Abstract
In yeast and mammalian cells, the spindle assembly checkpoint proteins Mad1p and Mad2p localize to the nuclear pore complex (NPC) during interphase. Deletion of MAD1 or MAD2 did not affect steady-state nucleocytoplasmic distribution of a classical nuclear localization signal-containing reporter, a nuclear export signal-containing reporter, or Ran localization. We utilized cells with conditional mutations in the yeast Ran GTPase pathway to examine the relationship between Ran and targeting of checkpoint regulators to the NPC. Mutations that disrupt the concentration of Ran in the nucleus displaced Mad2p but not Mad1p from the NPC. The displacement of Mad2p in M-phase cells was correlated with activation of the spindle checkpoint. Our observations demonstrate that Mad2p localization at NPCs is sensitive to nuclear levels of Ran and suggest that release of Mad2p from NPCs is closely linked with spindle assembly checkpoint activation in yeast. This is the first evidence indicating that Ran affects the localization of Mad2p to the NPC.
In all eukaryotes, mitosis is controlled through the abundance of B-type cyclins and Pds1p (also known as Securin) (13). B-type cyclins associate with and activate the cdc28 kinase in budding yeast during mitosis. Pds1p forms an inactive complex with the Esp1p protease (also known as Separase). Upon loss of Pds1p, Esp1p cleaves the cohesin Scc1p, which holds the sister chromatids together, allowing the onset of anaphase. Both B-type cyclins and Pds1 are ubiquitinated at the metaphase-anaphase transition by an E3 ligase called the anaphase-promoting complex (APC) (13). Premature APC activation is prevented in the presence of unattached kinetochores by a set of conserved spindle assembly checkpoint proteins, including Mad1p, Mad2p, Mad3p, Mps1p, Bub1p, and Bub3p (9). Some of these proteins have been localized to unattached kinetochores (Mad1p, Mad2p, Bub1p, and Bub3p), leading to a model in which a diffusible “delay anaphase” signal that prevents the activation of APC is generated by these proteins at unattached kinetochores (11, 22). After all of the chromosomes have achieved bipolar attachment and aligned on the mitotic spindle, the checkpoint is turned off, the APC becomes active, and anaphase ensues (9, 13).
Nuclear pore complexes (NPCs) are large, multiprotein structures that perforate the nuclear envelope (NE) and act as the conduits for nuclear-cytoplasmic trafficking (20). Recent studies have suggested a surprising link between the spindle checkpoint pathway and NPCs (19). In Saccharomyces cerevisiae, the Mad1p/Mad2p complex associates with the NPC through the Nup53p-containing complex of nucleoporins (7). In the same study, it was observed that upon checkpoint activation, Mad2p (but not the bulk of Mad1p) is released from the NPC and accumulates on kinetochores. These findings led to the proposal of a model wherein the association between Mad1p and the Nup53p complex sequesters Mad2p at the NPC when the spindle checkpoint is inactive (7). In a separate study, Gillett et al. used chromatin immunoprecipitation analysis to demonstrate that both Mad1p and Mad2p localize to kinetochores upon treatment with microtubule-destabilizing drugs. This finding does not demonstrate that Mad1p is quantitatively removed from NPCs but does suggest that at least some portion of Mad1p relocalizes to the kinetochores upon checkpoint activation (6).
Ran is an abundant, highly conserved Ras-like GTPase that acts as a critical regulator of nuclear-cytoplasmic trafficking (10). Ran has been implicated in the regulation of mitosis (18) and shown to directly control checkpoint activation in Xenopus egg extracts (3). We wished to examine the in vivo relationship between Ran and NPC-associated components of the spindle checkpoint pathway. Here we show that Mad2p localization to the NPC is closely linked to proper nuclear-cytoplasmic compartmentalization of the yeast Ran homologue, Gsp1p. The converse relationship was not found, since disruption of the MAD1 and MAD2 genes had no apparent effect on steady-state Gsp1p localization or classical nuclear localization signal (NLS) or nuclear export signal (NES) transport. Our data further indicate that there is a close relationship between Mad2p release from NPCs and spindle checkpoint activation.
MATERIALS AND METHODS
Yeast strains and plasmids.
