Abstract
Mitochondrial F1Fo-ATP synthase complexes do not exist as physically independent entities but rather form dimeric and possibly oligomeric complexes in the inner mitochondrial membrane. Stable dimerization of two F1Fo-monomeric complexes involves the physical association of two membrane-embedded Fo-sectors. Previously, formation of the ATP synthase dimeric-oligomeric network was demonstrated to play a critical role in modulating the morphology of the mitochondrial inner membrane. In Saccharomyces cerevisiae, subunit e (Su e) of the Fo-sector plays a central role in supporting ATP synthase dimerization. The Su e protein is anchored to the inner membrane via a hydrophobic region located at its N-terminal end. The hydrophilic C-terminal region of Su e resides in the intermembrane space and contains a conserved coiled-coil motif. In the present study, we focused on characterizing the importance of these regions for the function of Su e. We created a number of C-terminal-truncated derivatives of the Su e protein and expressed them in the Su e null yeast mutant. Mitochondria were isolated from the resulting transformant strains, and a number of functions of Su e were analyzed. Our results indicate that the N-terminal hydrophobic region plays important roles in the Su e-dependent processes of mitochondrial DNA maintenance, modulation of mitochondrial morphology, and stabilization of the dimer-specific Fo subunits, subunits g and k. Furthermore, we show that the C-terminal coiled-coil region of Su e functions to stabilize the dimeric form of detergent-solubilized ATP synthase complexes. Finally, we propose a model to explain how Su e supports the assembly of the ATP synthase dimers-oligomers in the mitochondrial membrane.
The mitochondrial F1Fo-ATP synthase complex plays a central role in the aerobic synthesis of adenosine 5′ triphosphate (ATP) in all eukaryotic cells (10, 12, 38, 41). The F1Fo-ATP synthase enzyme catalyzes the formation of this ATP from adenosine 5′ diphosphate (ADP) in a manner that is coupled to the transport of protons across the mitochondrial inner membrane from the intermembrane space to the matrix space. In general, two functionally distinct parts of the F1Fo-ATP synthase complex may be distinguished, the water-soluble F1-sector and the membrane-embedded Fo-sector. Both the Fo-sector and F1-sector are multisubunit complexes. The F1-sector, composed of nuclearly encoded subunits, performs the ATP synthesis and hydrolysis reactions. The Fo-sector, composed largely of hydrophobic subunits (some are nuclearly encoded and others are mitochondrially encoded), mediates the proton transport steps across the lipid bilayer (10, 12, 38, 41).
The Fo-sector of the bacterial ATP synthase complex is probably the simplest in form, as it is composed of only three known subunits, subunits a, b, and c. The mitochondrial Fo-sector is more complex in composition, however. The Saccharomyces cerevisiae Fo-sector, for example, is composed of at least nine different subunits. In addition to the counterparts of subunits a, b, and c (referred to as subunits 6, 4, and 9, respectively), the yeast Fo-sector has been described as containing subunits 8 (equivalent to bovine A6L), d, e, f, g, h, i and j, and k (12, 41).
In mitochondria, the F1Fo-ATP synthase complex does not exist as a separate entity in the membrane but rather forms dimeric complexes. Evidence for the presence of the dimeric ATP synthase complexes came initially from molecular sizing analysis performed on complexes, which were solubilized from mitochondrial membranes under mild detergent lysis conditions (e.g., by use of digitonin) (3, 4). More recently, chemical cross-linking analysis (16, 28, 37, 42) and fluorescence resonance energy transfer technology (14, 15) have been used to demonstrate the existence of interacting dimeric complexes in intact mitochondrial membrane systems in vitro and in vivo, respectively. Organization of the F1Fo-ATP synthase complex into dimeric complexes has been shown to involve the physical interaction of membrane-embedded subunits of neighboring Fo complexes (3, 4, 13, 16, 28, 35, 37, 42). Subunit 4 (Su 4) (equivalent to bovine subunit b), subunit e (Su e), and subunit g (Su g), all nuclearly encoded Fo-sector subunits, play a central role in the ATP synthase dimerization process in yeast mitochondria (3, 4, 27, 36, 37, 40). Su e and Su g are not unique to the yeast ATP synthase, as they are present in other mitochondrial F1Fo-ATP synthase complexes such as bovine complexes, in which context they are also referred to as Su e and Su g (11). To date, no equivalents of Su e and Su g in bacterial ATP synthase complexes have been reported. Unlike that of Su 4, the presence of Su e and Su g is not essential for the enzymatic activity of the ATP synthase complex (3, 4, 7, 27, 37, 40). In the absence of Su e and Su g, the digitonin-solubilized ATP synthase complex is present as only a monomeric species, indicating that Su e and Su g play an important role in dimeric complex stabilization (4, 28). Interestingly, the inner membrane of mitochondria from the Su e and Su g null mutant strains (Δsu e and Δsu g, respectively) does not display characteristic tubular mitochondrial shape and normal cristae morphology but instead exhibits aberrant shape and cristae structure (6, 28). Thus, the absence of the dimeric ATP synthase network has been correlated with disrupted inner membrane morphology (6, 28). Although this observation is consistent with an earlier hypothesis proposed by Allen (1), the molecular basis of how the ATP synthase complex modulates the inner membrane morphology is poorly understood.
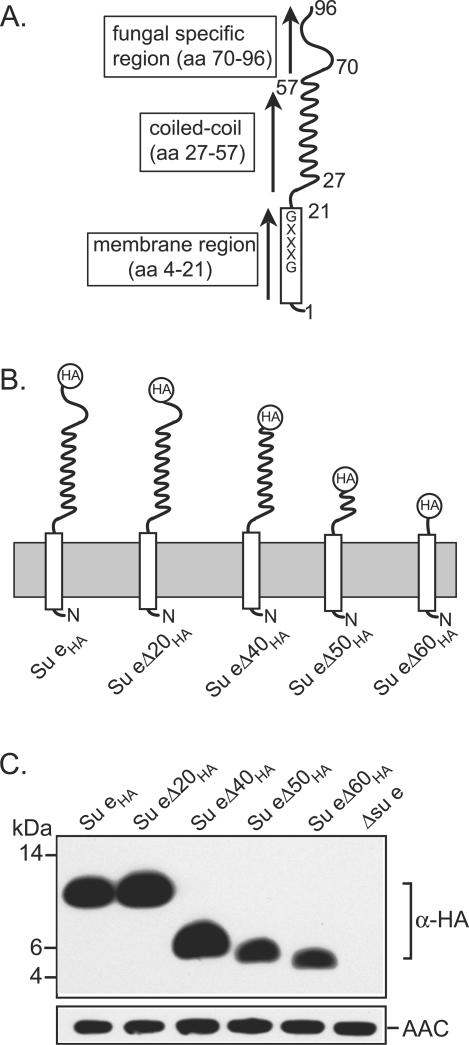
Our present work focuses on Su e and its role in the formation of F1Fo-ATP synthase dimers. Su e, a relatively small protein (96 amino acid residues in length), is an integral inner membrane protein (3, 5, 6, 9, 40). The stoichiometry of Su e in the Fo-sector has been best characterized in rat mitochondrion studies, from which a 2:1 ratio of Su e subunits to F1Fo-ATP synthase complex was reported (2). A single hydrophobic segment located at the extreme N-terminal region of the Su e protein (approximately residues 4 to 21) is thought to anchor the protein to the inner membrane, where the bulk of the protein, the C-terminal hydrophilic segment (residues 26 to 96), resides in the mitochondrial intermembrane space (Fig. 1A). The membrane anchor region displays a high degree of amino acid sequence conservation between Su e homologs (5). Thus, in addition to serving as a hydrophobic membrane anchor, it is possible that this region may exert an important role for the function of Su e. The C-terminal segment of the yeast Su e contains a region (residues 27 to 57) that displays the potential to form a coiled-coil structure (3). This motif may be important for the function of Su e, as it is conserved in all known members of the Su e protein family (3, 18). The extreme C-terminal region encompassed by the C-terminal approximately 40 amino acid residues appears to date to be restricted to fungal Su e homologs and displays little homology to the known mammalian or Drosophila Su e proteins (18).
