430 | KpLE2 transcriptional regulator | yjhU | < | < | yjhF | 3.1 | ||
| 127 | B | 4530030 | KpLE2 phage endoglucanase | sgcX | < | < | yjhP | 3.3 | ||
| 128 | A | 4538050 | N-acetylnuraminic acid channel protein | nanC | < | > | fimB | tyrosine recombinase, fimA regulator | 3.5 | |
| 129 | A | 4538964 | N-acetylnuraminic acid channel protein | nanC | < | > | fimB | tyrosine recombinase, fimA regulator | 3.4 | |
| 130 | B | 4540968 | fimE | > | > | fimA | major type 1 subunit fimbrin (pilin) | 3.4 | ||
| 131 | A | 4549330 | fructuronate transporter | gntP | < | > | uxuA | mannonate hydrolase | 5.0 | |
| 132 | D | 4576468 | 5-methylcytosine restriction enzyme | mcrC | < | mcrB | < | symE | 3.5 | |
| 133 | A | 4633370 | DNA-binding transcriptional activator | rob | < | > | *creA | conserved protein | 3.3 |
Genomic SELEX was performed for search of the binding sites of RNAP RpoH holoenzyme. By setting the cut-off level of 3.0, a total of 133 binding sites were identified (see Fig 2 for SELEX pattern), which are aligned along the map of E. coli K12 genome. A total of 107 sites are located within intergenic spacers: 41 wihin type-A spacers (shown under orange background); and 60 within type-B spacers (shown under green background). The constitutive promoters of RpoH were predicted based on the adjacent genes [note that only the genes next to the RpoH holoenzyme-binding sites are shown] and the gene orientation (shown by arrows in the column of transcription direction). A total of 26 RpoH holoenzyme-binding sites are located inside open reading frames as indicated by the gene symbols shown in RpoH column.
* The genes listed in RegulonDB as the regulated targets of RpoH.
Among a total of 322 RpoH promoters (or the transcription initiation sites) listed in RegulonDB database, 20 were identified setting the cut-off level at 3.0 (Table 3, marked by asterisk). The majority of RpoH promoters in the database were suggested to belong to the inducible promoters that are expressed only under the support of other positive regulatory factors. Otherwise these RpoH promoters might represent the inaccurate prediction as note above. Genomic SELEX analysis identified minimum 100 and maximum 140 RpoH constitutive promoters including 18 known RpoH dependent promoters (Table 2). The highest peak was 20-fold intensity that was detected on promoter region of the ybeD gene, which encodes a conserved protein of unknown function under regulation of RpoH (Fig 2) [49], followed by high-level peaks at the aspA-fxsA and the rlmJ-yhbY intergenic regions. The fxsA gene encodes an inner membrane protein, which is involved in sensitivity control to bacteriophage T7 [49]. The rlmE gene encodes 23S rRNA 2’-O-ribose U2552 metyltransferase, and has been proposed to carry RpoH-dependent promoter [50]. The regulatory target of RpoH sigma identified by Genomic SELEX expands to a set of genes related to varieties of stress-response genes beyond the HSP genes. In fact, the genes for response to environmental insults such as ethanol, alkaline pH, and hyperosmotic shock and the genes for proteolysis and cell division have been indicated under the control of RpoH. The set of RpoH-regulon genes thus identified in vivo, however, vary depending on the culture conditions.
The whole sets of constitutive promoters for the flagella-chemotaxis sigma RpoF
The bacterial flagellum is a complex organelle consisting of three distinctive structural parts, the basal body, the hook and the filament [51]. The synthesis, assembly and function of the flagellar and chemotaxis system require the expression of more than 50 genes, which are divided into three temporally regulated transcriptional classes based on the hierarchy of expression order: class-I (early), class-II (middle), and class-III (late) [52,53]. The class-1 (early) consists of a single operon including two genes, flhD and flhC, each encoding transcription factor FlhD and FlhC, respectively, which together form a complex, FlhD2-FlhC2 or FlhD4-FlhC2, that activates transcription of a set of class-2 (middle) genes, including both the rpoF sigma gene (renamed fliA) and the flgM gene encoding the anti-RpoF factor [51,52]. RpoF is the sigma factor for flagellar chemotaxis, which recognizes the promoters of motility and flagellar synthesis genes. The regulatory target of RpoF in Salmonella was identified to include a set of genes that were classified into the class-3 operons of the flagella regulon [54,55]. More than 30 genes have been proposed to carry promoters that are under the control of RpoF sigma, including a set of the structural genes for flagella formation, and the chemotaxis genes encoding sensor of environmental signals affecting the motility control [54,56]. With use the combination of ChIP-chip, ChIP-seq and RNA-seq systems, a more comprehensive screening was recently performed for identification of the regulatory targets of RpoF sigma in E. coli [57]. A total of 52 RpoF-binding sites were identified in vivo on the genome of exponentially growing E. coli K-12 MG1655 cells in a rich LB medium, with a considerable level of over-lapping with the hitherto identified target genes of the RpoF regulon. The total number of RpoF promoters (or the transcription initiation sites) listed in the current RegulonDB database is as many as 144, which have been identified in vivo using ChIP-chip and ChIP-RNA Seq analyses. Most of the targets predicted by the in vivo data, however, represent those indirectly affected upon knock-out of the rpoF gene or over-production of RpoF.
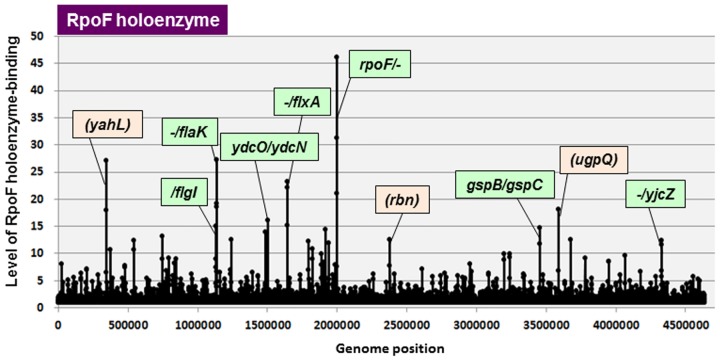
We then performed the Genomic SELEX screening in vitro for search of the direct target promoters, genes and operons under the control of RpoF using the reconstituted RNAP RpoF holoenzyme. By setting the cut-off level of 4.0 fold higher than the background of original library DNA, a total of 105 RpoF holoenzyme-binding peaks were identified (Fig 3 and Table 4), of which 37 (35%) are located within intergenic spacers and 68 (65%) are inside of genes (Table 2). One unique feature of RpoF holoenzyme is its high-level (65%) binding to inside of open reading frames of a number of genes. A high-level (60%) of RNAP binding was also identified for RpoD holoenzyme [27]. The identification of the promoter-like sequences inside these genes awaits further analysis. The spacers containing RpoF holoenzyme-binding sites were also classified into three types (Tables 2 and 4 for the whole list): 7 peaks are located within type-A spacer; 27 peaks are located within type-B spacers; and 3 peaks are located within type-C spacers. Based on the transcription direction of flanking genes, the total number of RpoF constitutive promoters was predicted to range between minimum 34 (7 type-A plus 27 type-B) and maximum 41 (7x2 type-A plus 27 type-B) (Table 2). The total number of RpoF promoters (or the transcription initiation sites) listed in the current RegulonDB database is as many as 144. Of which 14 were identified setting cut-off level at 4.0 (Table 4, marked by asterisk), indicating that these promoters are constitutive promoters and the majority of known promoters represent the inducible promoters that are expressed only under the support of positive regulatory factors.
Fig 3. SELEX-chip search for RNAP RpoF holoenzyme-binding sequences on the E. coli K-12 genome.
The y-axis represents the relative number of RpoF holoenzyme-bound DNA fragments whereas x-axis represents the position on the E. coli K-12 genome, in base pair. The adjacent gene on E. coli K-12 genome of peak position was indicated for high intensity peaks. The peaks located within spacer regions are shown in green color, while peaks located within open reading frames are shown in orange color. The list of RpoF holoenzyme-binding sites is described in Table 4.
Table 4. RpoF holoenzyme-binding sites on the E. coli K-12 genome.
