Abstract
The presence of plastids in diverse eukaryotic lineages that have lost the capacity for photosynthesis is well documented. The metabolic functions of such organelles, however, are poorly understood except in the case of the apicoplast in the Apicomplexa, a group of intracellular parasites including Plasmodium falciparum, and the plastid of the green alga Helicosporidium sp., a parasite for which the only host-free stage identified in nature so far is represented by cysts. As a first step in the reconstruction of plastid functions in a nonphotosynthetic, predominantly free-living organism, we searched for expressed sequence tags (ESTs) that correspond to nucleus-encoded plastid-targeted polypeptides in the green alga Prototheca wickerhamii. From 3,856 ESTs, we found that 71 unique sequences (235 ESTs) correspond to different nucleus-encoded putatively plastid-targeted polypeptides. The identified proteins predict that carbohydrate, amino acid, lipid, tetrapyrrole, and isoprenoid metabolism as well as de novo purine biosynthesis and oxidoreductive processes take place in the plastid of P. wickerhamii. Mg-protoporphyrin accumulation and, therefore, plastid-to-nucleus signaling might also occur in this nonphotosynthetic organism, as we identified a transcript which encodes subunit I of Mg-chelatase, the enzyme which catalyzes the first committed step in chlorophyll synthesis. Our data indicate a far more complex metabolism in P. wickerhamii's plastid compared with the metabolic pathways predicted to be located in the apicoplast of P. falciparum and the plastid of Helicosporidium sp.
The presence of plastids in eukaryotic cells is generally associated with the ability to perform photosynthesis; these light-harvesting organelles are the product of primary endosymbiosis in the chlorophyte, streptophyte, rhodophyte, and glaucophyte lineages, while in other groups, such as the apicomplexan parasites, heterokonts, euglenoids, and chlorarachniophytes, the plastid was acquired through secondary endosymbiosis. However, colorless plastids with various degrees of functional and structural degeneration, relative to their photosynthetically competent homologues, have been identified in several of these evolutionarily diverse eukaryotic lineages (2, 45, 61). The loss of plastid-encoded photosynthesis-related genes has been documented in plastids from achlorophyllic lineages among green algae (29, 51), land plants (63, 64), dictyochophytes (45), and euglenoids (16), but the functional role of these plastids is largely unknown.
The discovery of a relict plastid (the apicoplast) in several apicomplexan parasites, i.e., Plasmodium falciparum (62), Toxoplasma gondii (28, 32), and Eimeria tenella (7), whose impaired functioning leads to the delayed-death of the parasite (15), has greatly enhanced studies aimed to understand the roles of such plastids in cellular metabolism. In the malarial parasite P. falciparum, metabolic pathways such as fatty acid synthesis (59), non-mevalonate isopentenyl diphosphate synthesis (25), and tetrapyrrole biosynthesis (42) have been shown to take place in the apicoplast.
A slightly more complex metabolism was recently predicted for the as yet not visualized plastid of the nonphotosynthetic pathogenic green alga Helicosporidium sp. (11); so far, it is not clear whether this taxon, a member of the Trebouxiophyceae (50), is an obligate parasite which can in nature survive outside its host only as a cyst or if it has also retained a free-living stage (5, 51). In addition to the metabolic pathways already described to be present in the P. falciparum apicoplast, the plastid of Helicosporidium sp. likely harbors pathways related to the synthesis of several amino acids and probably uses different precursors for tetrapyrrole synthesis. However, these functions account only for a fraction of the metabolic pathways known to be located in different plastid types of photosynthetically competent land plants (reviewed in references 37 and 60). We therefore chose to investigate plastid functions in a nonphotosynthetic taxon that clearly has a free-living life style rather than an obligate parasitic one, as in apicomplexans, holoparasitic angiosperms, and possibly Helicosporidium sp.; obligate parasitism can obscure some of the important roles plastids assume in cellular metabolism as essential plastid-located metabolic pathways can be lost if the final product is imported from the parasitized host.
Prototheca wickerhamii (Trebouxiophyceae, Chlorophyta) is a nonphotosynthetic, predominantly free-living unicellular alga which is ubiquitous in soil and aqueous habitats. The green algal versus fungal nature of P. wickerhamii was the subject of debate until ultrastructural studies revealed the presence of a plastid with starch granules (30, 35, 36). Several Prototheca species, including P. wickerhamii, can act as opportunistic pathogens; P. wickerhamii is associated mainly with cutaneous and systemic infections in immunocompromised humans (26, 30, 53), while other Prototheca species have been shown to infect a wide range of animals (20, 23, 48).
Phylogenetic studies have confirmed that the closest relatives of P. wickerhamii are among the chlorococcales (21) and include Helicosporidium sp. (50) and that P. wickerhamii can be considered the achlorophyllic equivalent of the photosynthetic alga Auxenochlorella protothecoides (55). More than half of the plastid genome of P. wickerhamii was recently sequenced; this genome is very reduced relative to its counterpart in photosynthetic lineages, has no photosynthesis-related genes, and most of the coding functions identified are involved in gene expression (rRNA, tRNA, and ribosomal protein genes) (29).
As a first step in the reconstruction of the metabolic pathways that are associated with a nonphotosynthetic plastid in a free-living taxon, we searched for expressed sequence tags (ESTs) which correspond to nucleus-encoded plastid-targeted polypeptides in P. wickerhamii. The functions of the ESTs identified lead us to suggest that several metabolic pathways are located in this nonphotosynthetic plastid, and we discuss our findings in relation to the available information for other nonphotosynthetic plastids.
MATERIALS AND METHODS
Strain and growth conditions.
Prototheca wickerhamii strain SAG 263-11 was obtained from Sammlung von Algenkulturen at the University of Göttingen, Göttingen, Germany. Cells were grown in M16 medium (1% malt extract, 0.25% Bacto-tryptone, pH 7.5) with rotary shaking, without illumination, at 24°C. In the late exponential phase of growth (optical density at 750 nm = 0.8) cells were harvested by centrifugation at 3,000 × g at 4°C and resuspended in Trizol (Invitrogen, Carlsbad, Calif.).
Library construction and EST sequencing.
P. wickerhamii library construction and EST sequencing were part of the Protist EST Program (http://megasun.bch.umontreal.ca/pepdb/pep.html). Normalized and nonnormalized cDNA libraries were made by DNA Technologies, Inc., Gaithersburg, Md. Inserts were unidirectionally cloned between the EcoRV and NotI sites of the pcDNA3.1 vector (Invitrogen, Carlsbad, Calif.). ESTs were sequenced from the 5′ end at the National Research Council Institute for Marine Bioscience Joint Laboratory, Halifax, Canada. A total of 3,856 vector and quality trimmed ESTs were grouped into 1,401 unique sequences (i.e., clusters and singletons). The number of ESTs sequenced from the nonnormalized library accounts for 76% of the total number of sequenced ESTs.
The full P. wickerhamii EST data set (4,808 entries) is available in PEPdbPubat http://amoebidia.bcm.umontreal.ca/public/pepdb/agrm.php; the interactivemap of the P. wickerhamii metabolic pathways is available in PEPdb PGDBs (Pathway Genome DataBases) at: http://amoebidia.bcm.umontreal.ca:1555.
Identification and analysis of transcripts encoding plastid-targeted polypeptides.