FIG. 1.
Expression of Su eΔCHA derivatives. (A) Depiction of the Su e protein (96 amino acid residues in length). An open rectangle shape indicates the N-terminal hydrophobic membrane anchor region, and the curled line represents the putative coiled-coil structure. aa, amino acid; GXXXG, the conserved dimerization motif located in the membrane anchor region of Su e (see text for details). (B) The membrane topology of the C-terminal-truncated Su e derivatives. The position of the HA tag at the C terminus of the proteins is indicated. (C) Mitochondria were isolated from the Δsu e null mutant strain harboring the Su eHA derivatives as indicated and were analyzed by SDS-PAGE, Western blotting, and immune decoration with an HA-specific polyclonal antiserum. Loading levels of the mitochondria were controlled by decoration of a control protein, the ADP/ATP carrier (AAC). The mobilities of the protein standards (in kilodaltons) are indicated.
In the present study, we directly analyzed the role of the N-terminal membrane anchor and the intermembrane space-localized C-terminal regions of Su e for its function. To do so, we expressed a number of C-terminal-truncated derivatives of the yeast Su e in su e null mutant yeast cells. We have analyzed the ability of these truncated Su e derivatives to support a number of functions known to require the Su e protein. As described below, our findings highlight the importance of the N-terminal hydrophobic region for the function of the Su e protein.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Yeast strains used in this study were wild-type (WT) W303-1A (Mat a, leu2, trp1, ura3, his3, ade2), rho0 strain (W303-1A, rho0) (a kind gift from Alex Tzagoloff, Columbia University, N.Y.), the su e null mutant, Δsu e (W303-1A, leu2, trp1, ura3, ade2, ATP21::HIS3), and the su g null mutant, Δsu g (W303-1A, leu2, trp1, ura3, ade2, ATP20::HIS3) (4, 30). Mitochondria were isolated from the resulting yeast strains, which had been grown in YP (yeast extract-peptone) supplemented with 0.5% lactate and galactose (YP-Gal medium) (2%) or glycerol (3%, YPG medium) as indicated (17).
Cloning and expression of C-terminally truncated, HA-tagged Su e derivatives (Su eΔCHA).
The entire open reading frame (ORF) encoding Su e (ATP21/TIM11 gene) (39), or 3′ truncated derivatives thereof, was amplified by the PCR as a SpeI-PstI fragment, whereby the translational stop codon of the Su e open reading frame was omitted. The reverse primer used in each case contained the sequence information to encode the hemagglutinin (HA) epitope and a translational stop codon, in addition to a Su e-specific sequence for priming. The recombinant PCR products were cloned into yeast integration vector Yip351, which contained the galactose-inducible GAL10 promoter, and the LEU2 auxotrophic gene (20). The resulting plasmids were linearized at a unique BstEII restriction site located in the 5′ region of the LEU2 gene locus, ethanol precipitated, and transformed into the su e null mutant, Δsu e, by use of a protocol described previously (26). Leucine-positive transformants were selected and grown in galactose-containing medium. Mitochondria were isolated from each of the transformants, and the expression and mitochondrial localization of the Su eΔCHA derivatives were verified by Western blotting using an antibody specific for the HA epitope (Covance Research Products, Berkeley, Calif.). Electrophoretic separation of the Su eΔCHA derivatives was achieved using precast gels (10% acrylamide) from Invitrogen Research Corp. and using MES (morpholineethanesulfonic acid) as an electrophoresis buffer.
CN-PAGE analysis of F1Fo-ATP synthase complexes.
Mitochondria (200 μg of protein) were lysed with 36 μl of digitonin buffer (34 mM potassium acetate, 34 mM HEPES-KOH [pH 7.4], 11.4% glycerol, 1 mM phenylmethylsulfonyl fluoride) and various concentrations of digitonin (as indicated) for 30 min on ice and subjected to a clarifying spin (JA-25.50 rotor, Beckman Avanti J-25 system) (30 min, 30,000 × g). The supernatants were collected, and 2 μl of 50% glycerol was added to the samples prior to electrophoresis. Samples were analyzed by clear native (CN)-polyacrylamide gel electrophoresis (PAGE) using a 3.5 to 10% polyacrylamide gel (32). Following electrophoresis, Western blotting to nitrocellulose was performed and F1Fo-ATP synthase complexes were detected by decoration with an F1-sector-specific antibody. Thyroglobulin (669 kDa) was used as a molecular marker.
Rho0/Rho− cell conversion detection assay.
All yeast strains were grown on YPG (3% glycerol) plates, with the exception of the rho0 control strain, which was initially grown on YPAD (2% glucose supplemented with adenine) plates. Colonies were used to inoculate YP-Gal medium and allowed to grow overnight at 30°C. The optical density at 580 nm of the overnight cultures was measured, and equivalent numbers of cells from each culture were plated onto either YPG-0.1% galactose or YPD (2% glucose) plates. Following incubation at 30°C for 1 week, the colonies were counted and the percentage of rho+ cells (i.e., those able to grow on glycerol) was calculated.
Analysis of mitochondrial morphology.