All yeast plasmids and strains used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype | Source |
|---|---|---|
| Strain | ||
| PSY 580 | Wild type | 5 |
| ACY109 | prp20-1 | 15 |
| ACY60 | rna1-1 | 15 |
| BQY392 | MAD1-GFP::HIS5 ntf2::HIS3 (+pPS920) | This study |
| BQY393 | MAD1-GFP::HIS5 ntf2::HIS3 (+pPS919) | This study |
| KH141 | mad2::URA3 | 15 |
| YMB1299 | MAD1-GFP::HIS5 | 7 |
| BQY141 | nup53Δ KanMX | Research Genetics |
| BQY142 | Wild type | Research Genetics |
| BQY157 | mad1Δ KanMX | Research Genetics |
| BQY381 | mad2Δ KanMX | Research Genetics |
| BQY238 | ntf2::HIS3 (+pPS882) | This study |
| BQY239 | ntf2::HIS3 (+pPS920) | This study |
| BQY240 | ntf2::HIS3 (+pPS919) | This study |
| BQY 413 | mad2Δ KanMX ntf2::HIS3 (+pPS920) | This study |
| BQY412 | ntf2::HIS3 (+pPS920) | This study |
| BQY414 | prp20-1 mad2Δ KanMX | This study |
| Plasmid | ||
| pPS882 | CEN LEU2 NTF2 | 5 |
| pPS919 | CEN LEU2 ntf2-1 | 5 |
| pPS920 | CEN LEU2 ntf2-2 | 5 |
| pAC212 | 2μm URA3 pADH1 SV40-NLS PKI-NES 2×GFP | 8 |
| pAC410 | 2μm URA3 GSP1-GFP | 15 |
| pAC256 | 2μm URA3 PDS1-GFP | This study |
| pBQ181 | CEN TRP1 MAD2-GFP | This study |
| pBQ184 | 2μm URA3 pGAL SV40-NLS 2×GFP | This study |
| pBQ288 | CEN URA MAD2-GFP | This study |
Localization of GFP fusion proteins.
In all cases, a fully functional Mad2-green fluorescent protein (GFP) was expressed from a low-copy-number plasmid by using its native promoter, and Mad1-GFP was integrated at the MAD1 locus. All GFP fusion proteins were localized by directly viewing the GFP signal in living cells through a GFP-optimized filter with a Zeiss Axioscope epifluorescence microscope equipped with an Orca digital camera and analyzed with Openlab software (1) (see Fig. 1, 2D, 4, and 5), or they were viewed with a Zeiss LSM510 META confocal microscope equipped with a 100× Plan Apochromat (NA 1.4, oil, differential interference contrast [DIC]) objective and a 488-nm line Argon laser (25-mW nominal output; detection long pass [LP], 505 nm), and images were overlaid onto DIC images with Zeiss confocal microscopy software (version 3.2) (2) (see Fig. 2A and 3). To analyze G1-arrested cells, cells were arrested at 25°C with α-factor mating hormone (5 μM) and then shifted to 37°C for 3 h in the presence of α-factor.
FIG. 1.
The ntf2-2 mutant disrupts Mad2p but not Mad1p localization to the NPC. (A) Localization of Mad1-GFP (integrated MAD1-GFP) and Mad2-GFP (pBQ288) in NTF2 (BQY238), ntf2-2 (BQY239), and ntf2-1 (BQY240) cells at the permissive temperature (25°C) and after 3 h at the restrictive temperature (37°C). (B) Fraction of total cells containing Mad1-GFP and Mad2-GFP at the NPC at the permissive (25°C) and restrictive (37°C) temperatures. (C) Disruption of Mad2p localization by ntf2-2 mutation in G1-arrested cells. NTF2 and ntf2-2 cells expressing Mad2-GFP were arrested at 25°C with α-factor mating hormone and shifted to 37°C for 3 h.
FIG. 2.
