Abstract
Large chromosomal events such as translocations and segmental duplications enable rapid adaptation to new environments. Here we marshal genomic, genetic, meiotic mapping, and physical evidence to demonstrate that a chromosomal translocation and segmental duplication occurred during construction of a congenic strain pair in the fungal human pathogen Cryptococcus neoformans. Two chromosomes underwent telomere-telomere fusion, generating a dicentric chromosome that broke to produce a chromosomal translocation, forming two novel chromosomes sharing a large segmental duplication. The duplication spans 62,872 identical nucleotides and generated a second copy of 22 predicted genes, and we hypothesize that this event may have occurred during meiosis. Gene disruption studies of one embedded gene (SMG1) corroborate that this region is duplicated in an otherwise haploid genome. These findings resolve a genome project assembly anomaly and illustrate an example of rapid genome evolution in a fungal genome rich in repetitive elements.
Organisms evolve by natural selection, whereby individuals that harbor beneficial mutations are enriched in populations and those with deleterious mutations decline. While point mutations contribute, large-scale DNA transactions involving transposition, deletion, duplication, and translocation can enable more global genomic changes that can engender broad phenotypic changes. The creation of new genes through segmental duplication enables considerable evolutionary potential, and this mechanism is apparently common, even in humans (1, 11). These events therefore play a unique role in evolution by providing two copies of a gene: one to retain the original function and a second that is free to diverge. Ohno hypothesized that duplicated gene pairs might bear the signature of accelerated evolution in one of the two copies (26), and this hypothesis has recently been experimentally verified (19).
The fungi represent a unique opportunity in which entire relatively small genomes can be analyzed for unique genomic architectures that have punctuated their evolutionary history. For example, in Saccharomyces cerevisiae, an ancient whole-genome duplication event gave rise to over 60 regions of the genome that harbor duplicated genes in a syntenic arrangement (36). Comparison of the S. cerevisiae genome to related fungi, including Ashbya gossypii and Kluveryomyces waltii, confirmed that S. cerevisiae and closely related sensu stricto species derived from an ancestral organism in which the entire genome was duplicated ∼100 million years ago (6, 19). Many duplicated genes were subsequently lost, but others were retained and have fostered evolution as paralogs with divergent functions. Thus, whole-genome duplication can drive evolution.
In contrast, recent analysis of more distantly related fungi revealed that more limited segmental duplications are rampant in the genomes of many yeasts (7). For example, in response to selective pressure, a growth-defective S. cerevisiae ribosomal protein mutant yields faster-growing variants in which segmental duplications and translocations duplicate a conserved paralog that restores normal growth (20). Similar variants arise in response to selective pressures during fermentation and carbon source utilization (8, 17). Thus, either whole-genome duplication or segmental duplication can provide new genes as raw material for evolution. Furthermore, laboratory-induced changes in chromosome structure have been observed in Candida albicans that confer resistance to fluconazole or enable utilization of alternative metabolites (27, 30).
Although aneuploidy occurs in many fungi (3, 34), the specific mechanisms that give rise to duplications have not been detailed in pathogenic fungi. Using two genomic sequences, a genetic linkage map, and physical maps, we have discovered that a region of the haploid genome of a strain of the human pathogen Cryptococcus neoformans is segmentally duplicated. Our evidence supports a model showing that two chromosomes underwent telomere-telomere fusion, generating an intermediate dicentric chromosome that was subsequently broken to generate a chromosomal translocation, forming two novel chromosomes sharing a large segmental duplication. This unusual genomic event is also made apparent by comparing the two completed genomes, and it posed a formidable assembly anomaly. Similar telomere-telomere fusions, chromosome instability, and rearrangements have been described in S. cerevisiae telomere checkpoint mutants (25) and human cancer cells (5, 38) and even as the mechanism underlying the formation of human chromosome 2 (10, 16, 35, 37). This process is therefore a general mechanism of genome instability leading to chromosomal translocations and segmental duplications with implications for virulence, speciation, and oncogenesis. We hypothesize that this event occurred during meiosis, implying that sexual reproduction may imperil the integrity of the genome. Indeed, like for many other pathogenic fungi (18, 21), evidence for an active sexual cycle for C. neoformans in nature has only recently been reported (23).
