Abstract
Introns of Plasmodium falciparum var genes act as transcriptional silencing elements that help control antigenic variations. In transfected episomes, intron silencing of a drug-selectable marker under var promoter control is reversed by the spontaneous deletion of key intron regions. The resulting promoter activation does not affect the transcription of chromosomal var genes.
Antigenic variations in Plasmodium falciparum-parasitized erythrocytes result from switches in the major variable cytoadherence surface proteins encoded by members of the multicopy var gene family (1, 7, 8). Each parasite expresses one of 50 to 60 var copies in its genome while silencing the rest by a mechanism that is thought to involve var gene introns in cooperation with their corresponding gene promoters. Previous work with luciferase reporter constructs showed that cooperative silencing by these elements is S-phase dependent (3). Here we report the mapping of intron silencing elements in detail and show that the stable expression of an episomal var promoter does not affect the expression of endogenous chromosomal var genes.
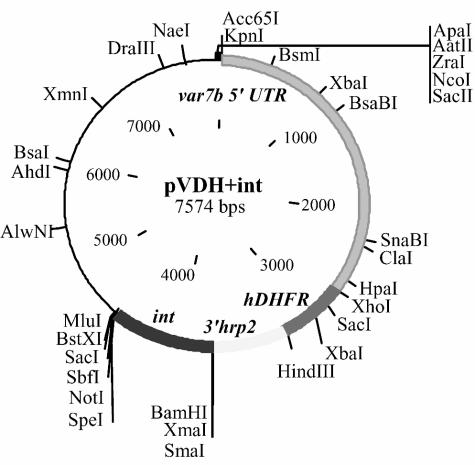
Our experiments first explored the effects of forced var promoter activity from plasmid pVDH+int, a modification of plasmid pVLH+int carrying the hDHFR (human dihydrofolate reductase) marker instead of a luciferase cassette (Fig. 1). Previous work showed that the activity of pVLH+int in transfected parasites was silenced by a cooperative interaction between the 5′ var7b flanking (promoter) region and the var intron (int) during the transition through S phase of the cell cycle (3). We therefore expected this same silencing mechanism to affect the ability of pVDH+int to confer drug resistance to WR99210, a powerful antifolate drug that transfected P. falciparum parasites can survive exposure to only by expressing hDHFR (4). At 25 days after transfection and selection of P. falciparum (FCB line) in the presence of WR99210 (5 nM), drug-resistant parasites were obtained; however, nontransfected parasites in control flasks did not survive. Rescue of the transfected episomes from the parasites and sequencing of the recovered plasmid DNA preparations identified a 460-bp deletion in the middle of the 821-bp int sequence. The remaining 361-bp sequence in the recovered plasmids (termed pVDH+intΔ2′) included all of var intron regions 1 and 3 and a 38-bp residual segment of intron region 2 described by Calderwood et al. (2).
FIG. 1.
Map of plasmid pVDH+int. The 3′ hrp2 flanking region provides proper termination for hDHFR expression; int is placed after this termination region.
The possibility that this deletion in the var intron had allowed plasmid pVDH+int-transfected parasites to escape hDHFR silencing and thereby survive WR99210 pressure led us to focus on the 460-bp region in further transfection and selection experiments. We replaced the full 821-bp var intron in plasmid pVDH+int with a segment containing var intron region 2 but missing almost all of regions 1 and 3 (intΔ1Δ3) (2). Parasites transfected with the resulting plasmid construct, pVDH+intΔ1Δ3, again yielded drug-resistant populations in the presence of 5 nM WR99210 selection, although only after a longer period (39 days) than with pVDH+int. Sequencing of plasmids rescued from these populations identified a 422-bp deletion in the original 571-bp intΔ1Δ3 segment, beginning 29 bp downstream of the start of region 2 (see Fig. S1 in the supplemental material). The plasmids were accordingly named pVDH+int149 for the short, 149-bp section remaining from the intΔ1Δ3 segment. Transcription of hDHFR was verified by reverse transcription (RT)-PCR, confirming that escape from the cooperative silencing mechanism could be attributed to the loss of the 422-bp deletion in the presence of drug pressure (data not shown).
Chromosomal var gene expression levels in transfected parasites and nontransfected control parasites showed no evidence that the presence of active var promoters on the episomal constructs affected the transcription of endogenous var genes. This conclusion followed from real-time RT-PCR experiments with RNAs from parasites stably transfected with pVDH+intΔ2′, pVDH+int149, and pVDH (counterpart of plasmid pVLH, with no intron) (3) and from nontransfected parasites (Table 1). Unaltered transcription of endogenous var genes in the presence of active episomal var promoters showed that stably transfected parasites can transcribe from more than one var promoter at a time. This result implies that episomal var promoters are not recognized by the allelic exclusion mechanism, perhaps because the chromatin structure that forms with the var promoter and the intron is altered when the sequences of region 2 are deleted. Alternatively, it is possible that only chromosomal copies are counted in the nuclear compartment, while episomal copies remain unrecognized.
TABLE 1.
