Abstract
Children from economically disadvantaged families experience worse cognitive, psychiatric, and medical outcomes compared to more affluent youth. Preclinical models suggest some of the adverse influence of disadvantage could be transmitted during gestation via maternal immune activation, but this hypothesis has not been tested in humans. It also remains unclear whether prenatal interventions can mitigate such effects. To fill these gaps, we conducted two studies. Study 1 characterized the socioeconomic conditions of 79 women during pregnancy. At delivery, placenta biopsies and umbilical blood were collected for transcriptional profiling. Maternal disadvantage was associated with a transcriptional profile indicative of higher immune activation and slower fetal maturation, particularly in pathways related to brain, heart, and immune development. Cord blood cells of disadvantaged newborns also showed indications of immaturity, as reflected in down-regulation of pathways that coordinate myeloid cell development. These associations were independent of fetal sex, and characteristics of mothers (age, race, adiposity, diabetes, pre-eclampsia) and babies (delivery method, gestational age). Study 2 performed the same transcriptional analyses in specimens from 20 women participating in CenteringPregnancy, a group-based psychosocial intervention, and 20 women in traditional prenatal care. In both placenta biopsies and cord blood, women in CenteringPregnancy showed up-regulation of transcripts found in Study 1 to be most down-regulated in conjunction with disadvantage. Collectively, these results suggest socioeconomic disparities in placental biology are evident at birth, and provide clues about the mechanistic origins of health disparities. They also suggest the possibility that psychosocial interventions could have mitigating influences.
Keywords: Early life stress, poverty, inflammation, placenta, neurodevelopment
INTRODUCTION
Children’s life outcomes differ as a function of their family’s economic conditions. The slope of this socioeconomic gradient varies across countries (Elgar et al. 2015) and, even in nations where it is steep, a sizeable minority of disadvantaged youth still achieve positive outcomes (Masten & Narayan 2012). Yet on the whole, disadvantaged children fare worse than their affluent peers, and these disparities are apparent in a variety of cognitive, psychiatric, and biomedical outcomes (Hertzman & Boyce 2010; Shonkoff & Garner 2012). With regard to cognition, childhood disadvantage forecasts slower language acquisition, worse executive function, and lower educational attainment (Duncan & Murnane 2011). Disadvantage also portends higher psychiatric risks (Reiss 2013). In the National Comorbidity Survey Replication Study, financial hardships during childhood presaged higher probability of first-onset anxiety, mood, behavioral, and substance disorders, and did so at all stages of the lifecourse (McLaughlin et al. 2011). In the realm of physical health, disadvantage is associated with the incidence and severity of obesity, diabetes, and asthma in childhood (Chen et al. 2002), and with increased vulnerability to cardiovascular disease, functional disability, and premature death in adulthood (Galobardes et al. 2008; Montez & Hayward 2014; Miller et al. 2011).
Mechanistic accounts of these disparities generally focus on characteristics of the postnatal environment, e.g., socioeconomic variations in children’s exposure to cognitive stimulation, sensitive caregiving, dietary imbalances, and environmental toxins. Although these characteristics undoubtedly contribute to socioeconomic disparities (Kundakovic & Champagne 2015; Hackman et al. 2010; Wright & Subramanian 2007; Schreier & Chen 2013), mounting evidence suggests that some of the relevant exposures could occur prenatally, and become “embedded” in aspects of physiology during sensitive windows of fetal development (Hertzman & Boyce 2010; Entringer et al. 2012). Indeed, socioeconomic disadvantage often co-occurs with psychological stress, depressive symptoms, cortisol dysregulation, poor nutrition, and toxin exposure (Wright 2011; Evans 2004). Experimental studies in animals indicate these exposures can affect the gestational milieu, with implications for offspring brain development, cognitive functioning, psychiatric disorders, and a host of allergic, metabolic, and cardiac diseases (Bale 2015; Hanson & Gluckman 2014; Prescott 2006; Pryce et al. 2005).
The placenta is likely to be a key route by which these exposures are transmitted from mother to offspring. It functions as a barrier that protects the fetus from maternal immunity and potential teratogens, and the interface where gases, nutrients, and waste are exchanged. These functions are dysregulated in animals subjected to experimental conditions that parallel human disadvantage, like psychological stress, glucocorticoid excess, and nutrient restriction (Bronson & Bale 2016; Braun et al. 2013; Coe & Lubach 2014). In many of these models, excessive activation of maternal immunity is a key pathway by which gestational manipulations predispose animals to altered patterns of neural, cognitive, and behavioral development (Bilbo 2013; Estes & McAllister 2016; Meyer 2013; Bale 2015; Nusslock & Miller 2015). These phenotypes are thought to emerge because maternal immune activity interferes with placental nutrient transfer, slowing maturation of fetal brain, heart, and liver (Arck & Hecher 2013; Dimasuay et al. 2016)
Despite these observations, studies have not yet examined how maternal socioeconomic conditions relate to placental immune activation in humans, or explored the implications for fetal maturation. Here, we attempted to begin to filling these gaps in knowledge by assessing the socioeconomic conditions of pregnant women and assembling transcriptional profiles of their placentas. Based on the findings in animal models outlined above (Bronson & Bale 2016; Hanson & Gluckman 2014; Arck & Hecher 2013; Braun et al. 2013; Coe & Lubach 2014), we hypothesized that maternal disadvantage would be associated with transcriptional indications of greater immune activation and slower tissue maturation in women’s placental biopsies. We also expected maternal disadvantage would be associated with transcriptional indications of slower leukocyte maturation in newborn cord blood cells.
In a small follow-up study, we also considered the possibility these disparities might be ameliorated through a prenatal intervention. CenteringPregnancy is group-based model of prenatal care, wherein 8–10 women of the same gestational age meet together on a weekly basis with a nurse or midwife. Patients receive all the obstetric components of traditional prenatal care, but the sessions also focus on building social support, and include discussions of nutrition, parenting, stress reduction, patient-provider communication, and other topics typically reserved for childbirth preparation classes (Hale et al. 2014). In multiple large-scale evaluations, Centering has improved birth outcomes, particularly among low-income minority women (Ickovics et al. 2007; Ickovics et al. 2016; Picklesimer et al. 2012). For example, in an initial randomized clinical trial of 1047 women, Ickovics reported that participation in Centering led to a 33% reduction in preterm birth compared with traditional prenatal care (Ickovics et al. 2007). These benefits were replicated in a follow-up randomized trial of 1148 women, which also found a 34% reduction in babies delivered small for gestational age (Ickovics et al. 2016). Given that Centering reduces the prevalence of adverse birth outcomes in low-income women, and simultaneously lowers distress, facilitates social connections, and improves lifestyle (Ickovics et al. 2011; Heberlein et al. 2016), we hypothesized it would ameliorate some of the transcriptional dysregulation associated with disadvantage.
PATIENTS & METHODS
Patients
Study 1 involved 100 women recruited from the obstetric clinics of NorthShore University Hospital in Evanston, Illinois. To participate, women had to be ≥ 18 years old, fluent in English, ≤ 26 weeks gestational age, and with a singleton pregnancy. To maximize generalizability, we included women regardless of whether they delivered vaginally or by Caesarean section. Exclusion criteria included fetal congenital anomaly, chromosomal abnormality, and treatment with oral corticosteroids during pregnancy or progesterone after 14 weeks’ gestation. All women gave written consent before participating, and the Institution Review Boards of Northwestern University and NorthShore University HealthSystem approved the protocol.