| No | Type | Map | Gene Function | Left | D | RpoF | D | Right | Gene Function | Intensity |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D | 25672 | ileS | > | lspA | > | fkpB | peptidyl-prolyl cis-trans isomerase | 8.1 | |
| 2 | B | 68342 | L-arabinose isomerase | araA | < | < | araB | 4.3 | ||
| 3 | D | 109556 | secM | > | secA | > | mutT | NTP pyrophosphohydrolase | 5.5 | |
| 4 | A | 131366 | predicted protein | yacH | < | > | acnB | aconitate hydratase | 4.6 | |
| 5 | B | 164658 | hrpB | > | > | mrcB | glycosyl transferase and transpeptidase | 6.4 | ||
| 6 | D | 198960 | rseP | > | bamA | > | skp | periplasmic chaperone | 4.7 | |
| 7 | B | 202068 | lpxD | > | > | fabZ | hydroxymyristol acyl carrier dehydratase | 7.1 | ||
| 8 | D | 298660 | conserved protein | yagQ | < | yagR | < | yagS | 4.4 | |
| 9 | D | 343660 | yahK | > | yahL | > | yahM | predicted protein | 27.1 | |
| 10 | D | 353230 | prpD | > | prpE | > | codB | cytosine transporter | 4.7 | |
| 11 | B | 463170 | ppiD | > | > | ybaV | conserved protein | 5.4 | ||
| 12 | D | 472938 | glnK | > | amtB | < | tesB | 4.0 | ||
| 13 | D | 477460 | ybaA | > | ylaB | < | ylaC | 7.8 | ||
| 14 | D | 482072 | predicted protein | tomB | < | acrB | < | acrA | 4.5 | |
| 15 | D | 543470 | conserved protein | ylbA | < | allC | < | allD | 12.3 | |
| 16 | D | 633930 | predicted oxidoreductase | ybdH | < | ybdL | < | ybdM | 5.4 | |
| 17 | D | 652632 | citrate lyase synthetase | citC | < | dpiB | > | dpiA | CitAB TCS response regulator | 5.0 |
| 18 | D | 747240 | predicted regulator | abrB | < | ybgO | < | ybgP | 13.1 | |
| 19 | D | 761630 | sucA | > | sucB | > | sucC | succinyl-CoA synthetase | 4.7 | |
| 20 | B | 794336 | ybhT | > | > | *modA | molybdate transporter subunit | 9.2 | ||
| 21 | B | 822944 | cardiolipin synthase 2 | ybhO | < | < | ybhP | 4.9 | ||
| 22 | B | 837732 | conserved protein | ybiI | < | < | ybiX | 5.7 | ||
| 23 | D | 844530 | rlmF | > | ybiO | < | glnQ | 9.0 | ||
| 24 | B | 874568 | yliE | > | > | yliF | predicted diguanylate cyclase | 4.3 | ||
| 25 | D | 878444 | bssR | > | yliI | < | yliJ | 5.3 | ||
| 26 | D | 893136 | ybjN | > | potF | > | potG | ABC superfamily putrescine transporter | 4.1 | |
| 27 | B | 956734 | ycaP | > | > | serC | 3-phosphoserine aminotransferase | 6.2 | ||
| 28 | D | 1039338 | appC | > | appB | > | yccB | hypothetical protein | 5.0 | |
| 29 | D | 1048250 | conserved protein | gfcC | < | gfcB | < | gfcA | 4.0 | |
| 30 | B | 1062060 | modulator of CbpA co-chaperone | cbpM | < | < | cbpA | 5.2 | ||
| 31 | B | 1129438 | anti-sigma factor for FliA (sigma 28) | *flgM | < | < | flgA | 15.0 | ||
| 32 | B | 1133868 | flgF | > | > | flgG | flagellar component | 6.7 | ||
| 33 | B | 1137530 | flgJ | > | > | *flgK | flagellar hook-filament junction protein 1 | 27.2 | ||
| 34 | D | 1158658 | ptsG | > | fhuE | > | hinT | purine nucleoside phosphoramidase | 6.5 | |
| 35 | B | 1193064 | tRNA methyltransferase | mnmA | < | < | nudJ | 5.3 | ||
| 36 | D | 1236068 | fadR | > | ycgB | > | dadA | D-amino acid dehydrogenase | 6.9 | |
| 37 | A | 1243852 | protein involved in flagellar function | *ycgR | < | > | ymgE | predicted inner membrane protein | 4.6 | |
| 38 | B | 1349272 | enoyl-[acyl-carrier-protein] reductase | fabI | < | < | ycjD | 5.6 | ||
| 39 | D | 1356550 | predicted protein | ymjA | < | puuP | < | puuA | 5.0 | |
| 40 | D | 1392064 | ycjZ | > | mppA | < | ynaI | 6.5 | ||
| 41 | A | 1434934 | outer membrane pore protein N | ompN | < | > | micC | ncRNA | 5.9 | |
| 42 | B | 1488236 | aldA | > | > | cybB | cytochrome b561 | 13.9 | ||
| 43 | D | 1504230 | predicted benzoate transporter | ydcO | < | ydcN | > | ydcP | predicted peptidase | 16.1 |
| 44 | B | 1644248 | ydfV | > | > | *flxA | Qin prophage; predicted protein | 23.2 | ||
| 45 | D | 1661452 | ynfF | > | ynfG | > | ynfH | oxidoreductase, membrane subunit | 5.5 | |
| 46 | D | 1734472 | sodB | > | ydhP | < | ynhF | 6.1 | ||
| 47 | D | 1794968 | integration host factor (IHF) | ihfA | < | pheT | < | pheS | 12.3 | |
| 48 | A | 1801758 | threonyl-tRNA synthetase | thrS | < | > | yniD | predicted protein | 5.3 | |
| 49 | B | 1815172 | conserved protein | *chbG | < | < | chbF | 4.2 | ||
| 50 | D | 1822736 | cho | > | *ves | < | spy | 10.8 | ||
| 51 | D | 1861032 | methionine sulfoxide reductase B | msrB | < | gapA | > | yeaD | conserved protein | 4.2 |
| 52 | B | 1887966 | acyl-CoA synthetase | fadD | < | < | yeaY | 9.9 | ||
| 53 | A | 1906860 | predicted protein | mgrB | < | > | yobH | predicted protein | 8.3 | |
| 54 | D | 1914836 | methionine-(R)-sulfoxide reductase | yebR | < | yebS | > | yebT | conserved protein | 14.4 |
| 55 | D | 1928332 | conserved protein | yebE | < | yebF | < | yebG | 4.3 | |
| 56 | D | 1938762 | myristoyl-ACP-dependent acyltransferase | lpxM | < | yebA | < | znuA | 11.9 | |
| 57 | D | 1971862 | purine-binding chemotaxis protein | cheW | < | cheA | < | motB | 4.3 | |
| 58 | D | 1988066 | predicted protein | *yecH | < | tyrP | < | yecA | 7.9 | |
| 59 | B | 1999848 | RNA polymerase sigma 28 | *fliA | < | < | fliC | 46.2 | ||
| 60 | D | 2233032 | yeiS | > | yeiT | > | yeiA | Dihydropyrimidine dehydrogenase | 4.8 | |
| 61 | D | 2262454 | fructose-specific PTS enzymes IIA | fruB | < | setB | < | yeiW | 6.2 | |
| 62 | D | 2379830 | N-acyltransferase | elaA | < | rbn | > | elaD | predicted enzyme | 12.5 |
| 63 | B | 2403570 | NADH:ubiquinone oxidoreductase | nuoA | < | < | lrhA | 3.5 | ||
| 64 | D | 2412454 | conserved inner membrane protein | yfbV | < | ackA | > | pta | phosphate acetyltransferase | 6.2 |
| 65 | D | 2456468 | phosphohistidine phosphatase | sixA | < | fadJ | < | fadI | 4.5 | |
| 66 | D | 2609062 | hyfH | > | hyfI | > | hyfJ | processing element hydrogenase 4 | 7.1 | |
| 67 | D | 2684772 | serine hydroxymethyltransferase | glyA | < | hmp | < | glnB | 5.8 | |
| 68 | D | 2745566 | 30S ribosomal subunit protein S16 | rpsP | < | ffh | > | ypjD | predicted inner membrane protein | 5.9 |
| 69 | D | 2868142 | L-isoaspartate carboxymethyltransfeerase | pcm | < | surE | < | truD | 5.8 | |
| 70 | D | 2916840 | barA | > | gudD | < | gudX | 5.4 | ||
| 71 | D | 2952958 | exonuclease V (RecBCD complex), | recD | < | recB | < | ptrA | 8.0 | |
| 72 | D | 2965066 | diacylglyceryl transferase | lgt | < | ptsP | < | rppH | 6.7 | |
| 73 | D | 3087058 | metK | > | galP | > | yggI | conserved protein | 6.4 | |
| 74 | D | 3124360 | glycolate oxidase iron-sulfur subunit | glcF | < | glcE | < | glcD | 4.0 | |
| 75 | C | 3146946 | yghZ | > | < | yqhA | 5.2 | |||
| 76 | B | 3197666 | deadenylyltransferase/adenylyltransferase | glnE | < | < | ygiF | 8.8 | ||
| 77 | B | 3237654 | alx | > | > | sstT | sodium:serine/threonine symporter | 9.9 | ||
| 78 | D | 3338440 | ABC superfamily toluene transporter | yrbF | < | yrbG | > | kdsD | D-arabinose 5-phosphate isomerase | 6.2 |
| 79 | D | 3344772 | hpf | > | ptsN | > | yhbJ | protein with NTP hydrolase domain | 5.9 | |
| 80 | C | 3388568 | aaeR | > | < | tldD | 4.7 | |||
| 81 | D | 3452166 | type II secretion divergon | gspB | < | gspA | > | gspC | general secretory pathway component | 11.8 |
| 82 | D | 3454168 | general secretory pathway component | gspA | < | gspC | > | gspD | general secretory pathway component | 14.7 |
| 83 | D | 3478072 | predicted protein | yheV | < | kefB | < | kefG | 6.1 | |
| 84 | D | 3515848 | predicted protein | damX | < | aroB | < | aroK | 4.5 | |
| 85 | C | 3528652 | hslO | > | < | yhgE | 4.0 | |||
| 86 | D | 3534454 | TCS sensory histidine kinase | envZ | < | ompR | > | *greB | transcription elongation factor | 4.3 |
| 87 | D | 3585866 | yhhA | > | ugpQ | < | ugpC | 18.1 | ||
| 88 | D | 3633746 | predicted protein | yhiJ | < | *yhiM | < | yhiN | 4.8 | |
| 89 | D | 3689064 | endo-1,4-D-glucanase | bcsZ | < | bcsB | < | bcsA | 4.6 | |
| 90 | B | 3779166 | lldD | > | > | yibK | predicted rRNA methylase | 9.2 | ||
| 91 | D | 3789732 | threonine 3-dehydrogenase | tdh | < | kbl | < | htrL | 4.9 | |
| 92 | A | 3826672 | glutamate transporter | gltS | < | > | yicE | predicted transporter | 5.4 | |
| 93 | D | 3829430 | yicE | > | yicH | < | yicI | 4.3 | ||
| 94 | B | 3906432 | phosphate transporter | pstB | < | < | pstA | 4.1 | ||
| 95 | D | 3945232 | trpT | > | *hdfR | > | yifE | conserved protein | 4.7 | |
| 96 | B | 3951430 | ilvE | > | > | ilvD | dihydroxyacid dehydratase | 8.6 | ||
| 97 | D | 4003446 | conserved protein | yigI | < | pldA | > | recQ | ATP-dependent DNA helicase | 4.5 |
| 98 | D | 4032158 | yigZ | > | trkH | > | hemG | protoporphyrin oxidase, flavoprotein | 6.0 | |
| 99 | B | 4068432 | predicted aldose-1-epimerase | yihR | < | < | yihS | 9.6 | ||
| 100 | D | 4178758 | rplJ | > | rplL | > | rpoB | RNA polymerase, beta subunit | 6.7 | |
| 101 | D | 4286472 | acetyl-CoA synthetase | acs | < | nrfA | > | nrfB | nitrite reductase | 5.8 |
| 102 | D | 4327160 | conserved protein | yjdM | < | *yjdA | > | yjcZ | conserved protein | 12.4 |
| 103 | D | 4338862 | biodegradative arginine decarboxylase | adiA | < | melR | > | melA | alpha-galactosidase, NAD(P)-binding | 4.5 |
| 104 | D | 4535430 | conserved protein | yjhX | < | yjhS | < | nanM | 4.7 | |
| 105 | A | 4589632 | predicted inner membrane protein | yjiY | < | > | *tsr | methyl-accepting chemotaxis protein I | 5.3 |
Genomic SELEX was performed for search of the binding sites of RNAP RpoF holoenzyme. By setting the cut-off level of 3.0, a total of 105 binding sites were identified (see Fig 3 for SELEX pattern), which are aligned along the map of E. coli K12 genome. A total of 37 sites are located within intergenic spacers: 7 wihin type-A spacers (shown under orange background); and 27 within type-B spacers (shown under green background). The constitutive promoters of RpoF were predicted based on the adjacent genes [note that only the genes next to the RpoF holoenzyme-binding sites are shown] and the gene orientation (shown by arrows in the column of transcription direction). A total of as many as 68 RpoF holoenzyme-binding sites are located inside open reading frames as indicated by the gene symbols shown in RpoF column.
