Abstract
Redox sensing is a ubiquitous mechanism regulating cellular activity. Fungal pathogens face reactive oxygen species produced by the host plant's oxidative burst in addition to endogenous reactive oxygen species produced during aerobic metabolism. An array of preformed and induced detoxifying enzymes, including superoxide dismutase, catalases, and peroxidases, could allow fungi to infect plants despite the oxidative burst. We isolated a gene (CHAP1) encoding a redox-regulated transcription factor in Cochliobolus heterostrophus, a fungal pathogen of maize. CHAP1 is a bZIP protein that possesses two cysteine-rich domains structurally and functionally related to Saccharomyces cerevisiae YAP1. Deletion of CHAP1 in C. heterostrophus resulted in decreased resistance to oxidative stress caused by hydrogen peroxide and menadione, but the virulence of chap1 mutants was unaffected. Upon activation by oxidizing agents or plant signals, a green fluorescent protein (GFP)-CHAP1 fusion protein became localized in the nucleus. Expression of genes encoding antioxidant proteins was induced in the wild type but not in chap1 mutants. Activation of CHAP1 occurred from the earliest stage of plant infection, in conidial germ tubes on the leaf surface, and persisted during infection. Late in the course of infection, after extensive necrotic lesions were formed, GFP-CHAP1 redistributed to the cytosol in hyphae growing on the leaf surface. Localization of CHAP1 to the nucleus may, through changes in the redox state of the cell, provide a mechanism linking extracellular cues to transcriptional regulation during the plant-pathogen interaction.
Aerobic organisms are exposed to reactive oxygen species formed by incomplete reduction of molecular oxygen during respiration and other metabolic processes. Fungal pathogens of plants must, in addition, deal with the oxidative burst generated by the host upon exposure to pathogens (45). In some interactions, the plant hypersensitive response is associated with blockage of the infection process (30). In contrast, some necrotrophic fungi may be insensitive to the hypersensitive response and even exploit plant cell death resulting from this response (20, 41).
It has been proposed that preformed and inducible detoxifying enzymes, including superoxide dismutase, catalases, and peroxidases, contribute to fungal reactive oxygen species tolerance (19, 23). The redox state of cells subjected to oxidative stress is altered, leading to activation of three major oxidant scavenging systems: glutathione, thioredoxin, and NADPH. Glutathione and thioredoxins are kept in their reduced state by NADPH and used as cofactors by a number of enzymes in various detoxification reactions. Altered redox potential also affects the sulfhydryl groups of cysteine residues. These groups are oxidized to form intra- or intermolecular disulfide bonds or produce protein-glutathione mixed disulfides. Oxidation of redox-sensitive proteins, in the normally reductive intracellular environment, can result in either gain or loss of function, providing a mechanism for redox signaling (9). Many mammalian proteins involved in signal transduction have critical thiols, and their redox-dependent modifications contribute directly to gene expression (12, 16).
The Saccharomyces cerevisiae transcription factor YAP1 was identified as a functional homolog of mammalian AP-1 (activator protein 1) on the basis of its ability to bind an AP-1 recognition element. Upon exposure to oxidative stress, YAP1 becomes localized to the nucleus, where it induces the expression of antioxidant proteins (8, 10, 11, 25, 27). YAP1 contains a basic leucine zipper (bZIP) domain similar to that of Jun, which is a component of mammalian AP-1 transcription factor complexes. Two cysteine-rich domains, carboxy terminal (c-CRD) and amino terminal (n-CRD), are critical for YAP1-mediated resistance to oxidative stress and for appropriate subcellular YAP1 localization. The activity of YAP1 is controlled by redox-sensitive nuclear export: redox signals cause formation of intramolecular disulfide bonds, resulting in changes in protein conformation that mask its nuclear export sequence (8, 10, 11, 25, 27). S. cerevisiae YAP2, a homolog of YAP1, is also activated during oxidative stress, inducing genes encoding products that help to stabilize and fold proteins in an oxidative environment (7).
Homologs of YAP1 have been found in Candida albicans (CAP1) (2), Schizosaccharomyces pombe (PAP1) (42), and Kluyveromyces lactis (KLYAP1) (5). Genetic analyses indicate that these proteins are involved in oxidative stress responses and cadmium resistance, as found for YAP1. CAP1 and PAP1, like YAP1, are involved in multidrug resistance (1, 2, 5, 34, 40, 42, 44). All fungal YAP1-like proteins are distinctive due to characteristic residues present in their basic DNA-binding domains (Fig. 1) (15). YAP1, YAP2, CAP1, PAP1, KLYAP1, and two uncharacterized genes from Neurospora crassa and Aspergillus nidulans constitute a subgroup within the YAP family of transcription factors that possess a conserved c-CRD.
FIG. 1.
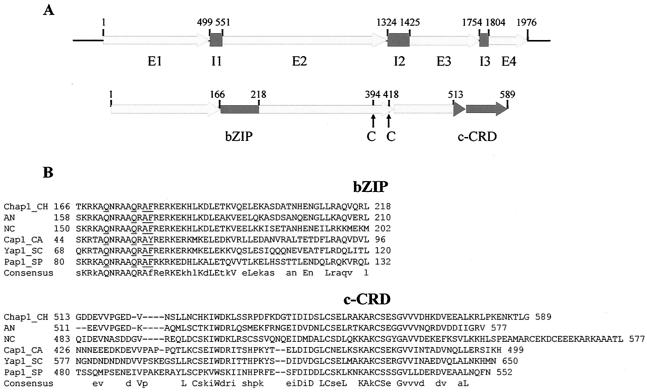
Structural organization of CHAP1. A. Upper map, organization of the gene: E1 to E4 indicate exons; I1 to I3 indicate introns. Lower map, predicted protein: bZIP, basic leucine zipper domain; c-CRD, C-terminal cysteine-rich domain; arrows indicate cysteine residues C394 and C418 belonging to the n-CRD (N-terminal cysteine-rich domain). For full details of the gene structure and domains, see GenBank accession no. AY486156. B. Alignment of bZIP and c-CRD domains of the YAP proteins. Consensus: uppercase letters indicate identity, lowercase letters indicate 50% or more similar residues. Residues characteristic of the YAP protein family are underlined. Abbreviations: Ch (CHAP1), Cochliobolus heterostrophus; An (AP-1-like protein, EAA62093), Aspergillus nidulans; Nc (AP-1-like protein, CAB91681), Neurospora crassa; Sp (Pap1, NP_593662), Schizosaccharomyces pombe; Ca (Cap1, EAL02784), Candida albicans; Sc (Yap1, NP_013707), Saccharomyces cerevisiae.
A number of YAP1-regulated genes have been identified, including GSH1, encoding γ-glutamylcysteine synthetase, catalyzing the first, rate-limiting step of glutathione synthesis (38, 46); GSH2, encoding glutathione synthetase, the second enzyme involved in glutathione biosynthesis (39); GLR1, encoding glutathione reductase (21); TRX2, encoding thioredoxin 2 (26, 33); TRR1, encoding thioredoxin reductase (28); FLR1, encoding a multidrug resistance transporter belonging to the major facilitator superfamily (1, 34); and YCF1, encoding an ABC transporter essential for cadmium tolerance (44). Numerous other YAP1-activated genes have been identified with microarray hybridization, genetic screens of target genes with a translational fusion library, and high-density membrane hybridization (13, 18).