MATERIALS AND METHODS
Strains and media.
The reference strains used in this study were the following serotype D strains of C. neoformans: NIH12 (MATα), NIH433 (MATa), B3501 (MATα), B3502 (MATa), JEC21 (MATα), and JEC20 (MATa) (15, 22). Strains were grown on yeast extract-peptone-dextrose medium.
Molecular techniques.
Standard methods were performed as described by Sambrook et al. (31). C. neoformans genomic DNA for Southern blot analysis was prepared as described by Pitkin et al. (29). Electrophoretic karyotypes were produced for all reference strains, and chromosomes were transferred from gels onto positively charged nylon membranes as described previously (24), with the following altered parameters: block 1, 75- to 150-s switch, 4.0 V/cm, 12°C for 40 h; and block 2, 200- to 400-s switch, 4.0 V/cm, 12°C for 60 h. These Southern blots of electrophoretically separated chromosomes were probed with sequences as indicated elsewhere (see Table S1 in the supplemental material). Probes for chromoblot analysis were labeled with digoxigenin (DIG), and the hybridizations were performed according to kit instructions supplied with the Roche DIG Luminescent Detection System (Roche, Mannheim, Germany) (24). Probes for genomic Southern blots were prepared with the Rediprime II Random Prime Labeling system (Amersham Biosciences). Sequences of primers and PCR products generated referred to in the text are presented elsewhere (see Table S1 in the supplemental material).
Gene disruption.
The smg1::URA5 mutant allele was made by PCR overlap as previously described (4). The left fragment was amplified with primers 130 and 132, and the right fragment was amplified with primers 184 and 136, with genomic DNA from strain JEC21 as a template. The URA5 fragment was amplified with primers 131 and 185. All three PCR amplicons were combined for the final overlap PCR with primers 130 and 136. The mutant allele was biolistically transformed into strain JEC43 as described previously (33), and transformants were selected on synthetic medium lacking uracil.
Nucleotide sequence accession numbers.
The novel telomeric sequences have been submitted to GenBank under accession no. AY772393 (chromosome 8) and AY772394 (chromosome 12). The sequences for the new telomeres, new chromosome numbering, and subsequent renaming of the JEC21 open reading frames (ORFs) have all been incorporated into the Cryptococcus neoformans var. neoformans strain JEC21 genome sequence.
RESULTS AND DISCUSSION
A JEC21 genome assembly anomaly.
Construction of a C. neoformans var. neoformans congenic strain pair (JEC20 and JEC21) over a decade ago provided a robust platform for molecular genetic studies of this important human pathogen (15, 22). However, during development of a meiotic linkage map, we discovered that these strains contain a chromosomal translocation compared to the parental isolates (24). Although karyotype variability had previously been observed in environmental isolates (28), during laboratory passage (12), in the host during persistent infection treated with long-term antifungal therapy (13), and following meiosis (2), the underlying molecular mechanisms were unknown. Given its prevalence, karyotypic variability may promote survival under adverse conditions.
Following electrophoresis by contour-clamped homogenous electric field (CHEF), Southern hybridizations of the separated chromosomes revealed a translocation entailing the exchange of genetic material between chromosomes 9 (linkage group 4) and 12 (linkage groups 14 and 18) of parental strain B3501A (Fig. 1). This event generated novel chromosome sizes in the congenic strains (linkage groups 14 and 18 on chromosome 8 and linkage group 4 on chromosome 12 of JEC21) (24). With additional molecular and genetic analyses, we reconciled the completed genomes of strains B3501A and JEC21, determining the molecular basis of this translocation.