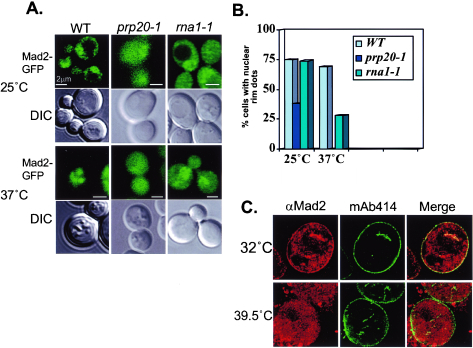
Mutations in the Ran GTPase pathway disrupt Mad2 localization to the NPC. (A) Localization of Mad2-GFP (pBQ181) in wild-type (PSY580), prp20-1 (ACY109), and rna1-1 (ACY60) cells at the permissive temperature (25°C) and after a 3-h shift to the restrictive temperature (37°C). (B) Fraction of total cells containing Mad2-GFP at the NPC at the permissive (25°C) and restrictive (37°C) temperatures. (C) Mad2p is displaced from pores in tsBN2 cells. An asynchronous culture of tsBN2 cells was shifted to the restrictive temperature (39.5°C) for 3 h. The cells were fixed and stained for immunofluorescence with rabbit anti-Mad2 antibodies (red) and mAb414 (green), a monoclonal antibody that recognizes a family of conserved nucleoporins.
FIG. 4.
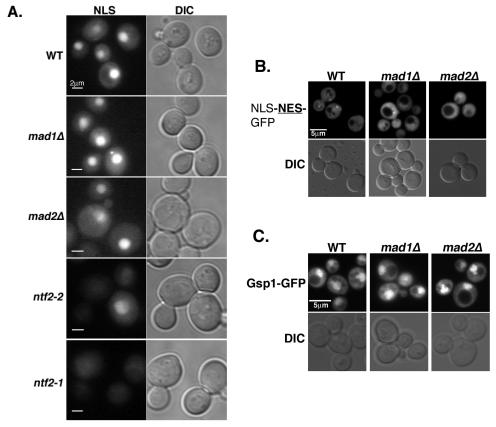
Mad2p does not affect classical nucleocytoplasmic transport. (A) NLS-mediated transport is not disrupted in mad1Δ or mad2Δ cells. Shown is cNLS-GFP-GFP (pBQ184) localization in WT (PSY580), ntf2-2 (BQY239), ntf2-1 (BQY240), mad1Δ (BQY157), and mad2Δ (BQY381) cells after 3 h at the restrictive temperature (37°C). (B) NES-mediated protein export is not disrupted in mad1Δ or mad2Δ cells. WT (BQY142), mad1Δ (BQY157), and mad2Δ (BQY381) cells expressing NLS-NES-GFP (pAC212) (8) are shown. (C) Gsp1p localization is maintained in mad1Δ and mad2Δ cells. Localization of Gsp1-GFP (pAC410) in WT (BQY142), mad1Δ (BQY157), and mad2Δ (BQY381) cells is shown.
FIG. 5.
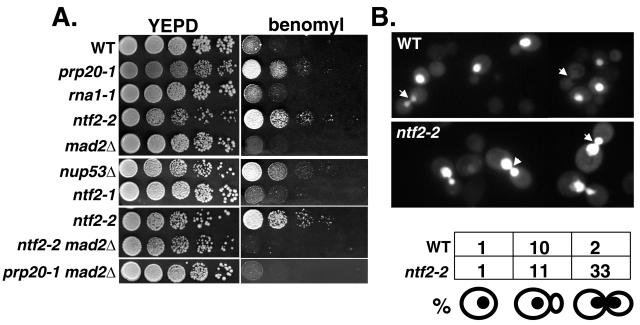
Displacement of Mad2p from the NPC correlates with checkpoint activation. (A) WT (BQY142), mad2Δ (BQY381), nup53Δ (BQY141), prp20-1 (ACY109), rna1-1 (ACY60), ntf2-1 (BQY240), ntf2-2 (top, BQY239; bottom, BQY412), ntf2-2 mad2Δ (BQY413), and prp20-1 mad2Δ (BQY414) cells grown on plates containing benomyl. Cells were grown overnight at 25°C, and 105, 104, 103, 100, and 10 cells were spotted onto yeast extract-peptone-dextrose (YEPD) medium and YEPD containing 20 μg of benomyl/ml. Plates were incubated at 25°C for 4 days. (B) Pds1p destruction is delayed in ntf2-2 cells at the restrictive temperature. Shown are micrographs of asynchronously growing WT (BQY142) and ntf2-2 (BQY412) cells expressing Pds1-GFP (pAC256) after shift to 37°C for 3 h. Arrowheads indicate cells in late mitosis. Shown below is the percentage of cells expressing Pds1-GFP at the point diagramed.