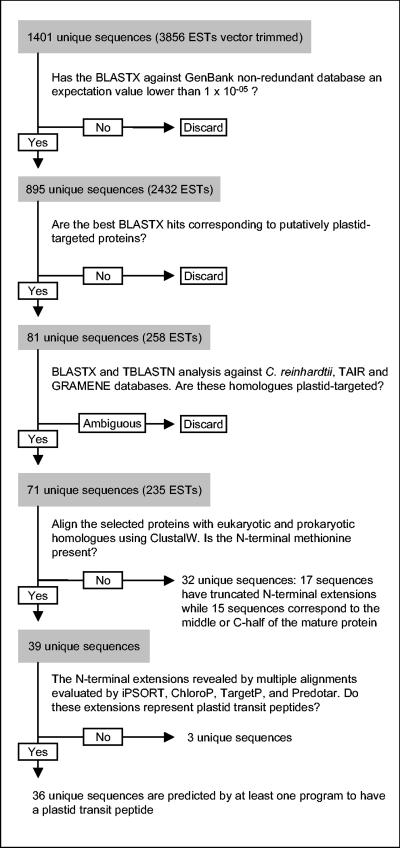
The identification of unique sequences that correspond to nucleus-encoded plastid-targeted proteins was performed in several steps (Fig. 1). All the unique sequences from the P. wickerhamii database were analyzed by BLASTX (1) against the nonredundant GenBank database, release 143.0. P. wickerhamii sequences having a BLASTX hit with an expectation value lower than 10−05 were selected as putative plastid-targeted polypeptides if the best BLASTX hit was for a protein that is known to be plastid targeted and annotated as such (chloroplast precursor) or the hit was for a protein involved in a process known to be localized in the plastid but not currently annotated as such. In the latter case, the presence of a transit peptide in the best-BLAST-score proteins was verified with TargetP (14) and iPSORT (4).
FIG. 1.
Flow diagram of the procedures used to identify and analyze putatively plastid-targeted polypeptides in P. wickerhamii.
The P. wickerhamii sequences identified as putatively plastid-targeted polypeptides were further analyzed for homologues by running BLASTX and TBLASTN analyses in the nonredundant and redundant Chlamydomonas reinhardtii (http://www.biology.duke.edu/chlamy), Arabidopsis thaliana (TAIR database, http://www.arabidopsis.org/index.jsp) and Oryza sativa (Gramene database, http://www.gramene.org) databases. The targeting peptide sequence was absent from all BLASTX and TBLASTN alignments, as expected, considering the low conservation of sequence among plastid-targeting peptides in plants (6). We also compared our data with putatively plastid-targeted proteins listed under Gene Ontology (GO) numbers 0009536 and 0009507 (proteins located in the plastid and chloroplast, respectively, see http://www.geneontology.org) in the TAIR and Gramene databases and with putatively apicoplast-targeted proteins from PlasmoDB (http://plasmodb.org).
The conceptual amino acid sequence of each cluster or singleton was aligned with several best-BLAST-hit proteins with ClustalX (52) or Multalin (10). For the identification of N-terminal extensions in P. wickerhamii proteins, eukaryotic cytosol-localized and/or prokaryotic/cyanobacterial homologues were also included in these alignments. The presence of a targeting peptide was then determined with iPSORT (4), ChloroP (13), TargetP (14), and Predotar (http://www.inra.fr/predotar). Additionally, we checked several databases (Brenda, http://www.brenda.uni-koeln.de, TAIR, and Gramene) to see whether multiple forms (isozymes) with potentially different cellular localizations (i.e., plastid and other cellular compartments) have been described for the proteins that we propose to be plastid targeted in P. wickerhamii.
Nucleotide sequence accession numbers.
A total of 258 ESTs and 81 unique sequences resulting from the first step of screening for putatively plastid-targeted proteins in P. wickerhamii (Fig. 1) have been deposited in the GenBank database (accession numbers CN587685 to CN587912, CO727030 to CO727059, CO729260, AY616038 to AY616113, and AY700206 to AY700210).
RESULTS
Nucleus-encoded plastid-targeted polypeptides in P. wickerhamii.
A total of 3,856 ESTs, with an average vector-trimmed length of 483 nucleotides, were grouped into 1,401 unique sequences (i.e., clusters or singletons); 895 unique sequences (2,432 ESTs) gave BLASTX (1) hits (expectation value cutoff of 10−05) against the GenBank nonredundant database (Fig. 1); 71 identifiable genes, or 8% of the unique sequences with a BLASTX hit, are predicted to encode different plastid-targeted polypeptides (Table 1). The average length of these unique sequences is 635 nucleotides; they resulted from clustering 235 ESTs with a mean length of 509 nucleotides. The polypeptides predicted in this study to be plastid targeted in P. wickerhamii represent a subset of the true number of plastid-targeted proteins for this taxon, as we sequenced a rather limited number of ESTs and we selected only EST clusters that have significant similarity to polypeptides known to be plastid located.
TABLE 1.
Nucleus-encoded proteins predicted to be plastid-targeted in P. wickerhamii
| Protein name (gene name) | Transit peptidea | Length (nt)b | E/Cc | Bit scored | e valuee | Accession no. (best hit)f | EC no. | Intracellular locationg |
|---|---|---|---|---|---|---|---|---|
| Chaperonins/heat shock proteins | ||||||||
| 60-kDa chaperonin alpha subunit (groEL) | A | 575 | 4 | 208 | 7e−53 | Q42694 | P# | |
| 60-kDa chaperonin beta subunit (groEL) | A | 711 | 14 | 207 | 1e−52 | NP_200461 | P# | |
| Heat shock 70 protein (chsp70) | C | 528 | 5 | 265 | 4e−70 | Q08080 | P# | |
| Transcription and translation | ||||||||
| Protein disulfide isomerase (pdi) | C | 526 | 3 | 158 | 7e−38 | AAD02069 | P#, ER# | |
| Poly(A) binding protein, RB47 homologue (pabp) | C | 345 | 1 | 137 | 6e−32 | AAC39368 | P# | |
| Catalytic subunit of ClpP5 protease (clpP) | A | 754 | 2 | 157 | 3e−37 | AAN18141 | P# | |
| Membrane protein/transport proteins | ||||||||
| Similar to Toc34, plastid protein import (toc34) | B | 720 | 1 | 105 | 1e−21 | CAB77551 | P# | |
| Similar to Tic22, plastid protein import apparatus (tic22) | B | 697 | 4 | 93 | 1e−18 | BAD35192 | P# | |
| SecA-type chloroplast protein transport factor (secA) | C | 719 | 1 | 274 | 2e−72 | NP_192089 | P# | |
| Similar to thylakoid lumenal 29.8-kDa protein | A | 739 | 2 | 147 | 3e−34 | NP_565149 | P# | |
| Putative plastidic ATP/ADP transporter (aatp1) | A | 643 | 1 | 149 | 6e−36 | XP_464574 | P# | |
| Hexose transporter (pGlcT) | B | 499 | 1 | 56 | 4e−07 | NP_909150 | P# | |
| Carbohydrate metabolism | ||||||||
| Phosphoglucomutase (pgm) | C | 562 | 1 | 263 | 2e−69 | AAN04961 | EC 5.4.2.2 | P, C |
| Phosphoglycerate dehydrogenase (serA) | B | 694 | 4 | 224 | 2e−57 | AAN12903 | EC 1.1.1.95 | P |
| Pyruvate kinase, similar to isozyme G (pyk) | A | 780 | 7 | 188 | 1e−46 | NP_917361 | EC 2.7.1.40 | P, C |
| Pyruvate dehydrogenase complex | ||||||||
| Dihydrolipoamide S-acetyltransferase (aceF) | A | 455 | 1 | 132 | 2e−30 | NP_487646 | EC 2.3.1.12 | P, M |
| Pyruvate dehydrogenase E1 beta subunit (aceA) | A | 681 | 9 | 276 | 3e−73 | AAB86804 | EC 1.2.4.1 | P, M |
| Starch synthesis | ||||||||
| Glucose-1-phosphate adenylyltransferase (glgC) | A | 1015 | 3 | 366 | e−100 | AAF75832 | EC 2.7.7.27 | P, C |