*The genes listed in RegulonDB as the regulated targets of RpoF.
The highest peak was 46-fold intensity detected on promoter region of rpoF itself (Fig 3), indicating the autoregulation as already suggested [58]. A high-level peak was also identified upstream of the flgK gene, which encodes flagellar hook-filament junction protein that connects the filament to the hook, and its transcription has been shown in vitro under the direct control of RpoF [59]. The flgM gene encodes the anti-sigma factor for RpoF [55]. FlgM forms a complex with RpoF, thereby inactivating its sigma function but protects its degradation by the Lon protease for preservation [60].
The whole set of constitutive promoters for extra-cytoplasmic stress response sigma RpoE
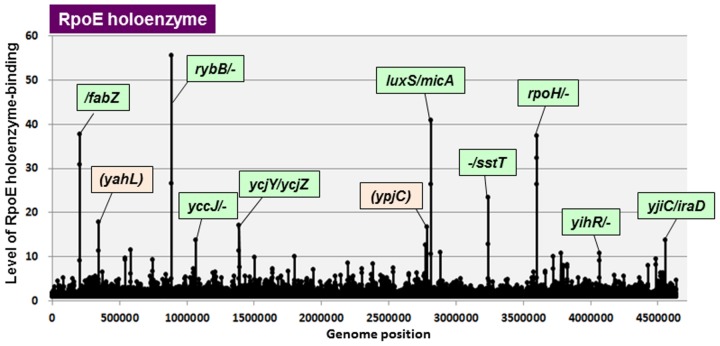
The bacterial cell envelope is a dynamic compartment, changing its structure and function in response to environmental conditions. Accordingly, the integrity of envelope is maintained through frequent modulation of its composition and components. The minor sigma factor RpoE plays a central role in this process, by controlling the selective expression of envelope components [61]. The regulatory targets have been estimated after proteome and transcriptome analyses in vivo [62–64]. The activity of RpoE is negatively regulated by a membrane-bound anti-sigma factor RseA, which sequesters RpoE under unstressed conditions. Within membrane, RseA is associated at its C-terminal domain with a periplasmic protein RseB, which senses misfolded proteins for release and activation of RpoE from RpoE-RseA complexes [62,65]. The total number of RpoE promoters (or the transcription initiation sites) listed in the current RegulonDB database is as many as 518, of which most are identified by computational analyses based on the consensus sequence of RpoE promoters without experimental analysis. After SELEX screening as noted below, most of these RpoE promoters must be inaccurate estimation due to the error in the consensus sequence.
The Genomic SELEX screening system was employed as a short-cut approach for identification of the RpoE regulon. Previously we purified RpoE and examined its promoter selectivity using an in vitro transcription system [66]. Using this purified RpoE, we performed SELEX screening. By setting the cut-off level of 4.0 fold against original library DNA, a total of 126 RpoE holoenzyme-binding peaks were identified (Fig 4 and Table 5), of which 84 (67%) are located within intergenic spacers and 42 (33%) are inside of open reading frames (Tables 2 and 5 for the whole set). Since the majority of hitherto identified promoters are located within spacers, detailed search for the constitutive promoters was focused on the total of 84 peaks within spacers. The spacers containing RpoE holoenzyme-binding sites were also classified into three types (Table 2): 29 peaks are located within type-A spacer; 48 peaks are located within type-B spacers; and 7 peaks are located within type-C spacers. Based on the transcription direction of flanking genes, the total number of RpoE constitutive promoters was predicted to range between minimum 77 (29 type-A plus 48 type-B) and maximum 106 (29x2 type-A plus 48 type-B) (Table 2). Within the set of constitutive promoters identified by setting the cut-off level at 4.0, a total of 19 known RpoE promoters were identified (Table 5, marked by asterisk). The majority of known promoters represent the inducible promoters that are expressed only under the support of positive regulatory factors.
Fig 4. SELEX-chip search for RNAP RpoE holoenzyme-binding sequences on the E. coli K-12 genome.
The y-axis represents the relative number of RpoE holoenzyme-bound DNA fragments whereas x-axis represents the position on the E.coli genome, in base pair. The adjacent gene on E. coli K-12 genome of peak position was indicated for high intensity peaks. The peaks located within spacer regions are shown in green color, while peaks located within open reading frames are shown in orange color. The list of RpoE holoenzyme-binding sites is described in Table 5.
Table 5. RpoE holoenzyme-binding sites on the E. coli K-12 genome.
| No. | Type | Map | Left Gene Function | Left | D | RpoE | D | Right | Right Gene Function | Intensity |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | D | 154236 | fimbrial-like adhesin protein | yadM | < | htrE | < | ecpD | 5.0 | |
| 2 | B | 164658 | hrpB | > | > | mrcB | glycosyl transferase and transpeptidase | 4.3 | ||
| 3 | B | 202068 | lpxD | > | > | fabZ | hydroxymyristol acyl carrier dehydratase | 30.9 | ||
| 4 | C | 262270 | thrW | > | < | ykfI | 4.6 | |||
| 5 | D | 270562 | CP4-6 prophage transcriptional regulator | perR | < | insI | < | insH | 4.6 | |
| 6 | B | 292358 | CP4-6 prophage; conserved protein | yagK | < | < | yagL | 4.7 | ||
| 7 | D | 302738 | xanthine dehydrogenase 2Fe-2S subunit | yagT | < | yagU | < | ykgJ | 5.3 | |
| 8 | A | 319448 | oxidoreductase with FAD-binding domain | *ykgC | < | > | ykgD | DNA-binding transcriptional regulator | 4.0 | |
| 9 | C | 323832 | ykgG | > | < | ykgH | 5.6 | |||
| 10 | B | 331456 | betT | > | > | yahA | DNA-binding transcriptional regulator | 4.9 | ||
| 11 | d | 343660 | yahK | > | yahL | > | *yahM | predicted protein | 17.8 | |
| 12 | A | 345542 | neutral amino-acid efflux system | yahN | < | > | yahO | predicted protein | 4.0 | |
| 13 | B | 477848 | conserved inner membrane protein | ylaB | < | < | ylaC | 4.5 | ||
| 14 | D | 543560 | conserved protein | *ylbA | < | allC | < | allD | 9.6 | |
| 15 | B | 581644 | nohB | > | > | appY | DLP12 DNA-binding transcriptional activator | 11.5 | ||
| 16 | B | 629042 | ybdB | > | > | cstA | carbon starvation protein | 4.3 | ||
| 17 | B | 728638 | ybfA | > | > | rhsC | rhsC element core protein RshC | 5.6 | ||
| 18 | D | 732870 | ybfA | > | rhsC | > | ybfB | predicted inner membrane protein | 4.1 | |
| 19 | D | 747240 | predicted regulator | abrB | < | *ybgO | < | ybgP | 9.3 | |
| 20 | A | 753948 | citrate synthase | gltA | < | > | sdhC | succinate dehydrogenase | 4.7 | |
| 21 | B | 773834 | ybgE | > | > | ybgC | predicted acyl-CoA thioesterase | 4.2 | ||
| 22 | B | 837732 | conserved protein | ybiI | < | < | ybiX | 4.1 | ||
| 23 | D | 838958 | conserved protein | ybiX | < | fiu | < | mcbA | 4.3 | |
| 24 | D | 844530 | rlmF | > | ybiO | < | glnQ | 6.0 | ||
| 25 | A | 862768 | pyruvate formate lyase activating enzyme | ybiY | < | > | fsaA | fructose-6-phosphate aldolase 1 | 4.4 | |
| 26 | B | 887366 | ncRNA | *rybB | < | < | ybjL | 55.5 | ||
| 27 | B | 986332 | outer membrane porin 1a (Ia;b;F) | ompF | < | < | asnS | 5.0 | ||
| 28 | D | 992744 | pepN | > | ssuB | < | ssuC | 5.3 | ||
| 29 | D | 1045134 | exopolysaccharide export protein | gfcE | < | gfcD | < | gfcC | 5.1 | |
| 30 | C | 1049830 | insB | > | < | cspH | 6.3 | |||
| 31 | A | 1050632 | CspA-family stress protein | *cspH | < | > | cspG | DNA-binding transcriptional regulator | 4.7 | |
| 32 | D | 1088072 | predicted glycosyl transferase | pgaC | < | pgaB | < | pgaA | 4.8 | |
| 33 | B | 1166772 | ndh | > | > | ycfJ | predicted protein | 4.0 | ||
| 34 | C | 1195868 | icd | > | < | ymfD | 6.4 | |||
| 35 | B | 1218154 | ycgG | > | > | *ymgF | predicted protein | 5.4 | ||
| 36 | D | 1259856 | ychH | > | ychM | < | prs | 5.3 | ||
| 37 | D | 1342036 | lipoprotein | osmB | < | *yciT | < | yciZ | 4.3 | |
| 38 | B | 1349272 | enoyl-[acyl-carrier-protein] reductaset | fabI | < | < | ycjD | 5.6 | ||
| 39 | B | 1384744 | ycjF | > | > | tyrR | DNA-binding transcriptional regulator | 5.0 | ||
| 40 | A | 1389946 | predicted hydrolase | ycjY | < | > | ycjZ | DNA-binding transcriptional regulator | 17.2 | |
| 41 | B | 1432738 | Rac prophage transcriptional regulator | ynaE | < | < | uspF | 5.6 | ||
| 42 | D | 1468142 | KpLE2 phage-like IS repressor InsA | insC | < | insI | > | ydbC | NAD(P)-binding oxidoreductase | 5.4 |
| 43 | D | 1500658 | tehB | > | *ydcL | > | ydcM | predicted transposase | 4.8 | |