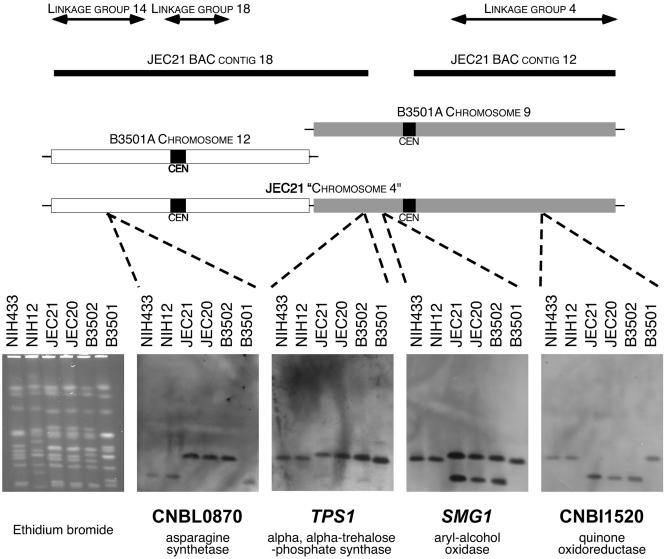
FIG. 1.
A duplication in the C. neoformans genome. Compiling complementary information from the meiotic map, the JEC21 BAC scaffolds, and the B3501A and JEC21 whole-genome sequence projects enabled the identification of a duplication in the JEC21 genome. Probing chromoblots with genes from the JEC21 chromosome 4 assembly revealed translocated (TPS1) and duplicated (SMG1) genomic regions. CEN, centromere.
The B3501A genome data set produced at the Stanford Genome Technology Center (SGTC) revealed 14 chromosomes, which agreed in size with those determined by CHEF analysis (chromosome 9 was 1.20 Mb according to CHEF analysis and 1.07 Mb according to genome analysis, and chromosome 12 was 0.94 Mb according to CHEF analysis and 0.89 Mb according to genome analysis). In contrast, the JEC21 genome sequence from The Institute for Genomic Research (TIGR) resolved 13 chromosomes; the sequences equivalent to chromosomes 9 and 12 of strain B3501A represented a single large contig of 2.04 Mb, designated chromosome 4. Alignment of the JEC21 and B3501A assemblies resembled a tandem fusion of chromosomes 9 and 12 (Fig. 1). Furthermore, during annotation of the JEC21 genome, each chromosome contained a large transposon cluster thought to represent the centromere. A single cluster is present on each chromosome except chromosome 4, which bears two and would therefore be predicted to be dicentric and unstable.
In C. neoformans, chromosomes are appended with tandem AGGGGGTT telomeric sequence arrays (9). Analysis of the fusion point of B3501A chromosomes 9 and 12 in the JEC21 chromosome 4 assembly revealed embedded copies of the canonical AGGGGGTT telomeric repeat on one strand. Interspersed are four partial copies of the class 1 transposable element Cnl1 (14), which is localized exclusively to subtelomeric regions with this cluster as the only exception in the genome.
The possibility that this large chromosome resulted from a misassembly in the fused telomeric region was discounted by probing high-density bacterial artificial chromosome (BAC) library filters, which revealed multiple clones spanning the fusion boundary (not shown). This finding agrees with physical maps based on BAC fingerprinting that scaffolded the sequence assembly (32).
Segmental duplication in a haploid genome.
Superimposing the BAC physical map contigs on the chromosome 4 assembly revealed that JEC21 BAC contig 18 covers the entire region representing chromosome 12 from B3501A and overlaps the chromosome 9 portion by >240 kb. In contrast, JEC21 BAC contig 12 corresponds to the opposite end of the chromosome 9 portion of the assembly, leaving a 116-kb gap that may contain a spurious join (Fig. 1).
Hybridizing CHEF blots with probes to opposite ends of the large assembly (B3501A predicted ORF CNBI1520 from chromosome 9 and CNBL0870 from chromosome 12) confirmed the JEC21 chromosome 4 assembly was incorrect (Fig. 1). This analysis revealed two smaller chromosomes corresponding in size to chromosomes 8 and 12 that were created by the translocation (24). To determine the precise point of the anomaly, probes from the BAC contig map break region were probed with CHEF chromosome blots. One such probe, TPS1 from B3501A chromosome 9, confirmed that a translocation was present, as it hybridized to the same chromosome as the B3501A chromosome 12 probe CNBI1520 rather than to the same chromosome as the B3501A chromosome 9 probe CNBL0870. However, while most probes identified either novel chromosome 8 or 12 from strains JEC20 and JEC21, three (SMG1, CNBI2940, and CNBI2900) hybridized to both, indicating that this region is duplicated (Fig. 1).