Cochliobolus heterostrophus is a foliar pathogen of maize, responsible for southern corn leaf blight (47). The conidia of the pathogen germinate and form appressoria; emerging penetration pegs then breach the leaf surface and start invasive growth inside the plant tissue. Spreading hyphae cause necrotic lesions, and mycelium growing on the surface of damaged plant tissue produces abundant conidia. We cloned a gene encoding the C. heterostrophus homolog of YAP1, designated CHAP1 (Cochliobolus heterostrophus AP-1-like), and studied its function with deletion mutants. We also isolated C. heterostrophus homologs of yeast YAP1 target genes and verified their regulation by CHAP1 during oxidative stress. Finally, we followed relocalization of a green fluorescent protein (GFP)-CHAP1 fusion protein into the nucleus upon exposure to oxidizing agents and during plant infection.
MATERIALS AND METHODS
Identification of CHAP1.
The degenerate oligonucleotide GCICKTTGAGCIGCICKRTTYTGRGC (where I is inosine, K is G/T, R is A/G, and Y is C/T) was end labeled and used to screen a cDNA library constructed in pADGAL4 (Stratagene) from a pool of RNA from fungus grown on synthetic media and infected plants. Positive clones were transferred from phages to Escherichia coli by in vivo excision and screened again. One of the second-round positive clones showed limited homology to the AP-1-like protein of Neurospora crassa, and this sequence was used to search the assembled 5× coverage of the C. heterostrophus genome (TMRI/Syngenta, San Diego, Calif.).
Fungal transformation vectors.
Vectors for replacement of CHAP1 by selection markers conferring resistance to hygromycin or bialaphos were constructed by ligation of upstream and downstream flanking regions to the relevant resistance cassette. Protoplasts of C. heterostrophus strain C4 (MAT1-2 Tox1+, ATCC 48331) were transformed (31).
The following fragments were assembled to create the CHAP1 disruption vector conferring hygromycin resistance: a left 616-bp EagI-XhoI flanking region including the 5′ untranslated region and part of the CHAP1 coding region; SalI-XbaI hygromycin resistance cassette from the vector pUC-ATPH (31); 798-bp NheI-HindIII right flanking region, including part of the coding region and 3′ untranslated region; and NotI-HindIII pBluescript cloning vector.
The GFP-CHAP1 transformation vector was constructed by joining the following fragments: 1.5-kb CHAP1 KpnI-NcoI promoter generated by PCR with primers chapp-sKpnI (cagggtaCCTCGTTGTCGCCAGTGTCC [uppercase letters indicate the gene homology region, and lowercase letters indicate the nonhomologous region with introduced restriction site]) and chapp-asNcoI (gaggccatggTGCGAAAGCTGGGGTGAAT); NcoI-NdeI GFP coding sequence (GFP GenBank accession number U84737); 2.4-kb CHAP1 NdeI-BglII genomic sequence, which includes coding region and part of 3′untranslated region generated by PCR with primers chap-sNdeI (AGCTTTCcatATGGCCGGCACAGGCAACGA) and chap-asBglII (ggaagaTCTGATAGCCAAGCACGCC); BamHI-XbaI hygromycin resistance cassette, and XbaI-KpnI pUC57-6D6 vector. The CHAP1 disruption vector conferring glufosinate (bialaphos) resistance was constructed from the following fragments: 955-bp EagI-PstI left flanking region including the 5′ untranslated region and part of the CHAP1 coding region; PstI-XbaI phosphinothricin acetyltransferase encoding gene (BAR) under regulation of the Aspergillus nidulans trpC promoter; 798-bp NheI-HindIII right flanking region including part of the CHAP1 coding sequence and 3′ untranslated region; and the NotI-HindIII pBluescript cloning vector (Stratagene). Hygromycin B (used at 50 μg/ml) was from Calbiochem, and bialaphos (used at 100 μg/ml, in minimal medium) was from Duchefa.
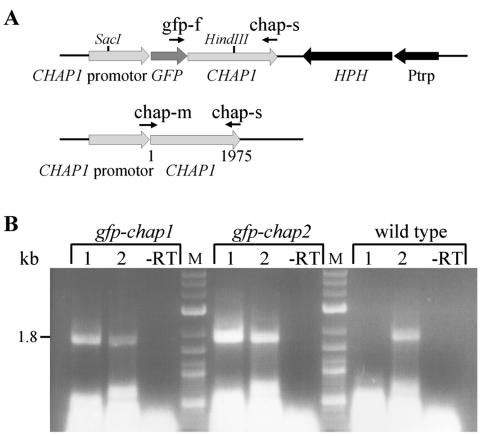
Reverse transcription-PCR with primers gfp-f (GGGATCACTCACGGCATGGA) and chap-s (CCAGTCAGATGAGAACAATTATCCAA) was applied to detect the expression of the GFP-CHAP1 coding sequence. Expression of the native CHAP1 copy was detected with primers chap-m (ACCCCAGCTTTCGCAATGGC) and chap-s. The sequence corresponding to the primer chap-m is present in the native CHAP1 copy but not in GFP-CHAP1 (see Fig. 6).
FIG. 6.
Construction of strains expressing the GFP-CHAP1 fusion protein. A. Construct map. GFP, GFP gene; Ptrp, constitutive promoter; HPH, hygromycin phosphotransferase gene; chap-m, chap-s, and gfp-f, primers used for transformant verification. Upper drawing: transformant; lower drawing: wild type. B. Confirmation of CHAP1-GFP expression by reverse transcription-PCR. The following reactions were performed with RNA of the wild type and transformants gfp-chap1 and gfp-chap2. Lanes 1, gfp-f and chap-s; lanes 2, chap-m and chap-s. Control reactions without reverse transcriptase (−RT) were performed with primers gfp-f and chap-s for transformants and chap-m and chap-s for the wild type. The predicted product size, 1.8 kb, is indicated at the left of the gel; note that this is smaller than the size indicated in A, 1,976 bp, due to splicing of introns.
Transformant verification.
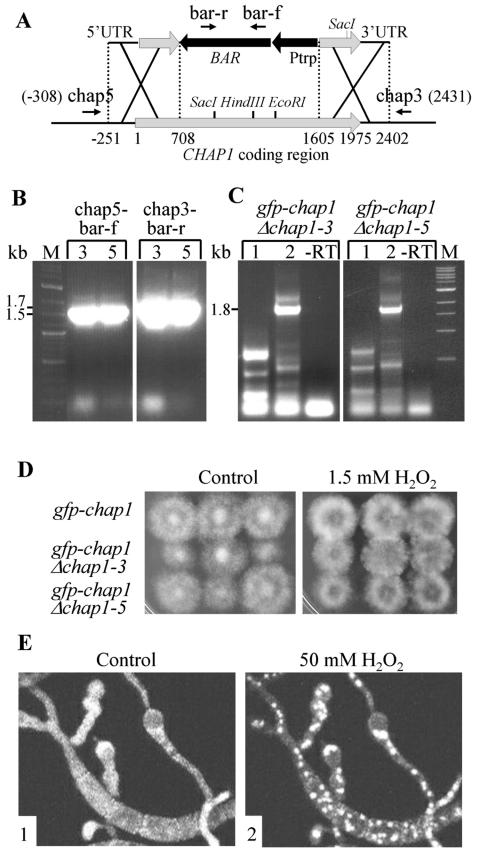
Disruption of CHAP1 was confirmed by PCR with genomic DNA as the template. The following primer pairs were used for verification of fungal lines transformed with the CHAP1 disruption vector carrying the hygromycin phosphotransferase gene: chap5 (TATTCGTCCGCCCTCCGCACG) and chap3 (CGTCTGATAGCCAAGCACGCC), chap5-ptrp (GGTCGTTCACTTACCTTGCTTG), and chap3-ttrp (GGTGTTCAGGATCTCGATAAG). To verify CHAP1 disruption in lines transformed with the vector carrying the phosphinothricin acetyltransferase gene (BAR), we used the following primer pairs (see Fig. 8): chap5-bar-f (CGGCCGTCTGCACCATCG) and chap3-bar-r (GGTACCGGCAGGCTGAAGTC).