| 44 | B | 1518232 | yncB | > | > | mcbR | DNA-binding transcriptional regulator | 4.5 | ||
| 45 | B | 1542352 | nitrate/nitrite transporter | narU | < | < | yddJ | 4.9 | ||
| 46 | B | 1585730 | fimbrial-like adhesin protein | ydeQ | < | < | ydeR | 4.4 | ||
| 47 | B | 1588350 | fimbrial-like adhesin protein | ydeS | < | < | hipA | 4.5 | ||
| 48 | A | 1630638 | predicted mannonate dehydrogenase | ydfI | < | > | *ydfK | Qin prophage transcriptional regulator | 5.1 | |
| 49 | B | 1638930 | Qin prophage; predicted S lysis protein | essQ | < | < | cspB | 7.0 | ||
| 50 | A | 1639660 | Qin prophage; cold shock protein | cspB | < | > | *cspF | Qin prophage; cold shock protein | 7.3 | |
| 51 | A | 1676254 | pyridine nucleotide transhydrogenase | pntA | < | > | ydgH | predicted protein | 5.0 | |
| 52 | B | 1751846 | predicted oxidoreductase | ydhV | < | < | ydhY | 6.6 | ||
| 53 | A | 1801758 | threonyl-tRNA synthetase | thrS | < | > | yniD | predicted protein | 10.1 | |
| 54 | D | 1852370 | predicted transporter | *ydjE | < | *ydjF | < | ydjG | 4.4 | |
| 55 | B | 1887966 | acyl-CoA synthetase | fadD | < | < | yeaY | 4.5 | ||
| 56 | A | 1923044 | conserved protein | yobA | < | > | holE | DNA polymerase III | 4.5 | |
| 57 | D | 1938762 | myristoyl-acyl carrier ACP acyltransferase | lpxM | < | *yebA | < | znuA | 7.1 | |
| 58 | A | 2060070 | DNA-binding transcriptional dual regulator | nac | < | > | asnV | Asn tRNA | 4.4 | |
| 59 | D | 2103368 | predicted acyl transferase | wbbJ | < | wbbI | < | rfc | 4.2 | |
| 60 | D | 2104870 | conserved protein | wbbI | < | rfc | < | glf | 5.7 | |
| 61 | D | 2144358 | uridine/cytidine kinase | udk | < | yegE | < | alkA | 5.8 | |
| 62 | B | 2196338 | metG | > | > | yehI | conserved protein | 8.6 | ||
| 63 | D | 2210570 | conserved protein | yehS | < | yehT | < | yehU | 5.2 | |
| 64 | D | 2214150 | yohO | > | yehW | < | yehX | 4.0 | ||
| 65 | D | 2302258 | yojO | > | eco | < | mqo | 7.2 | ||
| 66 | D | 2369270 | arnD | > | arnT | > | arnE | conserved protein | 5.9 | |
| 67 | A | 2405036 | DNA-binding transcriptional repressor | lrhA | < | > | yfbQ | predicted aminotransferase | 4.0 | |
| 68 | D | 2441138 | 3-oxoacyl-[acyl-carrier-protein] synthase I | fabB | < | mnmC | < | yfcL | 5.7 | |
| 69 | A | 2459264 | conserved protein | yfcZ | < | > | fadL | long-chain fatty acid OM transporter | 5.3 | |
| 70 | D | 2484966 | evgA | > | evgS | < | yfdE | 5.2 | ||
| 71 | D | 2490262 | predicted oxalyl-CoA decarboxylase | *oxc | < | frc | < | yfdX | 4.9 | |
| 72 | D | 2492854 | predicted protein | yfdX | < | *ypdI | < | yfdY | 4.1 | |
| 73 | A | 2507446 | glucokinase | glk | < | > | yfeO | predicted ion channel protein | 4.3 | |
| 74 | A | 2535342 | pyridoxal-pyridoxamine kinase | pdxK | < | > | *yfeK | predicted protein | 7.5 | |
| 75 | D | 2649866 | fused transglycosylase/transpeptidase | pbpC | < | yfhM | > | sseA | 3-mercaptopyruvate sulfurtransferase | 6.1 |
| 76 | A | 2696642 | conserved protein | yfhB | < | > | yfhH | DNA-binding transcriptional regulator | 4.7 | |
| 77 | A | 2708134 | RNA polymerase sigma 24 | rpoE | < | > | nadB | quinolinate synthase | 6.3 | |
| 78 | B | 2761570 | CP4-57 prophage protein | yfjK | < | < | yfjL | 5.6 | ||
| 79 | B | 2763272 | CP4-57 prophage protein | yfjL | < | < | yfjM | 3.2 | ||
| 80 | D | 2764730 | CP4-57 prophage protein | yfjM | < | rnlA | > | yfjO | CP4-57 prophage protein | 6.1 |
| 81 | B | 2773454 | yfjW | > | > | yfjX | CP4-57 prophage antirestriction protein | 6.1 | ||
| 82 | C | 2796040 | ygaP | > | < | stpA | 4.3 | |||
| 83 | A | 2812736 | S-ribosylhomocysteinase | luxS | < | > | *micA | ncRNA | 40.9 | |
| 84 | A | 2874372 | sulfate adenylyltransferase | cysD | < | > | *iap | aminopeptidase | 4.2 | |
| 85 | D | 2881556 | predicted protein | ygcK | < | ygcL | < | ygcB | 10.9 | |
| 86 | D | 2894072 | predicted flavoprotein | ygcQ | < | ygcR | < | ygcS | 4.0 | |
| 87 | D | 2903952 | conserved protein | ygcF | < | *ygcG | < | eno | 4.5 | |
| 88 | B | 2985164 | yqeG | > | > | yqeH | protein with bipartite regulator domain | 4.0 | ||
| 89 | A | 3004270 | DNA-binding transcriptional regulator | ygeV | < | > | *ygeW | conserved protein | 4.5 | |
| 90 | D | 3036652 | ssDNA exonuclease | recJ | < | *dsbC | < | xerD | 5.1 | |
| 91 | B | 3237758 | alx | > | > | sstT | sodium:serine/threonine symporter | 23.4 | ||
| 92 | D | 3246540 | yqjA | > | yqjB | > | yqjC | conserved protein | 4.4 | |
| 93 | A | 3276944 | DNA-binding transcriptional dual regulator | agaR | < | > | kbaZ | tagatose 6-phosphate aldolase 1 | 4.2 | |
| 94 | B | 3375554 | stringent starvation protein A | sspA | < | < | rpsI | 4.7 | ||
| 95 | B | 3387142 | p-hydroxybenzoic acid efflux system | aaeA | < | < | aaeX | 4.9 | ||
| 96 | C | 3528652 | hslO | > | < | yhgE | 4.1 | |||
| 97 | A | 3559934 | thiosulfate:cyanide sulfurtransferase | glpE | < | > | glpD | sn-glycerol-3-phosphate dehydrogenase | 4.0 | |
| 98 | B | 3576850 | DNA-binding transcriptional repressor | gntR | < | < | yhhW | 4.9 | ||
| 99 | A | 3579162 | ncRNA | ryhB | < | > | yhhY | predicted acetyltransferase | 6.5 | |
| 100 | B | 3599044 | RNA polymerase sigma 32 | *rpoH | < | < | ftsX | 37.4 | ||
| 101 | C | 3661646 | mdtF | > | < | gadW | 6.7 | |||
| 102 | D | 3666862 | glutamate decarboxylase A | gadA | < | yhjA | > | treF | cytoplasmic trehalase | 6.6 |
| 103 | B | 3720058 | insK | > | > | sokA | ncRNA | 7.3 | ||
| 104 | B | 3739660 | 4Fe-4S ferredoxin-type hydrogenase | ysaA | < | < | yiaJ | 4.2 | ||
| 105 | D | 3764054 | predicted glutathione S-transferase | yibF | < | rhsA | > | yibA | lyase containing HEAT-repeat | 4.0 |
| 106 | B | 3779166 | lldD | > | > | yibK | predicted rRNA methylase | 10.8 | ||
| 107 | B | 3828470 | yicE | > | > | yicH | conserved protein | 4.2 | ||
| 108 | D | 3829430 | yicE | > | yicH | < | yicI | 8.2 | ||
| 109 | D | 3862372 | yidL | > | yidP | < | yidE | 4.3 | ||
| 110 | B | 3906432 | phosphate transporter subunit | pstB | < | < | pstA | 5.2 | ||
| 111 | D | 3955270 | ilvA | > | ilvY | > | ilvC | ketol-acid reductoisomerase | 4.1 | |
| 112 | A | 3958552 | peptidyl-prolyl cis-trans isomerase C | ppiC | < | > | rep | DNA helicase/ssDNA-dependent ATPase | 4.8 | |
| 113 | B | 3978772 | rffM | > | > | yifK | predicted transporter | 4.0 | ||
| 114 | B | 4068538 | predicted aldose-1-epimerase | yihR | < | < | yihS | 10.7 | ||
| 115 | B | 4173964 | thrT | > | > | tufB | protein chain elongation factor EF-Tu | 5.7 | ||
| 116 | B | 4187734 | rpoC | > | > | yjaZ | heat shock protein | 4.0 | ||
| 117 | A | 4212172 | predicted acetyltransferase | yjaB | < | > | metA | homoserine O-transsuccinylase | 4.3 | |
| 118 | B | 4249366 | malM | > | > | ubiC | chorismate pyruvate lyase | 5.7 | ||
| 119 | B | 4364770 | C4-dicarboxylate antiporter | dcuA | < | < | aspA | 5.2 | ||
| 120 | A | 4368630 | predicted transporter | yjeH | < | > | groS | Cpn10 chaperonin GroES | 4.3 | |
| 121 | A | 4380530 | anaerobic fumarate reductase | frdA | < | > | poxA | predicted lysyl-tRNA synthetase | 4.0 | |
| 122 | B | 4427854 | fklB | > | > | cycA | D-alanine/D-serine/glycine transporter | 8.0 | ||
| 123 | D | 4486746 | yjgQ | > | yjgR | < | idnR | 9.4 | ||
| 124 | B | 4530030 | KpLE2 phage endoglucanase | sgcX | < | < | yjhP | 5.2 | ||
| 125 | B | 4533162 | conserved protein | yjhX | < | < | yjhS | 4.0 | ||
| 126 | A | 4538964 | N-acetylnuraminic acid outer membrane channel protein | nanC | < | > | fimB | tyrosine recombinase/fimA regulator | 6.4 | |
| 127 | B | 4638610 | yjjY | > | > | *yjtD | predicted rRNA methyltransferase | 4.6 |