Strains were grown in YP medium containing glycerol (2%) and ethanol (3%) and supplemented with 0.2% galactose (as indicated) to control expression of Su e derivatives. Cells were grown to mid-log phase and stained in growth medium with 33.3 ng of 3,3′-dihexyloxacarbocyanine iodide (DiOC6; Molecular Probes, Eugene, Oreg.)/ml. For collection of images, cells were examined using a DeltaVision system (Applied Precision Instruments, Issaquah, Wash.) that included an Olympus microscope with a 100× Plan Apochromat objective and a PXL charge-coupled device camera (Roper Industries, Princeton, N.J.). Images in the z axis were taken every 0.2 μm, and each image was deconvolved using DeltaVision software. For quantification of mitochondrial morphology phenotypes, cells were examined using a Zeiss Axioscop microscope with a 100× Plan Apochromat objective lens and an Endow 41017 filter (Chroma Technology Corp., Rockingham, Vt.).
Miscellaneous.
Protein determinations and sodium dodecyl sulfate (SDS)-PAGE were performed according to published methods (8, 22). The Western blot analysis and antibody decoration were performed using available Su e, Su g, and Su k antisera raised against peptides corresponding to the C-terminal region of each of the proteins (3, 4). The F1 antibody was raised in chicken following injection of purified yeast F1-sector and was obtained from David Mueller (The Chicago Medical School, Chicago, Ill.).
RESULTS
Expression of C-terminal-truncated derivatives of Su e.
To address the functional relevance of the N-terminal hydrophobic and C-terminal hydrophilic segments of the yeast Su e protein (Fig. 1A), a series of truncated derivatives of the Su e protein (Su eΔCHA) was constructed. The truncated derivatives correspond to a deletion of the C-terminal 20, 40, 50, or 60 amino acid residues of the Su e protein (Fig. 1B). Expression of the proteins was achieved by cloning the DNA fragments that encode the proteins into a yeast integration vector, Yip351, which contained a galactose-inducible promoter. The resulting plasmids were linearized and then integrated into the leu2 locus of the su e null yeast mutant, Δsu e. The Su e derivatives were expressed as C-terminally HA-tagged proteins, as our present Su e-specific antiserum was raised against the C-terminal 15 amino acid residues of Su e and hence would not have recognized the truncated Su e derivatives. As a control, we also expressed the full-length Su e protein with the C-terminal HA tag (Su eHA).
The Su eΔC20HA derivative shows a 20-amino-acid-residue deletion from the extreme C-terminal region, resulting in the deletion of the extreme nonconserved region of the yeast Su e protein (residues 77 to 96) (Fig. 1B). The amino acid residues 57 to 96 are deleted in the Su eΔC40HA derivative. These amino acid residues are located immediately C terminal to the predicted coiled-coil motif, which spans residues 27 to 57. The coiled-coil region is compromised and completely deleted in the Su eΔC50HA and Su eΔC60HA derivatives, respectively (Fig. 1B). Thus, the most extreme deletion, the Su eΔC60HA derivative, is composed of only the initial 36 amino acid residues of Su e, a region which encompasses the N-terminal hydrophobic membrane anchor.
The truncated derivatives were expressed in the yeast su e null mutant background. Mitochondria were isolated from the resulting transformants, and the presence of the Su e derivatives was verified by Western blotting using an HA-specific antiserum (Fig. 1C). For some unknown reason the Su eΔC20HA derivative displayed an electrophoretic mobility similar to that of the full-length HA-tagged Su e derivative, Su eHA. The mitochondrial localization of even the shortest derivative, Su eΔC60HA, indicates that the mitochondrial targeting information and submitochondrial sorting information for Su e must reside in its N-terminal 36 amino acid residues. This is worthy of note, as the Su e proteins lack classical N-terminal positively charged mitochondrial targeting sequences. The steady-state levels of the Su eΔC50HA and Su eΔC60HA derivatives appeared somewhat reduced in comparison to those of the other Su e truncation derivatives (Fig. 1C). It is possible that the presence of the coiled-coil structure of Su e is required to increase the stability of the Su e protein.
The membrane anchor of Su e is required for the stabilization of the Su g and Su k proteins and the maintenance of mitochondrial DNA (mt-DNA).
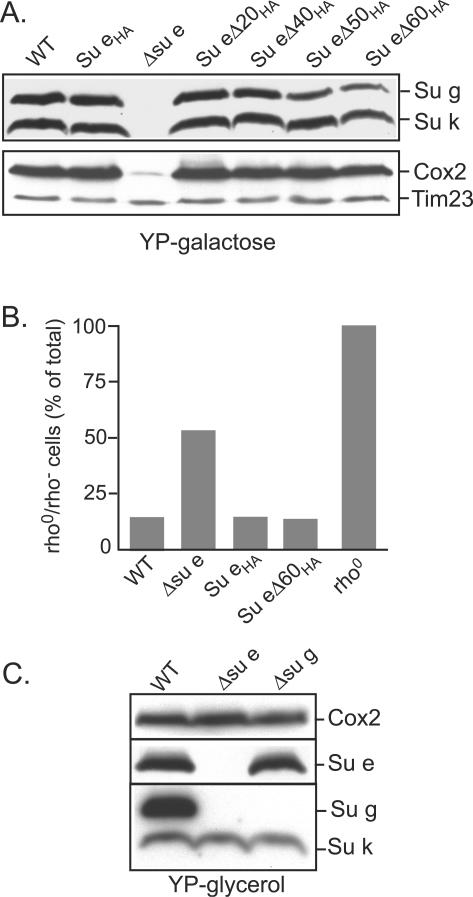
Following the successful expression and mitochondrial targeting of the Su eΔCHA derivatives, our subsequent experiments addressed the functional relevance of the N-terminal membrane region and the extreme C-terminal and coiled-coil regions of the Su e protein. To do so, we started by testing the steady-state levels of Su g and Su k proteins in the mitochondria harboring the truncated Su e derivatives. Su g and Su k are both subunits of the F1Fo-ATP synthase and, like Su e, are found in association with the dimeric form of the complex. As previously reported, the steady-state levels of both Su g and Su k are reduced in the absence of the Su e protein, i.e., in the Δsu e cells, when grown on a fermentable carbon source such as galactose (4). Western blot analysis of mitochondria isolated from the su e null mutant harboring the full-length (Su eHA) or C-terminal truncation derivatives Su eΔ20HA or Su eΔ40HA and grown in the presence of galactose indicated that the levels of Su g and Su k were similar to those in WT mitochondria, in contrast to the reduced levels observed in the su e null mutant (Fig. 2A). Su g and Su k were also observed in the Su eΔ50HA and Su eΔ60HA mitochondria; however, the levels of Su g were slightly reduced relative to those of the WT control mitochondria (Fig. 2A) and can be correlated with the observed reduction in the levels of the Su e derivatives in these mitochondria (Fig. 1C). Taken together, these data indicate that the presence of the N-terminal hydrophobic segment of Su e is necessary to ensure the stable accumulation of the Su g and Su k proteins.
FIG. 2.