Chromosomal var gene transcription levels in various transformed parasite linesa
| FCB lines transformed by constructs | Ratio (mean ± SD) of transcript levels in parasite lines after normalization for:
|
|
|---|---|---|
| C341 | C2143 | |
| pVDH, pVDH+intΔ2′ | 1.07 ± 0.89 | 0.81 ± 0.60 |
| pVDH, pVDH+int149 | 2.74 ± 1.14 | 1.38 ± 0.45 |
| pVDH, none | 1.53 ± 0.33 | 2.45 ± 0.97 |
| pVDH+intΔ2′, none | 1.29 ± 0.04 | 1.63 ± 0.45 |
| pVDH+int149, none | 0.86 ± 0.19 | 2.32 ± 0.64 |
Parasitized erythrocyte stages were separated on a six-step (40, 60, 70, 80, 85, and 90%) Percoll gradient in RPMI 1640-5% sorbitol by centrifugation at 12,000 × g for 20 min (room temperature). RNA was extracted from recovered parasites with a total RNA purification kit as recommended by the manufacturer (Invitrogen). Total RNA (1 to 3 μg) was treated with DNase for 15 min before RT (Invitrogen first-strand cDNA synthesis kit). Samples were primed with random hexamers. One sample in which reverse transcriptase was omitted served as a negative control to verify the absence of genomic DNA. Levels of transcription of var genes were determined by real-time quantitative RT-PCR with cDNA from transfected and nontransfected parasites and a Light Cycler (Roche Diagnostics, Indianapolis, Ind.). Five DNA standards containing 108 to 104 plasmid molecules were used for a standard curve. Amplification of the var cDNA DBLα domain with primers αAF [GCACG(A/C)AGTTTTGC] and αBR [GCCCATTC(G/C)TCGAACCA] (9) and amplification of C341 (disulfide-like protein) and C2143 (seryl tRNA synthetase) cDNA controls with the primers of Mamoun et al. (6) were performed in the same reaction tube. Five microliters of 10-fold-diluted cRNA or DNA standard was amplified in a 20-μl reaction mixture containing 9.6 μl of H2O, 2.4 μl of MgCl2 (25 mM), 0.5 μl of each primer (10 μM), and 2 μl of Fast Start DNA Master SYBR green I (Roche Diagnostics). PCR conditions were as follows: 10 min of denaturation at 95°C followed by 40 cycles of 95°C for 5 s, 48°C for 5 s, and 60°C for 25 s. Levels of expression from each transformed line were compared to each other and to that from untransformed FCB parasites and are presented as values normalized to those for C341 and C2143 control genes. All amplifications were repeated to ensure reproducibility (26 experiments for pVDH+intΔ2′, 24 experiments for pVDH+int149, and 15 experiments for untransformed FCB).
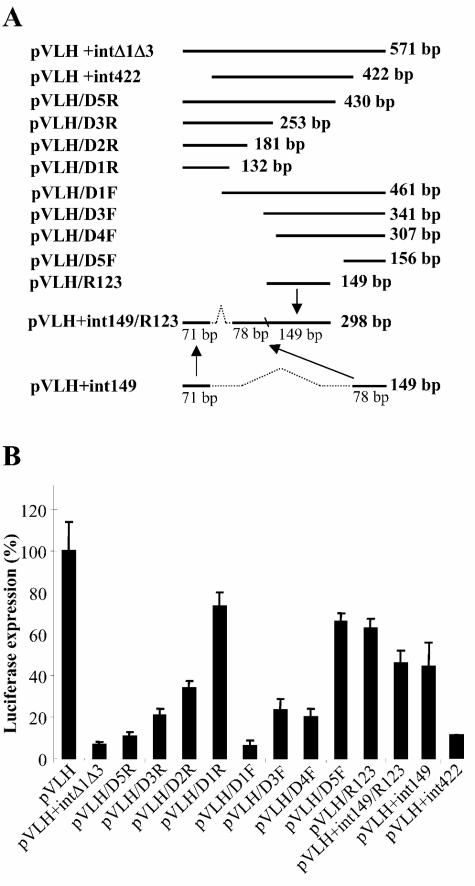
To further evaluate the silencing elements in var intron region 2, we used luciferase reporter experiments with plasmids in which the full-length intron of pVLH+int was replaced with various portions of region 2. A plasmid containing the 422-bp sequence that had been deleted from pVDH+intΔ1Δ3 (pVLH+int422) showed only 9 to 15% of the luciferase expression of control plasmid pVLH, with no intron. In contrast, luciferase expression from pVLH+int149, containing the 149-bp sequence from pVDH+int149, was much greater, about 45% of that of the control (Fig. 2).
FIG. 2.
Relative positions of var intron sequences from region 2 and their effect on luciferase activities from pVLH constructs. (A) Locations of segments from region 2 and map of the constructed int149/R123 segment containing three imperfect repeats. (B) Levels of luciferase expression from constructs containing different segments of region 2. Control plasmids pVLH and pVLH+intΔ1Δ3 demonstrated complete activity and full silencing of luciferase, respectively. All experiments were repeated at least twice in triplicate, and all Student's t tests yielded P values of <0.05 for comparisons of luciferase levels from pVLH+intΔ1Δ3 and the individual plasmids. Bars on the histogram indicate 95% confidence intervals.