Study 2 involved 40 women delivering at Greenville Memorial Hospital in Greenville, South Carolina. At admission to Labor and Delivery, resident physicians in Obstetrics and Gynecology identified women who had participated in CenteringPregnancy and screened them for eligibility. Criteria were identical to Study 1, with the additional stipulation that women had attended at least 5/10 Centering sessions. (The average number of sessions attended was 7.2, with a standard deviation of 1.1.) After enrolling a Centering participant, residents approached consecutively admitted women until they identified an eligible Control, defined as a patient who met Study 1 criteria and had not attended Centering. All women gave written consent and the Greenville Health System Institution Review Board approved the protocol.
Disadvantage
In Study 1, socioeconomic conditions were assessed during second trimester with a structured interview developed by the MacArthur Network on SES and Health. We calculated a composite disadvantage score (Miller et al. 2014a) that assigned one point for each of these indicators: household income below federal poverty threshold; savings less than one month of living expenses; receipt of TANF, WIC, SNAP, CHIP, SSI, or Medicaid; education less than two-year college degree; and inability to afford suitable housing. Reflecting the independence of these indicators, their average inter-correlation was .29 (range .08–.62). The sample had the full range of scores on the composite (0–5), but only 3 women scored 4 or 5. Accordingly, we compressed the composite into a four-level variable. In the analytic sample, the distribution of scores was 0 (57%), 1 (13.9%), 2 (11.4%), and ≥ 3 (17.7%).
Placenta Biopsy and Cord Blood Collection
Both studies used the same protocol to collect placenta biopsies and umbilical blood. Within two hours of childbirth, trained staff on Labor and Delivery units collected 0.5-cm3 biopsies of fetal chorionic villi, the inner placenta layer where nutrient exchange occurs. To minimize the effects of clonal variation, biopsies were collected from four separate cotyledons. Immediately following collection, the specimens were rinsed in PBS, stabilized in RNAlater (Qiagen), and frozen at −80 C. After completing the placenta biopsy, staff drew 2.5 ml of umbilical vein blood into a PAXgene Blood RNA Tube (Qiagen), and immediately froze the specimen at –80° C. At the end of the study, biopsies were thawed, then dissociated and homogenized on a gentleMACS Instrument (Miltenyi Biotec). Each woman’s biopsies were pooled into a single lysate and frozen at −80° C.
RNA Extraction and Transcriptional Profiling
From placental lysates and PAXgene specimens, total RNA was extracted using PCR-clean and RNAse-free techniques with a commercially available kit (Qiagen RNeasy). After extract purity and integrity had been verified on NanoDrop 1000 and Agilent TapeStation instruments, fluorescently-labeled cRNA targets were synthesized with a commercially available kit (Ambion TotalPrep), and hybridized to Illumina HT-12 v4 bead arrays, which were read on an Illumina iScan Station at the UCLA Neuroscience Genomics Core. This array covers 34,000+ transcripts including the vast majority of named human genes. Detailed descriptions of our methods are provided elsewhere (Miller et al. 2009; Cole et al. 2011).
Covariates
On an a priori basis, we selected a panel of covariates that pattern by socioeconomic status and forecast perinatal outcomes, and abstracted values from electronic medical records. The covariates were maternal age, race (coded as European descent vs. Other), body mass index (BMI) before pregnancy, gestational diabetes, pre-eclampsia, delivery method (coded as vaginal vs. Caesarean), fetal sex, and gestational age at delivery.
Statistical Analyses
Data analytic methods have been previously detailed (Cole et al. 2005; Miller et al. 2009; Miller et al. 2008). Briefly, transcript abundance was estimated using default algorithms in Illumina GenomeStudio, and raw data were quantile-normalized and log2-transformed. Linear models were used to estimate the magnitude of differential gene expression across the disadvantage composite (Study 1) or between intervention and comparison groups (Study 2). All models included the panel of a priori selected covariates. In Study 1, genes showing ≥ 1.25-fold differential expression over the range from lowest to highest disadvantage served as input into higher-order bioinformatics analyses, which examined transcription control pathways (Cole et al. 2005) cellular origins (Cole et al. 2011), and mesenchymal vs. epithelial polarization (Choi et al. 2010). Statistical testing was based on standard errors derived from bootstrap resampling of linear model residual vectors (controlling for correlations across genes). In Study 2, we limited analyses to a subset of genes that showed the largest differential expression in Study 1. This approach directly addressed the mitigation question, asking whether Centering was associated with a reversal of differences related to disadvantage. The number of analyzed transcripts was selected by optimizing the ratio of observations to parameters (with four genes, 160:21). Mixed-effect linear models with unstructured covariance matrices were used, treating the four genes as repeated measures.
STUDY 1 RESULTS
Preliminary Analyses
Placenta tissue and umbilical blood were collected in 88/100 deliveries. (The remainder involved precluding emergencies or unplanned deliveries elsewhere.) To avoid confounding effects of prematurity, we excluded 9 deliveries that occurred before 37 weeks, leaving an analytic sample of 79 full-term pregnancies. The sample’s characteristics are described in Table 1.
Table 1.
Characteristics of Study 1 analytic sample (N = 79).
| Characteristic | Mean (SD) or N (%) |
|---|---|
| Age, years | 30.72 (5.30) |
| Racial/ethnic group, White (non-Latina) | 43 (54.43%) |
| Racial/ethnic group, Black (non-Latina) | 20 (25.31%) |
| Racial/ethnic group, Latina (any race) | 12 (15.19%) |
| Pre-pregnancy body mass index (BMI) | 27.35 (6.94) |
| Normal weight (BMI ≤ 24.99) | 39 (49.40) |
| Overweight (BMI 25.00 – 29.99) | 18 (22.80) |
| Obese (BMI ≥ 30.00) | 22 (27.80) |
| Nulliparous | 35 (44.30%) |
| Pre-eclampsia | 7 (8.86%) |
| Diabetes | 9 (11.39%) |
| Caesarean delivery | 22 (27.85%) |
| Gestational age at delivery, weeks | 39.20 (1.13) |
| Fetal sex, female | 36 (45.60) |
| Disadvantage composite (0–3) | 0.90 (1.18) |
| Disadvantage score = 0 | 45 (57.00) |
| Disadvantage score = 1 | 11 (13.90) |
| Disadvantage score = 2 | 9 (11.40) |
| Disadvantage score = 3 or more | 14 (17.70) |
In preliminary analyses, we estimated associations between disadvantage and the panel of covariates, using Pearson’s correlations for continuous outcomes and Spearman’s correlations for categorical outcomes. To the extent they were disadvantaged, women tended to be younger (r = −.31, p = .005) and identify as African-American (r = −.62, p < .0001). They also tended to have higher pre-pregnancy BMI (r = .45, p = .001), and were more likely to have gestational diabetes (r = .22, p = .05) and pre-eclampsia (r = .27, p = .02). Disadvantage was not associated with delivery method (r = .12, p = .30), gestational age at delivery (r = −.18, p = .09), or fetal sex (r = −.07, p = .57). Regardless, these characteristics are still likely to influence transcriptome profiles, so we included all of them as covariates in subsequent analyses of Study 1 data. This strategy helps to minimize risks of residual confounding.