FIG. 8.
A GFP-CHAP1 fusion protein is functional in the absence of the resident CHAP1 copy. A. Deletion of the wild-type copy of CHAP1 in a GFP-CHAP1 reporter strain. BAR, phosphinothricin acetyltransferase gene; Ptrp, constitutive promoter; bar-r, bar-f, chap5, and chap3 are the primers used to confirm double-crossover integration. B. PCR verification of double-crossover integration at the CHAP1 locus in two transformants, 3 (each left lane) and 5 (each right lane). The primer pairs used for PCR were chap5 and bar-f and chap3 and bar-r. These primer pairs did not yield any product in reactions with template DNA from the wild type or the parent GFP-CHAP1 strain. Predicted product sizes are indicated at the left of the gel photo. C. Reverse transcription-PCR detection of GFP-CHAP1 but not native CHAP1 transcripts in transformants 3 and 5. Lanes 1 and 2 show reaction products with primer pairs chap-m and chap-s, and gfp-f and chap-s, respectively (described in Fig. 6A). Primer chap-m crosses the junction between genomic sequence and gfp and therefore can give a product only if the wild-type transcript is present (Fig. 6). Control reactions without reverse transcriptase (−RT) were done with primers gfp-f and chap-s. The 1.8-kb band present in lanes 2 is of the predicted size and corresponds to the transcript encoding the fusion protein (as in Fig. 6B). The lower-molecular-weight bands are nonspecific products. D. Complementation of the hydrogen peroxide-sensitive phenotype by the GFP-CHAP1 fusion protein. Left panel, control plate; right panel, 1.5 mM H2O2. The genotypes of the three strains tested are indicated at left. All strains grew equally well on both media. E. Nuclear localization of GFP-CHAP1 upon exposure to 50 mM H2O2 in the absence of native CHAP1. Panels: 1, conidia and germ tubes growing on a glass slide prior to hydrogen peroxide treatment; 2, 5 min after addition of 50 mM H2O2.
Nucleic acid isolation and reverse transcription-PCR.
Crude genomic DNA for PCR analysis was isolated from mycelium ground in liquid nitrogen (14). RNA was isolated from mycelia and leaf tissue ground in liquid nitrogen by a modified phenol-sodium dodecyl sulfate method (4) and purified by lithium acetate precipitation. For reverse transcription-PCR, 4 μg of total RNA was treated with RNase-free DNase (Qiagen 79254) in restriction enzyme buffer 4 (New England Biolabs) for 20 min at 30°C in a total volume of 10 μl and incubated 15 min at 65°C for DNase inactivation; 2 μl of the treated RNA was used as the template for reverse transcription and amplification (Access reverse transcription-PCR kit A1250; Promega).
Cloning of CHAP1 target genes.
The following primers were designed for PCR amplification of putative CHAP1 target genes: ChGLR1, 2071-s, GCCGAGACCAAGGAGTGTGATT, and 2071-as, CCGTGGGCTGCTTGTGCTCT; ChGSH1, 1353-s, CCCCATATCCGCTTCCCAAC, and 1353-as, CCAGACGAGGTTTGTTGCCC; ChTRXL, 4383-s, GCAAAATCATCGCCGTTACCTC, and 4383-as, TATCGCTTCGCCTTCACGTTG; ChTRX1, 4350-s, CCACTCTTCACCCCACGTTTCT, and 4350-as, CATGATTGCGGGAACGGTCC; ChTRR1, 4742-s, CGGCATCGCAGCAGGTGGTC, and 4742-as, GGGTTAGAGCGGTATTCGGGA; and ChGSH2, 4210-s, CCCAGTCTTTTCCCACGCTCT, and 4210-as, CCCTCCTCGCTCTGGTCGCC. PCRs were performed with C. heterostrophus genomic DNA as a template.
Assay of sensitivity to oxidizing agents, preparation of plant extracts, and host infection.
Hydrogen peroxide (0.5 to 2 mM) and menadione (2-methyl-1,4-naphthoquinone, Fluka) (20 to 50 μM, from a 100 mM ethanol stock) were incorporated into complete medium plates. Plates were inoculated with 1, 2, and 3 μl of conidial suspension (≈50 spores/μl) and incubated for 2 days at 30°C in darkness.
To follow GFP-CHAP1 relocalization, hydrogen peroxide (50 mM), menadione (1 mM prepared from a 100 mM stock in ethanol), diethyl maleate (10 mM), and maize and Arabidopsis thaliana plant extracts (25 to 100%) were applied to conidia germinated on glass slides. GFP-CHAP1 subcellular location was observed by confocal microscopy after 10 min of incubation. Plant extracts were prepared by grinding leaves in liquid nitrogen; leaf powder was defrosted immediately before use and mixed with water (unless otherwise indicated) to achieve the required percentage (100% is 1:1, wt/vol). The suspension was centrifuged at 14,000 rpm for 2 min, and the supernatant was used immediately. Hydrogen peroxide in maize extract was assayed with PeroXOquant (Pierce). To further fractionate the CHAP1 activating component, the 100% maize extract was heated for 5 min at 95°C and centrifuged 5 min to remove denatured proteins. The resulting supernatant was tested directly or further separated by phase partition against ethyl acetate. To test the activity of this organic extract, a sample was dried under vacuum and redissolved in water.
For CHAP1 activation in culture, fungal strains were grown in liquid complete medium with shaking (230 rpm) for 48 h at 30°C. Hydrogen peroxide was added to a final concentration of 20 mM, and samples were collected 30 and 60 min later. For induction by maize leaf extract, mycelium was filtered and resuspended in 25% plant extract. Samples were collected 1 h later.
To assay virulence, the first leaves of maize plants (Jubilee, 2 weeks old) were inoculated with drops of conidial suspension containing 0.05% Tween 80. Plants were incubated in moist chambers at 30°C with continuous illumination.
Microscopy.
Confocal microscopy was performed as described (29). For DAPI (4′,6′-diamidino-2-phenylindole) staining, germinated spores adhering to glass slides were fixed (6% paraformaldehyde in 100 mM piperazine ethanesulfonic acid (PIPES buffer, pH 6.5) for 20 min and then washed twice with Tris-buffered saline (TBS: 10 mM Tris, pH 7.5, 200 mM NaCl). Spores and germ tubes were stained with DAPI solution (1 μg/ml in TBS) for 5 min and washed twice with TBS.
Yeast YAP1 complementation.
S. cerevisiae Δyap1 strain BY4742-YML007W and the strain from which it was derived, BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), were obtained from Euroscarf (Frankfurt, Germany). The full-length CHAP1 cDNA (1.8 kb) was amplified with primers chap-m and chap-s and cloned into pTZ57R/T (Fermentas). The insert was reexcised with KpnI and SphI restriction enzymes, cloned into the yeast expression vector pYES2 (Invitrogen), and transformed into BY4742-YML007W and BY4742 strains, and colonies were selected on synthetic complete medium lacking uracil. As a control, these strains were also transformed with empty pYES2 vector. To induce CHAP1 expression under control of the GAL1 promoter, transformed yeast cells were grown on synthetic complete medium without uracil and with 1% galactose and 1% raffinose as a carbon source instead of glucose. About 2,000 yeast cells in a 10-μl volume were plated on peroxide-containing plates (see Fig. 2) and grown for 2 days at 30°C.