| Starch synthase isoform SS III (glgA) | C | 670 | 2 | 121 | 1e−26 | CAB40374 | EC 2.4.1.21 | P |
| 1,4-Alpha-glucan branching enzyme (glgB) | A | 639 | 2 | 57 | 3e−07 | AAP72266 | EC 2.4.1.18 | P |
| Fatty acid biosynthesis | ||||||||
| Acetyl-CoA carboxylase | EC 6.4.1.2 | P, C, ER | ||||||
| Biotin carboxylase subunit (accC) | A | 606 | 4 | 157 | 2e−37 | AAP99120 | ||
| Acetyl-CoA carboxylase, alpha subunit (accA) | B | 729 | 3 | 192 | 2e−47 | T06765 | ||
| Fatty acid synthase multienzyme complex | ||||||||
| Enoyl-[acyl-carrier protein] reductase (fabI) | B | 468 | 2 | 143 | 2e−33 | CAC41368 | EC 1.3.1.9 | P |
| 3-Oxoacyl-[acyl-carrier protein] synthase (fabB) | A | 654 | 4 | 79 | 1e−13 | CAA84023 | EC 2.3.1.41 | P |
| 3-Oxoacyl-[acyl-carrier protein] reductase (fabG) | B | 680 | 2 | 186 | 3e−46 | CAA45866 | EC 1.1.1.100 | P |
| Acyl-[acyl-carrier protein] desaturase (acpd) | A | 740 | 3 | 208 | 1e−52 | AAL26877 | EC 1.14.19.2 | P |
| Beta-hydroxyacyl-[acyl-carrier protein] dehydratase (fabZ) | A | 660 | 1 | 191 | 1e−47 | AAM78110 | EC 4.2.1- | P |
| [Acyl-carrier protein] S-malonyltransferase (fabD) | B | 687 | 2 | 226 | 3e−58 | AAM64515 | EC 2.3.1.39 | P |
| Galactolipid synthesis | ||||||||
| 1,2-Diacylglycerol 3-beta-galactosyltransferase (mgdA) | A | 637 | 3 | 81 | 2e−14 | BAB11980 | EC 2.4.1.46 | P |
| Carbohydrate-to-lipid interconversion | ||||||||
| Glycerol-3-phosphate O-acyltransferase (pslB) | A | 617 | 2 | 135 | 6e−31 | BAB39689 | EC 2.3.1.15 | P, M, C, ER |
| Aromatic amino acid metabolism | ||||||||
| 3-Deoxy-7-phosphoheptulonate synthase (aroF) | B | 762 | 6 | 302 | 5e−81 | AAF18536 | EC 2.5.1.54 | P, C |
| 3-Dehydroquinate synthase (aroB) | A | 696 | 2 | 203 | 3e−51 | AAL77575 | EC 4.2.3.4 | P |
| Prephenate dehydratase (pheA) | B | 447 | 1 | 142 | 3e−33 | AAS79603 | EC 4.2.1.51 | P# |
| Tryptophan synthase, beta chain (trpB) | B | 733 | 5 | 328 | 7e−89 | AAC25986 | EC 4.2.1.20 | P |
| Branched amino acid synthesis | ||||||||
| Acetolactate synthase, small subunit (ilvH) | A | 580 | 4 | 122 | 6e−27 | BAB09596 | EC 2.2.1.6 | P |
| Acetolactate synthase, large subunit (ilvI) | A | 658 | 1 | 227 | 2e−58 | AAC03784 | EC 2.2.1.6 | P |
| Ketol-acid reductoisomerase (ilvC) | B | 649 | 1 | 179 | 3e−44 | S30145 | EC 1.1.1.86 | P |
| 3-Isopropylmalate dehydrogenase (leuB) | A | 635 | 4 | 221 | 7e−57 | ZP_00110976 | EC 1.1.1.85 | P# |
| Serine metabolism and fixation of sulfur | ||||||||
| Glycine hydroxymethyltransferase (glyA) | A | 657 | 4 | 249 | 3e−65 | CAB79969 | EC 2.1.2.1 | P, M, C |
| Methylenetetrahydrofolate dehydrogenase and methenyl- tetrahydrofolate cyclohydrolase, bifunctional protein (folD) | A | 745 | 12 | 239 | 9e−61 | AAM62762 | EC 1.5.1.5 EC 3.5.4.9 | P, C |
| Cysteine synthase (cysM) | A | 727 | 3 | 253 | 2e−66 | BAA03542 | EC 2.5.1.47 | P, M, C |
| Cystathionine gamma-synthase (metB) | C | 594 | 2 | 256 | 3e−67 | AAF74982 | EC 2.5.1.48 | P |
| Aspartate group of amino acids | ||||||||
| Aspartate-semialdehyde dehydrogenase (asd) | A | 672 | 3 | 209 | 3e−53 | AAG33078 | EC 1.2.1.11 | P |
| Threonine synthase (thrC) | A | 590 | 1 | 198 | 2e−50 | NP_974637 | EC 4.2.3.1 | P |
| l-Aspartate oxidase (nadB) | C | 604 | 1 | 179 | 4e−44 | XP_464033 | EC 1.4.3.- | P# |
| Glutamate/glutathione metabolism | ||||||||
| Glutamate dehydrogenase (NADP dependant) (gdhA) | B | 643 | 9 | 307 | 1e−82 | CAA41636 | EC 1.4.1.4 | P |
| Glutamate-cysteine ligase (gshA) | A | 1313 | 9 | 426 | e−118 | O22493 | EC 6.3.2.2 | P, C |
| Ferredoxin-dependent glutamate synthase (gltB) | C | 639 | 2 | 206 | 3e−52 | AAF64387 | EC 1.4.7.1 | P |
| Acetylglutamate kinase (argB) | A | 688 | 8 | 213 | 4e−54 | NP_440676 | EC 2.7.2.8 | P# |
| Argininosuccinate lyase (argH) | A | 653 | 3 | 214 | 2e−54 | AAF43427 | EC 4.3.2.1 | P# |
| Histidine metabolism | ||||||||
| ATP phosphoribosyltransferase (hisG) | C | 259 | 3 | 107 | 1e−22 | AAB88880 | EC 2.4.2.17 | P# |
| Phosphoribosyl-AMP cyclohydrolase (hisI) | A | 807 | 3 | 124 | 3e−27 | AAM63514 | EC 3.5.4.19 | P# |
| Histidinol-phosphate transaminase (hisC) | B | 674 | 2 | 222 | 6e−57 | XP_467409 | EC 2.6.1.9 | P |
| Histidinol dehydrogenase (hisD) | C | 649 | 9 | 222 | 4e−57 | AAN28839 | EC 1.1.1.23 | P |
| Purine de novo synthesis | ||||||||
| Phosphoribosylformylglycinamidine cycloligase (purM) | A | 660 | 1 | 229 | 4e−59 | P52424 | EC 6.3.3.1 | P#, M# |
| Phosphoribosylaminoimidazole-succinocarboxamide synthase (purC) | A | 735 | 6 | 216 | 4e−55 | AAL48317 | EC 6.3.2.6 | P# |
| Adenylosuccinate lyase (purB) | B | 462 | 4 | 161 | 5e−39 | ZP_00263521 | EC 4.3.2.2 | P# |
| Phosphoribosylaminoimidazolecarboxamide formyl- transferase (purH) | A | 1181 | 5 | 423 | e−117 | AAM91661 | EC 2.1.2.3 | P# |
| Adenylosuccinate synthetase (purA) | A | 628 | 4 | 183 | 3e−45 | XP_469397 | EC 6.3.4.4 | P# |
| Sulfur metabolism and oxidoreductive processes | ||||||||
| Ferredoxin (fdx) | A | 450 | 2 | 109 | 2e−23 | P00252 | P# | |
| Similar to thioredoxin y (trxY) | A | 680 | 3 | 105 | 1e−21 | AAF63825 | EC 1.8.1.9 | P, C |
| Ferredoxin-NADP reductase (petH) | C | 411 | 3 | 214 | 5e−55 | AAB40978 | EC 1.18.1.2 | P |
| Thioredoxin (trxM) | B | 593 | 1 | 160 | 2e−38 | CAA56851 | EC 1.8.1.9 | P, C |
| Adenylyl-sulfate reductase (glutathione) (apr) | A | 666 | 1 | 254 | 1e−66 | AAC26855 | EC 1.8.4.9 | P |
| Isoprenoid metabolism | ||||||||
| Farnesyltranstransferase (crtE) | A | 587 | 6 | 159 | 3e−38 | AAS49033 | EC 2.5.1.29 | P |
| Hydroxymethylbutenyl 4-diphosphate synthase (gcpE) | C | 598 | 1 | 102 | 7e−21 | AAT70081 | P# | |
| Porphyrin pathway | ||||||||
| Glutamate-1-semialdehyde 2,1-aminomutase (hemL) | C | 249 | 1 | 108 | 3e−23 | Q55665 | EC 5.4.3.8 | P |
| Porphobilinogen synthase (hemB) | B | 410 | 2 | 103 | 9e−22 | CAA43833 | EC 4.2.1.24 | P# |
| Uroporphyrinogen-III synthase (hemD) | C | 694 | 1 | 149 | 5e−35 | CAC85287 | EC 4.2.1.75 | P# |
| Uroporphyrinogen decarboxylase (hemE) | B | 580 | 1 | 234 | 9e−61 | NP_850587 | EC 4.1.1.37 | P# |
| Mg-chelatase subunit I (chlI) | A | 600 | 1 | 234 | 1e−60 | Q94FT3 | EC 6.6.1.1 | P |
A, full-length transit-peptide identified; B, 5 start codon is missing (truncated transit peptide) or the N-terminal methionine could not be assigned unambiguously; C, the N-half of the protein was not identified (N-half-truncated clones); the amino acid sequence covers the middle or the C-half of the mature protein.