Placenta Transcriptome
Net of the covariates, linear mixed models identified 344 transcripts showing ≥ 1.25-fold differential expression across the range of disadvantage (online Table S1). The 111 up-regulated genes included multiple transcripts involved in immune activation (CXCL14, CCL13, IL2B, SPP1, STAT4, TNFRSF21, TNFSF10), prostaglandin synthesis (PTGES, PTGDS), and tissue remodeling (COL1A1, COL1A2, COL5A1 COL5A2, KRT6A, KRT6C, KRT24, KRT7). Prominent among the 233 transcripts down-regulated with disadvantage were pregnancy-specific glycoproteins (PSG5, PSG7, PSG11) and the chorionic gonadotropin beta/luteinizing hormone beta gene cluster (CGB, CGB1, CGB5, CGB7, CGB8, LGB), which all have roles in fetal immune tolerance (Martinez et al. 2013; Bansal et al. 2012). Other down-regulated transcripts included suppressors of cytokine signaling (SOCS2, A2M, PIK3AP1), and inhibitors of histamine and prostaglandin activity (PGT, ABP1), as well as mediators of fetal nutrient access, blood supply, and organ maturation (CGB, CGB1, CGB5, CGB8, MMP12), and transcripts found to be hypermethylated in association with nutrient deficiency (MEG3, PEG10, SLC38A2, and LEP) (Tobi et al. 2015). Also down-regulated were the CRH gene, which regulates the timing of parturition, and genes involved in maturation of the bones, heart, brain, and kidneys (FRZB, GPNMB, INSL4, LEP, NUCB2, INSIG1, GADD45G, SEMA3B, HOXB2, NDRG1, TEAD3, TFAP2A).
Role of Covariates
The project was not designed to examine the influence of the demographic and obstetrical covariates themselves. Nevertheless, for interested readers we present the full results of the linear mixed models in online Table S2. For each of the 34,000+ measured transcripts, coefficients reflecting the influence of demographic and obstetrical covariates are displayed. When examining these results, it is important to keep in mind that most of the covariates have limited variability. As a result, the coefficients are likely to be imprecise estimates of their true (population) effects.
To roughly quantify the influence of covariates, we compared results from crude models, where the only predictor was disadvantage, to those from covariate-adjusted models. For placental transcripts identified as up-regulated, the average crude association for disadvantage was .44 log2 expression units and the average covariate-adjusted association for disadvantage for these transcripts was .26 log2 units. These results imply that 40.4% of the total association between disadvantage and expression of these transcripts was attributable to the demographic and obstetrical covariates. Accordingly, the residual 59.6% was attributable to either direct effects of disadvantage and/or other (unmeasured) mediating pathways. Similarly, among the placental transcripts identified above as down-regulated, the average disadvantage coefficient in crude models was −.45 log2 units and the average covariate-adjusted coefficient for these transcripts was −.33. Thus, for the down-regulated transcripts, covariates explained 27.3% of the association with disadvantage, whereas the remaining 72.7% was attributable to direct influences of disadvantaged and/or other (unmeasured) mediating pathways.
Upstream Regulatory Pathways
As in past research (Cole et al., 2011; Miller et al., 2008), our goal was not to discover individual mRNAs that mediate health disparities, but instead to use the pool of differentially expressed genes to glean insights about the activity of upstream transcriptional networks. Accordingly, this pool of genes served as input for TELiS, a bioinformatics tool that quantifies the prevalence of transcription factor binding motifs (TFBMs) in promoters of differentially expressed genes (Cole et al. 2005). Results appear in Figure 1 and online Table S3. Consistent with hypotheses regarding immune activation, TELiS results linked disadvantage to up-regulation of multiple transcription pathways involved in macrophage and lymphocyte activity (AP-1, GATA-1, EGR2, EGR4, MAF, EBF1). Disadvantage also was associated with down-regulated activity of NF-κB, which as we discuss later may be a reflection of its role in embryonic development (Espín-Palazón & Traver 2016).
Figure 1. Placenta transcriptome.
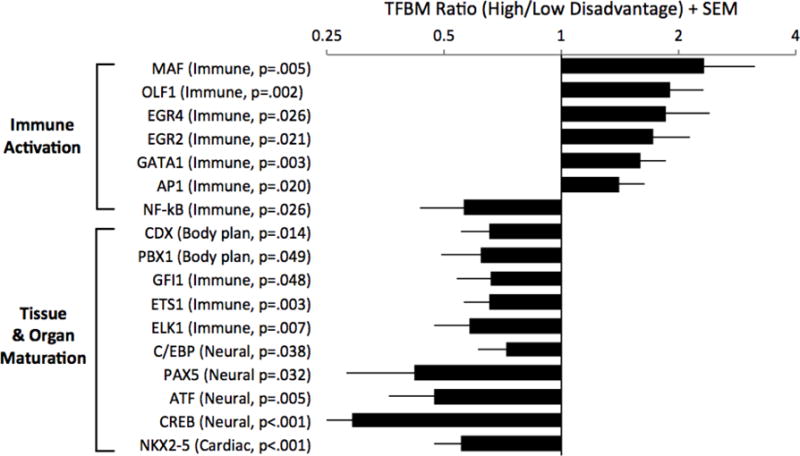
Genome-wide expression profiling was performed on biopsies of placental chorionic villi. Linear mixed models were used to estimate the magnitude of differential gene expression as a function of maternal socioeconomic disadvantage, adjusting for a panel of a priori selected covariates, with false discovery rate held at 5 percent. Genes showing ≥ 1.25-fold differential expression over the range from lowest to highest disadvantage served as input into higher-order bioinformatics analyses using the Transcription Factor Element Listening System. This platform quantified the prevalence of transcription factor binding motifs (TFBMs) in promoters of differentially expressed genes. TFBM ratios > 1 indicate specified transcriptional pathway is up-regulated with maternal disadvantage; ratios < 1 indicate converse.
Also consistent with hypotheses, results linked disadvantage to down-regulation of multiple transcription pathways involved in tissue development (Figure 1 and Table S3). These pathways included homeobox factors (CDX, PBX1), which control body plan and cell fate (Rezsohazy et al. 2015); members of the ETS family (ETS1, ELK1), which play roles in hematopoiesis (Ciau-Uitz et al. 2013), and NKX2-5, a key driver of cardiomyocyte development (McCulley & Black 2012). Of particular interest here were results indicating that disadvantage was associated with down-regulated activity of C/EBP-β, GFI1, PAX5, and CREB/ATF factors. These transcriptional pathways orchestrate brain maturation by promoting neurogenesis, dendrite formation, and synaptic plasticity (de la Torre-Ubieta & Bonni 2011; Blake & Ziman 2014).
Cellular Origins
Placental villi contain multiple leukocyte subsets as well as trophoblasts and fibroblasts. We used bioinformatic Transcript Origin Analyses in conjunction with reference datasets (Cole et al. 2011) to estimate how much each leukocyte population contributed to the transcriptional patterns described above. As shown in previous validation studies, this approach can accurately identify which specific cell type(s) mediate changes in gene expression within a heterogeneous pool of cell types (Cole et al. 2011). Results of these analyses identified monocytes and B-lymphocytes as major sources of the transcripts up-regulated in association with disadvantage (online Figure S1). No specific immune cell type was distinguished as an origin of down-regulated genes.
Fetal development entails a series of epithelial-mesenchymal transitions (EMTs) (Thiery et al. 2009). At the outset of pregnancy, EMTs mediate placental anchorage in the endometrium and remodel uterine circulation to perfuse the fetus. Later they facilitate tissue development by helping progenitor cells dislodge from epithelia, and acquire the specialized functions required to form organs. As these tissues mature, this process reverses to consolidate the cells into the well-structured epithelia characteristic of a mature organ. EMTs also play a role in mature tissue responses to injury. Consistent with a less fully mature or more injured tissue, TOA found disadvantage-related transcripts to be more predominately mesenchymal versus epithelial in status (online Figure S1).