FIG. 3.
Displacement of Mad2p correlates with nucleocytoplasmic Ran-GTP distribution. (A) Localization of Gsp1-GFP (pAC410) in WT (PSY580), ntf2-2 (BQY239), ntf2-1 (BQY240), rna1-1 (ACY109), and prp20-1 (ACY60) cells at the permissive temperature (25°C) and after 3 h at the restrictive temperature (37°C). (B) Fraction of total cells containing Ran-GFP concentrated in the nucleus (nuc), at the nuclear rim (rim), and diffusely localized (dif) at the permissive (25°C) and restrictive (37°C) temperatures.
Analysis of tsBN2 cells.
Cells from a hamster cell line that contains a temperature-sensitive allele of the vertebrate Ran nucleotide exchange factor (tsBN2 cells) were grown at 32°C and then shifted to 39.5°C for 3 h. The cells were permeabilized for immunofluorescence with 0.005% digitonin in transport buffer [110 mM KOAc, 20 mM HEPES, pH 7.3, 2 mM Mg(OAc)2, 0.5 mM EGTA, 2 mM dithiothreitol, 1 μg (each) of leupeptin, pepstatin, and aprotinin/ml] for 10 min, fixed with 3.7% formaldehyde in phosphate-buffered saline for 5 min, and permeabilized by using 0.2% Triton X-100 in phosphate-buffered saline for 5 min. The cells were incubated with polyclonal anti-MAD2 antibodies (1:200 dilution) (gift from R. Chen) and mAb414 antibodies (1:5,000) followed by appropriate fluorescent secondary antibodies. Slides were examined with a Zeiss Axioskop fluorescence microscope, and images were collected and analyzed with Openlab software.
RESULTS AND DISCUSSION
We analyzed Mad1p and Mad2p localization, Gsp1p localization, and checkpoint activation in cells with conditional mutations in the yeast Ran GTPase pathway (10): Ntf2p promotes the import of Gsp1-GDP from the cytosol into the nucleus, where Prp20p promotes nucleotide exchange to generate Gsp1-GTP (15). Gsp1-GTP binds to transport receptors and exits from the nucleus via NPCs. In the cytosol, Gsp1p's GTPase-activating protein, Rna1p, promotes hydrolysis of the Gsp1p-associated GTP. The Ran pathway mutants that we analyzed included ntf2-1, ntf2-2 (5, 15), prp20-1 (2), and rna1-1 (4).
To assay Mad1p and Mad2p localization in living cells, both proteins were fused at the carboxy terminus to GFP. In all cases, Mad2-GFP was expressed from its native promoter on a low-copy-number plasmid that fully complements the benomyl sensitivity of a mad2Δ strain (data not shown). Mad1-GFP was integrated into the genome at the Mad1 locus and has previously been shown to fully complement endogenous MAD1 function (7). We initially compared the localization of Mad1-GFP and Mad2-GFP in ntf2-1, ntf2-2, and wild-type (WT) cells. The ntf2-1 mutation disrupts dimerization, while the ntf2-2 mutation disrupts binding to Gsp1p (15). Mad1-GFP remained at the NPC under both permissive (25°C) and nonpermissive (37°C) conditions in all strains (Fig. 1A), indicating that these mutations did not disrupt its NPC targeting. However, Mad2-GFP was lost from NPCs in an allele-specific fashion: at 25°C, Mad2-GFP visibly accumulated at NPCs in 75% of WT cells and 66% of ntf2-1 cells, while only 36% of the ntf2-2 cells localized Mad2-GFP to NPCs (Fig. 1A and B). At 37°C, only 2% of ntf2-2 cells retained Mad2-GFP at the NPC, compared to 82% of WT cells and 70% of ntf2-1 cells. Notably, not only was the percentage of ntf2-2 cells that localized Mad2-GFP to the NPC significantly reduced in comparison to WT cells, but also the intensity of Mad2-GFP accumulation on NPCs was visibly lower in the subset of ntf2-2 cells where NPC accumulation could be observed than in WT cells. Mad2-GFP was retained within the nucleoplasm of ntf2-2 cells at 37°C, indicating that its failure to bind the NPC was not a simple result of its exclusion from the nucleus.