Stabilization of Su g, Su k, and the mt-DNA requires the N-terminal hydrophobic region of Su e. (A) The Δsu e null strain, isogenic WT (W303-1A), and the Δsu e null mutant harboring the truncated Su e derivatives, as indicated, were grown on YP-Gal medium. Isolated mitochondria (150 μg of protein) were analyzed by SDS-PAGE and Western blotting. Levels of the Su g, Su k, and Cox2 proteins were analyzed using subunit-specific antisera. Loading levels of the mitochondria were controlled by decoration of a control protein, Tim23. (B) Δsu e, the Δsu e strains expressing Su eHA, Su eΔ60HA derivatives, and the WT control strain maintained on YPG (glycerol) medium were transferred to YP-Gal (galactose) medium and grown overnight. The number of cells that became rho0/rho− (i.e., exhibited loss of mt-DNA) was determined as described in the Materials and Methods. A control rho0 strain that had been maintained in YP-Gal medium was analyzed in parallel. For each strain type, the number of cells that exhibited loss of mt-DNA was expressed as a percentage of the total cells analyzed (% rho0/rho−). (C) The Δsu e and Δsu g null strains and their isogenic WT strain were maintained on YPG medium, and mitochondria were isolated. The isolated mitochondria (150 μg of protein) were further analyzed as described above for panel A.
As recently reported, the su e null mutant strain displays an increased frequency in spontaneous rho0/rho− cell conversion (5, 28). Consistently, mitochondria isolated from Δsu e cells that had been grown under fermentable carbon source (galactose) conditions contained significantly reduced levels of subunit 2 of the cytochrome oxidase complex (Cox2) (Fig. 2A). Cox2 is encoded on the mt-DNA; therefore, rho0/rho− cells, which exhibit a loss of the mt-DNA, may have lost their capacity to synthesize Cox2. Mitochondria isolated from the Δsu e yeast strain expressing the truncated derivatives of Su e and grown in galactose-containing medium contained WT levels of Cox2, however (Fig. 2A). This observation suggests that the N-terminal hydrophobic region of Su e is sufficient to stabilize the mt-DNA. Consistently, expression of the Su eΔ60HA derivative in the Δsu e strain prevented the high frequency of spontaneous rho0/rho− cell formation which was observed in the Δsu e strain in the absence of an expressed Su e derivative (Fig. 2B). Taking these results together, we conclude that the presence of Su e, and in particular that of its N-terminal hydrophobic region, is required to ensure the stable maintenance of the mt-DNA and hence the coding capacity of Cox2 in the Δsu e yeast cells.
The mitochondria isolated from Δsu e cells grown in the presence of galactose contained reduced levels of Su g and Su k proteins (Fig. 2A), indicating that Su e plays a role in the stabilization of these proteins. However, the levels of Su k, like those of Cox2, were reduced in the Δsu e cells only when grown under conditions that included fermentable, but not nonfermentable, carbon sources (Fig. 2A versus C). We conclude therefore that the presence of Atp6, Atp8, and/or Atp9, the mitochondrially encoded Fo subunits, rather than the immediate presence of Su e may be required for the stabilization of Su k in the Δsu e mitochondria. The presence of the assembled Fo-sector has been previously shown to be required for the stable accumulation of Su k (4). In contrast, the levels of Su g were decreased in the mitochondria of Δsu e cells irrespective of whether they were grown on fermentable or nonfermentable carbon sources (Fig. 2A versus C). Thus, unlike the Su k protein, Su g displays a direct dependence on the presence of Su e (in particular, the N-terminal hydrophobic region of Su e) for its stabilization. However, Su e does not depend on the presence of Su g for its stabilization, as the levels of Su e were found to be unaffected in the su g null mutant mitochondria (Fig. 2C).
CN-PAGE analysis of mitochondria harboring the Su eΔCHA derivatives.
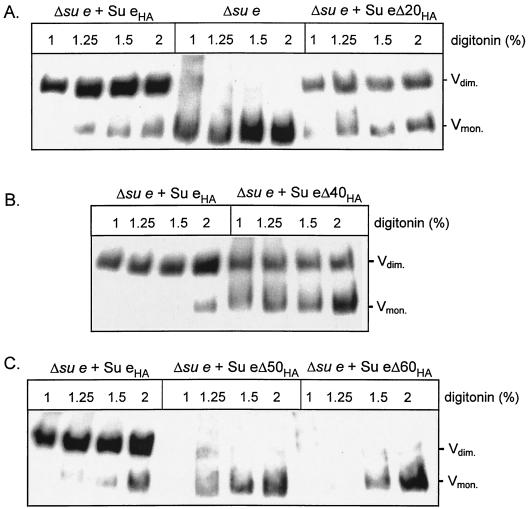
When solubilized from WT mitochondria, the ATP synthase complex can be isolated as a dimeric complex. Formation of a detergent-stable dimeric complex depends on the presence of Su e, as only monomeric complexes are observed upon digitonin solubilization of Δsu e mitochondria. The assembly state of the ATP synthase complexes was therefore analyzed in mitochondria isolated from the su e null mutant strain, together with those from the su e null strains harboring the Su eΔCHA derivatives. Mitochondrial protein complexes were solubilized with a series of different concentrations of digitonin, and the complexes were resolved by CN gel electrophoresis (CN-PAGE) (Fig. 3). CN-PAGE was selected rather than the previously used blue native PAGE, since we routinely observed that the solubilized dimeric ATP synthase was more stable in the absence of Coomassie blue (results not shown).
FIG. 3.
CN gel analysis of the dimeric state of the F1Fo-ATP synthase complexes. Mitochondria were isolated from the Δsu e null strain harboring the Su eΔCHA-tagged derivatives as indicated. The Δsu e null strain had been grown on YPG medium, whereas the other strains were grown on YP-Gal. Isolated mitochondria (200 μg of protein) were solubilized with digitonin at the concentrations indicated and, following a clarifying centrifugation, were directly analyzed on CN-PAGE as described in Materials and Methods. Following electrophoresis, the native gels were Western blotted and immune decoration was performed using a yeast F1-specific antisera. The positions of the dimeric (Vdim.) and monomeric (Vmon.) F1Fo-ATP synthase complexes are indicated.
As previously described, in the absence of Su e, the ATP synthase complex is solubilized with digitonin from the mitochondrial membranes predominantly in its monomeric form (4) (Fig. 3A). The functionality of the expressed Su eHA and Su eΔ20HA derivatives was indicated by the ability of these derivatives to support the stabilization of the dimeric form of the ATP synthase in the su e null mutant mitochondria (Fig. 3A). The ratio of dimeric to monomeric ATP synthase in the mitochondria harboring the full-length Su eHA derivative was similar to that in the WT untagged control for each of the digitonin concentrations (results not shown). The presence of dimeric complexes was also observed for mitochondria harboring the Su eΔ20HA derivative. However, the dimeric complexes supported by the Su eΔ20HA derivative did not appear to be as stable as those formed in the presence of the full-length HA-tagged derivative. Increasing digitonin concentrations resulted in a larger proportion of the monomeric form of the complex in the Su eΔ20HA mitochondria (Fig. 3A).