Cord Blood Transcriptome
Net of covariates, linear mixed models identified 610 transcripts from cord blood cells that were differentially expressed by ≥ 1.25-fold across the range of disadvantage (online Table S4). Among the 379 up-regulated transcripts were genes involved with response to hypoxia (HBG2, HBE1, HBB, HBM, HBBP1) and anti-viral/cell-mediated immunity (IL18, IFI27, ISG20, FOX04, CD8A, CD5). The 231 down-regulated genes included transcripts involved in monocyte chemotaxis and activation (CCL3, TLR4, IL1B, IL1R2, IL8) and transcription factors involved in myeloid effector functions (MAFB, EGR1, EGR2, CEBPA). Also down-regulated were transcripts involved in B-cell activation and differentiation (CD19, FCRLA, CD72, CD24, CD52), and antigen presentation (HLADQB1, HLADRB3, HLADRB4, HLADRB6, HLADOB).
Role of Covariates
For readers interested in associations between demographic and obstetrical covariates and the umbilical transcriptome, online Table S2 displays the complete results of linear mixed models. As with the placenta results, readers should keep in mind that most of the covariates have limited variability and, as a consequence, the coefficients are likely to be imprecise estimates of their true (population) effects. To roughly quantify the nature of these effects, we again compared results from crude and adjusted models. Among the umbilical transcripts identified as up-regulated, the average crude association for disadvantage was .44 log2 units and the average covariate-adjusted coefficient for these transcripts was .42 log2 units. These patterns imply that less than three percent of disadvantage’s total association with these transcripts was attributable to demographic and obstetrical covariates. For the transcripts identified as down-regulated, the average crude coefficient was −.43 and the average covariate-adjusted coefficient for these transcripts was −.38, implying that covariates explained 11.8% of these relationships.
Upstream Regulatory Pathways
TELiS analysis of umbilical blood data (Figure 2, online Table S5) linked disadvantage with up-regulation of c-Myb, which mobilizes hematopoietic stem and progenitor cells, and a panel of transcription factors indicative of up-regulation of lymphoid lineage precursor cells (E2F, ETS1, ELK1, EGR1, EGR3) (Gómez-Martín et al. 2010). Results also linked disadvantage to a down-regulation of transcription factors involved in myeloid cell differentiation and mature effector activities (i.e., C/EBP-β, EVI1, TAL1, AP-1, STAT1, STAT3, NF-κB).
Figure 2. Cord blood transcriptome.
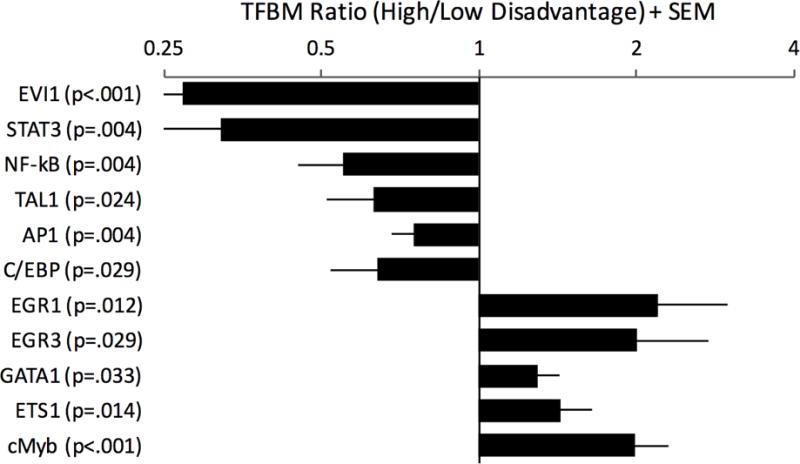
Genome-wide expression profiling was performed on bulk cord blood cells (i.e., un-separated) from newborns. Linear mixed models were used to estimate the magnitude of differential gene expression as a function of maternal socioeconomic disadvantage, adjusting for a panel of a priori selected covariates, with false discovery rate held at 5 percent. Genes showing ≥ 1.25-fold differential expression over the range from lowest to highest disadvantage served as input into higher-order bioinformatics analyses using the Transcription Factor Element Listening System. This platform quantified the prevalence of transcription factor binding motifs (TFBMs) in promoters of differentially expressed genes. TFBM ratios > 1 indicate specified transcriptional pathway is up-regulated with maternal disadvantage; ratios < 1 indicate converse.
Cellular Origins
Umbilical blood contains multiple types of leukocytes. As with the placenta analyses, we used Transcript Origin Analyses in conjunction with leukocyte reference datasets (Cole et al. 2011) to estimate how much each of these cell types contributed to the transcriptional patterns described above. The results identified monocytes, dendritic cells, and macrophages as primary sources of downregulated cord blood transcripts (online Figure S2). B-lymphocytes were identified as a source of both upregulated and downregulated transcripts. One potential explanation for this biphasic pattern is that disadvantage is associated with a shift in B-lymphocyte differentiation patterns, e.g., in the balance of precursor vs. mature B-lymphocytes or naive vs. activated phenotypes).
STUDY 2 RESULTS
Study 2 collected placenta biopsies and cord blood from 20 women in CenteringPregnancy, and 20 comparison women in traditional prenatal care who delivered babies in the same clinical setting over the same time period. As Table 2 illustrates, women in this study were generally low in socioeconomic status, with 35% lacking a high-school diploma, and only 5% having completed a Bachelor’s degree. The groups had similar demographic and obstetrical profiles except that women in Centering were slightly older. There also were hints of group differences in fetal sex, gestational age, diabetes, and preeclampsia (p’s < .20). To minimize the risks of confounding, we included all five of these covariates (maternal age, fetal sex, gestational age, diabetes, and preeclampsia) along with Centering status in statistical models.
Table 2.
Characteristics of Study 2 sample according to intervention status (N=40).
| Centering Group Mean (SD) or N (%) |
Comparison Group Mean (SD) or N (%) |
Difference (p value) |
|
|---|---|---|---|
| Age, years | 24.80 (4.36) | 28.45 (6.19) | .04 |
| Racial/ethnic group, White | 9 (45.0 %) | 10 (50.0 %) | .75 |
| Racial/ethnic group, Black | 4 (20.0 %) | 4 (20.0 %) | .99 |
| Racial/ethnic group, Latina | 7 (35.0 %) | 5 (25.0%) | .49 |
| Pre-pregnancy BMI | 30.51 (6.25) | 28.66 (6.01) | .35 |
| Nulliparous | 6 (30.0 %) | 5 (25.0 %) | .72 |
| Pre-eclampsia | 0 (0.0 %) | 2 (10.0%) | .15 |
| Diabetes | 0 (0.0 %) | 2 (10.0%) | .15 |
| Caesarean delivery | 6 (30.0 %) | 5 (25.0 %) | .72 |
| Gestational age, weeks | 39.57 (1.05) | 39.01 (1.37) | .16 |
| Fetal sex, female | 11 (55.0 %) | 8 (40.0 %) | .34 |
| Didn’t complete high school | 6 (30.0 %) | 8 (40.0 %) | .51 |
| High school diploma, but no Bachelor’s degree | 14 (70.0 %) | 10 (50.0%) | .74 |
As noted in the Statistical Analyses section, analyses of transcriptional activity focused on a set of four placenta transcripts that were most strongly associated with disadvantage in Study 1. These transcripts were CGB1, CCK, LHB, and KRTAP26-1, and all of them were down-regulated in association with disadvantage in Study 1. Consistent with hypotheses, women in Centering showed higher average expression of these mRNAs relative to Controls, both in crude (F = 12.56, p = .001, d = .63; Figure 3a) and adjusted mixed-model analyses (F = 11.10, p = .002, d = .59).