At 37°C, ntf2-2 cells arrest in G2/M phase as large budded cells with duplicated DNA, whereas ntf2-1 cells show a loss of viability that is not associated with arrest in any particular phase of the cell cycle (15). It was therefore plausible that Mad2p may be released from Mad1p at the NPC near the G2/M transition and that the loss of Mad2-GFP from NPCs in ntf2-2 cells might result indirectly from cell cycle arrest. To test this possibility, we arrested WT and ntf2-2 cells in G1 phase with α-factor, followed by a shift to 37°C for 3 h in the continued presence of α-factor (Fig. 1C). Both WT and ntf2-2 cells retained a shmoo morphology throughout the experiment, confirming that they remained appropriately arrested. Under these circumstances, Mad2-GFP remained at the NPC in the WT cells, but it was displaced from the NPC in the ntf2-2 cells (Fig. 1C). This finding shows that Mad2p is released from the NPC in ntf2-2 cells at the restrictive temperature, regardless of cell cycle stage.
We next analyzed Mad2-GFP localization at the permissive (25°C) and restrictive (37°C) temperatures in prp20-1 and rna1-1 cells. There were fewer prp20-1 cells with Mad2-GFP at the NPC than both WT and rna1-1 cells at 25 and 37°C (Fig. 2). At 37°C, the residual Mad2-GFP was completely lost from the NPC in prp20-1 cells. In rna1-1 cells, the reduction of Mad2-GFP association with NPCs at 37°C was less dramatic: we found no NPC-associated Mad2-GFP within 72% of cells, while 28% of cells still showed a concentration of Mad2-GFP at their NEs. As with ntf2-2 cells, Mad2-GFP still concentrated within the nucleoplasm in the majority of prp20-1 and rna1-1 cells at 37°C, indicating that a loss of NPC association was not an indirect result of reduced levels of Mad2-GFP or nuclear exclusion. Together with observations of ntf2-2 cells, these observations suggested that disruption of the Ran pathway profoundly alters the localization of Mad2p to the NPC.
We examined the localization of Mad2p by immunofluorescence in a hamster cell line that contains a temperature-sensitive allele of the vertebrate Ran nucleotide exchange factor, RCC1 (tsBN2 cells) (12). A subpopulation of Mad2 clearly localizes to the nuclear rim in tsBN2 cells at the permissive temperature (32°C) (Fig. 2C). This population of Mad2p was no longer localized to NPCs 3 h after the shift to the restrictive temperature (39.5°C). As in yeast, a large fraction of Mad2p remained within the tsBN2 nucleus at the restrictive temperature. This finding strongly suggests that the Ran pathway is required for the retention of the Mad2 protein on the NPC during interphase in vertebrate cells, suggesting that the role of Ran in Mad2p localization may be highly conserved.
We considered two possible explanations of our findings in yeast. First, localization of Mad2p to the NPC may require ongoing nuclear transport. This idea is not consistent with the presence of Mad2-GFP at the NPC in ntf2-1 cells at 37°C, because it has been previously demonstrated that ntf2-1 cells have lower transport capacity than ntf2-2 cells under nonpermissive conditions (15) (see Fig. 4A). Alternatively, the localization of Mad2p may be directly sensitive to the localization of Gsp1p or its nucleotide binding state. To examine the behavior of Gsp1p in the mutant strains, we analyzed the localization of a Gsp1-GFP fusion protein in each of the mutants at 25 and 37°C (Fig. 3).