Such different levels of silencing from the 422- and 149-bp intron sequences led us to map the silencing effects in more detail. We generated a series of fragments from intron region 2 in both the forward and the reverse directions, cloned these into plasmid pVLH, and tested them for silencing of luciferase expression (Fig. 2A). Silencing comparable to that obtained with full region 2 was obtained with three constructs, pVLH+intΔ1Δ3, pVLH/D5R, and pVLH/D1F; other constructs showed partial silencing ranging from 20 to 80%, depending on the length of the inserted fragment (Fig. 2B). We also tested a stretch of three imperfect repeats (49, 48, and 52 bp in the center of region 2) (Fig. 2A; see also Fig. S1 in the supplemental material) for silencing activity. Repeats of this type are frequently found in var introns and contain some of the Inr-like elements previously noted by Calderwood et al. (2). The plasmid containing these three repeats (pVLH/R123) showed less silencing than constructs such as pVLH/D3R and pVLH/D4F, which contained different intron sequences of comparable or smaller size (Fig. 2B). Taken together, these results suggest that cooperative silencing involves structural complexities of the intron, extending across hundreds of nucleotides of region 2. Interestingly, a var gene conserved in many P. falciparum isolates, varCOMMON, contains an intron that is naturally missing region 2. The varCOMMON gene has been found to be constitutively transcribed in an unusual stage-specific pattern (5, 10), consistent with the idea that elements of region 2 have a key role in var gene silencing.
The var introns have their own promoter activity and produce var-associated “sterile” transcripts that are thought to be involved in gene regulation (2, 8). We have tested whether this promoter activity resides within the 422-bp subsequence of region 2 that was deleted from pVDH+intΔ2′. Substitution of this subsequence for the 5′ var7b flanking region upstream of luc in pVLH yielded a plasmid that produced the same luciferase activity in transfected parasites as p2LH-1, a promoter construct that contains all of region 2 (2) (data not shown). These results locate promoter activity within the same intron subsequence as silencing activity, lending further support for a role of silent transcripts in epigenetic control.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Baruch, D. I., B. L. Pasloske, H. B. Singh, X. Bi, X. C. Ma, M. Feldman, T. F. Taraschi, and R. J. Howard. 1995. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82:77-87. [DOI] [PubMed] [Google Scholar]
- 2.Calderwood, M. S., L. Gannoun-Zaki, T. E. Wellems, and K. W. Deitsch. 2003. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 278:34125-34132. [DOI] [PubMed] [Google Scholar]
- 3.Deitsch, K. W., M. S. Calderwood, and T. E. Wellems. 2001. Malaria. Cooperative silencing elements in var genes. Nature 412:875-876. [DOI] [PubMed] [Google Scholar]
- 4.Fidock, D. A., and T. E. Wellems. 1997. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. USA 94:10931-10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyes, S. A., Z. Christodoulou, A. Raza, P. Horrocks, R. Pinches, J. A. Rowe, and C. I. Newbold. 2003. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol. Microbiol. 48:1339-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamoun, C. B., I. Y. Gluzman, C. Hott, S. K. MacMillan, A. S. Amarakone, D. L. Anderson, J. M. R. Carlton, J. B. Dame, D. Chakrabarti, R. K. Martin, B. H. Brownstein, and D. E. Goldberg. 2001. Coordinated programme of gene expression during asexual intraerythrocytic development of the human malaria parasite Plasmodium falciparum revealed by microarray analysis. Mol. Microbiol. 39:26-36. [DOI] [PubMed] [Google Scholar]
- 7.Smith, J. D., C. E. Chitnis, A. G. Craig, D. J. Roberts, D. E. Hudson-Taylor, D. S. Peterson, R. Pinches, C. I. Newbold, and L. H. Miller. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su, X., V. M. Heatwole, S. P. Wertheimer, F. Guinet, J. V. Herrfeldt, D. S. Peterson, J. V. J. Ravetch, and T. E. Wellems. 1995. A large and diverse gene family (var) encodes 200-350 kD proteins implicated in the antigenic variation and cytoadherence of Plasmodium falciparum-infected erythrocytes. Cell 82:89-100. [DOI] [PubMed] [Google Scholar]
- 9.Taylor, H. M., S. A. Kyes, D. Harris, N. Kriek, and C. I. Newbold. 2000. A study of var gene transcription in vitro using universal var gene primers. Mol. Biochem. Parasitol. 105:13-23. [DOI] [PubMed] [Google Scholar]
- 10.Winter, G., Q. J. Chen, K. Flick, P. Kremsner, V. Fernandez, and M. Wahlgren. 2003. The 3D7var5.2 (varCOMMON) type var gene family is commonly expressed in non-placental Plasmodium falciparum malaria. Mol. Biochem. Parasitol. 127:179-191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.