Figure 3. CenteringPregnancy Intervention.
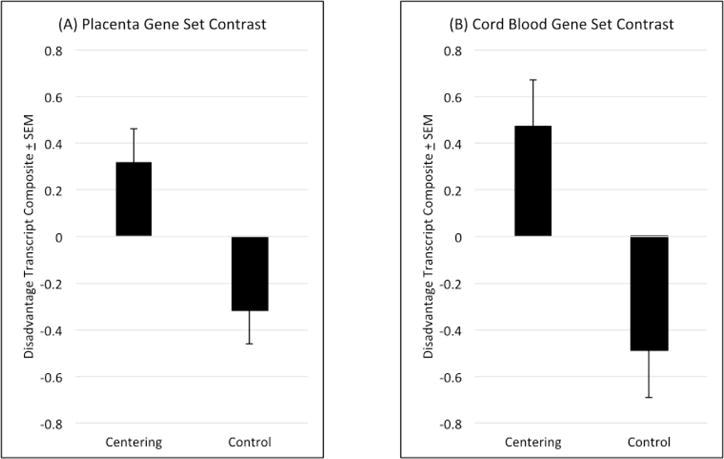
Relative to comparison subjects, women who participated in the intervention displayed higher expression of transcripts that were down-regulated in concert with disadvantage in Study 1. These patterns were apparent in both (A) biopsies of the placenta’s chorionic villous layer and (B) bulk cord blood cells (i.e., un-separated) from newborns.
We next conducted parallel analyses of the 4 cord blood transcripts most strongly associated with disadvantage in Study 1. They were GH1, CSH1, CSHL1, and CSH2; and again all were down-regulated in connection with disadvantage in Study 1. Again, consistent with hypotheses, women in Centering showed higher average expression of these transcripts relative to Controls in mixed models (crude: F = 11.75, p = .002, d = .96; covariate-adjusted: F = 19.75, p < .001, d = 1.27; Figure 3b).
DISCUSSION
Economic hardship in childhood is associated with a variety of adverse outcomes across the lifecourse, spanning the cognitive, psychiatric, and biomedical domains. The prevailing mechanistic accounts of these disparities focus on variations in the postnatal environment. However, results of the studies presented here indicate that molecular correlates of maternal disadvantage are present in mRNA from the placenta’s chorionic villous layer and newborn umbilical blood cells. These patterns are consistent with the possibility that some of the biological substrates of lifecourse disparities originate in utero. Because of the study’s observational design, it is unclear whether these associations reflect a causal influence of disadvantage. However, experimental studies in animals demonstrate the biological plausibility of a causal effect, showing that gestational exposure to stressors associated with disadvantage can leave molecular footprints detectable in the nervous, immune, and metabolic systems of offspring (Bale 2015; Hanson & Gluckman 2014; Prescott 2006; Pryce et al. 2005; Coe & Lubach 2014).
The findings also provide clues about the mechanistic origins of socioeconomic disparities. Several themes emerged, all paralleling experimental models of pregnancy stress (Blois et al. 2005; Friebe et al. 2011; Howerton et al. 2013; Hanson & Gluckman 2014). First, disadvantage was associated with multiple indications of perturbed immune homeostasis in the placenta. The patterns suggest a scenario where disadvantage impairs maternal immune tolerance of the fetal allograft, as reflected in decreased expression of pregnancy-specific glycoproteins, chorionic gonadotropins, and other anti-inflammatory mediators (Martinez et al. 2013; Bansal et al. 2012). This diminution of tolerance is permissive of immune activity in the chorionic villi, as reflected in up-regulation of the AP-1, GATA-1, EGR2, EGR4, and MAF transcriptional pathways, which collectively are hallmarks of macrophage and lymphocyte activation (Gómez-Martín et al. 2010; Natoli et al. 2011; Naito et al. 2011; Geissmann et al. 2010). Consistent with this interpretation, bioinformatic analyses suggested these differential transcription profiles were orchestrated by monocytes and macrophages, B-lymphocytes, and to a lesser extent, dendritic cells. The villi are an immune privileged site, with a pivotal role in nutrient exchange. To the extent that immune activity disrupts nutrient transfer (Dimasuay et al. 2016), it could underlie a second prominent theme in the results, the down-regulation of transcriptional pathways that orchestrate tissue maturation. Indeed, disadvantage was associated with bioinformatic indications of reduced activity of transcription factors that control body plan, and promote maturation of the heart, brain, and immune system (Rezsohazy et al. 2015; Ciau-Uitz et al. 2013; de la Torre-Ubieta & Bonni 2011; Blake & Ziman 2014; McCulley & Black 2012). Cellular origin analyses substantiated these results, revealing patterns characteristic of mesenchymal cells engaged in organ development.
Whether these transcriptional patterns are sufficient to functionally alter trajectories of organ development is not yet clear. But socioeconomic disparities in phenotypic development are apparent in early childhood, particularly in the brain. By 6 months, socioeconomic differences in the functional connectivity of default-mode and sensori-motor networks are apparent (Gao et al. 2015). By school age, there are well-established socioeconomic disparities in amygdala and hippocampal volume, as well as cortical grey matter (Hanson et al. 2013; Luby et al. 2013). The placenta gene expression findings reported here could be gestational manifestations of these later phenotypic variations in brain maturation. Consistent with the developmental relevance of our observations in the placenta, analyses revealed transcriptional correlates of disadvantage in umbilical blood cells, suggesting that newborns enter the world with distinct immunologic profiles, which pattern by socioeconomic conditions. The transcription profile of disadvantaged newborns was consistent with a relative immaturity of myeloid cells, as reflected in down-regulation of pathways that orchestrate functions like phagocytosis, antigen processing and presentation, and tissue remodeling (Gómez-Martín et al. 2010; Ciau-Uitz et al. 2013). These newborns also showed up-regulation of pathways that control mobilization of lymphoid progenitors, and commitment and differentiation of B-lymphocytes (Medvedovic et al. 2011; Phelan et al. 2010). Research is needed to clarify the functional significance of these variations for subsequent immune functioning and health outcomes.
Broadly speaking, the perturbed immune homeostasis seen here is consistent with results of earlier research on transcriptional correlates of socioeconomic disadvantage (Chen et al. 2009; Powell et al. 2013; Miller et al. 2009; Levine et al. 2015). But in several regards there were substantive differences in the pattern of results. First, in placenta biopsies disadvantage was generally associated with transcriptional indications of immune activation, as reflected in up-regulation of the AP-1, GATA-1, EGR2, EGR4, and MAF pathways (Gómez-Martín et al. 2010; Natoli et al. 2011; Naito et al. 2011; Geissmann et al. 2010). However, these analyses simultaneously indicated down-regulated activity of NF-κB, a pivotal pro-inflammatory transcription factor, which has shown up-regulation in previous studies of disadvantage and adversity more generally (Cole 2014; Chen et al. 2009; Powell et al. 2013; Miller et al. 2009; Cole et al. 2011; Miller et al. 2014b; Cole et al. 2007). Future research will be required to determine why the NF-κB up-regulation previously associated with social adversity in the blood cells of more developmentally mature humans is not observed here in the context of placental villi. However, it seems plausible that in trophoblasts, fibroblasts, and other cells of the placenta, NF-κB plays different functional roles from those of mature leukocyte populations observed in previous studies. Indeed, emerging research shows that during embryonic development, NF-κB signaling is crucial in maturation of the liver and brain, as well as limbs, muscle, and skin (Espín-Palazón & Traver 2016). If that is (primarily) what our measure of placental NF-κB activity reflects, the pattern of results could be understood as convergent with this study’s other findings connecting disadvantage with transcriptional indications of slower tissue maturation. A second inconsistency with previous research is the pattern observed in umbilical blood cells. Here, disadvantage was associated with a bioinformatic profile suggesting relative down-regulation of myeloid cells functions and up-regulation of lymphoid cell functions. These patterns contrast markedly with the “Conserved Transcriptional Response to Adversity (CTRA),” seen in peripheral leukocytes of more developmentally mature humans exposed to various forms of social adversity (Cole 2014; Cole 2010; Irwin & Cole 2011). Again, future research will be required to identify the basis for the observed differences, but we speculate that they might reflect (a) disparities between the cellular makeup of umbilical versus antecubital blood, and/or (b) disparities in these cells’ developmental stage and historical exposure to microbial and hormonal stimulation. Either way, our results suggest that in umbilical vein cells from newborns, adversity does not manifest in the CTRA phenotype of increased pro-inflammatory and decreased anti-viral activity observed later in development. Instead, it appears to be associated with a profile indicative of relative hematopoietic immaturity, which is consistent with results of studies in non-human primates. When exposed to stress during the prenatal period, newborn monkeys display reductions in lymphocyte proliferation, natural killer activity, and cytokine production (Coe & Lubach 2005). If substantiated in future research, these findings would suggest significant developmental heterogeneity in the transcriptional response to adversity.