As previously reported (15), ntf2-1 cells concentrate Gsp1-GFP within their nuclei at both temperatures (Fig. 3A), although there is an increase in cytoplasmic Gsp1-GFP in these cells compared to WT cells. This may account for the decreased level of Mad2-GFP at the pore observed in this mutant (Fig. 1). By contrast, ntf2-2 cells exhibit extensive loss of Gsp1-GFP compartmentalization at 37°C. In the prp20-1 cells, Gsp1-GFP localization was abnormal even at 25°C: Gsp1-GFP localized to the nuclear periphery instead of filling the entire nuclear compartment (Fig. 3B). The localization of Gsp1-GFP was further disrupted in prp20-1 cells at 37°C, with a loss of Gsp1-GFP compartmentalization similar to that of ntf2-2 cells. Gsp1-GFP localization was also abnormal at 25°C in rna1-1 cells, although this defect was not as severe as that in prp20-1 cells. Approximately 25% of rna1-1 cells showed some punctuate nuclear rim staining at 25°C (Fig. 3B). For reasons that are unclear, Gsp1-GFP protein levels were consistently lower in rna1-1 cells at 37°C than at 25°C (Fig. 3C). The residual Gsp1-GFP was no longer concentrated within the nucleus of rna1-1 cells at 37°C, although it was enriched on the nuclear envelope in 35% of cells (Fig. 3).
In summary, the two strains with mutations in the Ran pathway that showed complete displacement of Mad2-GFP from the NPC (the ntf2-2 and prp20-1 mutants) at the restrictive temperature also showed a loss of Gsp1p compartmentalization under the same conditions. rna1-1 cells exhibited an intermediate phenotype, with a residual population of Gsp1-GFP at the NE and incomplete dissociation of Mad2-GFP from NPCs. By contrast, the single mutant that maintained nuclear Gsp1-GFP localization at 37°C (ntf2-1) also maintained Mad2p on NPCs. Together, these findings suggest that Gsp1p concentration within the nucleus is closely correlated with Mad2p targeting to the NPC. Interphase tsBN2 cells similarly showed both a loss of Ran compartmentalization (16) and release of Mad2 from the NPC at 39.5°C (Fig. 2C), suggesting that similar mechanisms may be operative in metazoan cells.
We considered two possible roles for the regulated localization of Mad2p to the NPC: Mad2p might promote nuclear-cytoplasmic trafficking or its sequestration to NPCs might control its activity in the mitotic checkpoint. It has previously been shown that deletion of MAD1 inhibits import by the Kap121p transport receptor (7). However, deletion of MAD2 had no apparent effect in the same assay. To determine more generally whether the deletion of MAD1 or MAD2 alters classical nucleocytoplasmic transport, we examined whether cells with deletions in the MAD1 (mad1Δ) or MAD2 (mad2Δ) genes maintain correct localization of a model substrate bearing a classical nuclear localization sequence (cNLS-GFP) (Fig. 4A). For comparison, the localization of the reporter was also examined in ntf2-1 and ntf2-2 cells at the nonpermissive temperature (37°C). ntf2-1 and ntf2-2 cells showed extensive and partial loss of cNLS-GFP nuclear localization, respectively. By contrast, both the mad1Δ and mad2Δ cells were indistinguishable from the WT controls, arguing that the deletion of MAD1 or MAD2 has an insignificant effect on NLS-mediated protein import.
To investigate whether Mad1p or Mad2p has a role in protein export, we examined a similar model substrate bearing a leucine-rich nuclear export sequence (NLS-NES-GFP) in mad1Δ and mad2Δ cells (Fig. 4B). When nuclear export is compromised, this reporter is redistributed to the nucleus due to the presence of a functional NLS within the protein (8). The steady-state distribution of NLS-NES-GFP between the cytoplasm and nucleus in mad1Δ and mad2Δ cells is essentially indistinguishable from the distribution in WT cells, arguing that these cells retain high levels of nuclear export capacity. Together with the observations from cNLS-GFP import, these data suggest that steady-state nucleocytoplasmic trafficking is not altered in the absence of Mad1p and Mad2p. Moreover, GFP-Gsp1p remained localized within the nucleus of both mad1Δ and mad2Δ cells (Fig. 4C), arguing against the notion that Mad2p or Mad1p is required for the maintenance of Gsp1p-GTP within the nucleus. For all of these reasons, we feel that it is very unlikely that Mad1p or Mad2p plays a direct role in regulation of bulk trafficking through the NPC.