FIG. 2.
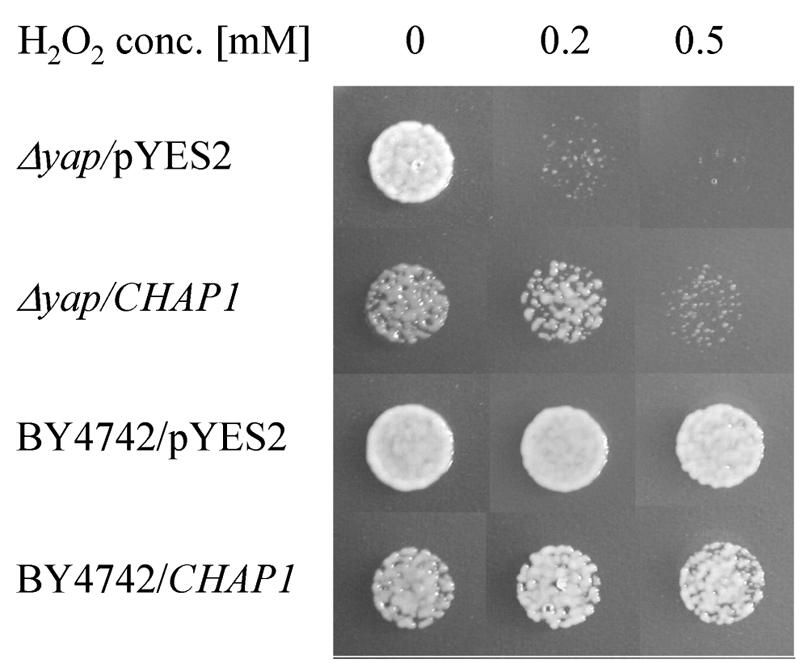
Partial complementation of yeast Δyap1 hydrogen peroxide sensitivity by C. heterostrophus CHAP1 cDNA. Yeast strains were tested for growth in the presence of 0.2 and 0.5 mM hydrogen peroxide. The Δyap1/pYES2 and Δyap1/CHAP1 Δyap1 mutant yeast strains were transformed with empty expression vector pYES2 and a vector containing CHAP1 cDNA, respectively. BY4742/pYES2 and BY4742/CHAP1 are the parental wild-type yeast strain with the same genetic background as the mutant transformed with empty pYES2 and pYES2 containing CHAP1, respectively.
Nucleotide sequence accession number.
The C. heterostrophus CHAP1 sequence has been deposited in GenBank with accession number AY486156.
RESULTS
Identification of an AP-1 homolog in C. heterostrophus.
A cDNA library was screened with a degenerate oligonucleotide corresponding to a region of basic amino acids that is the most conserved motif in known fungal AP-1-like proteins. One of the resulting clones, designated CHAP1, had limited homology to the AP-1-like protein of Neurospora crassa (3), and this sequence was used to search the C. heterostrophus genome. The genomic sequence of CHAP1 has a predicted 1,976-bp coding region. A full-length cDNA clone was obtained by reverse transcription-PCR. The cDNA sequence confirmed the locations of three introns (53, 102, and 51 bp) and encodes a protein of 590 amino acids. The predicted CHAP1 protein has a basic leucine zipper domain with residues characteristic of the YAP1-like protein family: Q171, Q176, A178, and F179 (15) and two cysteine-rich domains, c-CRD and n-CRD (Fig. 1) (10, 43). The homology of CHAP1 to other YAP proteins is low outside the bZIP and c-CRD regions. The BLAST search of the C. heterostrophus genome with the amino acid sequence corresponding to the bZIP domain of CHAP1 revealed that there are at least two additional proteins sharing the residues that are a signature of the YAP protein family.
Complementation of a yeast yap1 mutant.
To determine whether CHAP1 could replace yeast YAP1 in a deletion mutant, we transformed a Δyap1 strain with the expression vector pYES2 harboring a full-length CHAP1 cDNA clone. In addition, pYES2-CHAP1 was transformed into the parental wild-type yeast strain BY4742. As a control, the Δyap1 and BY4742 strains were transformed with empty pYES2 vector. All the strains were grown on galactose medium to activate the GAL4 promoter of the expression vector. In the absence of oxidizing agents, yeast strains harboring CHAP1 were less viable than those transformed with empty vector (Fig. 2). In the presence of 0.2 mM hydrogen peroxide, growth of the Δyap1 mutant was almost completely inhibited, whereas growth of the Δyap1/CHAP1 strain was only slightly affected compared to the BY4742/CHAP1 strain. At 0.5 mM hydrogen peroxide the growth of the Δyap1/CHAP1 strain was further inhibited, whereas growth of the BY4742/CHAP1 and BY4742/pYES2 strains was unaffected (Fig. 2). These findings indicate that CHAP1 is able to complement yeast Δyap1, at least partially. Neither the toxic effect of CHAP1 expression nor complementation could be observed when the strains were grown on glucose instead of galactose (data not shown).
Deletion of CHAP1.
To investigate the function of CHAP1, the gene was deleted by double crossover integration, replacing the entire coding region with an expression cassette conferring resistance to hygromycin B. The disruption construct was transformed into C. heterostrophus. More than 20 hygromycin-resistant transformants were obtained, and homologous integration and loss of the wild-type copy were confirmed by PCR amplification with primer pairs chap5 and chap3; chap5 and ptrp; and chap3 and ttrp (see Materials and Methods). Deletion mutants were indistinguishable from the wild type with respect to colony morphology, sporulation, and conidial germination. Two independently isolated transformants were selected for further characterization.
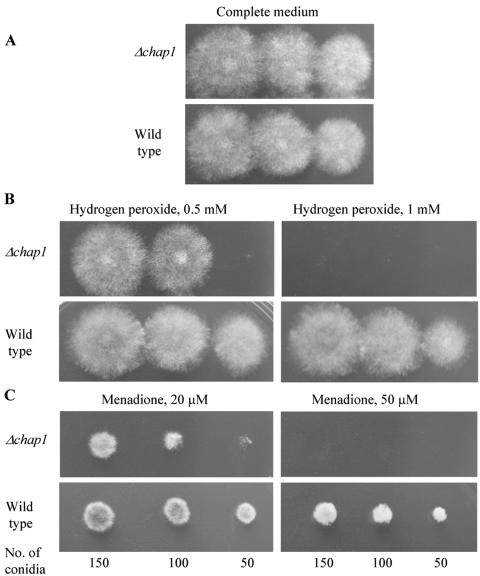
Drops of conidial suspension of the wild-type and Δchap1 strains were inoculated on complete medium plates containing different concentrations of hydrogen peroxide. Conidia of both the mutant and the wild type began to germinate at the same time on plates containing 0.5, 1, or 2 mM hydrogen peroxide (not shown). Elongation of the germ tubes of the mutant, however, was inhibited shortly after germination. Subsequent colony growth was markedly inhibited in the mutant (Fig. 3A and B).
FIG. 3.