Length in nucleotides of the EST cluster or singleton.
Number of ESTs/ cluster (one for singletons).
Best BLASTX bit score.
Best BLASTX e value.
Accession number of the best BLASTX hit in the GenBank database.
Intracellular localization in land plants and algae of proteins that are homologues of the P. wickerhamii proteins. These data were compiled from the Brenda (http://www.brenda.uni-koeln.de/), TAIR (http://www.arabidopsis.org/index.jsp), and Gramene (http://www.gramene.org) databases and from journal articles about the best-BLAST proteins. When such data were not available in the Brenda database, only the remaining three sources were used (marked with #). P, plastid targeted; M, mitochondrion targeted; C, cytosolic; ER, endoplasmic reticulum (microsomal fraction).
Plastid transit peptide in P. wickerhamii.
For 39 unique sequences we identified the N-terminal methionine and the presence of a transit peptide was evaluated by iPSORT (4), ChloroP (13), TargetP (14), and Predotar (http://www.inra.fr/predotar). ChloroP predicted the presence of a plastid transit peptide in 85% of these polypeptides followed by TargetP with 77%, iPSORT with 62%, and Predotar with 44%. In more than 64% of these unique sequences, the presence of the plastid transit peptide was confirmed by at least three of the four programs (for more details of the prediction of the plastid transit peptide, see the supplemental material). The cleavage site of the plastid transit peptide predicted by TargetP and/or ChloroP was in all situations located upstream of the amino acid that corresponds, in multiple alignments, to the N-terminal methionine from prokaryotic/cyanobacterial or eukaryotic cytosol-localized homologues; therefore, all predicted transit peptides correspond to N-terminal extensions.
For three of the enzymes that have full-length N-terminal extensions, i.e., the 1,4-alpha-glucan branching enzyme, 1,2-diacylglycerol 3-beta-galactosyltransferase, and phosphoribosyl formylglycinamidine cycloligase, all four programs failed to identify a plastid-targeting peptide, although some of the programs indicated the presence of a mitochondrion-targeting peptide. However, in green algae and land plants, the only known localization of the 1,4-alpha-glucan branching enzyme (the starch-branching enzyme) is in the plastid (24). The enzyme 1,2-diacylglycerol 3-beta-galactosyltransferase, type A, catalyzes the synthesis of the major plastid membrane lipid monogalactosyldiacylglycerol and is known to be embedded in the plastid inner membrane of algae and land plants (3, 34). For phosphoribosylformylglycinamidine cycloligase, which catalyzes the third step in the de novo purine pathway, it has been demonstrated that the same transit peptide can direct this enzyme to both the plastid and the mitochondrial compartments (17). As we identified three other enzymes involved in the de novo purine pathway with an intact N terminus (Table 1 and supplemental data) and with a plastid-targeting peptide predicted by at least three programs, we propose that this enzyme is also plastid targeted in P. wickerhamii.
For 17 of the remaining 32 unique sequences identified as plastid targeted based on high similarity to homologues from photosynthetic algae and land plants, the N-terminal methionine could not be assigned unambiguously or the 5′ start codon is missing (truncated transit peptide) (Table 1 and supplemental material). Thirteen of the proteins that belong to this category are known to be only plastid located; the four proteins that are reported to have counterparts with alternative intracellular localizations were identified as putatively plastid targeted based on (i) BLAST analysis, which gave significantly better scores for plastid-targeted isozymes (the e value was at least 20 orders of magnitude smaller for the plastid-targeted isozymes) or failed to reveal plant proteins other than plastid targeted, and (ii) structural features, i.e., dissimilar oligomeric status of the isozymes. Moreover, all four polypeptides have N-terminal extensions revealed by alignments with eukaryotic cytosol-localized or prokaryotic/cyanobacterial homologues. The protein that has similarity to Toc33 represents the only exception. Toc33/Toc34 proteins (translocon at the outer chloroplastic envelope membrane) from land plants have their hydrophilic N-terminally located GTPase domain exposed to the cytosol. Similar to most proteins of the plastid outer membrane, they do not contain an N-terminal transit sequence and do not use the general import pathway (38). Most likely the Toc33 homologue that we identified in P. wickerhamii has the same structural characteristics.
The remaining 15 unique sequences identified as encoding putatively plastid-targeted proteins in P. wickerhamii are made up of sequences that correspond to the middle or C-half of the mature protein (Table 1 and supplemental material). Apart from phosphoglucomutase, for which two forms (plastid targeted and cytosolic) are described, and protein disulfide isomerase, which might have dual localization, plastid and endoplasmic reticulum, all the other proteins in this category are known to be only plastid located. The phosphoglucomutase form we identified in the P. wickerhamii library is likely to be plastid located as the best BLASTX hits in GenBank are for the plastid isozyme of land plants and the alignment of 187 amino acids from P. wickerhamii with plastid and cytosolic land plant isozymes, and green algal (C. reinhardtii) and cyanobacterial enzymes revealed that P. wickerhamii lacks the three conserved insertions that are specific to the cytosolic isozyme from land plants. The protein disulfide isomerase that we propose to be plastid targeted in P. wickerhamii has the highest similarity with sequences from the algal species C. reinhardtii, Volvox carteri, and Helicosporidium sp. The N-terminal extension of protein disulfide isomerase from these taxa is predicted by TargetP (14) and iPSORT (4) to represent a signal peptide, indicating endoplasmic reticulum localization. Moreover, the C. reinhardtii and V. carteri sequences also possess the C-terminal endoplasmic reticulum retention signal KDEL (for Helicosporidium sp. the C-half of the protein is not available in the databases). However, as several studies of the C. reinhardtii protein disulfide isomerase clearly indicated this enzyme as a regulator of plastid translational activation (27, 54), it is likely that this protein has a dual localization, i.e., it is plastid and endoplasmic reticulum targeted (54).
DISCUSSION
Nonphotosynthetic metabolic pathways located in the plastid.