Alternatively, it has previously been shown that deletion of the NUP53 gene renders cells resistant to the microtubule (MT)-destabilizing drug benomyl (7). These observations would be consistent with the possibility that Mad2p release from NPC potentiates its activity in the spindle checkpoint, thereby rendering the cells better able to cope with MT destabilization. This hypothesis predicts that Mad2p release by other means should also promote checkpoint activation. To test this prediction, we examined whether ntf2-2, prp20-1, and rna1-1 mutants showed resistance to benomyl (Fig. 5A), as nup53Δ cells do (7). It was previously found that the ntf2-2 mutation conferred benomyl sensitivity (15). In the course of performing these experiments, we determined that the benomyl sensitivity observed in those experiments was due to an additional background mutation present in the ntf2-2 strain used at that time. We grew each of the yeast strains at 25°C on plates containing 20 μg of benomyl/ml. ntf2-2 and prp20-1 cells exhibited benomyl resistance, although ntf2-1 cells did not, consistent with the idea that NPC release potentiates spindle checkpoint activation under conditions where Gsp1p-GTP levels are low (Fig. 5A).
Ran-GTP has been shown to have roles in MT dynamics in fission yeast (17) and to promote MT polymerization in Xenopus egg extracts (14). While some contribution of MT dynamics toward the benomyl resistance phenotype cannot be fully eliminated, we do not believe that benomyl resistance of ntf2-2 and prp20-1 cells resulted simply from MT stabilization for several reasons. First, ntf2-2 cells arrest with morphologically normal short metaphase spindles, suggesting that their MT dynamics are relatively normal (15). Second, ntf2-2 mad2Δ double mutants are viable and undergo normal mitosis with WT spindle morphology in the absence of benomyl (15). If the ntf2-2 mutation altered spindle assembly, ntf2-2 mad2Δ cells should show inviability and increased chromosome missegregation. Moreover, similar to the benomyl resistance observed for nup53Δ cells (7), the benomyl resistance of ntf2-2 and prp20-1 cells was entirely dependent upon the presence of Mad2p, since the ntf2-2 mad2Δ and prp20-1 mad2Δ double mutants were as sensitive to benomyl as mad2Δ cells (Fig. 5B). This finding suggests that changes in Mad2p activity in these cells at the permissive temperature underlie the benomyl resistance observed.
Interestingly, rna1-1 cells were not benomyl resistant at 25°C (Fig. 5A). It is possible that the NPC association of Mad2p was not as perturbed in this mutant as in ntf2-2 or prp20-1 cells. This idea is consistent with the frequent persistence of Mad2-GFP on NPCs of rna1-1 cells at 37°C (Fig. 2B and C). It is also possible that the nucleotide binding state of Gsp1p is important for checkpoint activation: defects in Ntf2p or Prp20p should cause accumulation of GDP-bound Gsp1p, whereas loss of Rna1p function should cause accumulation of GTP-bound Gsp1p. Increased Ran-GTP levels disrupt the localization of Mad2p and other spindle checkpoint regulators to kinetochores in Xenopus egg extracts (3), and Gsp1p-GTP might act similarly in yeast. These two possibilities are obviously not mutually exclusive, and it is imaginable that the nucleotide binding state of Gsp1p may influence both the release of Mad2p from the NPC and its activity in the spindle checkpoint.
Finally, we noted that ntf2-2 cells arrest at the G2/M transition when shifted to 37°C. To determine whether this arrest is related to activation of the spindle checkpoint, we analyzed the stability of Pds1p in living WT and ntf2-2 cells at 37°C by using a Pds1p-GFP fusion protein (Pds1-GFP) (21). Pds1-GFP is degraded in a manner that is similar to published epitope-tagged Pds1 (1) (data not shown). In wild-type cells, Pds1-GFP fluorescence was reduced or completely lost in large budded cells (Fig. 5B, arrows), and a smaller percentage of cells exhibited Pds1-GFP. However, more ntf2-2 cells retain Pds1-GFP fluorescence at 37°C, with 33% of cells arresting as large budded cells with bright Pds1-GFP fluorescence, indicating that APC is inactive and Pds1p is stabilized under these conditions.
Taken together, we have shown that the loss of nuclear Ran-GTP is closely linked to release of Mad2p from the NPC and to activation of the spindle assembly checkpoint. These results argue that Ran interacts with the spindle checkpoint pathway in yeast as well as in vertebrates, suggesting that Ran's function as a mitotic regulator may be conserved across all eukaryotes, and this report is the first time that Ran has been shown to affect the localization of Mad2p to the nuclear pore complex.