Sensitivity of the wild-type and chap1-deleted mutant to oxidizing agents. Hydrogen peroxide (B) and menadione (C) were incorporated into complete medium at the indicated concentrations (A is complete medium alone). Plates were inoculated with, from left to right in each panel, 3, 2, and 1 μl of conidial suspension (≈50 spores/μl) and grown for 3 days at 30°C.
We examined the resistance of a Δchap1 mutant to another oxidizing agent, the superoxide-generating compound menadione (2-methyl-1,4-naphthoquinone). Growth of the wild type on complete medium with 20 or 50 μM menadione was inhibited less than that of a Δchap mutant (Fig. 3C).
CHAP1 target genes.
S. cerevisiae YAP1 is activated in response to oxidative stress and then regulates the expression of genes involved in detoxification. We cloned C. heterostrophus homologs of several YAP1-dependent genes in order to investigate whether their expression is regulated by CHAP1 in response to oxidative stress. Genes encoding the following proteins were identified, with the indicated similarity to their yeast homologs, in the C. heterostrophus genome: thioredoxin reductase (ChTRR1, 77%), γ-glutamylcysteine synthetase (ChGSH1, 65%), glutathione reductase (ChGLR1, 69%), glutathione synthetase (ChGSH2, 70%), and two thioredoxins (ChTRX1 and ChTRX2). ChTRX2 is highly similar to both thioredoxins of S. cerevisiae (78% to TRX1 and 80% to TRX2), in contrast to ChTRX1, which is only 43% similar to both TRX1 and TRX2. There is 42% similarity between the two C. heterostrophus thioredoxins.
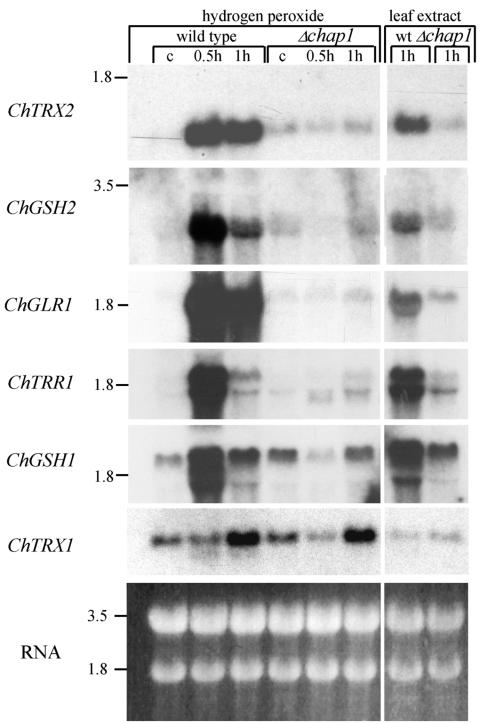
We used hydrogen peroxide (20 mM) as an oxidizing agent to activate CHAP1 in mycelium grown in liquid culture; samples were collected 30 and 60 min after addition of hydrogen peroxide. RNA blot analyses were performed with probes corresponding to putative CHAP1 target genes. All the genes tested except ChTRX1 are upregulated in response to hydrogen peroxide, and their expression depends on CHAP1 (Fig. 4). This implies that the same mechanism of oxidative stress defense functions in S. cerevisiae and in C. heterostrophus and that structurally similar members of the YAP family are responsible for it. The thioredoxin gene ChTRX1 was upregulated upon exposure to hydrogen peroxide in a CHAP1-independent manner. Its activation was delayed in comparison to that of other genes tested and occurred to a lesser extent (Fig. 4). Perhaps an additional redox state sensor is involved in ChTRX1 activation, as in S. cerevisiae, whose two thioredoxins are differentially regulated (17).
FIG. 4.
Induction of putative CHAP1 target genes in response to oxidative stress and plant extract in wild-type and chap1 deletion strains. Gene abbreviations: ChTRX1 and ChTRX2, thioredoxins 1 and 2, respectively; ChGSH1, γ-glutamylcysteine synthetase; ChGSH2, glutathione synthetase large chain; ChTRR1, thioredoxin reductase; ChGLR1, glutathione reductase. RNA loading was verified by ethidium bromide staining. Size markers (in kilobases) at the left indicate the rRNA bands. C, control prior to treatment; 0.5 h and 1 h indicate the time after addition of hydrogen peroxide or leaf extract.
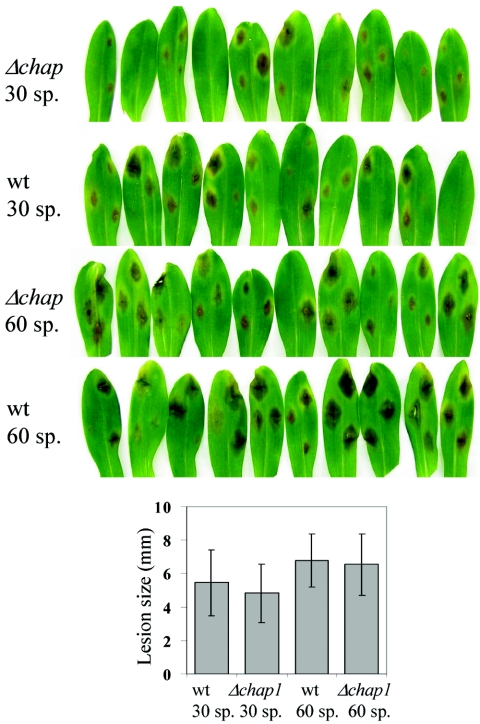
Despite the increased sensitivity of the chap1 deletion mutants to oxidizing agents, as well as the loss of induction of a set of target genes, inoculation of maize leaves by drops of conidial suspension (30 and 60 conidia per drop) of the wild-type and mutant strains resulted in similar disease symptoms. The average lesion size was the same within the variability for the wild type and the mutant (Fig. 5).
FIG. 5.
Disease symptoms on maize leaves caused by wild-type and Δchap1 fungal strains. A. First leaves of 2-week-old seedlings were inoculated with drops containing 30 and 60 spores, and the plants were incubated at 30°C for 2 days. B. The bar graph compares average lesion size. Bars indicate standard deviation, and each sample includes 20 to 28 lesion spots.
Nuclear localization of the GFP-CHAP1 fusion protein upon activation.
To investigate the mode of CHAP1 activation, we followed the intracellular response of CHAP1 to oxidizing conditions. A construct was designed encoding an N-terminal fusion of GFP to the CHAP1 coding sequence under regulation of the CHAP1 promoter (Fig. 6A). Fungal transformation was performed, and integration of the CHAP1 promoter-GFP-CHAP1 sequence was confirmed by PCR with genomic DNA as the template (data not shown) and by reverse transcription-PCR (Fig. 6B). An endogenous CHAP1 was transcribed in the transformants as well as the fused GFP-CHAP1 coding sequence, indicating that the construct was not integrated into the CHAP1 coding region but elsewhere in the genome.