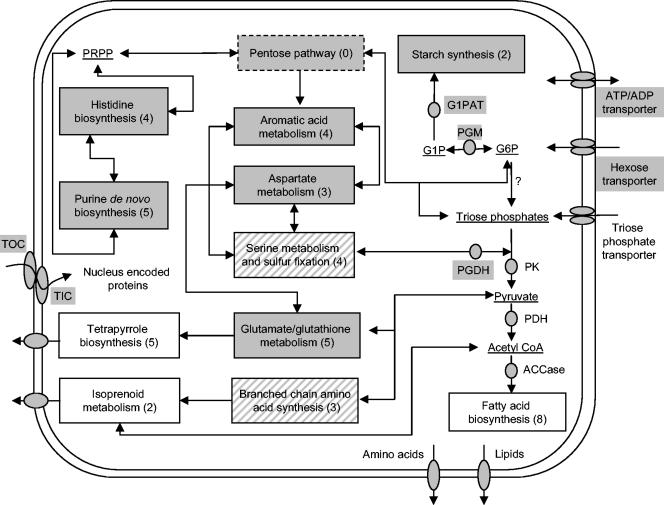
Based on the putative plastid-targeted enzymes identified in P. wickerhamii, carbohydrate, lipid, amino acid metabolism, de novo purine biosynthesis, oxidoreductive processes, isoprenoid metabolism, and porphyrin (tetrapyrrole) synthesis (Fig. 2, Table 1) seem to take place in the plastid of this alga.
FIG. 2.
Overview of the metabolism and pathways predicted to be located in the plastids of P. wickerhamii, Helicosporidium sp., and P. falciparum based on available data. The conversion of imported trioses to acetyl-coenzyme A seems to be present in all three organisms, while that of glucose-6-phosphate to starch is present only in P. wickerhamii. Enzymes are represented by solid circles, and substrates are underlined. Other pathways (indicated in boxes) predicted to be present in the plastids of P. wickerhamii, Helicosporidium sp., and P. falciparum are shown on a white background, in P. wickerhamii and Helicosporidium sp. but not in P. falciparum are on a striped background, and only in P. wickerhamii are on a grey background. ACCase, acetyl-coenzyme A carboxylase; G1P, glucose-1-phosphate; G1PAT, glucose-1-phosphate adenylyltransferase; G6P, glucose-6-phosphate; PDH, pyruvate dehydrogenase complex; PGDH, phosphoglycerate dehydrogenase; PK, pyruvate kinase; PRPP, phosphoribosyl pyrophosphate; TIC, translocon at the inner envelope membrane of chloroplasts; TOC, translocon at the outer envelope membrane of chloroplasts. The number of identified unique sequences that are predicted to encode plastid-targeted enzymes in P. wickerhamii and are associated with the various metabolic processes are indicated in parentheses; the pentose phosphate pathway and triose phosphate transporters are probably present in P. wickerhamii, although no transcripts associated with these functions were detected.
Carbohydrate metabolism.
The identification of several clusters encoding homologues of enzymes related to the glycolytic pathway (phosphoglucomutase, phosphoglycerate dehydrogenase, and pyruvate kinase) including two members of the pyruvate dehydrogenase complex, suggests that the imported carbohydrates can be converted to acetyl-coenzyme A in the plastid of P. wickerhamii. A similar process is known to take place in the leucoplasts of photosynthetically competent land plants (37, 40). The pyruvate dehydrogenase complex is also present in Helicosporidium sp. (11). and P. falciparum (41), implying that these parasites can use pyruvate to generate acetyl-coenzyme A, which is used for fatty acid synthesis (Fig. 2).
The presence of a plastid transit peptide on the small subunit of glucose-1-phosphate adenylyltransferase (ADP-glucose pyrophosphorylase) indicates that in P. wickerhamii, as in dicotyledenous plants, the synthesis of ADP-glucose from glucose-1-phosphate takes place in the plastid (37). The identification of P. wickerhamii's homologues of starch synthase isoform SS III and of the 1,4-alpha-glucan branching enzyme provides evidence for additional steps in the conversion of ADP-glucose to starch.
Lipid metabolism.
In land plants, type II fatty acid biosynthesis is strictly limited to the plastid compartment (37, 39); this pathway is also apicoplast located in P. falciparum (41, 59) and predicted to be plastid located in Helicosporidium sp. (11). In P. wickerhamii, we identified several plastid-targeted components of the heterotetrameric acetyl-coenzyme A carboxylase and polypeptides of the fatty acid synthase multienzyme complex. Several pathways related to lipid and galactolipid metabolism might also be functional, as we found homologues of the plastid-located glycerol-3-phosphate O-acyltransferase and 1,2-diacylglycerol 3-beta-galactosyltransferase (MGDG synthase type A) in our cDNA libraries.
Amino acid metabolism.
Leucine, serine, and lysine biosynthesis was proposed to be plastid located in Helicosporidium sp. (11). while amino acid biosynthesis is not present in the P. falciparum apicoplast (41). In the P. wickerhamii libraries we identified a wide array of transcripts encoding putative plastid-located enzymes that are involved in the biosynthesis of several amino acids (Fig. 2, Table 1). However, due to the limited data available, it is difficult to asses how many steps of these biosynthetic pathways are actually located in the plastid of P. wickerhamii. The presence of aromatic amino acid metabolism (including the shikimate pathway) is supported by several enzymes involved in chorismate (3-deoxy-7-phosphoheptulonate synthase and 3-dehydroquinate synthase), phenylalanine (prephenate dehydratase), and tryptophan (the beta chain of the tryptophan synthase) synthesis. Several other unique sequences encode enzymes that catalyze the synthesis of branched amino acids from pyruvate, histidine from phosphoribosyl-pyrophosphate, and serine from the glycolytic intermediate phosphoglycerate. Aspartate-4-semialdehyde, the product of the reaction catalyzed by aspartate-semialdehyde dehydrogenase, is a common intermediate for threonine, lysine, and methionine biosynthesis, while cysteine synthase and cystathionine gamma-synthase can use intermediates of serine and aspartate/threonine biosynthesis and represent a link to sulfur metabolism. Also related to sulfur metabolism is glutamate-cysteine ligase, which catalyzes the first step of glutathione biosynthesis. Finally, we identified a ferredoxin-dependent glutamate synthase, which represents a key enzyme of nitrogen assimilation and amino acid biosynthesis.
Purine biosynthesis.
We identified five out of the 11 enzymes involved in the synthesis of AMP from phosphoribosyl-pyrophosphate, all clusters being represented by a rather high number of ESTs. Purine biosynthesis is not present in P. falciparum (see below) and transcripts encoding enzymes involved in this pathway were not identified yet in Helicosporidium sp. Interestingly, the tendency to terminate this energy-consuming pathway can be encountered in phylogenetically unrelated organisms. For example, most of the prokaryotic intracellular parasites (i.e., Chlamydia, Treponema, Rickettsia, and Mycoplasma) and several protozoan parasites (i.e., Apicomplexa [Plasmodium and Toxoplasma]; Parabasala [Trichomonas]; Diplomonada [Giardia]; Euglenozoa [Leishmania]) studied so far are lacking the de novo purine biosynthetic pathway.
In P. falciparum the primary flux of purine nucleotide synthesis is via hypoxanthine. Hypoxanthine derived from the host is converted to IMP by the enzyme hypoxanthine guanine phosphoribosyltransferase. IMP serves as the precursor for both AMP and GMP, which will be further converted to triphosphates (12). In contrast, P. wickerhamii, which is usually a free-living organism, has to synthesize its own pool of purine nucleotides; the de novo purine pathway is present and, as in land plants, at least some enzymes are plastid localized.
Oxidoreductive processes.
A few of the proteins involved in a variety of redox reactions are predicted to be plastid targeted in Helicosporidium sp. (11), while in P. falciparum, a plant-type ferredoxin-NADP reductase and a ferredoxin were identified as apicoplast targeted (56); therefore, some processes related to the generation of reducing power are expected to be present in the degenerate organelle of both parasites. The enzymatic activity of several proteins identified in the present study (e.g., ADP-glucose pyrophosphorylase, acetyl-coenzyme A carboxylase, and ferredoxin-dependent glutamate synthase) has been shown to be regulated by thioredoxins (22); it is therefore not unexpected that we found homologues of thioredoxin m and the recently discovered thioredoxin y (31). In the absence of light-driven electron transport, the power required for thioredoxin reduction (i.e., reduced ferredoxin) is probably provided by NADPH through the ferredoxin-NADP reductase/ferredoxin-thioredoxin reductase system. The electron flow in the reverse direction to that which occurs in the chloroplast is thought to supply the reductant for other plastid-localized enzymes such as nitrite reductase and lipid desaturases (37, 46) and for biosynthetic processes localized in the apicoplast (41, 56).