Consistent with other studies suggesting that early disadvantage can be mitigated (Campbell et al. 2014; Miller et al. 2014a), we found that participation in CenteringPregnancy was associated with favorable gene expression profiles in placenta biopsies and umbilical cells. These patterns suggest the possibility that Centering ameliorated some of the transcriptional disparities most strongly connected to disadvantage in Study 1. However, given the observational design of the study, the small number of participants, the potential for Type 1 error, and the fallibility of covariance procedures in rendering groups truly equivalent, these results should be interpreted cautiously. To ascertain the causal status of our observations, a randomized, controlled trial of Centering with the same outcomes is needed. If efficacious, this trial would also provide the opportunity to address questions about mechanisms of action. Centering reduces distress, improves lifestyle, and bolsters social support in high-risk women (Heberlein et al. 2016; Ickovics et al. 2011). Any of these changes might ameliorate immune activation in the placenta (Straub et al. 2014) and their role should be explored in follow-up studies.
In the meantime, further research is needed to substantiate our findings regarding disadvantage and elucidate their underpinnings and significance. After confirming the patterns seen here, research should identify the cellular actors involved, and clarify whether disadvantage is associated with their distribution, transcriptional patterns, or both. Research also should identify factors that connect disadvantage with transcriptional activity. Our study considered a host of demographic (age, racial/ethnic background) and obstetrical (BMI, diabetes, pre-eclampsia, delivery method, fetal sex, gestational age at delivery) factors. Collectively, they explained 3–40 percent of the associations between disadvantage and differential gene expression in both placenta biopsies and umbilical blood, which implies that other mechanistic pathways are substantial contributors to these relationships. Thus, future research should consider the role of other presumptive mediators, including variations in nutrition, glucocorticoid activity, toxin exposure, and intrauterine infection. Interestingly, many functions of the placenta are regulated by a circadian clock (Waddell et al. 2012). Given the marked socioeconomic variations in sleep and activity cycles (Laposky et al. 2016), clock-related disruptions are a potential mechanism to consider in follow-up research. Substance use might also play a role, although is unlikely to be a factor here. Only two women drank alcohol during pregnancy (both < 1 drink weekly) and two others smoked (both < 5 cigarettes daily.) A more thorough assessment of characteristics and complications of delivery is also warranted; we did not have the statistical power or range of variation in risk factors to do that here. Finally, longitudinal research that tracks newborns’ health into childhood is needed, so the phenotypic relevance of the differences we observed can be evaluated.
Despite these uncertainties, these results suggest that multiple dimensions of newborn gene expression are dysregulated following gestation in adverse socioeconomic conditions. The results also provide encouraging, albeit preliminary, suggestions that some of those alternations might be ameliorated by a prenatal intervention that emphasizes the importance of women’s psychosocial environment alongside more conventional obstetric characteristics.
Supplementary Material
Highlights.
In pregnant women, socioeconomic disadvantage was associated with a placenta transcriptional profile indicative of higher immune activation and slower fetal maturation
These patterns were particularly evident in pathways related to brain, heart, and immune development
Cord blood cells of disadvantaged newborns also showed indications of immaturity, as reflected in down-regulation of pathways that coordinate myeloid cell development
A small follow-up study found preliminary support for the hypothesis that group-based prenatal care, focusing on women’s psychosocial status, could ameliorate some of these socioeconomic differences
Acknowledgments
Supported by NIH grants HL122328, DA027827, and NorthShore University HealthSystem. The funders had no role in study design, statistical analysis, data interpretation, or manuscript preparation.
Abbreviations
- BMI
body mass index
- EMT
epithelial-mesenchymal transition
- IL
interleukin
- NF-κB
nuclear factor kappa B
- TFBM
transcription factor binding motif
- TNFα
tumor necrosis factor alpha
- TELiS
transcription element listening system
- TOA
transcript origin analysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
None of the authors reports a (1) biomedical financial interest or (2) potential conflict of interest related to this study.