Genomic SELEX was performed for search of the binding sites of RNAP RpoE holoenzyme. By setting the cut-off level of 4.0, a total of 126 binding sites were identified (see Fig 4 for SELEX pattern), which are aligned along the map of E. coli K12 genome. A total of 84 sites are located within intergenic spacers: 29 wihin type-A spacers (shown under orange background); and 48 within type-B spacers (shown under green background). The constitutive promoters of RpoE were predicted based on the adjacent genes [note that only the genes next to the RpoE holoenzyme-binding sites are shown] and the gene orientation (shown by arrows in the column of transcription direction). A total of 42 RpoE holoenzyme-binding sites are located inside open reading frames as indicated by the gene symbols shown in RpoE column.
*The genes listed in RegulonDB as the regulated targets of RpoE.
Genomic SELEX analysis identified minimum 77 and maximum 106 for the RpoE constitutive promoters. The highest peak (55-fold intensity) was detected in the promoter region of rybB, which encodes a small regulatory RNA for expression control of some outer membrane proteins. The rybB promoter is known to be regulated by RpoE (Fig 4) [67]. The second highest peak was located on the luxS-micA intergenic region. The micA gene again encodes a small regulatory RNA that regulates expression of many genes including outer membrane proteins [68,69]. The micA promoter is also established under the control of RpoE. These sRNAs control the repair of damages in the outer membrane that took place in response to envelope stress [70,71]. High-intensity peaks were detected on some other known RpoE-dependent promoters such as rpoH, pgrR and ycjY [72,73]. Among the total of 136 binding sites of RpoE holoenzyme, 36 overlaps with that of RpoH holoenzyme. Most of these overlapping sites are related to the genes that are expressed under envelope stresss or heat-shock stress.
The intracellular levels of all seven sigma factors in E. coli K-12 W3110
In this study, we determined the constitutive promoters for the four minor sigma factors, RpoS, RpoH, RpoF and RpoE, from E. coli K-12 W3110. These promoters are recognized by the RNAP holoenzyme containing each sigma in the absence of other supporting factors. Using the mixed reconstitution in vitro of RNAP holoenzyme in the presence of all seven sigma factors, we estimated the binding affinity of each sigma to the common core enzyme, the order being RpoD (highest) > RpoN > RpoF > RpoH > FecI > RpoE > RpoS (lowest) [74]. Once we get the knowledge of intracellular concentrations of these sigma factors, it should be possible to predict the expression levels of the regulatory target genes and operons under the control of the constitutive promoters of each sigma factor. Including these four minor sigma factors, we then determined the intracellular concentrations of all seven sigma subunits. For this purpose, antibodies were made against each of the purified sigma factors that were also used for SELEX screening.
E. coli K-12 W3110 type-A was cultivated with shaking at 37°C in LB medium, and the whole cell lysates were prepared in both exponential growing phase and the stationary phase. By using the quantitative immuno-blotting method and the purified sigma proteins as the reference controls, we measured the concentrations of all seven sigma subunits. The measurement was carried out for two independent cultures, and the immuno-blot analysis was repeated for all the samples. The intracellular concentration of RpoD sigma is maintained at a constant level (on average, 160 fmol/μg total protein) throughout the transition from the exponential growth phase to the stationary phase (Fig 5) in good agreement with the previous estimation [1,75]. Based on the relative level of RNAP core enzyme subunits, the number of RpoD sigma was estimated to be 800 molecules per genome [note that the number of RNAP core enzyme is 2,000 molecules per genome]. The level of RpoD sigma in E. coli K-12 MG1655 was about half the level of W3110 (data not shown). The second abundant sigma was RpoF (70 fmol/μg protein at log phase and 80 fmol/μg protein at stationary phase) (Fig 5), but the unneccesary RpoF is stored in an inactive form by forming complexes with the anti-RpoF sigma, FlgM [76]. RpoN is also always present at a constant level (approximately 40 fmol/μg protein) in both log and stationary phases (Fig 5). The levels of other four sigma, RpoS, RpoH, RpoE and FecI, are very low during the steady-state growth (Fig 5A), but upon entry into the stationary phase, the level of RpoS increased markedly up to the level (about 100 fmol/μg protein) higher than other five minor sigma factors (Fig 5B). Taking the intracellular concentrations and the binding affinity of sigma to RNAP core enzyme as noted above, we are now able to estimate the level of each RNAP holoenzyme. Noteworthy is that the total number of all seven sigma factors is approximately as many as that of the core enzyme, but the RNAP involved in the transcription cycle or the elongation of RNA chains is considered to lack sigma subunit, the RNAP not involved in transcription should be stored as the holoenzyme forms.
Fig 5. Intracellular concentrations of seven sigma factors in E. coli K-12 W3110 type-A strain.
E. coli K-12 W3110 type-A strain was grown in LB medium at 37°C with shaking. Cells were harvested at various times and cell lysates were subjected to the quantitative immuno-blot analysis of all seven sigma factors as described in Materials and Methods. [A] The sigma levels at exponential growth phase; [B] the sigma levels in the stationary phase.
Discussion
Seven species of the sigma subunit exist in E. coli K-12, the widely used model E. coli strain. Here we identified the whole set of constitutive promoters for four minor sigma factors, RpoS, RpoH, RpoF and RpoE, by using the Genomic SELEX system. Up to the present time, the binding sites of RNAP and TF have been identified in vivo using the high-throughput systems such as ChIP-chip, ChIP-seq and RNA-seq systems. Even using these modern techniques, however, it is in principle impossible to obtain the whole set of binding sites for both RNAP and TFs because of the competition with other DNA-binding proteins in binding to DNA targets [8,9,24] [note that E. coli contains more than 500 DNA-binding proteins [77], and because the binding of RNAP and TFs often depends on the supporting factors for binding to targets [8,9,25].
The computational approaches in silico have also been employed to identify the target binding sequences of RNAP and TFs, relying on the consensus sequences predicted based on the known target sequences listed in the databases such as RegulonDB [20,21] and EcoCyc [22,23] (Table 2) (Details of the promoter list and the evidence are in Supplemental Information: S1 Table for RpoS; S2 Table for RpoH; S3 Table for RpoF; and S4 Table for RpoE). In particular, more than 80% of RpoE-dependent promoters were predicted in silico (Table 5; and S4 Table). The consensus sequences, however, often include the inaccurate non-target sequences due to the lack of experiments for confirmation or some regulators recognize wide-varieties of the binding sequences [8,9,27,78]. Another serious problem associated with in vivo approaches is the difference in genetic background of E. coli strains used. Up to the present, the complete genome sequence has been determined for more than 1,000 different E. coli strains, allowing the prediction of about 3,000 core genes for all strains but at least one third of the total genes on the E. coli genome are different among the hitherto sequenced E. coli genome [79]. The difference in genetic background exists even in the RNAP and TF genes and between not only different strains but also different stocks of the same E. coli strain. For instance, the difference in the gene encoding the stationary-phase sigma RpoS was first identified between laboratory stocks of a single and the same E. coli K-12 W3110 strain [80]. The widely used databases such as RegulonDB [20,21] and EcoCyc [22,23] include huge collections of useful data of transcription in vivo, but care should be taken to use these data for theoretical prediction of transcription regulation, in particular, as to the bacterial strains and culture conditions used in each experiment.