Solubilization of the mitochondria harboring the Su eΔ40HA derivative with low digitonin concentrations also demonstrated the presence of assembled dimeric ATP synthase complexes. The stability of the Su eΔ40HA-supported dimeric ATP synthase complexes was, however, significantly reduced (Fig. 3B). Solubilization of the Su eΔ40HA mitochondria with the highest digitonin concentration resulted in approximately 80% of the ATP synthase complex present as the monomeric form on the CN-PAGE gel (approximately 20:80 dimer/monomer ratio). This was in contrast to the full-length Su eHA mitochondria, for which an approximately 90:10 dimer/monomer ratio was observed under the same detergent conditions (Fig. 3B). Although the computer prediction analysis indicates that the coiled-coil motif (residues 27 to 57) is retained in the Su eΔ40HA derivative, it is possible that deleting the 40 amino acid residues (residues 57 to 97) immediately C terminal to this predicted motif may exert some adverse influence on the stability of the coiled-coil structure, which in turn may affect the stability of the dimeric ATP synthase.
CN-PAGE analysis of the detergent-solubilized ATP synthase from the mitochondria harboring the Su eΔ50HA or Su eΔ60HA derivatives indicated the presence of primarily the monomeric form of the enzyme (Fig. 3C). Furthermore, we observed that the solubilization of the ATP synthase complex in the Su eΔ50HA or Su eΔ60HA mitochondria was reproducibly less efficient at lower concentrations of digitonin. It is possible that the reduced levels of the Su eΔ50HA or Su eΔ60HA proteins may also contribute to the observed reduction in the level of stable dimeric ATP synthase. However, taking these results together with those obtained with the Su eΔ20HA or Su eΔ40HA mitochondria, for which despite normal levels of these proteins a clear instability of the dimeric complexes was also observed, it appears that the C-terminal elements of Su e play a role in the stabilization-formation of dimeric ATP synthase complexes. We therefore conclude that the C-terminal region of Su e, and in particular the coiled-coil motif, is required to support and/or maintain the dimerization of the F1Fo-ATP synthase under detergent solubilization and CN-PAGE electrophoresis conditions.
The N-terminal hydrophobic region of Su e supports normal mitochondrial morphology.
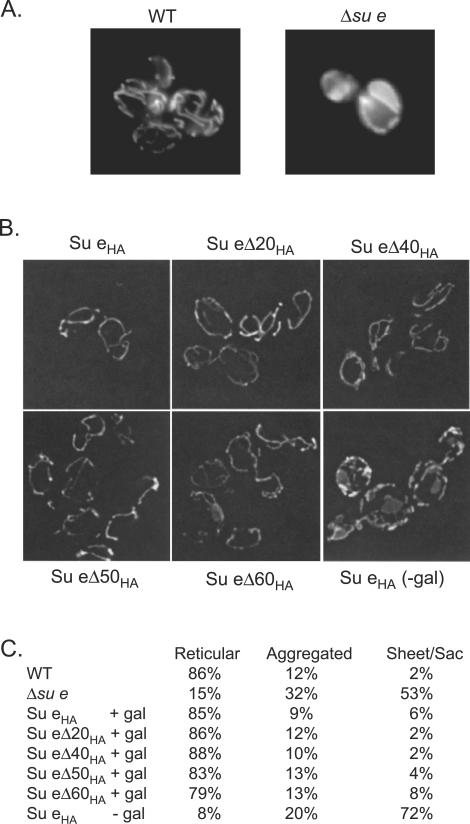
Mitochondrial yeast mutants deficient in either Su e or Su g fail to develop normal mitochondrial morphology (6, 28). Transmission electron microscope studies have shown that the inner membrane of mitochondria isolated from the Δsu e and Δsu g mutants have onion-like membrane morphology (6, 28). Hence, it was concluded that the presence of the Su e- and Su g-mediated ATP synthase dimers-oligomers plays a critical role in the modulation of the cristae morphology.
In the present study, mitochondrial morphology was analyzed using standard fluorescence microscopy (Fig. 4). As previously described, WT mitochondria form a branched tubular network extending throughout the cell and located below the cell surface (19, 34). In contrast, the mitochondrial membranes of the Δsu e null mutant appeared “collapsed” or “sheet like,” and the tubular network system was disrupted (Fig. 4A). The role of the N-terminal membrane anchor and C-terminal regions of Su e in supporting the development of the normal mitochondrial morphology was addressed for the Δsu e mutant cells harboring the various Su eΔCHA derivatives. When Su eHA or the C-terminally truncated derivatives (Su eΔC20HA, Su eΔC40HA, Su eΔC50HA, or Su eΔC60HA) were expressed in the su e null mutant background, the branched tubular network morphology was restored (Fig. 4B and C). The mitochondrial network in all the Su eΔCHA cells appeared to be very similar to that observed with the WT or Δsu e cells expressing Su eHA. Analysis of the cells expressing the Su eΔC60HA derivative did indicate, however, that the tubular network, although present, appeared slightly thinner and sometimes fragmented (results not shown). This slight increase in aberrant mitochondrial morphology in the Su eΔC60HA mitochondria could be the result of lower steady-state levels of this derivative.
FIG. 4.
Expression of Su eΔCHA-tagged derivatives can restore normal mitochondrial morphology to the Δsu e null mutant strain. Mitochondrial morphology was analyzed in WT and Δsu e cells (A) or Δsu e cells expressing the HA-tagged derivatives (under control of a galactose-inducible promoter) (B), which had been grown in the presence or absence (−gal) of galactose as indicated. The morphology of the mitochondria was observed using fluorescence microscopy and following staining of the mitochondrial membranes by use of DiOC6 as described in Materials and Methods. (C) The percentages of cells (n > 200 for each cell type) displaying the normal reticular, aggregated, and sheet-like morphologies were quantified.
We therefore conclude that the presence of the C-terminal hydrophilic region of Su e, encompassing the most extreme C-terminal end and the coiled-coil region, does not play an essential role in the establishment of the mitochondrial tubular network system within the cell. Rather, it appears that the expression of the N-terminal hydrophobic segment of Su e is sufficient to restore the tubular morphology in the Δsu e null mutant cells. A more detailed analysis at the electron microscope level would be required, however, before we can conclude that the architecture of the cristae membrane is normal in the absence of the coiled-coil region of Su e and in particular in the case of the Su eΔ60HA derivative, for which a slight narrowing and fragmentation of the membrane system was observed.