Isoprenoid metabolism and porphyrin (tetrapyrrole) synthesis.
Several steps of the isoprenoid metabolism, including the nonmevalonate pathway, and probably all steps leading to the formation of Mg-protoporhyrin IX from glutamate appear to take place in the plastid of P. wickerhamii. Several steps of tetrapyrrole synthesis are also apicoplast located in P. falciparum (41-43) and plastid located in Helicosporidium sp. (11). However, it seems that the precursor of this pathway, δ-aminolevulinic acid, has a distinct origin in the different species, i.e., plastid in the two algal taxa versus mitochondrial in the apicomplexan parasite. In P. falciparum, δ-aminolevulinic acid, the first intermediate of tetrapyrrole biosynthesis, is synthesized in the mitochondrion from succinyl-coenzyme A and glycine (the Shemin pathway) and then transported into the apicoplast (41, 43). In cyanobacteria and photosynthetic plants (9), tetrapyrrole biosynthesis occurs in the plastid from glutamate (the C5 pathway); the enzyme which catalyzes the last step in the formation of δ-aminolevulinic acid from glutamate is the plastid-targeted glutamate-1-semialdehyde 2,1-aminomutase. This enzyme was identified in Helicosporidium sp. (11); in P. wickerhamii we also found a glutamate-1-semialdehyde 2,1-aminomutase homologue along with three other putative plastid-targeted enzymes which are involved in tetrapyrrole biosynthesis. Most likely, therefore, in P. wickerhamii and in Helicosporidium sp., tetrapyrrole biosynthesis is similar to that of cyanobacteria and photosynthetic plants (9).
Retrograde (plastid-to-nucleus) signaling pathway mediated by tetrapyrroles might be still functional.
At first glance, Mg-protoporphyrin IX synthesis in a colorless alga, suggested by the presence of a transcript of the chlI gene, which encodes subunit I of Mg-chelatase (Table 1), is surprising. Recent studies, however, have indicated that the accumulation of Mg-protoporphyrin IX, the first committed precursor of chlorophyll, is both necessary and sufficient for regulation by retrograde signaling of a large number of nuclear genes encoding plastid products (18, 49). Whether Mg-protoporphyrin IX indeed accumulates in the P. wickerhamii plastid remains to be experimentally demonstrated. Nonetheless, taking into account the multiple pathways proposed here to be plastid located in P. wickerhamii, plastid-to-nucleus signaling might also be required in this nonphotosynthetic taxon.
Interestingly, chlI is plastid encoded in most green algae, including Chlorella vulgaris, the closest relative of P. wickerhamii with a sequenced plastid genome (58), as well as in rhodophytes, glaucophytes, and cryptophytes. So far, the only evidence within the green algal group that chlI can be/was transferred to the nucleus comes from C. reinhardtii where chlI is not plastid encoded (33), and several ESTs corresponding to subunit I of Mg-chelatase are present in the C. reinhardtii database (http://www.biology.duke.edu/chlamy).
Leucoplast-like function or cryptic plastid?
In contrast to the metabolism predicted to be located in the P. falciparum apicoplast and in the Helicosporidium sp. plastid, two phylogenetically divergent parasites, the repertoire of nucleus-encoded plastid-targeted polypeptides predicted for P. wickerhamii reveals a more complex network of pathways (Fig. 2). The metabolism of P. wickerhamii's plastid is therefore rather similar to that encountered in the plastid of photosynthetically competent plants except that processes directly related to photosynthesis, i.e., carbon fixation and photophosphorylation, seem to be lost from the P. wickerhamii plastid.
Clearly, some of the functions proposed to be located in the P. wickerhamii plastid are present in the same cellular compartment of P. falciparum and Helicosporidium sp. Triose phosphates conversion to acetyl-coenzyme A, fatty acid synthesis, several steps in isoprenoid and tetrapyrrole biosynthesis, oxidoreductive processes, and translation and chaperone activity seem to be present in the plastids of all three species.
Amino acid synthesis was demonstrated to be present in the plastid of Helicosporidium sp. but not in the apicoplast of P. falciparum. This feature, along with the different origin of the precursor of tetrapyrrole biosynthesis, led to the suggestion that the Helicosporidium sp. plastid is metabolically more diverse (11).
When plastid metabolism from P. wickerhamii and Helicosporidium sp. is compared, however, carbohydrate, amino acid, and de novo purine metabolism point out a higher complexity of plastid metabolism in P. wickerhamii. Carbohydrate metabolism is clearly more elaborate in P. wickerhamii, as this taxon deposits starch granules in the plastid, which can be easily visualized by electron microscopy. In Helicosporidium sp. this method has failed to reveal the presence of a plastid. Amino acid metabolism in P. wickerhamii is probably more complex, as we identified putative plastid-targeted enzymes that are involved in the biosynthesis and interconversion of more than 10 amino acids. In Helicosporidium sp., only pathways related to the biosynthesis of the amino acids leucine, serine, and lysine have been described to be located in its plastid. Finally, the de novo purine biosynthesis pathway in P. wickerhamii is represented by several putative plastid-targeted polypeptides while in Helicosporidium sp. none of the enzymes involved in this pathway was identified. This is clearly a very interesting finding, as de novo purine biosynthesis represents an energy-consuming pathway, not present in most obligate parasites.
Putative plastid-targeted enzymes reported here and elsewhere support a gradient of decreasing complexity of plastid metabolic pathways in P. wickerhamii, Helicosporidium sp., and P. falciparum. P. wickerhamii is primarily a ubiquitous free-living soil alga, Helicosporidum sp. is a parasite for which the only host-free stage identified in nature so far is represented by cysts (5), while P. falciparum is completely dependent on its hosts for all life cycle stages (12, 41). Two evolutionary factors have been suggested to explain the greater plastid metabolic complexity of Helicosporidium sp. compared to P. falciparum: the need for Helicosporidium sp. to maintain greater metabolic autonomy because of its host-free cyst stage and the more recent autotrophic ancestry of Helicosporidium sp. (11). We note that the P. falciparum apicoplast is of secondary endosymbiotic origin (2, 41, 62) and this may also have contributed in some unknown way to its simplified plastid metabolism relative to Helicosporidium sp. (and P. wickerhamii). However, even though Helicosporidium sp. is a close relative of P. wickerhamii (50, 51) and their plastids are both of primary origin, it appears that the plastid of P. wickerhamii has the greater metabolic complexity of the two, supporting the connection between the degree of parasitism and plastid metabolic complexity. On the other hand, the difference in the diversity of plastid metabolic functions identified to be plastid located in the two nonphotosynthetic green algae might be biased by the number of ESTs characterized so far from the two taxa; we identified 71 putative plastid-targeted proteins in P. wickerhamii by analyzing 3,856 ESTs, while for Helicosporidium sp. the survey of 1,360 ESTs allowed the identification of 20 such proteins (11). Although available data from P. wickerhamii, Helicosporidium sp., and P. falciparum suggest a connection between the degree of parasitism and plastid metabolic complexity, more data are needed from Helicosporidium sp. and from additional nonphotosynthetic primary and secondary plastid-harboring-species before this connection can be confirmed.
Plastid-located metabolic pathways in P. wickerhamii can reveal enzymes that might be drug targeted.
The reconstructed metabolic network proposed here to be plastid located in P. wickerhamii could suggest a new approach in the treatment of protothecosis. Intriguingly, antiprokaryotic and antifungal medications are still widely used in humans infected with Prototheca, though it is largely ineffective or large doses have to be used (26, 53), while for other animals no treatment presently exists (20, 23, 48). Other drugs that target enzymes of pathways that are lacking in mammals (i.e., the shikimate and nonmevalonate pathway, one-carbon pool metabolism, and type II fatty acid synthesis) (8, 41, 57) might be considered along with prophytotoxins such as hydantocidin and ribofuranosyl triazolone, which require plant-specific bioactivation and inhibit the plastid-targeted adenylosuccinate synthetase (19, 44, 47).