In this study, we performed the SELEX screening for the constitutive promoters that are recognized in vitro by four minor sigma factors, RpoS, RpoH, RpoF and RpoE, but in the absence of repressors, activators and other DNA-binding proteins. It should be noted that all the proteins and promoters used in this study are prepared from a single and the same E. coli K-12 W3110. Here we also determined the intracellular concentrations of all seven sigma factors in both growing and stationary-phase cells of E. coli K-12 W3110. These data altogether will be used for our ultimate purpose of the prediction of genome expression under a given condition. The list of constitutive promoters for the minor sigma factors will be deposited into TEC database (Transcriptional profile of Escherichia coli database: https://shigen.nig.ac.jp/ecoli/tec/top/) [9].
Materials and methods
Bacterial strains and plasmids
E. coli K12 W3350 type-A containing the full-set of seven sigma factors [80] was used for purification of RNA polymerase and the template DNA for Genomic SELEX screening of RpoS, RpoH, RpoF and RpoE promoters. E. coli BL21(DE3) was used for the expression and purification of sigma and core enzyme subunit proteins. Expression plasmids for the core enzyme subunits (pRpoA, pRpoB and pRpoC) and all seven sigma subunits (pRpoD, pRpoN, pRpoS, pRpoH, pRpoF, pRpoE and pFecI) were constructed by ligating the respective coding sequences, which were prepared by PCR amplification of the E. coli K12 W3350 type-A genome DNA as template, into pET21 expression vector essentially according to the standard procedure used for expression of all sigma and all transcription factors in this laboratory [74,81].
Purification of core RNA polymerase
RNAP was purified from log-phase cells of E. coli K-12 W3350 by the standard procedure [29]. Separation of the native core from holoenzymes was performed by passing the purified RNAP through P11-phosphocellulose column in the presence of 50% glycerol. To remove trace amounts of the core enzyme-associated sigma factors, the purified RNAP in the storage buffer containing 50% glycerol was dialyzed against the same buffer but containing 5% glycerol and fractionated by P11-phosphocellulose column chromatography in the presence of 5% glycerol. The level of remaining sigma factors was less than 0.1%, if any, as checked of SDS-PAGE gels by both protein-staining with a silver reagent and immuno-staining with antibodies against each of seven sigma factors.
Purification of core and sigma subunits
The core enzyme subunits (RpoA, RpoB, RpoC and RpoZ) were expressed using the respective expression plasmids and purified by two cycles of column chromatography through DEAE (DE52) and P11-phosphocellulose [29]. Sigma subunits were expressed and purified by ion-exchange column chromatography through DE52 and P11 followed by Sephacryl S-300 gel filtration column. The purified sigma and core subunit proteins were more than 99% pure as judged by both protein-staining and immuno-staining of SDS-PAGE gels.
Preparation of antibodies
Antibodies against sigma factors and core enzyme subunits were produced in rabbits by injecting purified sigma proteins [75,76]. Antibodies against each RNA polymerase protein were produced in two rabbits, and after examination of antibody activity using immune-blot analysis, the batch of higher activity was used in this study. Anti-RpoD, anti-RpoS, anti-RpoN, anti-RpoH, anti-RpoF, anti-RpoE, anti-FecI and anti-RpoC used in this study did not cross-react with each other. These antibodies were produced in the Nippon Institute for Biological Science (One, Tokyo) and the Animal Laboratory of Mitsubishi Chemical Medience Co. (Uto, Kumamoto, Japan).
Genomic SELEX screening of RNA polymerase holoenzyme binding sequences
The Genomic SELEX screening was carried out under the standard procedure [26,27]. This method was developed by improvement of the original SELEX methods [30–32]. A mixture of DNA fragments of the E. coli K-12 W3110 genome was prepared after sonication of purified genome DNA, and cloned into a multi-copy plasmid pBR322 at EcoRV site. In each SELEX screening, the DNA mixture was regenerated by PCR using a pair of primers with the flanking sequences of pBR322 EcoRV. For SELEX screening, 5 pmol of the mixture of DNA fragments and 10 pmol of RNA polymerase holoenzyme were mixed in a binding buffer (10 mM Tris-HCl, pH 7.8 at 4°C, 3 mM magnesium acetate, 150 mM NaCl, and 1.25 mg/ml bovine serum albumin) and incubated for 30 min at 37°C. The DNA-RNA polymerase mixture was treated with anti-RpoC antibody and DNA fragments recovered from the complexes were PCR-amplified and subjected to next cycle of SELEX for enrichment of RNA polymerase-bound DNA fragments.
For SELEX-chip analysis, DNA samples were isolated from the DNA-protein complexes at the final state of SELEX, PCR-amplified and labeled with Cy5 while the original DNA library was labeled with Cy3. The fluorescent labeled DNA mixtures were hybridized to a DNA microarray consisting of 43,450 species of 60 b-long DNA probe, which are designed to cover the entire E. coli K-12 MG1655 genome at 105 bp interval (Oxford Gene Technology, Oxford, UK) [14]. The fluorescent intensity of test sample at each probe was normalized with that of the corresponding peak of original library. After normalization of each pattern, the Cy5/Cy3 ratio was measured and plotted along the E. coli K-12 MG1655 genome. The gene organization is almost identical between two well-characterized E. coli K-12 strains except for a long-range inversion between the rrnD and rrnE operons.
Immuno-blot analysis for determination of sigma levels
For the measurement of sigma factors in E. coli K-12 W3110, a quantitative Western blot analysis was employed with the anti-sigma antibodies as employed in the previous studies [66,75,76]. In brief, cell lysates were treated with a SDS (sodium dodecyl sulfate) sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 1% 2-mercaptoethanol, 10% glycerol, and 0.025% bromophenol blue) and separated on SDS–7.5 or 10% polyacrylamide gels. Proteins in gels were directly electro-blotted onto polyvinylidene difluoride membranes (Nippon Genetics). Blots were blocked overnight at 48C in 3% BSA in PBS (phosphate-buffered saline), probed with the polyclonal antibodies against each sigma factor, washed with 0.5% Tween 20 in PBS, and incubated with goat anti-rabbit immunoglobulin G conjugated with hydroxyperoxidase (Cappel). The blots were developed with 3,3’-diaminobenzidine tetrahydrochloride (Dojindo). Staining intensity was measured with a PDI image analyzer system equipped with a white light scanner. The standard curve for the calculation of each sigma level was prepared from the immuno-blot patterns of increasing concentrations of each sigma factor. Under the standard Western-blot conditions herein employed, the linearity was detected over a 10-fold range at least between 2 and 20 ng sigma proteins. The determination of test sigma proteins subunits was first performed using several different volumes of the cell lysates. Using the optimum volumes of cell lysates to give the sigma concentrations within the linear range of standard curves, we finally repeated the determination of individual sigma factors.
Supporting information
Promoters listed in RegulonDB are classified into those not identified as the constitutive promoters (A) and the constitutive promoters identified by SELEX screening (B). Evidence for each promoter are as described in RegulonDB (see Table 5): (Group-A) Promoters were experimentally identified by using HTTIM (high-throughput transcription initiation mapping), TIM (transcription initiation mapping), FP (footprinting), or IDA (inferred by direct promoter assay; (Class-B) Promoters were predicted based on AIPP (automated inference of promoter position), ICWHO (inferred computationally without human oversight), HIPP (human inference of promoter position), NTAS (non-traceable author statement), TASES (traceable author statement to experimental support), TAS (traceable author statement), IMP (inferred from mutant) or IEP (inferred from expression pattern).
(PDF)
Promoters listed in RegulonDB are classified into those not identified as the constitutive promoters (A) and the constitutive promoters identified by SELEX screening (B). Evidence for each promoter are as described in S1 Table.
(PDF)
Promoters listed in RegulonDB are classified into those not identified as the constitutive promoters (A) and the constitutive promoters identified by SELEX screening (B). Evidence for each promoter are as described in S1 Table.
(PDF)
Promoters listed in RegulonDB are classified into those not identified as the constitutive promoters (A) and the constitutive promoters identified by SELEX screening (B). Evidence for each promoter are as described in S1 Table.
(PDF)
Acknowledgments
We thank Nobuyuki Fujita, Ayako Kori and Kayoko Yamada for expression and purification of RNAP proteins.
Data Availability
The data described in this report has been deposited to TEC (Transcription Profile of Escherchia coli) database (https://shigen.nig.ac.jp/ecoli/tec/). The data of each minor sigma will be shown by setting the gene symbol, rpoS, rpoH, rpoF or rpoE, respectively [https://shigen.nig.ac.jp/ecoli/tec/tfmap]; for details, follow the instructions.
Funding Statement
This work was supported by National Institute of Genetics to YY; MEXT Grants-in-Aid for Scientific Research (A) (21241047), (B) (18310133), and (C) (25430173) to AI; MEXT Grant-in-Aid for Young Scientists (B) (24710214) to TS; Research Fund from IFO (Institute for Fermentation, Osaka) to TS; funding from the MEXT Cooperative Research Program of Network Joint Research Center for Materials and Devices to AI and KT; and funding to AI and TS from the MEXT-Supported Program for the Strategic Research Foundation at Private Universities 208-2012 (S0801037) to Hosei University.