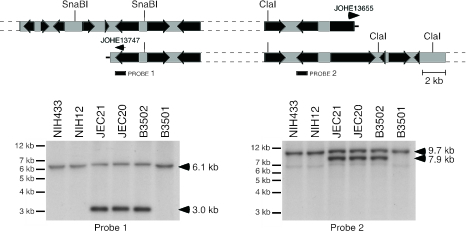
To map the duplicated genomic region, Southern hybridizations were performed, using probes for randomly chosen predicted genes in the anomalous region. The presence of a nearby chromosomal terminus serves as a common restriction fragment end, creating additional fragments that do not correspond to those predicted from the genome sequence. Accordingly, CNBI3110 and CNBI2900 gene-specific probes produced additional hybridization signals, allowing more precise mapping of the left and right ends of the duplication (Fig. 2). This revealed that the duplication spans a region of ∼60 kb, including the three duplicated genes detected based on chromoblot analysis.
FIG. 2.
Fine-mapping novel JEC21 telomeres by Southern blotting. Following restriction digestion (SnaBI or ClaI), the location of each duplication boundary/telomeric end was established by Southern blotting. Filled black arrows represent predicted genes, and the probe used for each terminus is indicated. Small arrows represent primers used in conjunction with telomeric primer JOHE13427 to amplify the novel chromosome ends.
In C. neoformans, linear extrachromosomal DNA fragments are appended with tandem arrays of the telomeric octanucleotide repeat AGGGGGTT (9), and the stability of novel chromosomal ends requires telomerase activity. The predicted new telomeres were amplified using CNBI3110- and CNBI2900-specific primers (Fig. 2) in conjunction with a generic telomeric primer. This approach yielded unambiguous products for both predicted end points, and sequencing determined the specific nucleotide at which each new chromosome terminates at telomeric repeats. The duplication is exactly 62,872 bp in length and contains 22 genes with diverse predicted functions (see Table S2 in the supplemental material).
When these data are incorporated into the whole-genome sequence, the larger incorrect assembly (JEC21 chromosome 4) is divided into two smaller contigs of 1.194 and 0.907 Mb, defining these as the 8th and 12th largest chromosomes, respectively, in accord with those observed on CHEF gels (1.20 Mb for JEC21 chromosome 8, 0.96 Mb for JEC21 chromosome 12).
Comparison of the region duplicated in strain JEC21 with the corresponding genomic region reveals it to be single copy in B3501A and monomorphic between the two. The genetic map could not detect the translocation as there are no polymorphisms in this region; rather, CHEF blot hybridizations were required. While the translocation is 257 kb in length, due to the duplication event, only 195 kb was lost from the JEC21 equivalent of B3501A chromosome 9, a size change that alters its designation to chromosome 12.
SMG1 is duplicated.
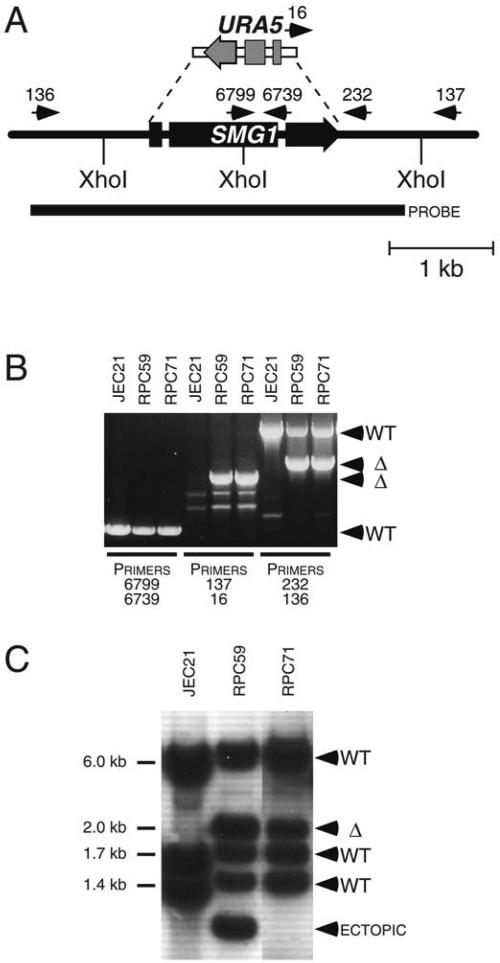
The SMG1 gene was identified in an independent genetic screen, but disruption experiments support the hypothesis that SMG1 lies in the JEC21 duplication. BLAST searches and Southern hybridizations analyzing the SMG1 locus initially suggested that a single copy was present in the JEC21 genome (not shown). In gene disruption experiments (Fig. 3A), genomic DNA from 140 transformants was analyzed by PCR. Using SMG1 internal primers, a 0.6-kb amplicon was present in all transformants, suggesting that each strain retains the wild-type SMG1 gene (Fig. 3B). Using primers that amplify different size fragments from the SMG1 wild-type or smg1 mutant locus, PCRs of strains RPC59 and RPC71 yielded products consistent with the presence of both wild-type and mutant SMG1 alleles. Southern hybridization also confirmed that both alleles are present (Fig. 3C), providing corroborative evidence that a segmental duplication exists in the JEC21 genome.
