Abstract
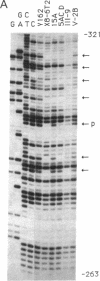
The 5' region of the gene encoding human X chromosome-linked phosphoglycerate kinase 1 (PGK1) is a promoter-containing CpG island known to be methylated at 119 of 121 CpG dinucleotides in a 450-base-pair region on the inactive human X chromosome in the hamster-human cell line X8-6T2. Here we report the use of polymerase chain reaction-aided genomic sequencing to determine the complete methylation pattern of this region in clones derived from X8-6T2 cells after treatment with the methylation inhibitor 5-azacytidine. We find (i) a clone showing full expression of human phosphoglycerate kinase is fully unmethylated in this region; (ii) clones not expressing human phosphoglycerate kinase remain methylated at approximately 50% of CpG sites, with a pattern of interspersed methylated (M) and unmethylated (U) sites different for each clone; (iii) singles, defined as M-U-M or U-M-U, are common; and (iv) a few CpG sites are partially methylated. The data are interpreted according to a model of multiple, autonomous CpG sites, and estimates are made for two key parameters, maintenance efficiency (Em approximately 99.9% per site per generation) and de novo methylation efficiency (Ed approximately 5%). These parameter values and the hypothesis that several independent sites must be unmethylated for transcription can explain the stable maintenance of X chromosome inactivation. We also consider how the active region is kept free of methylation and suggest that transcription inhibits methylation by decreasing Em so that methylation cannot be maintained. Thus, multiple CpG sites, independent with respect to a dynamic methylation system, can stabilize two alternative states of methylation and transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot F., Acs G., Christman J. K. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2'-deoxycytidine. J Biol Chem. 1982 Feb 25;257(4):2041–2048. [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Drahovský D., Morris N. R. Mechanism of action of rat liver DNA methylase. I. Interaction with double-stranded methyl-acceptor DNA. J Mol Biol. 1971 May 14;57(3):475–489. doi: 10.1016/0022-2836(71)90104-5. [DOI] [PubMed] [Google Scholar]
- Flatau E., Gonzales F. A., Michalowsky L. A., Jones P. A. DNA methylation in 5-aza-2'-deoxycytidine-resistant variants of C3H 10T1/2 C18 cells. Mol Cell Biol. 1984 Oct;4(10):2098–2102. doi: 10.1128/mcb.4.10.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S. G., Chapman V. M. Mechanisms of X-chromosome regulation. Annu Rev Genet. 1988;22:199–233. doi: 10.1146/annurev.ge.22.120188.001215. [DOI] [PubMed] [Google Scholar]
- Grant S. G., Worton R. G. Differential activation of the hprt gene on the inactive X chromosome in primary and transformed Chinese hamster cells. Mol Cell Biol. 1989 Apr;9(4):1635–1641. doi: 10.1128/mcb.9.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Ellis N. A., Gartler S. M. Demethylation of specific sites in the 5' region of the inactive X-linked human phosphoglycerate kinase gene correlates with the appearance of nuclease sensitivity and gene expression. Mol Cell Biol. 1988 Nov;8(11):4692–4699. doi: 10.1128/mcb.8.11.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R. S., Gartler S. M. 5-Azacytidine-induced reactivation of the human X chromosome-linked PGK1 gene is associated with a large region of cytosine demethylation in the 5' CpG island. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4174–4178. doi: 10.1073/pnas.87.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Holliday R. The inheritance of epigenetic defects. Science. 1987 Oct 9;238(4824):163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Kaslow D. C., Migeon B. R. DNA methylation stabilizes X chromosome inactivation in eutherians but not in marsupials: evidence for multistep maintenance of mammalian X dosage compensation. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6210–6214. doi: 10.1073/pnas.84.17.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith D. H., Singer-Sam J., Riggs A. D. Active X chromosome DNA is unmethylated at eight CCGG sites clustered in a guanine-plus-cytosine-rich island at the 5' end of the gene for phosphoglycerate kinase. Mol Cell Biol. 1986 Nov;6(11):4122–4125. doi: 10.1128/mcb.6.11.