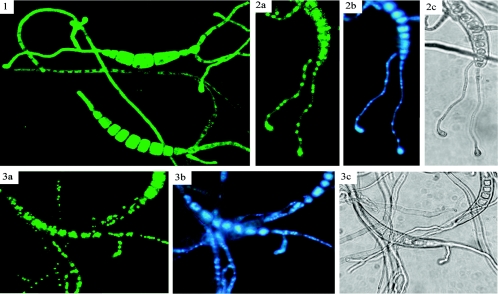
Conidia of GFP-CHAP1 transformants were placed on glass slides and allowed to germinate (4 h at 30°C). Germ tubes and conidia were uniformly fluorescent during germination and appressorium formation on glass slides (Fig. 7, panel 1). Hydrogen peroxide (50 mM) was added, and conidia were observed by confocal microscopy 15 min later. Fluorescence was restricted to subcellular compartments in treated conidia and germ tubes. GFP fluorescence and DAPI staining colocalized to the same compartment, the nucleus (Fig. 7). Upon addition of hydrogen peroxide, bubbling was observed almost immediately, indicating catalase activity. Bubbling and CHAP1 activation occur almost simultaneously, perhaps a manifestation of two strategies to cope with stress caused by hydrogen peroxide. Catalase activity makes the effective concentration of hydrogen peroxide available at the surface of the fungal cell unstable. Consistent and widespread activation of CHAP1 was observed only upon application of at least 15 mM hydrogen peroxide. About 10-fold lower hydrogen peroxide concentrations were inhibitory when conidia were plated on solid medium (Fig. 3). It is likely that conidia germinating on agar plates containing hydrogen peroxide encounter a continuous supply of H2O2, in contrast to the mycelium growing on a glass slide to which 20- to 30-μl drops of H2O2-containing solution were added. It seems that catalase secreted from the growing hyphae was sufficient to partially degrade H2O2 in the first moments after addition.
FIG. 7.
Nuclear localization of CHAP1-GFP fusion protein upon activation. 1. Conidia germinating on glass surface prior to hydrogen peroxide treatment; 2 and 3, germinated conidia treated with 50 mM hydrogen peroxide after 15 min of incubation. 2a and 3a, visualization of GFP-CHAP1 fusion protein by GFP fluorescence; 2b and 3b, DAPI staining of fungal nuclei; 2c and 3c, light microscope image. GFP fluorescence was visualized with an MRC-1024 laser confocal scanning microscope (Bio-Rad, Hempstead, United Kingdom) with a Nikon Plan Fluor 40/1.30 objective (Tokyo, Japan). Image processing was performed with Confocal Assistant 4.02 software (Bio-Rad, Hampstead, United Kingdom). DAPI fluorescence was visualized with a Zeiss Axioskop fluorescence microscope.
To test the function of the GFP-CHAP1 fusion protein in the absence of the native protein, we disrupted the endogenous CHAP1 copy in a GFP-CHAP1-expressing strain with resistance to bialaphos as the selection marker. Double-crossover integration was confirmed by PCR on genomic DNA of the transformants, and reverse transcription-PCR confirmed the absence of wild-type CHAP1 transcripts in these bialaphos-resistant lines (Fig. 8A and B). The growth of these transformants (GFP-CHAP1 Δchap1) on plates containing 1.5 mM hydrogen peroxide was indistinguishable from that of the parental strain expressing both GFP-CHAP1 and wild-type CHAP1 (Fig. 8D). CHAP1 activation by hydrogen peroxide was tested with a GFP-CHAP1 Δchap1 transformant (Fig. 8E), and similar results were obtained, indicating that the fusion protein becomes localized to the nucleus under oxidative stress in the absence of a wild-type copy.
Activation of CHAP1 by oxidizing agents and plant extracts.
CHAP1 activation (nuclear localization) was examined in response to other oxidizing agents: the superoxide-generating compound menadione (2-methyl-1,4-naphthoquinone) and the glutathione-depleting compound diethyl maleate. Both menadione (1 mM) and diethyl maleate (10 mM) caused rapid CHAP1 activation in treated hyphae at a rate comparable to the response to hydrogen peroxide. Maize leaf extract (25, 50, and 100%) was added to conidia germinated on glass slides. Within a few minutes after application of the 100% extract, nearly all GFP fluorescence relocalized into nuclei (the rate of response was similar to that observed in activation by hydrogen peroxide); 50% extract was less effective than 100%, and 25% extract caused only slight activation. Plant extract induced expression of CHAP1-dependent genes encoding antioxidant proteins in the wild type but not in a Δchap1 mutant (Fig. 4).
To test whether hydrogen peroxide is the inductive factor in plant extracts, catalase (5,000 U/ml) was added to 100% plant extract prepared in 50 mM potassium phosphate buffer, pH 7. As a control, the same quantity of catalase was added to 50 mM hydrogen peroxide. Both reaction mixtures were allowed to stand 10 min and then added to germinated conidia. Neither buffer alone nor catalase-treated hydrogen peroxide activated CHAP1, whereas CHAP1 was activated by catalase-treated and untreated leaf extract as well as by hydrogen peroxide without catalase treatment. Independent evidence that hydrogen peroxide is not the major inductive factor present in plant extract comes from measurement of the hydrogen peroxide content. The hydrogen peroxide concentration in 100% plant extract, determined by spectrophotometric assay, was about 160 μM. This concentration was completely ineffective in activating CHAP1 nuclear localization. Furthermore, catalase treatment of the extract reduced the measured hydrogen peroxide content to nearly background levels. Thus, it appears that plant extracts contain CHAP1-activating factors other than hydrogen peroxide, the presence of which does not depend on active plant defense. To determine whether the CHAP1-activating substance is specific to maize, we tested a similar extract prepared from Arabidopsis thaliana leaves. It caused a response comparable to that of maize extract. Apparently, the activator(s) of CHAP1 is not plant species specific.
Initial characterization of active components in the plant extract.
To provide an initial characterization of the active components in the plant extract, we used nuclear localization of GFP-CHAP1 for activity-guided fractionation. First, we filtered the extract through an ultrafiltration membrane with a cutoff of 10,000 Da. The active component was able to cross the membrane. When the extract was heated for 5 min at 95°C and centrifuged at 14,000 rpm for 2 min to precipitate denatured proteins, the resulting supernatant contained the activator. The supernatant after heat treatment and centrifugation was then extracted with an equal volume of ethyl acetate. Upon separation of the phases by centrifugation, the active compound(s) partitioned into the organic phase. No detectable active components were extracted from the leaf powder by chloroform-methanol, while ethanol did efficiently extract the activator(s).
Activation of CHAP1 during plant infection.
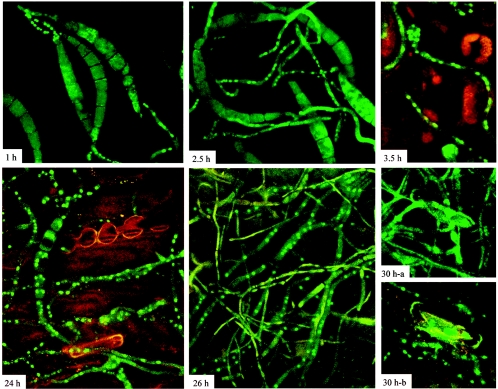
Two-week-old maize plants were inoculated with suspensions of conidia from GFP-CHAP1 transformants. Germ tubes emerged from conidia within an hour, and the fusion protein began migrating into the nuclei of the germ tubes on the leaf surface prior to penetration. This migration was specific to the plant surface, as germ tubes on glass slides remained uniformly fluorescent. During the subsequent 2 to 3 h, the germ tubes elongated and CHAP1 continued to be localized in nuclei. After about 3 h, the hyphal tips formed appressoria, and CHAP1 became localized to their nuclei. During hyphal penetration of the plant cuticle and epidermis and development inside the leaf, CHAP1 remained in the hyphal nuclei. After about 24 h, when well-developed necrotic lesions were visible at inoculation sites, bundles of uniformly fluorescent hyphae appeared. During further progression of the disease, the pattern of CHAP1 inactivation (reappearance in the cytoplasm) became more discernible: hyphae on leaf surfaces were uniformly fluorescent, whereas inside the leaf CHAP1 was localized in the nuclei (Fig. 9).