FIG. 3.
The SMG1 gene is duplicated in strain JEC21. (A) The SMG1 gene was replaced with URA5 to create an smg1::URA5 mutant. Arrows indicate primer positions. Primers 136 and 137 correspond to regions outside the smg1::URA5 allele. (B) Genomic DNA from the indicated strains was used as PCR templates with the indicated primers. Primers 137 and 16 amplify a 1.1-kb product from strains in which the smg1::URA5 allele precisely replaced the SMG1 locus. WT, wild type. Δ, deletion mutant. (C) Genomic DNA from the indicated strains was XhoI digested and analyzed by Southern blotting with an SMG1 gene probe.
Translocation during congenic strain pair production.
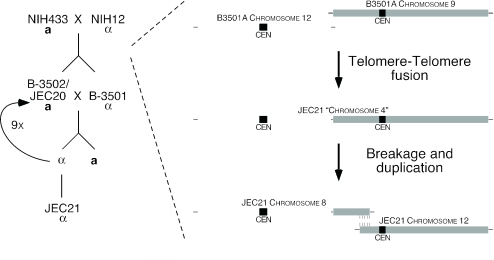
We have identified a significant genomic event that remained undetected by the genome project. When did the fusion occur? As the congenic strains were produced by crossing parental strains NIH12 and NIH433, neither of which contains the duplication (Fig. 1), the proposed chromosomal events must have occurred during either meiosis or laboratory passage. During one of these initial steps in the isolation of the JEC20/JEC21 congenic pair, chromosomes 9 and 12 underwent telomere-telomere fusion (Fig. 4). Furthermore, PCR analysis of three independent subcultures of the F1 MATa and MATα progeny (B3501 and B3502) obtained from different laboratories—in one case from Kwon-Chung laboratory lyophilized stocks—revealed the same novel telomeric sequences, indicating that this result is not an artifact of passage in our laboratory (data not shown). Analysis of the fusion point does not reveal a head-to-head arrangement of the telomeric repeats; rather the telomeric sequence from chromosome 12 is absent at the junction point. This is in contrast to the well-characterized telomere-telomere fusion that formed human chromosome 2, where such head-to-head telomeric repeats are found embedded at the fusion point (16). This structure in C. neoformans therefore suggests that this translocation was caused by recombination, possibly within subtelomeric Cnl1 elements, rather than nonhomologous end joining. The unstable dicentric chromosome formed by this fusion was then broken, subsequently forming a segmental duplication of 62,872 bp in a region that shares 100% identity with the corresponding region in strain B3501A. This also contrasts with human chromosome 2, where instead of chromosome breakage resolving the dicentric structure, one of the centromeres (2q) was instead suppressed (10). In C. neoformans, the canonical telomeric repeat AGGGGGTT was then appended to the termini of these newly formed novel chromosomes. The original JEC21 genome chromosome 4 assembly (21 October 2003 release) therefore represents the transitional dicentric chromosome.
FIG. 4.