Supplementary Material
Acknowledgments
We thank Aurora Nedelcu, University of New Brunswick at Fredericton, for helpful suggestions used in the writing of the paper. We are also grateful to Liisa Koski, Université de Montréal, for implementing the interactive map of the P. wickerhamii metabolic pathways, and Cristina Iancu, California Institute of Technology, and Martin Mallet, Dalhousie University, for editorial suggestions.
This work is part of the Protist EST Program (PEP) and was supported by Genome Canada and the Atlantic Canada Opportunities Agency (Atlantic Innovation Fund). T.B. was the recipient of a PEP Postdoctoral Fellowship, and C. E. P. was funded by the Faculty of Graduate Studies, Dalhousie University, and a Natural Sciences and Engineering Research Council of Canada Discovery Grant to R.W.L.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Archibald, J. M., and P. J. Keeling. 2002. Recycled plastids: a ‘green movement’ in eukaryotic evolution. Trends Genet. 18:577-584. [DOI] [PubMed] [Google Scholar]
- 3.Awai, K., E. Marechal, M. A. Block, D. Brun, T. Masuda, H. Shimada, K. Takamiya, H. Ohta, and J. Joyard. 2001. Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98:10960-10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannai, H., Y. Tamada, O. Maruyama, K. Nakai, and S. Miyano. 2002. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18:298-305. [DOI] [PubMed] [Google Scholar]
- 5.Boucias, D. G., J. J. Becnel, S. E. White, and M. Bott. 2001. In vivo and in vitro development of the protist Helicosporidium sp. J. Eukaryot. Microbiol. 4:460-470. [DOI] [PubMed] [Google Scholar]
- 6.Bruce, B. D. 2001. The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim. Biophys. Acta 1541:2-21. [DOI] [PubMed] [Google Scholar]
- 7.Cai, X., A. L. Fuller, L. R. McDougald, and G. Zhu. 2003. Apicoplast genome of the coccidian Eimeria tenella. Gene 321:39-46. [DOI] [PubMed] [Google Scholar]
- 8.Coggins, J. R., C. Abell, L. B. Evans, M. Frederickson, D. A. Robinson, A. W. Roszak, and A. P. Lapthorn. 2003. Experiences with the shikimate-pathway enzymes as targets for rational drug design. Biochem. Soc. Trans. 31:548-552. [DOI] [PubMed] [Google Scholar]
- 9.Cornah, J. E., M. J. Terry, and A. G. Smith. 2003. Green or red: what stops the traffic in the tetrapyrrole pathway? Trends Plant Sci. 8:224-230. [DOI] [PubMed] [Google Scholar]
- 10.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Koning, A. P., and P. J. Keeling. 2004. Nucleus-encoded genes for plastid-targeted proteins in Helicosporidium: functional diversity of a cryptic plastid in a parasitic alga. Eukaryot. Cell 3:1198-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.el Kouni, M. H. 2003. Potential chemotherapeutic targets in the purine metabolism of parasites. Pharmacol. Ther. 99:283-309. [DOI] [PubMed] [Google Scholar]
- 13.Emanuelsson, O., H. Nielsen, and G. von Heijne. 1999. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 15.Fichera, M. E., and D. S. Roos. 1997. A plastid organelle as a drug target in apicomplexan parasites. Nature 390:407-409. [DOI] [PubMed] [Google Scholar]
- 16.Gockel, G., and W. Hachtel. 2000. Complete gene map of the plastid genome of the nonphotosynthetic euglenoid flagellate Astasia longa. Protist 151:347-351. [DOI] [PubMed] [Google Scholar]
- 17.Goggin, D. E., R. Lipscombe, E. Fedorova, A. H. Millar, A. Mann, C. A. Atkins, and P. M. Smith. 2003. Dual intracellular localization and targeting of aminoimidazole ribonucleotide synthetase in cowpea. Plant Physiol. 131:1033-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray, J. C. 2003. Chloroplast-to-nucleus signalling: a role for Mg-protoporphyrin. Trends Genet. 19:526-529. [DOI] [PubMed] [Google Scholar]
- 19.Hanessian, S., P. P. Lu, J. Y. Sanceau, P. Chemla, K. Gohda, R. Fonne-Pfister, L. Prade, S. W. Cowan-Jacob. 1999. An enzyme-bound bisubstrate hybrid inhibitor of adenylosuccinate synthetase. Angew. Chem. Int. Ed. Engl. 38:3159-3162. [DOI] [PubMed] [Google Scholar]
- 20.Hollingsworth, S. R. 2000. Canine protothecosis. Vet. Clin. North Am. Small Anim. Pract. 30:1091-1101. [DOI] [PubMed] [Google Scholar]
- 21.Huss, V. A., and M. L. Sogin. 1990. Phylogenetic position of some Chlorella species within the chlorococcales based upon complete small-subunit ribosomal RNA sequences. J. Mol. Evol. 31:432-442. [DOI] [PubMed] [Google Scholar]
- 22.Jacquot, J. P., E. Gelhaye, N. Rouhier, C. Corbier, C. Didierjean, and A. Aubry. 2002. Thioredoxins and related proteins in photosynthetic organisms: molecular basis for thiol dependent regulation. Biochem. Pharmacol. 64:1065-1069. [DOI] [PubMed] [Google Scholar]
- 23.Janosi, S., F. Ratz, G. Szigeti, M. Kulcsar, J. Kerenyi, T. Lauko, F. Katona, and G. Huszenicza. 2001. Review of the microbiological, pathological, and clinical aspects of bovine mastitis caused by the alga Prototheca zopfii. Vet. Q. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 24.Jobling, S. A., G. P. Schwall, R. J. Westcott, C. M. Sidebottom, M. Debet, M. J. Gidley, R. Jeffcoat, and R. Safford. 1999. A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: cloning and characterisation of multiple forms of SBE A. Plant J. 18:163-171. [DOI] [PubMed] [Google Scholar]
- 25.Jomaa, H., J. Wiesner, S. Sanderbrand, B. Altincicek, C. Weidemeyer, M. Hintz, I. Turbachova, M. Eberl, J. Zeidler, H. K. Lichtenthaler, D. Soldati, and E. Beck. 1999. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science 285:1573-1576. [DOI] [PubMed] [Google Scholar]
- 26.Kantrow, S. M., and A. S. Boyd. 2003. Protothecosis. Dermatol. Clin. 21:249-255. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J., and S. P. Mayfield. 2002. The active site of the thioredoxin-like domain of chloroplast protein disulfide isomerase, RB60, catalyzes the redox-regulated binding of chloroplast poly(A)-binding protein, RB47, to the 5′ untranslated region of psbA mRNA. Plant Cell Physiol. 43:1238-1243. [DOI] [PubMed] [Google Scholar]
- 28.Kohler, S., C. F. Delwiche, P. W. Denny, L. G. Tilney, P. Webster, R. J. Wilson, J. D. Palmer, and D. S. Roos. 1997. A plastid of probable green algal origin in Apicomplexan parasites. Science 75:1485-1489. [DOI] [PubMed] [Google Scholar]
- 29.Knauf, U., and W. Hachtel. 2002. The genes encoding subunits of ATP synthase are conserved in the reduced plastid genome of the heterotrophic alga Prototheca wickerhamii. Mol. Genet. Genomics 267:492-497. [DOI] [PubMed] [Google Scholar]
- 30.Lee, W.-S., M. D Lagios, and R. Leonards. 1975. Wound infection by Prototheca wickerhamii, a saprophytic alga pathogenic for man. J. Clin. Microbiol. 2:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaire, S. D., V. Collin, E. Keryer, A. Quesada, and M. Miginiac-Maslow. 2003. Characterization of thioredoxin y, a new type of thioredoxin identified in the genome of Chlamydomonas reinhardtii. FEBS Lett. 543:87-92. [DOI] [PubMed] [Google Scholar]