FIG. 9.
Subcellular localization of CHAP1 during plant infection. Numbers indicate time after inoculation: 1 h, conidia start germination, CHAP1 is activated in emerging germ tubes, and germ tubes elongate (2.5 h) and form appressoria (3.5 h); 24 h and 26 h, extensive mycelial network interlaces plant tissue, necrotic lesion is formed at the site of inoculation, fluorescence is localized in the nucleus in most of the hyphae; 30 h-a, hyphae found in the outermost layers of the infected leaf: GFP fluorescence is distributed in the cytosol; 30 h-b, hyphae found inside the plant tissue: in these hyphae CHAP1 is still activated. Confocal image acquisition was done as described for Fig. 7; the autofluorescence of the leaf, contributed primarily by chlorophyll, was detected with the Cy5-optimized channel.
DISCUSSION
Redox regulation is a ubiquitous mechanism for controlling cellular activity (16). Direct modification of transcription factors or signal transduction by redox-sensing proteins alters gene expression in response to an oxidizing environment. We isolated a C. heterostrophus homolog of the yeast YAP1 transcription factor. CHAP1 has the same structural features as YAP1, including a bZIP domain and two cysteine-rich regions. An oxidizing environment causes activation of CHAP1 and its relocalization into the nucleus. It then regulates a set of genes encoding antioxidant proteins. We found that this set overlaps the set of YAP1-dependent genes of S. cerevisiae.
Despite the marked homology of both AP-1-like proteins in their DNA-binding domains, no YAP1 recognition sequence could be identified in the promoters of the CHAP1-dependent genes tested. Nevertheless, CHAP1 could partially complement the hydrogen peroxide-sensitive phenotype of a yeast yap1 mutant. It appears that there is sufficient structural homology to allow at least partial replacement of YAP1 function in S. cerevisiae. It is possible that in C. heterostrophus, CHAP1 forms a heterodimer with another bZIP protein and the resulting complex has a different DNA binding specificity, as was shown for the mammalian bZIP protein family (24). Expression of CHAP1 under the strong GAL4 promoter in S. cerevisiae led to reduced growth. The reason for this toxicity is not clear, but it was independent of oxidative stress and of the presence of the resident yeast YAP1 copy (Fig. 2). CHAP1 overexpression might have allowed binding to nonspecific sequences, interfering with metabolism and growth.
Activation of CHAP1 nuclear relocalization during plant infection might be induced by penetration of the activator into the cell directly or through membrane channels. Another route of activation could be interaction of the activating substance with the extracellular domains of membrane receptor proteins, followed by transduction of the redox signal into the cell by thiol-disulfide exchange (6, 16, 36). Several lines of evidence indicate that existing molecules other than reactive oxygen species are involved in CHAP1 activation on the plant. First, plant extracts prepared from maize leaves that have never been in contact with the pathogen caused intense CHAP1 activation. As the extract was made from quickly frozen plant tissue, it is unlikely that any wounding or defense responses were activated during its preparation. Second, pretreatment of the plant extract with catalase did not affect its activating property. The activator(s) is not plant species specific and activates CHAP1 as rapidly as hydrogen peroxide does.
Our initial fractionation experiments revealed that the active component is heat stable and has a relatively low molecular weight. The active molecule is sufficiently polar to dissolve in water or in ethanol, and it partitions from water into ethyl acetate. In view of the recent report (35) that indole-3-acetic acid activates downstream targets of the yeast CHAP1 ortholog, YAP1, indole-3-acetic acid might be a candidate inducer. We therefore tested whether indole-3-acetic acid activates CHAP1 nuclear relocalization. Up to the highest concentration tested, 500 μM, indole-3-acetic acid did not activate CHAP1. Methylglyoxal, a toxic by-product of glycolyis, was recently shown to activate yeast YAP1 (32). Methylglyoxal could be produced intracellularly in the fungus and also by the plant; its activity as a plant-derived inducer can be tested with CHAP1 nuclear relocalization as an assay. Oxidizing agents, cadmium, indole-3-acetic acid, and methylglyoxal, are unrelated compounds, all of which activate yeast YAP1. This raises the possibility that YAP1, characterized as a redox-responsive transcription factor, may have a more general sensory role than initially thought. In C. heterostrophus, it seems likely that a novel plant compound is responsible for the induction of CHAP1 relocalization, and its full chemical characterization is currently in progress.
Activation of CHAP1 on the plant (Fig. 9) suggests a role in infection. Nevertheless, deletion of CHAP1 had no significant effect on virulence. Apparently, other C. heterostrophus YAP-like proteins are not able to compensate for the defect caused by CHAP1 disruption: transcription of the CHAP1-dependent antioxidant genes is not induced in absence of CHAP1. The ability of the fungus to overcome the absence of CHAP1 during plant infection might therefore result from alternative protective systems that compensate for the loss of CHAP1. In S. cerevisiae, glutathione and catalase provide overlapping defenses against hydrogen peroxide (22). The genome of C. heterostrophus encodes three catalase-encoding (CAT) genes (37). Mutants deficient in CAT3 had enhanced sensitivity to hydrogen peroxide compared with the wild type and with mutants deficient in CAT1, CAT2, or both. Nevertheless, all the catalase-deficient mutants had normal virulence on maize.
One interpretation of normal virulence in both CHAP1 mutants (this study) and catalase mutants (37) is that either of these distinct mechanisms of defense against oxidative stress is sufficient. Another possibility is that C. heterostrophus does not encounter damaging levels of reactive oxygen species during infection under the conditions that have been tested. CHAP1 relocalization was induced before penetration into the leaf and certainly before massive death of host cells. This suggests that the activation of CHAP1 may act as an “early warning” system, relaying a signal for impending exposure to oxidative stress. The antioxidant transcriptional program would be induced and ready, even if this stress does not actually occur. Some defense mechanisms, particularly against as universal a challenge as oxidative stress, may have evolved before specialization of fungi to their hosts. The homologs of CHAP1 as well as of the catalase genes might have essential roles in the virulence of other plant pathogens.
Transcriptional regulation mediated by nuclear localization is well known in animals and plants and in S. cerevisiae. Nuclear relocalization of CHAP1 at the earliest stages of infection is among the most rapid known molecular consequences of growth of a plant pathogen on its host. Further study should uncover both the protective and signaling roles of redox-sensitive transcription factors in fungus-plant interactions.
Acknowledgments
We are grateful to Jonathan Gressel for critical reading of the manuscript. We are grateful to Torrey Mesa Research Institute/Syngenta for access to the Cochliobolus sequence database (sequenced by Celera Genomics for TMRI/Syngenta).
This work was supported in part by ISF grant 233002 from the Israel Academy of Sciences.