Translocation and formation of a segmental duplication during congenic strain construction. (Left) Construction of a C. neoformans strain pair. The outlined mating scheme incorporates more recent discoveries that strains JEC20 and B3502 are identical. (Right) Proposed model for genomic events giving rise to a segmental duplication in the otherwise haploid C. neoformans genome. First, either during laboratory passage or meiosis in the construction of the JEC20/JEC21 congenic strain pair, the parental chromosomes 9 (filled) and 12 (open) underwent telomere-telomere fusion, most likely via recombination. This produced an unstable dicentric structure, represented by the misassembled chromosome 4 in the TIGR genome sequence, which subsequently broke, duplicating a 61-kb genomic region in the process of forming a translocation. CEN, centromere.
In S. cerevisiae, the occurrence of telomere-telomere fusion is increased in cell cycle-telomere checkpoint control mutants (25). In mec1 tel1 mutant strains, telomere-telomere fusions via nonhomologous end joining are rampant and breakage of the resulting dicentric chromosomes rapidly and dramatically alters the entire organization of the genome; similar mechanisms may have driven ascomycete speciation. Furthermore, such events are common in tumors, where the cell cycle is perforce unregulated, with telomere-telomere fusion driven by nonhomologous end joining (38). However, in the example discovered here, the mechanism driving segmental duplications may have been meiotic recombination between subtelomeric transposable elements. If so, this mechanism would be different from that of S. cerevisiae mec1 tel1 mutants, where nonhomologous end joining mediates telomere-telomere fusion (25). Of course, we cannot exclude the possibility that this chromosomal rearrangement transpired mitotically. Further studies of environmental, clinical, and laboratory isolates will be required to determine when and at what frequency similar large-scale genomic events occur in C. neoformans.
The generation of segmental aneuploidy offers a rapid alternative response to drastic changes in the environment or host. Further studies of this evolutionary mechanism will help elucidate how this evolutionary force might contribute to the development of drug resistance, limited fecundity, the ability to occupy novel evolutionary niches, sexual isolation, and speciation.
Supplementary Material
Acknowledgments
We thank Richard Hyman and Brendan Loftus for discussions, James W. Kronstad for BAC map information, Chris Parker for chromoblots, and Timothy Y. James and Robert E. Marra for comments on the manuscript.
This work was supported in part by NIAID grants AI25783, AI44975, AI50113, and AI50128; the Burroughs Wellcome Fund; and the HHMI. We acknowledge C. neoformans genome projects at the Stanford Genome Center and TIGR, funded by NIAID/NIH cooperative agreements AI47087 and AI48594.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bailey, J. A., A. M. Yavor, H. F. Massa, B. J. Trask, and E. E. Eichler. 2001. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 11:1005-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boekhout, T., and A. van Belkum. 1997. Variability of karyotypes and RAPD types in genetically related strains of Cryptococcus neoformans. Curr. Genet. 32:203-208. [DOI] [PubMed] [Google Scholar]
- 3.Chibana, H., J. L. Beckerman, and P. T. Magee. 2000. Fine-resolution physical mapping of genomic diversity in Candida albicans. Genome Res. 10:1865-1877. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 5.DePinho, R. A., and K. Polyak. 2004. Cancer chromosomes in crisis. Nat. Genet. 36:932-934. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 7.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 8.Dunham, M. J., H. Badrane, T. Ferea, J. Adams, P. O. Brown, F. Rosenzweig, and D. Botstein. 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:16144-16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edman, J. C. 1992. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol. Cell. Biol. 12:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, Y., E. Linardopoulou, C. Friedman, E. Williams, and B. J. Trask. 2002. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13-2q14.1 and paralogous regions on other human chromosomes. Genome Res. 12:1651-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortna, A., Y. Kim, E. MacLaren, K. Marshall, G. Hahn, L. Meltesen, M. Brenton, R. Hink, S. Burgers, T. Hernandez-Boussard, A. Karimpour-Fard, D. Glueck, L. McGavran, R. Berry, J. Pollack, and J. M. Sikela. 2004. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2:937-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzot, S. P., J. Mukherjee, R. Cherniak, L.-C. Chen, J. S. Hamdan, and A. Casadevall. 1998. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect. Immun. 66:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fries, B. C., F. Chen, B. P. Currie, and A. Casadevall. 1996. Karyotype instability in Cryptococcus neoformans infection. J. Clin. Microbiol. 34:1531-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodwin, T. J., and R. T. Poulter. 2001. The diversity of retrotransposons in the yeast Cryptococcus neoformans. Yeast 18:865-880. [DOI] [PubMed] [Google Scholar]