Acknowledgments
B.Q. is a recipient of a Pharmacology Research Associate Training fellowship.
We are grateful to Anita Corbett, Oliver Kerscher, and Rick Wozniak for plasmids and strains used in this study and insightful discussions. We are also grateful to Ferhan Ayaydin for help with confocal microscopy.
REFERENCES
- 1.Agarwal, R., and O. Cohen-Fix. 2002. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 16:1371-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhtar, N., H. Hagan, J. E. Lopilato, and A. H. Corbett. 2001. Functional analysis of the yeast Ran exchange factor Prp20p: in vivo evidence for the RanGTP gradient model. Mol. Genet. Genomics 265:851-864. [DOI] [PubMed] [Google Scholar]
- 3.Arnaoutov, A., and M. Dasso. 2003. The Ran GTPase regulates kinetochore function. Dev. Cell 5:99-111. [DOI] [PubMed] [Google Scholar]
- 4.Corbett, A. H., D. M. Koepp, G. Schlenstedt, M. S. Lee, A. K. Hopper, and P. A. Silver. 1995. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J. Cell Biol. 130:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbett, A. H., and P. A. Silver. 1996. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J. Biol. Chem. 271:18477-18484. [DOI] [PubMed] [Google Scholar]
- 6.Gillett, E. S., C. W. Espelin, and P. K. Sorger. 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164:535-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iouk, T., O. Kerscher, R. J. Scott, M. A. Basrai, and R. W. Wozniak. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159:807-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones, A. L., B. B. Quimby, J. K. Hood, P. Ferrigno, P. H. Keshava, P. A. Silver, and A. H. Corbett. 2000. SAC3 may link nuclear protein export to cell cycle progression. Proc. Natl. Acad. Sci. USA 97:3224-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37:251-282. [DOI] [PubMed] [Google Scholar]
- 10.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 3:731-741. [DOI] [PubMed] [Google Scholar]
- 12.Nishitani, H., M. Ohtsubo, K. Yamashita, H. Iida, J. Pines, H. Yasudo, Y. Shibata, T. Hunter, and T. Nishimoto. 1991. Loss of RCC1, a nuclear DNA-binding protein, uncouples the completion of DNA replication from the activation of cdc2 protein kinase and mitosis. EMBO J. 10:1555-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters, J. M. 1999. Subunits and substrates of the anaphase-promoting complex. Exp. Cell Res. 248:339-349. [DOI] [PubMed] [Google Scholar]
- 14.Quimby, B. B., and M. Dasso. 2003. The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol. 15:338-344. [DOI] [PubMed] [Google Scholar]
- 15.Quimby, B. B., C. A. Wilson, and A. H. Corbett. 2000. The interaction between Ran and NTF2 is required for cell cycle progression. Mol. Biol. Cell 11:2617-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren, M., G. Drivas, P. D'Eustachio, and M. G. Rush. 1993. Ran/TC4: a small nuclear GTP-binding protein that regulates DNA synthesis. J. Cell Biol. 120:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salus, S. S., J. Demeter, and S. Sazer. 2002. The Ran GTPase system in fission yeast affects microtubules and cytokinesis in cells that are competent for nucleocytoplasmic protein transport. Mol. Cell. Biol. 22:8491-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sazer, S., and M. Dasso. 2000. The Ran decathlon: multiple roles of Ran. J. Cell Sci. 113:1111-1118. [DOI] [PubMed] [Google Scholar]
- 19.Stukenberg, P. T., and I. G. Macara. 2003. The kinetochore NUPtials. Nat. Cell Biol. 5:945-947. [DOI] [PubMed] [Google Scholar]
- 20.Suntharalingam, M., and S. R. Wente. 2003. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4:775-789. [DOI] [PubMed] [Google Scholar]
- 21.Yellman, C. M., and D. J. Burke. 2004. Assaying the spindle checkpoint in the budding yeast Saccharomyces cerevisiae. Methods Mol. Biol. 280:275-290. [DOI] [PubMed] [Google Scholar]
- 22.Yu, H. 2002. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14:706-714. [DOI] [PubMed] [Google Scholar]