REFERENCES
- 1.Alarco, A. M., I. Balan, D. Talibi, N. Mainville, and M. Raymond. 1997. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 272:19304-19313. [DOI] [PubMed] [Google Scholar]
- 2.Alarco, A. M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kinston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
- 5.Billard, P., H. Dumond, and M. Bolotin-Fukuhara. 1997. Characterization of an AP-1-like transcription factor that mediates an oxidative stress response in Kluyveromyces lactis. Mol. Gen. Genet. 257:62-70. [DOI] [PubMed] [Google Scholar]
- 6.Ciriolo, M. R., M. Paci, M. Sette, A. De Martino, A. Bozzi, and G. Rotilio. 1993. Transduction of reducing power across the plasma membrane by reduced glutathione. A 1H-NMR spin-echo study of intact human erythrocytes. Eur. J. Biochem. 215:711-718. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, B. A., Y. Pilpel, R. D. Mitra, and G. M. Church. 2002. Discrimination between paralogs using microarray analysis: application to the Yap1p and Yap2p transcriptional networks. Mol. Biol. Cell 13:1608-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, S. T., E. A. Epping, S. M. Steggerda, and W. S. Moye-Rowley. 1999. Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 19:8302-8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danon, A. 2002. Redox reactions of regulatory proteins: do kinetics promote specificity? Trends Biochem. Sci. 27:197-203. [DOI] [PubMed] [Google Scholar]
- 10.Delaunay, A., A. D. Isnard, and M. B. Toledano. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaunay, A., D. Pflieger, M. B. Barrault, J. Vinh, and M. B. Toledano. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471-481. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson, D. A., and H. J. Forman. 2002. Cellular glutathione and thiols metabolism. Biochem. Pharmacol. 64:1019-1026. [DOI] [PubMed] [Google Scholar]
- 13.Dumond, H., N. Danielou, M. Pinto, and M. Bolotin-Fukuhara. 2000. A large-scale study of Yap1p-dependent genes in normal aerobic and H2O2-stress conditions: the role of Yap1p in cell proliferation control in yeast. Mol. Microbiol. 36:830-845. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, K., C. Johnstone, and C. Thompson. 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19:1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes, L., C. Rodrigues-Pousada, and K. Struhl. 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17:6982-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filomeni, G., G. Rotilio, and M. R. Ciriolo. 2002. Cell signalling and the glutathione redox system. Biochem. Pharmacol. 64:1057-1064. [DOI] [PubMed] [Google Scholar]
- 17.Garrido, E. O., and C. M. Grant. 2002. Role of thioredoxins in the response of Saccharomyces cerevisiae to oxidative stress induced by hydroperoxides. Mol. Microbiol. 43:993-1003. [DOI] [PubMed] [Google Scholar]
- 18.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gil-ad, N. L., N. Bar-Nun, T. Noy, and A. M. Mayer. 2000. Enzymes of Botrytis cinerea capable of breaking down hydrogen peroxide. FEMS Microbiol. Lett. 190:121-126. [DOI] [PubMed] [Google Scholar]
- 20.Govrin, E. M., and A. Levine. 2000. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10:751-757. [DOI] [PubMed] [Google Scholar]
- 21.Grant, C. M., F. H. Maciver, and I. W. Dawes. 1996. Stationary-phase induction of GLR1 expression is mediated by the yAP-1 transcriptional regulatory protein in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 22:739-746. [DOI] [PubMed] [Google Scholar]
- 22.Grant, C. M., G. Perrone, and I. W. Dawes. 1998. Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 253:893-898. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki, L., and J. Aguirre. 2001. Multiple catalase genes are differentially regulated in Aspergillus nidulans. J. Bacteriol. 183:1434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerppola, T. K., and T. Curran. 1994. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene 9:3149-3158. [PubMed] [Google Scholar]
- 25.Kuge, S., M. Arita, A. Murayama, K. Maeta, S. Izawa, Y. Inoue, and A. Nomoto. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21:6139-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuge, S., and N. Jones. 1994. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuge, S., N. Jones, and A. Nomoto. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040-16046. [DOI] [PubMed] [Google Scholar]
- 29.Lev, S., and B. A. Horwitz. 2003. A mitogen-activated protein kinase pathway modulates the expression of two cellulase genes in Cochliobolus heterostrophus during plant infection. Plant Cell 15:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine, A., R. I. Pennell, M. E. Alvarez, R. Palmer, and C. Lamb. 1996. Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 6:427-437. [DOI] [PubMed] [Google Scholar]
- 31.Lu, S., L. Lyngholm, G. Yang, C. Bronson, O. C. Yoder, and B. G. Turgeon. 1994. Tagged mutations at the Tox1 locus of Cochliobolus heterostrophus by restriction enzyme-mediated integration. Proc. Natl. Acad. Sci. USA 91:12649-12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeta, K., S. Izawa, S. Okazaki, S. Kuge, and Y. Inoue. 2004. Activity of the Yap1 transcription factor in Saccharomyces cerevisiae is modulated by methylglyoxal, a metabolite derived from glycolysis. Mol. Cell. Biol. 24:8753-8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen, D. T., A. M. Alarco, and M. Raymond. 2001. Multiple Yap1p-binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J. Biol. Chem. 276:1138-1145. [DOI] [PubMed] [Google Scholar]
- 35.Prusty, R., P. Grisafi, and G. R. Fink. 2004. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:4153-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reglinski, J., S. Hoey, W. E. Smith, and R. D. Sturrock. 1988. Cellular response to oxidative stress at sulfhydryl group receptor sites on the erythrocyte membrane. J. Biol. Chem. 263:12360-12366. [PubMed] [Google Scholar]
- 37.Robbertse, B., O. C. Yoder, A. Nguyen, C. L. Schoch, and B. G. Turgeon. 2003. Deletion of all Cochliobolus heterostrophus monofunctional catalase-encoding genes reveals a role for one in sensitivity to oxidative stress but none with a role in virulence. Mol. Plant-Microbe Interact. 16:1013-1021. [DOI] [PubMed] [Google Scholar]
- 38.Stephen, D. W., S. L. Rivers, and D. J. Jamieson. 1995. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol. Microbiol. 16:415-423. [DOI] [PubMed] [Google Scholar]
- 39.Sugiyama, K., S. Izawa, and Y. Inoue. 2000. The Yap1p-dependent induction of glutathione synthesis in heat shock response of Saccharomyces cerevisiae. J. Biol. Chem. 275:15535-15540. [DOI] [PubMed] [Google Scholar]
- 40.Tenreiro, S., A. R. Fernandes, and I. Sa-Correia. 2001. Transcriptional activation of FLR1 gene during Saccharomyces cerevisiae adaptation to growth with benomyl: role of Yap1p and Pdr3p. Biochem. Biophys. Res. Commun. 280:216-222. [DOI] [PubMed] [Google Scholar]
- 41.Tiedemann, A. V. 1997. Evidence for a primary role of active oxygen species in induction of host cell death during infection of bean leaves with Botrytis cinerea. Physiol. Mol. Plant Pathol. 50:151-166. [Google Scholar]
- 42.Toone, W. M., S. Kuge, M. Samuels, B. A. Morgan, T. Toda, and N. Jones. 1998. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12:1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toone, W. M., B. A. Morgan, and N. Jones. 2001. Redox control of AP-1-like factors in yeast and beyond. Oncogene 20:2336-2346. [DOI] [PubMed] [Google Scholar]
- 44.Wemmie, J. A., M. S. Szczypka, D. J. Thiele, and W. S. Moye-Rowley. 1994. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J. Biol. Chem. 269:32592-32597. [PubMed] [Google Scholar]
- 45.Wojtaszek, P. 1997. Oxidative burst: an early plant response to pathogen infection. Biochem. J. 322:681-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, A. L., and W. S. Moye-Rowley. 1994. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 14:5832-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoder, O. C. 1988. Cochliobolus heterostrophus, cause of southern corn leaf blight. Adv. Plant Pathol. 6:93-112. [Google Scholar]