References
- Arck PC, Hecher K. Fetomaternal immune cross-talk and its consequences for maternal and offspring’s health. Nat Med. 2013;19:548–556. doi: 10.1038/nm.3160. [DOI] [PubMed] [Google Scholar]
- Bale TL. Epigenetic and transgenerational reprogramming of brain development. Nat Rev Neurosci. 2015;16:332–344. doi: 10.1038/nrn3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal AS, Bora SA, Saso S, Smith JR, Johnson MR, Thum MY. Mechanism of human chorionic gonadotrophin-mediated immunomodulation in pregnancy. Expert Rev Clin Immunol. 2012;8:747–753. doi: 10.1586/eci.12.77. [DOI] [PubMed] [Google Scholar]
- Bilbo SD, Frank A. Beach award: programming of neuroendocrine function by early-life experience: a critical role for the immune system. Horm Behav. 2013;63:684–691. doi: 10.1016/j.yhbeh.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Ziman MR. Pax genes: regulators of lineage specification and progenitor cell maintenance. Development. 2014;141:737–751. doi: 10.1242/dev.091785. [DOI] [PubMed] [Google Scholar]
- Blois S, Tometten M, Kandil J, Hagen E, Klapp BF, Margni RA, Arck PC. Intercellular adhesion molecule-1/LFA-1 cross talk is a proximate mediator capable of disrupting immune integration and tolerance mechanism at the feto-maternal interface in murine pregnancies. J Immunol. 2005;174:1820–1829. doi: 10.4049/jimmunol.174.4.1820. [DOI] [PubMed] [Google Scholar]
- Braun T, Challis JR, Newnham JP, Sloboda DM. Early-life glucocorticoid exposure: the hypothalamic-pituitary-adrenal axis, placental function, and long-term disease risk. Endocr Rev. 2013;34:885–916. doi: 10.1210/er.2013-1012. [DOI] [PubMed] [Google Scholar]
- Bronson SL, Bale TL. The placenta as a mediator of stress effects on neurodevelopmental reprogramming. Neuropsychopharmacology. 2016;41:207–218. doi: 10.1038/npp.2015.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell F, Conti G, Heckman JJ, Moon SH, Pinto R, Pungello E, Pan Y. Early childhood investments substantially boost adult health. Science. 2014;343:1478–1485. doi: 10.1126/science.1248429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Miller GE, Walker HA, Arevalo JM, Sung CY, Cole SW. Genome-wide transcriptional profiling linked to social class in asthma. Thorax. 2009;64:38–43. doi: 10.1136/thx.2007.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Matthews K, Boyce WT. Socioeconomic differences in children’s health: How and why do these relationships change with age? Psychol Bull. 2002;128:295–329. doi: 10.1037/0033-2909.128.2.295. [DOI] [PubMed] [Google Scholar]
- Choi YL, Bocanegra M, Kwon MJ, Shin YK, Nam SJ, Yang JH, Kao J, Godwin AK, Pollack JR. LYN is a mediator of epithelial-mesenchymal transition and a target of dasatinib in breast cancer. Cancer Res. 2010;70:2296–2306. doi: 10.1158/0008-5472.CAN-09-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciau-Uitz A, Wang L, Patient R, Liu F. ETS transcription factors in hematopoietic stem cell development. Blood Cells Mol Dis. 2013;51:248–255. doi: 10.1016/j.bcmd.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Coe CL, Lubach GR. Prenatal origins of individual variation in behavior and immunity. Neurosci Biobehav Rev. 2005;29:39–49. doi: 10.1016/j.neubiorev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Coe CL, Lubach GR. Vital and vulnerable functions of the primate placenta critical for infant health and brain development. Front Neuroendocrinol. 2014;35:439–446. doi: 10.1016/j.yfrne.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Sung CS, Rose RM, Cacioppo JT. Social regulation of gene expression: Inflammation and the human transcriptional response to loneliness. Genome Biol. 2007;8:r89. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW. Elevating the perspective on human stress genomics. Psychoneuroendocrinology. 2010;35:955–962. doi: 10.1016/j.psyneuen.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW. Human social genomics. PLoS Genet. 2014;10:e1004601. doi: 10.1371/journal.pgen.1004601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci U S A. 2011;108:3080–3085. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 2005;21:803–810. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Bonni A. Transcriptional regulation of neuronal polarity and morphogenesis in the mammalian brain. Neuron. 2011;72:22–40. doi: 10.1016/j.neuron.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimasuay KG, Boeuf P, Powell TL, Jansson E. Placental responses to changes in the maternal environment determine fetal growth. Front Physiol. 2016;7:12. doi: 10.3389/fphys.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan GJ, Murnane J. Whither Opportunity? Rising Inequality, Schools, and Children’s Life Chances. New York: Russell Sage Foundation; 2011. [Google Scholar]
- Elgar FJ, Pförtner TK, Moor I, De Clercq B, Stevens GW, Currie C. Socioeconomic inequalities in adolescent health 2002–2010: a time-series analysis of 34 countries participating in the Health Behaviour in School-aged Children study. Lancet. 2015;385:2088–2095. doi: 10.1016/S0140-6736(14)61460-4. [DOI] [PubMed] [Google Scholar]
- Entringer S, Buss C, Wadhwa PD. Prenatal stress, telomere biology, and fetal programming of health and disease risk. Sci Signal. 2012;5:pt12. doi: 10.1126/scisignal.2003580. [DOI] [PubMed] [Google Scholar]
- Espín-Palazón R, Traver D. The NF-κB family: Key players during embryonic development and HSC emergence. Exp Hematol. 2016;44:519–527. doi: 10.1016/j.exphem.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Estes ML, McAllister AK. Maternal immune activation: Implications for neuropsychiatric disorders. Science. 2016;353:772–777. doi: 10.1126/science.aag3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans GW. The environment of childhood poverty. Am Psychol. 2004;59:77–92. doi: 10.1037/0003-066X.59.2.77. [DOI] [PubMed] [Google Scholar]
- Friebe A, Douglas AJ, Solano E, Blois SM, Hagen E, Klapp BF, Clark DA, Arck PC. Neutralization of LPS or blockage of TLR4 signaling prevents stress-triggered fetal loss in murine pregnancy. J Mol Med (Berl) 2011;89:689–699. doi: 10.1007/s00109-011-0743-5. [DOI] [PubMed] [Google Scholar]
- Galobardes B, Lynch JW, Smith GD. Is the association between childhood socioeconomic circumstances and cause-specific mortality established? Update of a systematic review. J Epidemiol Community Health. 2008;62:387–390. doi: 10.1136/jech.2007.065508. [DOI] [PubMed] [Google Scholar]
- Gao W, Alcauter S, Elton A, Hernandez-Castiloo CR, Smith JK, Ramirez J, Lin W. Functional network development during the first year: Relative sequence and socioeconomic correlations. Cerebral Cortex. 2015;25:2919–2928. doi: 10.1093/cercor/bhu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Martín D, Díaz-Zamudio M, Galindo-Campos M, Alcocer-Varela J. Early growth response transcription factors and the modulation of immune response: implications towards autoimmunity. Autoimmun Rev. 2010;9:454–458. doi: 10.1016/j.autrev.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Hackman DA, Farah MJ, Meaney MJ. Socioeconomic status and the brain: mechanistic insights from human and animal research. Nat Rev Neurosci. 2010;11:651–659. doi: 10.1038/nrn2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale N, Picklesimer AH, Billings DL, Covington-Kolb S. The impact of Centering Pregnancy Group Prenatal Care on postpartum family planning. Am J Obstet Gynecol. 2014;210:50.e1–7. doi: 10.1016/j.ajog.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Hanson JL, Hair N, Shen DG, Shi F, Gilmore JH, Wolfe BL, Pollak SD. Family poverty affects the rate of human infant brain growth. PLoS One. 2013;8:e80954. doi: 10.1371/journal.pone.0080954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Gluckman PD. Early developmental conditioning of later health and disease: physiology or pathophysiology. Physiol Rev. 2014;94:1027–1076. doi: 10.1152/physrev.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein EC, Picklesimer AH, Billings DL, Covington-Kolb S, Farber N, Frongillo EA. The comparative effects of group prenatal care on psychosocial outcomes. Arch Womens Ment Health. 2016;19:259–269. doi: 10.1007/s00737-015-0564-6. [DOI] [PubMed] [Google Scholar]
- Hertzman C, Boyce T. How experience gets under the skin to create gradients in developmental health. Annu Rev Public Health. 2010;31:329–347. doi: 10.1146/annurev.publhealth.012809.103538. [DOI] [PubMed] [Google Scholar]