References
- 1.Kawakami K., Saitoh T. and Ishihama A. (1979) Biosynthesis of RNA polymerase in Escherichia coli: Growth-dependent variations in the synthesis rate, content and distribution of RNA polymerase. Mol. Gen. Genet., 174, 107–116. [DOI] [PubMed] [Google Scholar]
- 2.Ishihama A. (1999) Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells, 4, 135–143. [DOI] [PubMed] [Google Scholar]
- 3.Ishihama A. (2000) Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol., 54, 499–518. doi: 10.1146/annurev.micro.54.1.499 [DOI] [PubMed] [Google Scholar]
- 4.Ishihama A. (2010) Prokaryotic genome regulation: Multi-factor promoters, multi-target regulators and hierarchic networks. FEMS Microbiol. Rev., 34, 628–645. doi: 10.1111/j.1574-6976.2010.00227.x [DOI] [PubMed] [Google Scholar]
- 5.Ishihama A. (2012) Procaryotic genome regulation: A revolutionary paradigm. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci., 88, 485–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruber T.M. and Gross C.A. (2003) Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol., 57, 441–466. doi: 10.1146/annurev.micro.57.030502.090913 [DOI] [PubMed] [Google Scholar]
- 7.Perez-Rueda E. and Collado-Vides J. (2000) The repertoire of DNA-binding transcription regulators in Escherichia coli K-12. Nucleic Acids Res., 28, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishihama A. (2016) A Revolutionary Paradigm of Bacterial Genome Regulation In: Stress and Environmental Control of Gene Expression in Bacteria. Ed: De Bruijin Frans J. (John Wiley & Sons; ), Chapter 2–3, pp. 23–35. [Google Scholar]
- 9.Ishihama A., Shimada T. and Yamazaki Y. (2016) Transcription profile of Escherichia coli: Genomic SELEX search for regulatory targets of transcription factors. Nucleic Acids Res., 44, 2058–2074 doi: 10.1093/nar/gkw051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richmond C.S., Glasner J.D., Mau R., Jin H and Blattner R. (1999) Genome-wide expression profiling in Escherichia coli. Nucleic Acids Res., 27, 3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oshima T., Aiba H., Masuda Y., Kanaya S., Sugiura M., Wanner B.L., Mori H. and Mizuno T. (2002) Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol., 46, 281–291. [DOI] [PubMed] [Google Scholar]
- 12.Herring C.D., Raffaelle M., Allen T.E., Kanin E.I., Landick R., Ansari A.Z. and Palsson B.O. (2005) Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol., 187, 6166–6174. doi: 10.1128/JB.187.17.6166-6174.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grainger D.C., Hurd D., Goldberg M.D. and Busby S.J.W. (2006) Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res., 34, 4642–4652. doi: 10.1093/nar/gkl542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimada T., Ishihama A., Busby S.J.W. and Grainger D.C. (2008) The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res., 36, 3950–3955. doi: 10.1093/nar/gkn339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas B.J., Chin M., Nusbaum C., Birren B.W. and Livny J. (2012) How deep is deep enough for RNA-seq profiling of bacterial transcriptomes? BMC Genomics 13: 734 doi: 10.1186/1471-2164-13-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway T., Creecy J.P., Maddox S.M., Grissom J.E., Conkie T.L., Shadid T.M., Teramoto J., San Miquel P., Shimada T., Ishihama A., Mori H. and Wanner B.L. (2014) Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. mBio, 5: e1442–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peano C., Wolf J., Demol J., Rossi E., Petiti L., De Bellis G., Geiselmann J., Egli T., Lacour S. and Landini P. (2015) Characterization of the Escherichia coli σ(S) core regulon by Chromatin Immunoprecipitation-sequencing (ChIP-seq) analysis. Sci. Rep., 5: 10469 doi: 10.1038/srep10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panyukov V.V. and Ozoline O.N. (2013) Promoters of Escherichia coli versus promoter islands: function and structure comparison. PLoS ONE, 8: e62601 doi: 10.1371/journal.pone.0062601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomason M.K., Bischler T., Eisenbart S.K., Forstner K.U., Zhang A., Neselt K., Sharma C.M. and Stortz G. (2015) Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol., 197, 18–28. doi: 10.1128/JB.02096-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salgado H., Gama-Castro S., Martinez-Flores I., Diaz-Peredo E., Sanchez-Solano F., Peralta-Gil M., Garcita-Alonso D., Jimenez-Jacinto V., Santos-Zavaleta A., Bonavides-Martinez C. and Collado-Vides J. (2006) RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions. Nucleic Acids Res., 32, D303–D306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salgado H., Peralta-Gil M., Gama-Castro S., Santos-Zavaleta A., Muniz-Raccado L., Garcia-Sotelo J.S., Weiss V., Solano-Lira H., Martinez-Flores I., Medina-Revira A., Salgado-Osrio G., Porron-Sotelo L., Huerta A.M., Bonavides-Martinez C., Balderas-Martinez Y.I., Pannier L., Olvera M., Labastida A., Jimenez-Jacinto V., Vega-Alvarado L., del Moral-Chavez V., Hernandez-Alvarez A., Morett E. and Collado-Vides J. (2013) RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards an dmore. Nucleic Acids Res., 41, D203–D213. doi: 10.1093/nar/gks1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham S., Paley S., Paulsen I.T., Peralta-Gil M. and Karp P.D. (2005) EcoCyc: a compehensive database resource for Escherichia coli. Nucleic Acids Res., 44, D334–D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karp P.D., Weaver D., Paley S., Fulcher C., Kubo A., Kothari A., Krummenacker M., Subharaveti P., Weerasinghe D., Gama-Castro S., Huerta A.M., Muniz-Rascado L., Bonavides-Martinez C., Weiss V, Peralta-Gil M., Santos-Zavaleta A., Schrder I., Mackie A., Gunsalus R., Collado-Vides J., Kaseler I.M. and Paulsen I. (2014) The EcoCyc database. EcoSal Plus [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimada T., Bridier A., Briandet R. and Ishihama A. (2012) Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with universal silencer H-NS. Mol. Microbiol., 82, 37–397. [DOI] [PubMed] [Google Scholar]
- 25.Ogasawara H., Yamada K., Kori A., Yamamoto K. and Ishihama A. (2010) Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology, 156, 2470–2483. doi: 10.1099/mic.0.039131-0 [DOI] [PubMed] [Google Scholar]
- 26.Shimada T., Fujita N., Maeda M. and Ishihama A. (2005) Systematic search for the Cra-binding promoters using genomic SELEX systems. Genes Cells, 10, 907–918. doi: 10.1111/j.1365-2443.2005.00888.x [DOI] [PubMed] [Google Scholar]
- 27.Shimada T., Yamazaki Y., Tanaka K. and Ishihama A. (2014) The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PLoS ONE 9: e90447 doi: 10.1371/journal.pone.0090447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochs M., Veitinger S., Kim I., Welz D., Angerer A. and Braun V. (1995) Regulation of citrate-dependent iron transport of Escherichia coli: fecR is required for transcription activation by FecI. Mol. Microbiol., 15, 119–132. [DOI] [PubMed] [Google Scholar]
- 29.Fujita N. and Ishihama A. (1996) Reconstitution of RNA polymerase. Meth Enzymol 273: RNA Polymerase and Associated Factors, Part A, Adhya, S. ed., pp. 121–130, Academic Press, New York, 1996. [DOI] [PubMed] [Google Scholar]
- 30.Singer B.S., Shtatland T., Brown D. and Gold L. (1997) Libraries for genomic SELEX. Nucleic Acids Res., 25, 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellington A.D. and Szostak J.W. (1990) In vitro selection of DNA molecules that bind specific ligands. Nature, 346, 818–822. doi: 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- 32.Tuerk C. and Gold L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 33.Ishihama A. (1997) Adaptation of gene expression in stationary phase bacteria. Curr. Opn. Genet. Devel., 7, 582–588. [DOI] [PubMed] [Google Scholar]
- 34.Loewen P.C. and Hengge-Aronis R. (1994) The role of the sigma factor S (KatF) in bacterial global regulation. Annu. Rev. Microbiol., 48, 53–80. doi: 10.1146/annurev.mi.48.100194.000413 [DOI] [PubMed] [Google Scholar]
- 35.Collet A., Cosette P., Beloin C., Ghigo J.M., Rihouey C., Lerouge P., Junter G.A. and Jouenne T. (2008) Impact of rpoS deletion on the proteome of Escherichia coli grown planktonically and as biofilm. J. Proteome Res., 7, 4659–4669. doi: 10.1021/pr8001723 [DOI] [PubMed] [Google Scholar]
- 36.Lacour S. and Landini P. (2004) Sigma S-dependent gene expression at the onset of stataionary phase in Escherichia coli: function of sigma S-dependent genes and identification of their promoter sequences. J. Bacteriol., 186, 7186–7195. doi: 10.1128/JB.186.21.7186-7195.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patten C.L., Kirchhof M.G., Schertzberg M.R., Morton R.A. and Schellhorn H.E. (2004) Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics, 272, 580–591. doi: 10.1007/s00438-004-1089-2 [DOI] [PubMed] [Google Scholar]
- 38.Weber H., Polen T., Heuveling J., Wendisch V.F. and Hengge R. (2005) Genomewide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol., 187, 1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimada T., Makinoshima H., Ogawa N., Maeda M. and Ishihama A. (2004) Classification and strength measurement of stationary-phase promoters by use of a newly developed promoter cloning vector. J. Bacteriol. 186, 7112–7122. doi: 10.1128/JB.186.21.7112-7122.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong S.H., Wang X., O’Connor H.F., Benedik M.J. and Wood T.K. (2012) Bacterial persistence increases as environmental fitness decreases. Microbial. Biotechnol., 5, 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glover W.A., Yang Y. and Zhang Y. (2009) Insights into the molecular basis of L-form formation and survival in Escherichia coli. PLoS ONE 4:e7316 doi: 10.1371/journal.pone.0007316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yura T., Nagai H. and Mori H. (1993) Regulation of the heat-shock response in bacteria. Annu. Rev. Microbiol., 47, 321–350. doi: 10.1146/annurev.mi.47.100193.001541 [DOI] [PubMed] [Google Scholar]
- 43.Guisbert E., Yura T., Rhodius V.A. and Gross C.A. (2008) Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microb. Molec. Biol. Rev., 72, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemaux P.G., Herendeen S.L., Bloch P.L. and Neidhardt F.C. (1978) Transient rates of synthesis of individual polypeptides in E. coli following temperature shifts. Cell, 13, 427434. [DOI] [PubMed] [Google Scholar]