- 15.Heitman, J., B. Allen, J. A. Alspaugh, and K. J. Kwon-Chung. 1999. On the origins of congenic MATalpha and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet. Biol. 28:1-5. [DOI] [PubMed] [Google Scholar]
- 16.IJdo, J. W., A. Baldini, D. C. Ward, S. T. Reeders, and R. A. Wells. 1991. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. USA 88:9051-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Infante, J. J., K. M. Dombek, L. Rebordinos, J. M. Cantoral, and E. T. Young. 2003. Genome-wide amplifications caused by chromosomal rearrangements play a major role in the adaptive evolution of natural yeast. Genetics 165:1745-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, A. 2003. The biology of mating in Candida albicans. Nat. Rev. Microbiol. 1:106-116. [DOI] [PubMed] [Google Scholar]
- 19.Kellis, M., B. W. Birren, and E. S. Lander. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617-624. [DOI] [PubMed] [Google Scholar]
- 20.Koszul, R., S. Caburet, B. Dujon, and G. Fischer. 2004. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 23:234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koufopanou, V., A. Burt, and J. W. Taylor. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 94:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litvintseva, A. P., R. E. Marra, K. Nielsen, J. Heitman, R. Vilgalys, and T. G. Mitchell. 2003. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot. Cell 2:1162-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marra, R. E., J. C. Huang, E. Fung, K. Nielsen, J. Heitman, R. Vilgalys, and T. G. Mitchell. 2004. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans). Genetics 167:619-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mieczkowski, P. A., J. O. Mieczkowska, M. Dominska, and T. D. Petes. 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc. Natl. Acad. Sci. USA 100:10854-10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohno, S. 1970. Evolution by gene duplication. Allen and Unwin, London, United Kingdom.
- 27.Perepnikhatka, V., F. J. Fischer, M. Niimi, R. A. Baker, R. D. Cannon, Y.-K. Wang, F. Sherman, and E. Rustchenko. 1999. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J. Bacteriol. 181:4041-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perfect, J. R., N. Ketabchi, G. M. Cox, C. W. Ingram, and C. L. Beiser. 1993. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J. Clin. Microbiol. 31:3305-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitkin, J. W., D. G. Panaccione, and J. D. Walton. 1996. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142:1557-1565. [DOI] [PubMed] [Google Scholar]
- 30.Rustchenko, E. P., D. H. Howard, and F. Sherman. 1997. Variation in assimilating functions occurs in spontaneous Candida albicans mutants having chromosomal alterations. Microbiology 143:1765-1778. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schein, J. E., K. L. Tangen, R. Chiu, H. Shin, K. B. Lengeler, W. K. MacDonald, I. Bosdet, J. Heitman, S. J. Jones, M. A. Marra, and J. W. Kronstad. 2002. Physical maps for genome analysis of serotype A and D strains of the fungal pathogen Cryptococcus neoformans. Genome Res. 12:1445-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Lee, T., A. Testa, A. Robold, J. van't Klooster, and F. Govers. 2004. High-density genetic linkage maps of Phytophthora infestans reveal trisomic progeny and chromosomal rearrangements. Genetics 167:1643-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wienberg, J., A. Jauch, H. J. Ludecke, G. Senger, B. Horsthemke, U. Claussen, T. Cremer, N. Arnold, and C. Lengauer. 1994. The origin of human chromosome 2 analyzed by comparative chromosome mapping with a DNA microlibrary. Chromosome Res. 2:405-410. [DOI] [PubMed] [Google Scholar]
- 36.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 37.Yunis, J. J., and O. Prakash. 1982. The origin of man: a chromosomal pictorial legacy. Science 215:1525-1530. [DOI] [PubMed] [Google Scholar]
- 38.Zheng, M. H., P. Siu, J. M. Papadimitriou, D. J. Wood, and A. R. Murch. 1999. Telomeric fusion is a major cytogenetic aberration of giant cell tumors of bone. Pathology 31:373-378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.