DISCUSSION
Dimerization of F1Fo-ATP synthase complexes in the mitochondrial inner membrane involves the physical interaction of neighboring Fo-sectors, anchored in the membrane, in a manner which is dependent on the Fo-sector subunit, Su e. How this dimeric network is formed from the ATP synthase monomers is not fully understood and requires a better understanding of the molecular organization and function of Su e within the assembled Fo-sector. The major goal of this study was to gain more information on the regions within the Su e protein that are important for its function. We have focused on the N-terminal, hydrophobic region of Su e, which embeds the protein in the mitochondrial inner membrane, and also on the C-terminal hydrophilic region of Su e, which is localized in the intermembrane space of mitochondria. A series of C-terminal truncation derivatives of Su e bearing a HA tag (Su eΔCHA derivatives) were generated and expressed in the yeast su e null mutant (Δsu e) background. The function of the Su e-truncated derivatives was tested by analyzing their ability to complement phenotypes generated in the absence of the authentic Su e protein. The data presented here indicates that the presence of the N-terminal hydrophobic region of Su e is required for a number of Su e-dependent processes.
The Su eΔCHA derivatives, including the Su eΔC60HA derivative, which represents only the most N-terminal 36 residues of the Su e protein, were expressed and correctly localized to the mitochondria. Hence, we conclude that the extreme N-terminal region of Su e is sufficient for targeting the Su e protein to the mitochondria. The N-terminal 36 residues of Su e do not contain a classical mitochondrial targeting sequence, which is typically a positively charged, amphipathic helix (43) and would therefore appear to comprise a nonclassical, and possibly novel, targeting mechanism to the mitochondrial inner membrane. Preliminary evidence for the involvement of Tim8 and Tim10 protein in the import pathway of Su e has been previously presented. Interestingly, the insertion of Su e into the inner membrane was reported not to require the Tim22 machinery (24), however, and thus it is presently unclear how Su e is integrated into the mitochondrial inner membrane.
The presence of Su e is required for the stabilization of Su g in the mitochondrial membrane. Strongly reduced levels of Su g were observed in Δsu e cells grown with either fermentable or nonfermentable carbon sources. Thus, the requirement of Su e for Su g stabilization appears to be a direct one and not to be due to an indirect effect caused by the loss of mt-DNA when cells are grown under fermentable conditions. Stabilization of Su g may be mediated by a direct interaction between the Su e and Su g proteins. Our findings reported here indicate that the N-terminal membrane anchor region (residues 1 to 36) is sufficient to stabilize the Su g protein. Steady-state levels of Su g close to WT levels were observed in the mitochondria harboring only the N-terminal membrane region of Su e (Su eΔC60HA). These findings would suggest that the N-terminal hydrophobic region of Su e (residues 4 to 21) and/or the short stretch of hydrophilic amino acids directly following it (residues 22 to 36), which are not deleted in the Su eΔC60HA construct, directly interact with and thereby stabilize the Su g protein. All Su e and Su g protein family members contain a conserved GXXXG motif. The GXXXG motif, where G stands for glycine and X for any hydrophobic amino acid residue, which typically resides in membrane-spanning segments, often represents dimerization (homo- or heterodimerization) motifs, the best characterized case being the homodimerization of glycophorin A protein (25, 31, 33). A recent study by Velours and colleagues has indicated the importance of the GXXXG motif of Su e. Interestingly, mutations in the GXXXG motif in Su e protein did not prevent the homodimerization of Su e (and even enhanced it) but did result in a strong reduction of Su g levels (5). We therefore conclude that the physical interaction of Su e and Su g, which is required for the stability of Su g, is mediated through the N-terminal region of Su e in a manner which is supported by the conserved GXXXG motif in Su e.
The su e null mutant (Δsu e) exhibits an increased propensity to form rho0/rho− cells, i.e., spontaneous loss of mt-DNA, relative to WT cells. Analysis of the Su eΔC60HA cells indicated that the N-terminal hydrophobic region of Su e was necessary and sufficient to maintain the mt-DNA. The loss of the mtDNA is a phenotype commonly observed with yeast mutants with an aberrant ATP synthase and defective in coupling proton transport to ATP synthesis (23). It is presently unknown, however, whether the formation of rho0/rho− cells is the cause or consequence of an aberrant ATP synthase. Therefore we conclude that the presence of Su e, in particular the N-terminal hydrophobic segment, may be important for the correct assembly of a tightly coupled F1Fo-ATP synthase complex. Interestingly, an increased formation of rho0/rho− cells has also been observed in mitochondrial morphology mutants such as the fzo1 and mdm33 mutants (21, 29), suggesting that defective morphology may also be correlated with the loss of mt-DNA.
Taken together, our findings emphasize the importance of the membrane anchor region of Su e for a number of functions of Su e. Although conserved in all Su e homologs, the importance of the C-terminal coiled-coil region remains unclear. Our findings indicate that the coiled-coil region of Su e may facilitate a Su e-Su e interaction and thereby stabilize the ATP synthase dimers, particularly under detergent solubilization conditions, as evidenced by the CN-PAGE analysis. The ability of Su e to stabilize the ATP synthase dimers depends largely on the presence of the C-terminal hydrophilic region of Su e. In the absence of the coiled-coil structure (in the Su eΔC50HA and Su eΔC60HA cells), the majority of the digitonin-solubilized complexes were present as their monomeric species. Even deletions of regions C terminal to the coiled-coil (i.e., in the Su eΔC20HA and Su eΔC40HA mitochondria) resulted in a destabilization of the ATP synthase dimers. Taking both of these observations together, we conclude that the extreme C-terminal region, in particular the coiled-coil segment of Su e, is necessary to stabilize the ATP synthase dimerization under conditions of detergent solubilization (Fig. 5).
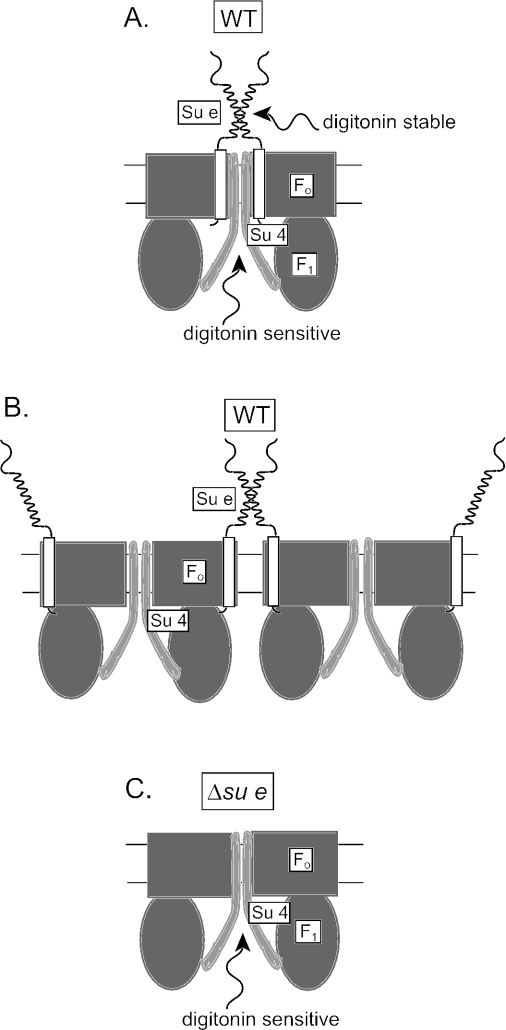
FIG. 5.