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey J. R., Riggs A. D. Determinator-inhibitor pairs as a mechanism for threshold setting in development: a possible function for pseudogenes. Proc Natl Acad Sci U S A. 1986 Feb;83(3):679–683. doi: 10.1073/pnas.83.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means A. L., Farnham P. J. Transcription initiation from the dihydrofolate reductase promoter is positioned by HIP1 binding at the initiation site. Mol Cell Biol. 1990 Feb;10(2):653–661. doi: 10.1128/mcb.10.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon B. R., Jan de Beur S., Axelman J. Frequent derepression of G6PD and HPRT on the marsupial inactive X chromosome associated with cell proliferation in vitro. Exp Cell Res. 1989 Jun;182(2):597–609. doi: 10.1016/0014-4827(89)90262-0. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
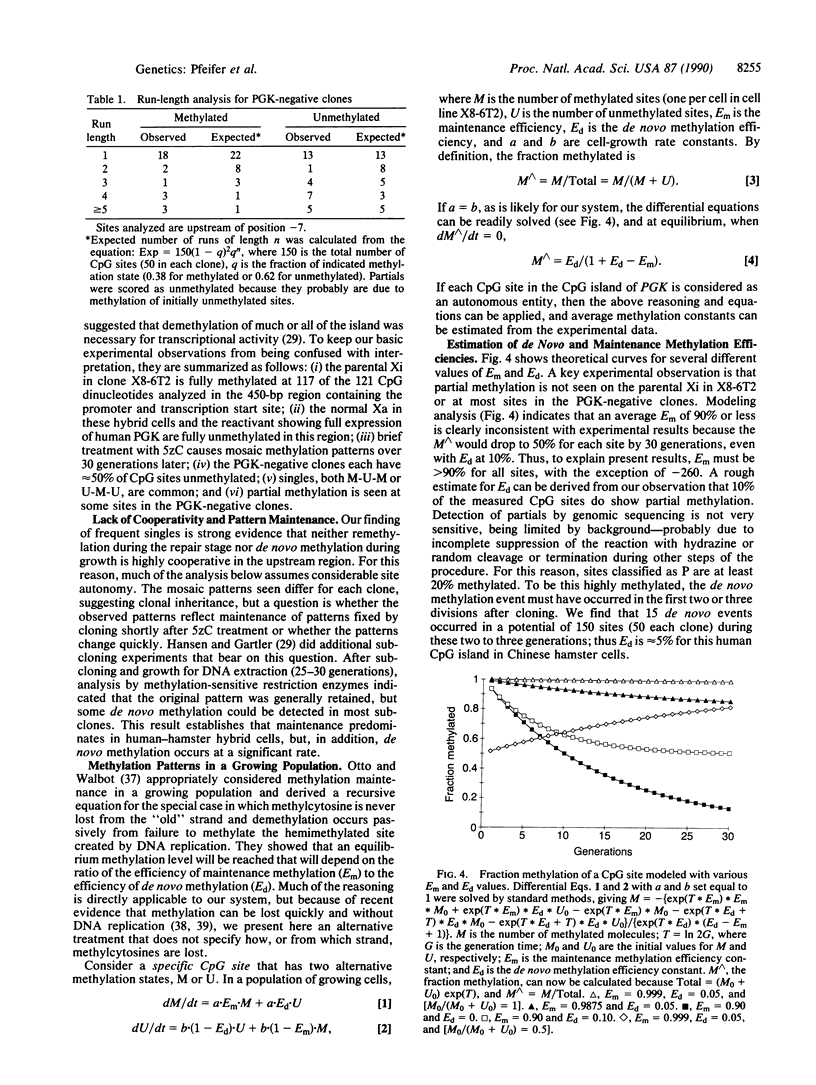
- Otto S. P., Walbot V. DNA methylation in eukaryotes: kinetics of demethylation and de novo methylation during the life cycle. Genetics. 1990 Feb;124(2):429–437. doi: 10.1093/genetics/124.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Grünwald S., Boehm T. L., Drahovsky D. Isolation and characterization of DNA cytosine 5-methyltransferase from human placenta. Biochim Biophys Acta. 1983 Aug 2;740(3):323–330. doi: 10.1016/0167-4781(83)90141-0. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Tanguay R. L., Steigerwald S. D., Riggs A. D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990 Aug;4(8):1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Razin A., Szyf M., Kafri T., Roll M., Giloh H., Scarpa S., Carotti D., Cantoni G. L. Replacement of 5-methylcytosine by cytosine: a possible mechanism for transient DNA demethylation during differentiation. Proc Natl Acad Sci U S A. 1986 May;83(9):2827–2831. doi: 10.1073/pnas.83.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D. DNA methylation and cell memory. Cell Biophys. 1989 Aug-Oct;15(1-2):1–13. doi: 10.1007/BF02991574. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Jones P. A. 5-methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Saluz H. P., Jiricny J., Jost J. P. Genomic sequencing reveals a positive correlation between the kinetics of strand-specific DNA demethylation of the overlapping estradiol/glucocorticoid-receptor binding sites and the rate of avian vitellogenin mRNA synthesis. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess E., Tomassetti A., Hernaiz-Driever P., Pfeifer G. P. Structure of mouse DNA (cytosine-5-)-methyltransferase. Eur J Biochem. 1988 Oct 15;177(1):29–34. doi: 10.1111/j.1432-1033.1988.tb14341.x. [DOI] [PubMed] [Google Scholar]
- Steigerwald S. D., Pfeifer G. P., Riggs A. D. Ligation-mediated PCR improves the sensitivity of methylation analysis by restriction enzymes and detection of specific DNA strand breaks. Nucleic Acids Res. 1990 Mar 25;18(6):1435–1439. doi: 10.1093/nar/18.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J Mol Biol. 1982 Dec 15;162(3):679–692. doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- Toniolo D., Martini G., Migeon B. R., Dono R. Expression of the G6PD locus on the human X chromosome is associated with demethylation of three CpG islands within 100 kb of DNA. EMBO J. 1988 Feb;7(2):401–406. doi: 10.1002/j.1460-2075.1988.tb02827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih F., Stewart A. F., Schütz G. A novel and rapid method to generate single stranded DNA probes for genomic footprinting. Nucleic Acids Res. 1988 Feb 25;16(4):1628–1628. doi: 10.1093/nar/16.4.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S. F., Jolly D. J., Lunnen K. D., Friedmann T., Migeon B. R. Methylation of the hypoxanthine phosphoribosyltransferase locus on the human X chromosome: implications for X-chromosome inactivation. Proc Natl Acad Sci U S A. 1984 May;81(9):2806–2810. doi: 10.1073/pnas.81.9.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen P. H., Patel P., Chinault A. C., Mohandas T., Shapiro L. J. Differential methylation of hypoxanthine phosphoribosyltransferase genes on active and inactive human X chromosomes. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1759–1763. doi: 10.1073/pnas.81.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker K. E., Riggs A. D., Smith S. S. Purification of human DNA (cytosine-5-)-methyltransferase. J Cell Biochem. 1985;29(4):337–349. doi: 10.1002/jcb.240290407. [DOI] [PubMed] [Google Scholar]