- Howerton CL, Morgan CP, Fischer DB, Bale TL. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc Natl Acad Sci U S A. 2013;110:5169–5174. doi: 10.1073/pnas.1300065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ickovics JR, Earnshaw V, Lewis JB, Kershaw TS, Magriples U, Stasko E, Rising SS, Cassells A, Cunningham S, Bernstein P, Tobin JN. Cluster randomized controlled trial of group prenatal care: Perinatal outcomes among adolescents in New York City health centers. Am J Public Health. 2016;106:359–365. doi: 10.2105/AJPH.2015.302960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ickovics JR, Kershaw TS, Westdahl C, Magriples U, Massey Z, Reynolds H, Rising SS. Group prenatal care and perinatal outcomes: a randomized controlled trial. Obstet Gynecol. 2007;110:330–339. doi: 10.1097/01.AOG.0000275284.24298.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ickovics JR, Reed E, Magriples U, Westdahl C, Schindler Rising S, Kershaw TS. Effects of group prenatal care on psychosocial risk in pregnancy: results from a randomised controlled trial. Psychol Health. 2011;26:235–250. doi: 10.1080/08870446.2011.531577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol. 2011;11:625–632. doi: 10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Champagne FA. Early-life experience, epigenetics, and the developing brain. Neuropsychopharmacology. 2015;40:141–153. doi: 10.1038/npp.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposky AD, Van Cauter E, Diez-Roux AV. Reducing health disparities: the role of sleep deficiency and sleep disorders. Sleep Med. 2016;18:3–6. doi: 10.1016/j.sleep.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Cole SW, Weir DR, Crimmins EM. Childhood and later life stressors and increased inflammatory gene expression at older ages. Soc Sci Med. 2015;130:16–22. doi: 10.1016/j.socscimed.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby J, Belden A, Botteron K, Marrus N, Harms MP, Babb C, Nishino T, Barch D. The effects of poverty on childhood brain development: the mediating effect of caregiving and stressful life events. JAMA Pediatr. 2013;167:1135–1142. doi: 10.1001/jamapediatrics.2013.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FF, Cervi L, Knubel CP, Panzetta-Dutari GM, Motran CC. The role of pregnancy-specific glycoprotein 1a (PSG1a) in regulating the innate and adaptive immune response. Am J Reprod Immunol. 2013;69:383–394. doi: 10.1111/aji.12089. [DOI] [PubMed] [Google Scholar]
- Masten AS, Narayan AJ. Child development in the context of disaster, war, and terrorism: pathways of risk and resilience. Annu Rev Psychol. 2012;63:227–257. doi: 10.1146/annurev-psych-120710-100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin KA, Breslau J, Green JG, Lakoma MD, Sampson NA, Zaslavsky AM, Kessler RC. Childhood socio-economic status and the onset, persistence, and severity of DSM-IV mental disorders in a US national sample. Soc Sci Med. 2011;73:1088–1096. doi: 10.1016/j.socscimed.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol. 2011;111:179–206. doi: 10.1016/B978-0-12-385991-4.00005-2. [DOI] [PubMed] [Google Scholar]
- Meyer U. Developmental neuroinflammation and schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2013;42:20–34. doi: 10.1016/j.pnpbp.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Miller GE, Chen E, Sze J, Marin T, Arevalo JMG, Doll R, Ma R, Cole SW. A genomic fingerprint of chronic stress in humans: Blunted glucocorticoid and increased NF-κB signaling. Biol Psychiatry. 2008;64:266–272. doi: 10.1016/j.biopsych.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Fok A, Walker H, Lim A, Nicholls EP, Cole SW, Kobor MS. Low early-life social class leaves a biological residue manifest by decreased glucocorticoid and increased pro-inflammatory signaling. Proc Natl Acad Sci U S A. 2009;106:14716–14721. doi: 10.1073/pnas.0902971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Brody GH, Yu T, Chen E. A family-oriented psychosocial intervention reduces inflammation in low-SES African American youth. Proc Natl Acad Sci U S A. 2014a;111:11287–11292. doi: 10.1073/pnas.1406578111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Chen E, Parker KJ. Psychological stress in childhood and susceptibility to the chronic diseases of aging: Moving toward a model of behavioral and biological mechanisms. Psychol Bull. 2011;137:959–997. doi: 10.1037/a0024768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GE, Murphy ML, Cashman R, Ma R, Ma J, Arevalo JM, Kobor MS, Cole SW. Greater inflammatory activity and blunted glucocorticoid signaling in monocytes of chronically stressed caregivers. Brain Behav Immun. 2014b;41:191–199. doi: 10.1016/j.bbi.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montez JK, Hayward MD. Cumulative childhood adversity, educational attainment, and active life expectancy among U.S. adults. Demography. 2014;51:413–435. doi: 10.1007/s13524-013-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Tanaka H, Naoe Y, Taniuchi I. Transcriptional control of T-cell development. Int Immunol. 2011;23:661–668. doi: 10.1093/intimm/dxr078. [DOI] [PubMed] [Google Scholar]
- Natoli G, Ghisletti S, Barozzi I. The genomic landscapes of inflammation. Genes Dev. 2011;25:101–106. doi: 10.1101/gad.2018811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslock R, Miller GE. Early-life adversity and physical and emotional health across the lifespan: a neuroimmune network hypothesis. Biol Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan JD, Shroyer NF, Cook T, Gebelein B, Grimes HL. Gfi1-cells and circuits: unraveling transcriptional networks of development and disease. Curr Opin Hematol. 2010;17:300–307. doi: 10.1097/MOH.0b013e32833a06f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picklesimer AH, Billings D, Hale N, Blackhurst D, Covington-Kolb S. The effect of CenteringPregnancy group prenatal care on preterm birth in a low-income population. Am J Obstet Gynecol. 2012;206:415.e1–7. doi: 10.1016/j.ajog.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Powell ND, Sloan EK, Bailey MT, Arevalo JM, Miller GE, Chen E, Kobor MS, Reader BF, Sheridan JF, Cole SW. Social stress up-regulates inflammatory gene expression in the leukocyte transcriptome via beta-adrenergic induction of myelopoiesis. Proc Natl Acad Sci U S A. 2013;110:16574–16579. doi: 10.1073/pnas.1310655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott SL. The development of respiratory inflammation in children. Paediatr Respir Rev. 2006;7:89–96. doi: 10.1016/j.prrv.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Pryce CR, Ruedi-Bettschen D, Dettling AC, Weston A, Russig H, Ferger B, Feldon J. Long-term effects of early-life environmental manipulations in rodents and primates: Potential animal models in depression research. Neurosci Biobehav Rev. 2005;29:649–674. doi: 10.1016/j.neubiorev.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Reiss F. Socioeconomic inequalities and mental health problems in children and adolescents: a systematic review. Soc Sci Med. 2013;90:24–31. doi: 10.1016/j.socscimed.2013.04.026. [DOI] [PubMed] [Google Scholar]
- Rezsohazy R, Saurin AJ, Maurel-Zaffran C, Graba Y. Cellular and molecular insights into Hox protein action. Development. 2015;142:1212–1227. doi: 10.1242/dev.109785. [DOI] [PubMed] [Google Scholar]
- Schreier HM, Chen E. Socioeconomic status and the health of youth: a multilevel, multidomain approach to conceptualizing pathways. Psychol Bull. 2013;139:606–654. doi: 10.1037/a0029416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonkoff JP, Garner AS. The lifelong effects of early childhood adversity and toxic stress. Pediatrics. 2012;129:e232–46. doi: 10.1542/peds.2011-2663. [DOI] [PubMed] [Google Scholar]
- Straub H, Qadir S, Miller G, Borders A. Stress and stress reduction. Clin Obstet Gynecol. 2014;57:579–606. doi: 10.1097/GRF.0000000000000038. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Tobi EW, Slieker RC, Stein AD, Suchiman HE, Slagboom PE, van Zwet EW, Heijmans BT, Lumey LH. Early gestation as the critical time-window for changes in the prenatal environment to affect the adult human blood methylome. Int J Epidemiol. 2015;44:1211–1223. doi: 10.1093/ije/dyv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell BJ, Wharfe MD, Crew RC, Mark PJ. A rhythmic placenta? Circadian variation, clock genes and placental function. Placenta. 2012;33:533–539. doi: 10.1016/j.placenta.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Wright RJ. Epidemiology of stress and asthma: from constricting communities and fragile families to epigenetics. Immunol Allergy Clin North Am. 2011;31:19–39. doi: 10.1016/j.iac.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RJ, Subramanian SV. Advancing a multilevel framework for epidemiologic research on asthma disparities. Chest. 2007;132:757S–769S. doi: 10.1378/chest.07-1904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