Model of F1Fo-ATP synthase dimerization and the role of Su e and Su 4. (A and B) Two alternative models for the role of Su e in stabilizing the neighboring ATP synthase dimers. (A) In this model, Su e-Su e interaction would serve to further stabilize (in a digitonin-stable fashion) two neighboring Fo-sectors whose Su 4 proteins are interacting (in a detergent-sensitive fashion). (B) In this model, the Su e-Su e interaction would serve to stabilize the contact of two Fo-sectors from neighboring Su 4-mediated F1Fo dimers. The predicted stoichiometry of Su e is 2 Su e subunits per Fo-sector. For simplicity, the models drawn depict one Su e per Fo-sector and the dimeric-specific Su g protein has been omitted. It is entirely plausible that one Su e protein may serve to stabilize neighboring Su 4-mediated Fo-dimers (as shown in panel A) and that the other Su e protein per Fo-sector could be involved in a Su e-Su e interaction between adjacent Fo-sectors, as depicted in panel B. By doing so, an oligomeric arrangement of ATP synthase complexes in the lipid bilayer may be achieved. (C) Model depicting the arrangement of F1Fo complexes in the absence of Su e. The Su 4-Su 4 interaction between neighboring Fo-sectors occurs in theabsence of Su e, but these dimers are sensitive to detergent solubilization. Hence, monomeric ATP synthase complexes are observed upon native-gel analysis following detergent solubilization of Δsu e mitochondria. For further explanation, see text.
As detailed in Fig. 5, ATP synthase dimers would be stabilized during digitonin solubilization (and CN-PAGE analysis) if supported by a Su e-Su e interaction in a manner that relied on the coiled-coil structural feature of the Su e protein. Cross-linking of Su 4-Su 4 dimers was observed in the absence of the Su e protein (37), indicating that the interaction of two neighboring Fo-sectors brings two Su 4 subunits in close proximity and does not rely on Su e (28). Consistently, the Su 4-Su 4 cross-linking was not adversely affected in the presence of the C-terminal-truncated derivatives of Su e (results not shown). However, in the absence of Su e (or the C-terminal hydrophilic region), the Su 4-Su 4 interactions appear not to be detergent stable, as the CN-PAGE analysis indicated the presence of only monomeric F1Fo complexes upon digitonin solubilization of Δsu e mitochondria (Fig. 5C). It is therefore possible that the coiled-coil Su e-Su e interaction may serve to strengthen the interaction between the Su 4-Su 4 interacting Fo-sectors (Fig. 5A) or alternatively to join neighboring Su 4-linked dimers into a tetrameric or possibly a larger oligomeric network (Fig. 5B).
Finally, our data indicate that the N-terminal anchor region of Su e, and not the coiled-coil region or extreme C-terminal end of Su e, is essential for the modulation of the mitochondrial tubular morphology. We therefore conclude that the formation of a possible ATP synthase oligomeric network mediated through a Su e-Su e coiled-coil interaction (as depicted in Fig. 5B) is not a determining factor for modulation of mitochondrial morphology. Rather, it appears that the importance of Su e in this process may be result from indirect activity; i.e., it may be that it is required for stabilization of another factor, Su g, or possibly another unknown protein which would be directly required to modulate the morphology. Clearly, the presence of the N-terminal membrane anchor region of Su e is sufficient to ensure the stabilization and function of this morphology factor.
Acknowledgments
We are grateful to Mary Dienhart for technical support and Sonika Saddar for helpful discussions.
This research was supported by funding from the National Institutes of Health (NIH) RO1GM61573 to R.A.S. and RO1GM46803 to R.E.J. and by NIH Predoctoral Training Grant 5T32GM07445 to C.D.D.
REFERENCES
- 1.Allen, R. D. 1995. Membrane tubulation and proton pumps. Protoplasma 189:1-8. [Google Scholar]
- 2.Arakaki, N., Y. Ueyama, M. Hirose, T. Himeda, H. Shibata, S. Futaki, K. Kitagawa, and T. Higuti. 2001. Stoichiometry of subunit e in rat liver mitochondrial H(+)-ATP synthase and membrane topology of its putative Ca(2+)-dependent regulatory region. Biochim. Biophys. Acta 1504:220-228. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, I., M. F. Bauer, M. Brunner, W. Neupert, and R. A. Stuart. 1997. Yeast mitochondrial F1Fo-ATPase: the novel subunit e is identical to Tim11. FEBS Lett. 411:195-200. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, I., K. Pfeiffer, W. Neupert, R. A. Stuart, and H. Schägger. 1998. Yeast mitochondrial F1Fo-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 17:7170-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arselin, G., M. F. Giraud, A. Dautant, J. Vaillier, D. Brethes, B. Coulary-Salin, J. Schaeffer, and J. Velours. 2003. The GxxxG motif of the transmembrane domain of subunit e is involved in the dimerization/oligomerization of the yeast ATP synthase complex in the mitochondrial membrane. Eur. J. Biochem. 270:1875-1884. [DOI] [PubMed] [Google Scholar]
- 6.Arselin, G., J. Vaillier, B. Salin, J. Schaeffer, M. F. Giraud, A. Dautant, D. Brethes, and J. Velours. 2004. The modulation in subunits e and g amounts of yeast ATP synthase modifies mitochondrial cristae morphology. J. Biol. Chem. 279:40392-40399. [DOI] [PubMed] [Google Scholar]
- 7.Boyle, G. M., X. Roucou, P. Nagley, R. J. Devenish, and M. Prescott. 1999. Identification of subunit g of yeast mitochondrial F1Fo-ATP synthase, a protein required for maximal activity of cytochrome c oxidase. Eur. J. Biochem. 262:315-323. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Brunner, S., V. Everard-Gigot, and R. A. Stuart. 2002. Su e of the yeast F1Fo-ATP synthase forms homodimers. J. Biol. Chem. 277:48484-48489. [DOI] [PubMed] [Google Scholar]
- 10.Capaldi, R. A., and R. Aggeler. 2002. Mechanism of the F1Fo-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 27:154-160. [DOI] [PubMed] [Google Scholar]
- 11.Collinson, I. R., M. J. Runswick, S. K. Buchanan, I. M. Fearnley, J. M. Skehel, M. J. van Raaij, D. E. Griffiths, and J. E. Walker. 1994. Fo membrane domain of ATP synthase from bovine heart mitochondria: purification, subunit composition, and reconstitution with F1-ATPase. Biochemistry 33:7971-7978. [DOI] [PubMed] [Google Scholar]
- 12.Devenish, R. J., M. Prescott, X. Roucou, and P. Nagley. 2000. Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochim Biophys Acta 1458:428-442. [DOI] [PubMed] [Google Scholar]
- 13.Dienhart, M., K. Pfeiffer, H. Schägger, and R. A. Stuart. 2002. Formation of the yeast F1Fo-ATP synthase dimeric complex does not require the ATPase inhibitor protein, Inh1. J. Biol. Chem. 277:39289-39295. [DOI] [PubMed] [Google Scholar]
- 14.Gavin, P. D., R. J. Devenish, and M. Prescott. 2003. FRET reveals changes in the F1-stator stalk interaction during activity of F1Fo-ATP synthase. Biochim. Biophys. Acta 1607:167-179. [DOI] [PubMed] [Google Scholar]
- 15.Gavin, P. D., M. Prescott, S. E. Luff, and R. J. Devenish. 2004. Cross-linking ATP synthase complexes in vivo eliminates mitochondrial cristae. J. Cell Sci. 117:L2333-L2343. [DOI] [PubMed] [Google Scholar]
- 16.Giraud, M. F., P. Paumard, V. Soubannier, J. Vaillier, G. Arselin, B. Salin, J. Schaeffer, D. Brethes, J. P. di Rago, and J. Velours. 2002. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim. Biophys. Acta 11555:174-180. [DOI] [PubMed] [Google Scholar]
- 17.Herrmann, J. M., H. Fölsch, W. Neupert, and R. A. Stuart. 1994. Isolation of yeast mitochondria and study of mitochondrial translation, p. 538-544. In J. E. Celis (ed.), Cell biology: a laboratory handbook. Academic Press, San Diego, Calif.