- 32.MacFadden, G. I., M. Reith, J. Munholland, and N. Lang-Unnasch. 1996. Plastid in human parasites. Nature 381:482. [DOI] [PubMed] [Google Scholar]
- 33.Maul, J. E., J. W. Lilly, L. Cui, C. W. de Pamphilis, W. Miller, E. H. Harris, and D. B. Stern. 2002. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14:2659-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miege, C., E. Marechal, M. Shimojima, K. Awai, M. A. Block, H. Ohta, K. Takamiya, R. Douce, and J. Joyard. 1999. Biochemical and topological properties of type A MGDG synthase, a spinach chloroplast envelope enzyme catalyzing the synthesis of both prokaryotic and eukaryotic MGDG. Eur. J. Biochem. 265:990-1001. [DOI] [PubMed] [Google Scholar]
- 35.Nadakavukaren, M. J., and D. A. McCracken. 1973. Prototheca: an alga or a fungus? J. Phycol. 9:113-116. [Google Scholar]
- 36.Nadakavukaren, M. J., and D. A. McCracken. 1977. An ultrastructural survey of the genus Prototheca with special reference to plastids. Mycopathologia 61:117-119. [DOI] [PubMed] [Google Scholar]
- 37.Neuhaus, H. E., and M. J. Emes. 2000. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:111-140. [DOI] [PubMed] [Google Scholar]
- 38.Obadou, S., R. Tien, J. Soll, and E. Schleiff. 2003. Membrane insertion of the chloroplast outer envelope protein, Toc34: constrains for insertion and topology. J. Cell Sci. 116:837-846. [DOI] [PubMed] [Google Scholar]
- 39.Ohlrogge, J. B., and J. G. Jaworski. 1997. Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:109-136. [DOI] [PubMed] [Google Scholar]
- 40.Qi, Q., K. F. Kleppinger-Sparace, and S. A. Sparace. 1995. The utilization of glycolytic intermediates as precursors for fatty acid biosynthesis by pea root plastids. Plant Physiol. 107:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ralph, S. A., G. G. van Dooren, R. F. Waller, M. J. Crawford, M. J. Fraunholz, B. J. Foth, C. J. Tonkin, D. S. Roos, and G. I. Mc Fadden. 2004. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2:203-216. [DOI] [PubMed] [Google Scholar]
- 42.Sato, S., and R. J. Wilson. 2002. The genome of Plasmodium falciparum encodes an active delta-aminolevulinic acid dehydratase. Curr. Genet. 40:391-398. [DOI] [PubMed] [Google Scholar]
- 43.Sato, S., B. Clough, L. Coates, and R. J. M. I. Wilson. 2004. Enzymes for heme biosynthesis are found in both the mitochondrion and plastid of the malaria parasite Plasmodium falciparum. Protist 155:117-125. [DOI] [PubMed] [Google Scholar]
- 44.Schmitzer, P. R., P. R. Graupner, E. L. Chapin, S. C. Fields, J. R. Gilbert, J. A. Gray, C. L. Peacock, and B. C. Gerwick. 2000. Ribofuranosyl triazolone: a natural product herbicide with activity on adenylosuccinate synthetase following phosphorylation J. Nat. Prod. 63:777-781. [DOI] [PubMed] [Google Scholar]
- 45.Sekiguchi, H., M. Moriya, T. Nakayama, I. Inouye. 2002. Vestigial chloroplasts in heterotrophic stramenopiles Pteridomonas danica and Ciliophrys infusionum (Dictyochophyceae). Protist 153:157-167. [DOI] [PubMed] [Google Scholar]
- 46.Shanklin, J., and E. B. Cahoon. 1998. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:611-641. [DOI] [PubMed] [Google Scholar]
- 47.Siehl, D. L., M. V. Subramanian, E. W. Walters, S. F. Lee, R. J. Anderson, and A. G. Toschi. 1996. Adenylosuccinate synthetase: site of action of hydantocidin, a microbial phytotoxin. Plant Physiol. 110:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spalton, D. E. 1985. Bovine mastitis caused by Prototheca zopfii: a case study. Vet. Rec. 116:347. [DOI] [PubMed] [Google Scholar]
- 49.Strand, A., T. Asami, J. Alonso, J. R. Ecker, and J. Chory. 2003. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin IX. Nature 421:79-83. [DOI] [PubMed] [Google Scholar]
- 50.Tartar, A., D. G. Boucias, B. J. Adams, and J. J. Becnel. 2002. Phylogenetic analysis identifies the invertebrate pathogen Helicosporidium sp. as a green alga (Chlorophyta). Int. J. Syst. Evol. Microbiol. 52:273-279. [DOI] [PubMed] [Google Scholar]
- 51.Tartar, A., and D. G. Boucias. 2004. The non-photosynthetic, pathogenic green alga Helicosporidium sp. has retained a modified, functional plastid genome. FEMS Microbiol. Lett. 233:153-157. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torres, H. A., G. P. Bodey, J. J. Tarrand, and D. P. Kontoyiannis. 2003. Protothecosis in patients with cancer: case series and literature review. Clin. Microbiol. Infect. 9:786-792. [DOI] [PubMed] [Google Scholar]
- 54.Trebitsh, T., E. Meiri, O. Ostersetzer, Z. Adam, and A. Danon. 2001. The protein disulfide isomerase-like RB60 is partitioned between stroma and thylakoids in Chlamydomonas reinhardtii chloroplasts. J. Biol. Chem. 276:4564-4569. [DOI] [PubMed] [Google Scholar]
- 55.Ueno, R., N. Urano, and M. Suzuki. 2003. Phylogeny of the non-photosynthetic green micro-algal genus Prototheca (Trebouxiophyceae, Chlorophyta) and related taxa inferred from SSU and LSU ribosomal DNA partial sequence data. FEMS Microbiol. Lett. 223:275-280. [DOI] [PubMed] [Google Scholar]
- 56.Vollmer, M., N. Thomsen, S. Wiek, and F. Seeber. 2001. Apicomplexan parasites possess distinct nuclear-encoded, but apicoplast-localized, plant-type ferredoxin-NADP+ reductase and ferredoxin. J. Biol. Chem. 276:5483-5490. [DOI] [PubMed] [Google Scholar]
- 57.Wakabayashi, K., and P. Boger. 2002. Target sites for herbicides: entering the 21st century. Pest Manag. Sci. 58:1149-1154. [DOI] [PubMed] [Google Scholar]
- 58.Wakasugi, T., T. Nagai, M. Kapoor, M. Sugita, M. Ito, S. Ito, J. Tsudzuki, K. Nakashima, T. Tsudzuki, Y. Suzuki, A. Hamada, T. Ohta, A. Inamura, K. Yoshinaga, and M. Sugiura. 1997. Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division. Proc. Natl. Acad. Sci. USA 94:5967-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waller, R. F., P. J. Keeling, R. G. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A. F. Cowman, G. S. Besra, D. S. Roos, and G. I. McFadden. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 95:12352-12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weeden, N. F. 1981. Genetic and biochemical implications of the endosymbiotic origin of the chloroplast. J. Mol. Evol. 17:133-139. [DOI] [PubMed] [Google Scholar]
- 61.Williams, B. A., and P. J. Keeling. 2003. Cryptic organelles in parasitic protists and fungi. Adv. Parasitol. 54:9-68. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, R. J., P. W. Denny, P. R. Preiser, K. Rangachari, K. Roberts, A. Roy, A. Whyte, M. Strath, D. J. Moore, P. W. Moore, and D. H. Williamson. 1996. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 261:155-172. [DOI] [PubMed] [Google Scholar]
- 63.Wolfe, K. H., C. W. Morden, and J. D. Palmer. 1992. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 89:10648-10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolfe, A. D., and C. W. dePamphilis. 1998. The effect of relaxed functional constraints on the photosynthetic gene rbcL in photosynthetic and nonphotosynthetic parasitic plants. Mol. Biol. Evol. 15:1243-1258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.