- 45.Yamamori T., Ito K. and Yura T. (1978) Transient regulation of protein synthesis in Escherichia coli upon shift-up of growth temperature. J. Bacteriol., 134, 1133–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung S.E. and Blattner F.R. (1993) Characterization of twenty-six new heat shock genes of Escherichia coli. J. Bacteriol., 175, 5242–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao K., Liu M. and Burgess R.R. (2005) The global transcriptional response of Escherichia coli to induced sigma 32 protein involves sigma 32 regulon activation followed by inactivation and degradation of sigma 32 in vivo. J. Biol. Chem., 280,17758–17768. doi: 10.1074/jbc.M500393200 [DOI] [PubMed] [Google Scholar]
- 48.Fujita N., Nomura T. and Ishihama A. (1987) Promoter selectivity of Escherichia coli RNA polymerase: Purification and properties of holoenzyme containing the heat-shock sigma subunit. J. Biol. Chem., 262, 1855–1859. [PubMed] [Google Scholar]
- 49.Nonaka G., Blankschien M., Herman C., Gross C.A. and Rhodius V.A. (2006) Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev., 20, 1776–1789. doi: 10.1101/gad.1428206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herman C., Thevenet D., D’Ari R. and Bouloc P. (1995) Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. USA 92: 3516–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macnab R.M. (1992) Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26, 131–158. doi: 10.1146/annurev.ge.26.120192.001023 [DOI] [PubMed] [Google Scholar]
- 52.Kutsukake K. and lino T. (1994) Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol., 176, 3598–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macnab R.M. (2003) How bacteria assemble flagella. Annu. Rev. Microbiol., 57, 77–100. doi: 10.1146/annurev.micro.57.030502.090832 [DOI] [PubMed] [Google Scholar]
- 54.Ohnishi K., Kutsukake K., Suzuki H. and Iino T. (1990) Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet., 221, 139–147. [DOI] [PubMed] [Google Scholar]
- 55.Ide N. and Kutsukake K. (1997) Identification of a novel Escherichia coli gene whose expression is dependent on the flagellum-specific sigma factor, FliA, but dispensable for motility development. Gene, 199, 19–23. [DOI] [PubMed] [Google Scholar]
- 56.Arnosti D.N. and Chamberlin M.J. (1989) Secondary s factor controls transcription of flagella and chemotaxis genes in Escherichia coli. Proc. Natl. Acad. Sci. USA, 86, 830–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fitzgerald D.M., Bonocora R.P. and Wade J.T. (2014) Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet., 10: e1004649 doi: 10.1371/journal.pgen.1004649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X. and Matsumura P. (1996) Differential regulation of multiple overlapping promoters in flagellar class II operons in Escherichia coli. Mol. Microbiol. 21, 613–620. [DOI] [PubMed] [Google Scholar]
- 59.Kundu T.K., Kusano S. and Ishihama A. (1997) Promoter selectivity of Escherichia coli RNA polymerase sigma-F holoenzyme involved in transcription of flagellar and chemotaxis genes. J. Bacteriol., 179, 4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barembruch C. and Hengge R. (2007) Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol., 65, 76–89. doi: 10.1111/j.1365-2958.2007.05770.x [DOI] [PubMed] [Google Scholar]
- 61.Ravio T.L. and Silhavy T.J. (2001) Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol., 55, 5911–624. [DOI] [PubMed] [Google Scholar]
- 62.De Las Penas A., Connolly L. and Gross C.A. (1997) σE is an essential sigma factor in Escherichia coli. J, Bacteriol., 179, 6862–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dartigalongue C., Missiakas D. and Raina S. (2001) Characterization of the Escherichia coli σE regulon. J. Biol. Chem., 276, 20666–20875. [DOI] [PubMed] [Google Scholar]
- 64.Rezuchova B., Miticka H., Homerova D., Roberts M. and Kormanec J. (2003) New members of the Escherichia coli σE regulon identified by a two-plasmid system. FEMS Microb. Lett., 225, 1–7. [DOI] [PubMed] [Google Scholar]
- 65.Missiakas D., Betton J.M., Mayer C., Georgopolos C. and Raina S. (1997) Modulation of the Escherichia cols σE (RpoE) heat shock transcription factor activity by the RseA, RseB, and RseC protein. Mol. Microbiol., 24, 335–372. [DOI] [PubMed] [Google Scholar]
- 66.Maeda H., Jishage M., Nomura T., Fujita N. and Ishihama A. (2000) Two cytoplasmic function sigma subunits, σE and σFecI, of Escherichia coli: Promoter selectivity and intracellular levels. J. Bacteriol., 182, 1181–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johansen J., Rasmussen A.A., Overgaard M. and Valentin-Hansen P. (2006) Conserved small non-coding RNAs that belong to the sigma-E regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol., 364, 1–8. doi: 10.1016/j.jmb.2006.09.004 [DOI] [PubMed] [Google Scholar]
- 68.Rasmussen A.A., Eriksen M., Gilany K., Udesen C., Franch T., Petersen C. and Valentin-Hansen P. (2005) Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol., 58, 1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x [DOI] [PubMed] [Google Scholar]
- 69.Udekwu K.I., Darfeuille F., Vogel J., Reimegard J., Holmqvist E. and Wagner E.G. (2005) Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev., 19, 2355–2366. doi: 10.1101/gad.354405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gogol E.B., Rhodius V.A., Papenfort K., Vogel J. and Gross C.A. (2011) Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. USA, 108, 12875–12880. doi: 10.1073/pnas.1109379108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagamitsu H., Murata M., Kosaka T., Kawaguchi J., Mori H. and Yamada M. (2013) Crucial roles of MicA and RybB as vital factors for s-dependent cell lysis in Escherichia coli long-term stationary phase. J. Mol. Microbiol. Biotechnol., 23, 227–232. doi: 10.1159/000350370 [DOI] [PubMed] [Google Scholar]
- 72.Wang Q.P. and Kaguni J.M. (1989) A novel sigma factor is involved in expression of the rpoH gene of Escherichia coli. J. Bacteriol., 171, 4248–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shimada T., Yamazaki K. and Ishihama A. (2013) Novel regulator PgrR for switch control of peptidoglycan recycling in Escherichia coli. Genes Cells, 18, 123–134. doi: 10.1111/gtc.12026 [DOI] [PubMed] [Google Scholar]
- 74.Maeda H., Fujita N. and Ishihama A. (2000) Competition among seven Escherichia coli s subunits: relative binding affinity to the core RNA polymerase. Nucleic Acids Res., 28, 3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jishage M. and Ishihama A. (1995) Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: Intracellular levels of σ70 and σ38. J. Bacteriol., 177, 6832–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jishage M., Iwata A., Ueda S. and Ishihama A. (1996) Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: Intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol., 178, 5447–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ishihama A. (1999) The Nucleoid: an Overview. EcoSal Plus, Chapter 2.6, Edited by Bëák A., Curtiss R., Kaper J. B., Karp P. D., Neidhardt F. C., Nystrëë T., Slauch J. M., Squires C. L., and Ussery D., ASM Press, Washington, DC [Google Scholar]
- 78.Shimada T., Fujita N., Maeda M. and Ishihama A. (2005) Systematic search for the Cra-binding promoters using genomic SELEX. Genes Cells 10, 907–918. doi: 10.1111/j.1365-2443.2005.00888.x [DOI] [PubMed] [Google Scholar]
- 79.Land M., Hauser L., Jun S.R., Nookaew I., Leuze M.R., Ahn T.H., Karpinets T., Lund O., Kora G., Wassenaar T., Poudel S. and Ussery D.W. (2015) Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics 15, 141–161. doi: 10.1007/s10142-015-0433-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jishage M. and Ishihama A. (1997) Variation in RNA polymerase sigma subunit composition within different strains of Escherichia coli W3110. J. Bacteriol., 179, 959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Igarashi K. and Ishihama A. (1991) Bipartite functional map of the E. coli RNA polymerase α subunit: Involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell, 65,1015–1022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Promoters listed in RegulonDB are classified into those not identified as the constitutive promoters (A) and the constitutive promoters identified by SELEX screening (B). Evidence for each promoter are as described in RegulonDB (see Table 5): (Group-A) Promoters were experimentally identified by using HTTIM (high-throughput transcription initiation mapping), TIM (transcription initiation mapping), FP (footprinting), or IDA (inferred by direct promoter assay; (Class-B) Promoters were predicted based on AIPP (automated inference of promoter position), ICWHO (inferred computationally without human oversight), HIPP (human inference of promoter position), NTAS (non-traceable author statement), TASES (traceable author statement to experimental support), TAS (traceable author statement), IMP (inferred from mutant) or IEP (inferred from expression pattern).
(PDF)
Promoters listed in RegulonDB are classified into those not identified as the constitutive promoters (A) and the constitutive promoters identified by SELEX screening (B). Evidence for each promoter are as described in S1 Table.
(PDF)
Promoters listed in RegulonDB are classified into those not identified as the constitutive promoters (A) and the constitutive promoters identified by SELEX screening (B). Evidence for each promoter are as described in S1 Table.
(PDF)
Promoters listed in RegulonDB are classified into those not identified as the constitutive promoters (A) and the constitutive promoters identified by SELEX screening (B). Evidence for each promoter are as described in S1 Table.
(PDF)
Data Availability Statement
The data described in this report has been deposited to TEC (Transcription Profile of Escherchia coli) database (https://shigen.nig.ac.jp/ecoli/tec/). The data of each minor sigma will be shown by setting the gene symbol, rpoS, rpoH, rpoF or rpoE, respectively [https://shigen.nig.ac.jp/ecoli/tec/tfmap]; for details, follow the instructions.