- 18.Hong, S., and P. L. Pedersen. 2003. Subunit e of mitochondrial ATP synthase: a bioinformatic analysis reveals a phosphopeptide binding motif supporting a multifunctional regulatory role and identifies a related human brain protein with the same motif. Proteins 51:155-161. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, R. E., A. E. Hobbs, K. L. Cerveny, and H. Sesaki. 2000. Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsci. Res. Tech. 51:573-583. [DOI] [PubMed] [Google Scholar]
- 20.Jia, L., M. Dienhart, M. Schramp, M. McCauley, K. Hell, and R. A. Stuart. 2003. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22:6438-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaur, R., I. Castano, and B. P. Cormack. 2004. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 48:1600-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lai-Zhang, J., Y. Xiao, and D. M. Mueller. 1999. Epistatic interactions of deletion mutants in the genes encoding the F1-ATPase in yeast Saccharomyces cerevisiae. EMBO J. 18:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leuenberger, D., N. A. Bally, G. Schatz, and C. M. Koehler. 1999. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 18:4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleiger, G., R. Grothe, P. Mallick, and D. Eisenberg. 2002. GXXXG and AXXXA: common alpha-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry 41:5990-5997. [DOI] [PubMed] [Google Scholar]
- 26.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 27.Paul, M. F., J. Velours, G. Arselin de Chateaubodeau, M. Aigle, and B. Guerin. 1989. The role of subunit 4, a nuclear-encoded protein of the Fo sector of yeast mitochondrial ATP synthase, in the assembly of the whole complex. Eur. J. Biochem. 185:163-171. [DOI] [PubMed] [Google Scholar]
- 28.Paumard, P., J. Vaillier, B. Coulary, J. Schaeffer, V. Soubannier, D. M. Mueller, D. Brethes, J. P. di Rago, and J. Velours. 2002. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rapaport, D., M. Brunner, W. Neupert, and B. Westermann. 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273:20150-20155. [DOI] [PubMed] [Google Scholar]
- 30.Rothstein, R. J., and F. Sherman. 1980. Genes affecting the expression of cytochrome c in yeast: genetic mapping and genetic interactions. Genetics 94:871-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russ, W. P., and D. M. Engelman. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296:911-919. [DOI] [PubMed] [Google Scholar]
- 32.Schägger, H. 2001. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65:231-244. [DOI] [PubMed] [Google Scholar]
- 33.Senes, A., I. Ubarretxena-Belandia, and D. M. Engelman. 2001. The Cα-H.O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl. Acad. Sci. USA 98:9056-9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw, J. M., and J. Nunnari. 2002. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soubannier, V., F. Rusconi, J. Vaillier, G. Arselin, S. Chaignepain, P. V. Graves, J. M. Schmitter, J. L. Zhang, D. Mueller, and J. Velours. 1999. The second stalk of the yeast ATP synthase complex: identification of subunits showing cross-links with known positions of subunit 4 (subunit b). Biochemistry 38:15017-15024. [DOI] [PubMed] [Google Scholar]
- 36.Soubannier, V., J. Vaillier, P. Paumard, B. Coulary, J. Schaeffer, and J. Velours. 2002. In the absence of the first membrane-spanning segment of subunit 4(b), the yeast ATP synthase is functional but does not dimerize or oligomerize. J. Biol. Chem. 277:10739-10745. [DOI] [PubMed] [Google Scholar]
- 37.Spannagel, C., J. Vaillier, G. Arselin, P. V. Graves, X. Grandier-Vazeille, and J. Velours. 1998. Evidence of a subunit 4 (subunit b) dimer in favor of the proximity of ATP synthase complexes in yeast inner mitochondrial membrane. Biochim. Biophys. Acta 1414:260-264. [DOI] [PubMed] [Google Scholar]
- 38.Stock, D., A. G. W. Leslie, and J. E. Walker. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286:1700-1705. [DOI] [PubMed] [Google Scholar]
- 39.Tokatlidis, K., T. Junne, S. Moes, G. Schatz, B. S. Glick, and N. Kronidou. 1996. Translocation arrest of an intramitochondrial sorting signal next to Tim11 at the inner-membrane import site. Nature 384:585-588. [DOI] [PubMed] [Google Scholar]
- 40.Vaillier, J., G. Arselin, P. V. Graves, N. Camougrand, and J. Velours. 1999. Isolation of supernumerary yeast ATP synthase subunits e and i. Characterization of subunit i and disruption of its structural gene ATP18. J. Biol. Chem. 274:543-548. [DOI] [PubMed] [Google Scholar]
- 41.Velours, J., and Arselin, G. 2000. The Saccharomyces cerevisiae ATP synthase. J. Bioenerg. Biomembr. 32:383-390. [DOI] [PubMed] [Google Scholar]
- 42.Velours, J., P. Paumard, V. Soubannier, C. Spannagel, J. Vaillier, G. Arselin, and P. V. Graves. 2000. Organisation of the yeast ATP synthase F(0):a study based on cysteine mutants, thiol modification and cross-linking reagents. Biochim. Biophys. Acta 1458:443-456. [DOI] [PubMed] [Google Scholar]
- 43.von Heijne, G. 1986. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 5:1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]