Abstract
Thioredoxin reductase (TRR1) is an important component of the thioredoxin oxidative stress resistance pathway. Here we show that it is induced during oxidative and nitrosative stress and is preferentially localized to the mitochondria in Cryptococcus neoformans. The C. neoformans TRR1 gene encodes the low-molecular-weight isoform of the thioredoxin reductase enzyme, which shares little homology with that of its mammalian host. By replacing the endogenous TRR1 promoter with an inducible copper transporter promoter, we showed that Trr1 appears to be essential for viability of this pathogenic fungus, making it a potential antifungal target.
In addition to its importance in defense against oxidative stress, the thioredoxin system of bacteria, yeast, and mammals is involved in regulating DNA synthesis, gene transcription, cell growth, and apoptosis (1, 12). Thioredoxin reductase, a flavoenzyme homodimer which binds flavin adenine dinucleotide and NADPH, reduces the oxidoreductase thioredoxin and is found in two forms throughout all five kingdoms. The high-molecular-weight isoform, which most likely evolved from glutathione reductase rather than the prokaryotic thioredoxin reductase (17), is present in mammals and some parasites, while the low-molecular-weight isoform is found in most bacteria, plants, and fungi (6). These two isoforms are thought to have independently evolved to having similar substrate specificity profiles. Though these isoforms have similar functions, they have very distinct protein structures; therefore, thioredoxin reductase may be a selective drug target for those pathogens harboring the low-molecular-weight form.
Through BLAST homology results determined using Saccharomyces cerevisiae Trr1, we have identified one thioredoxin reductase gene in C. neoformans. It encodes an enzyme of the low-molecular-weight form and is more similar to TRR1 than to TRR2 of S. cerevisiae (61% compared to 58% identity), since it lacks the N-terminal mitochondrial signal sequence found in TRR2. We have also sequenced the cDNA of the TRR1 gene from strain H99, a C. neoformans var. grubii clinical isolate, and have submitted the coding sequences to GenBank (accession number AY796185). The TRR1 gene of C. neoformans contains seven exons and encodes a 343-amino-acid protein.
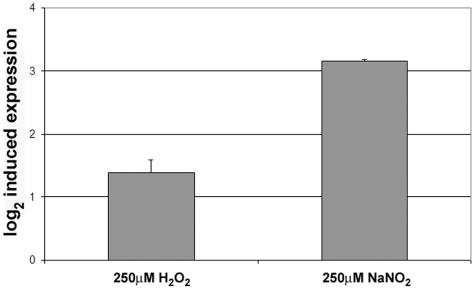
Thioredoxin reductase has been studied in many organisms, including bacteria and fungi. Increased transcription of the thioredoxin system genes in Staphylococcus aureus is observed with diamide, menadione, and t-butyl hydroperoxide but not hydrogen peroxide stress (19). Both TRR1 and TRR2 are induced in response to H2O2 stress in S. cerevisiae (5, 16), while TRR1 is induced with menadione stress in S. pombe (7) and with peroxide stress in Kluyveromyces lactis (18). In Candida albicans, TRR1 has been shown to be induced during H2O2 stress and osmotic stress and during exposure to whole blood (3, 4). By performing real-time PCR using actin as an internal control, we show that in C. neoformans, TRR1 is induced during both oxidative stress from H2O2 and nitrosative stress from nitric oxide (Fig. 1). Cells grown in yeast nitrogen base (pH 4) were treated for 2 h with the appropriate stress. RNA was isolated, cDNA was generated, and real-time PCR was performed as described previously (11). Though TRR1 is induced more than 2-fold (1 log2) with oxidative stress, we observe greater than 8-fold (3 log2) induction during nitrosative stress. This may imply that the thioredoxin system is the primary defense against nitric oxide stress, while multiple systems may respond to hydrogen peroxide stress.
FIG. 1.
TRR1 induction during oxidative and nitrosative stress in H99. Cells grown in yeast nitrogen base (pH 4) were treated with 250 μM hydrogen peroxide or sodium nitrite for 2 h.
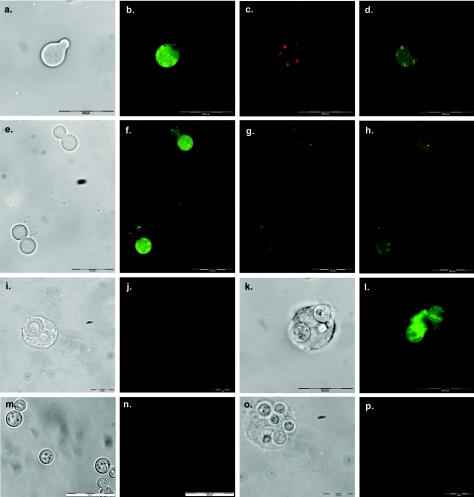
By fusing yeast enhanced green fluorescent protein (GFP) (GenBank accession U73901) to the 3′ end of the thioredoxin reductase gene, we are able to show localization and expression of this enzyme in H99. Control wild-type cells not expressing GFP show a slight ring of autofluorescence around the cell surface (Fig. 2m to n). During logarithmic growth in rich medium, Trr1 is localized throughout the cell (Fig. 2a to h). It is preferentially localized to the mitochondria, but is also seen in the cytoplasm. In budding cells, Trr1 is more generally localized in the mother cell but is seen in the mitochondria in both mother and daughter cells. As C. neoformans, unlike S. cerevisiae, does not encode a separate mitochondrial thioredoxin system, it appears that the single thioredoxin reductase enzyme functions as both cytoplasmic and mitochondrial forms. By incubating this strain of H99 expressing the Trr1-GFP fusion protein with macrophages, we can also show that Trr1 is expressed when C. neoformans is phagocytosed (Fig. 2i to l) whereas wild-type H99 that has been phagocytosed by macrophages shows minimal fluorescence (Fig. 2o to p). This helps support the hypothesis that thioredoxin reductase is important for the virulence of this fungal pathogen.
FIG. 2.
Localization and expression of Trr1-GFP fusion protein in H99. Phase-contrast results (panels a and e), Trr1-GFP localization (b and f), mitochondrial localization using 1 μM MitoTracker (panels c and g), and merged images of panels b and c (panel d) or panels f and g (panel h) are shown. The results of expression of Trr1-GFP when incubated with RAW macrophages as determined by phase contrast (panels i and k) and Trr1-GFP expression (panels j and l) are shown. The results obtained with wild-type control cells alone (panels m and n) and in macrophages (panels o and p) with minimal autofluorescence are shown.
Multiple attempts were made to delete the TRR1 gene from H99, but double-homologous recombination was not achieved. After a few hundred isolates were screened, we hypothesized that TRR1 was essential for vegetative growth. To reduce the expression of TRR1, we chose to replace the endogenous promoter of this gene with an inducible copper transporter promoter. The promoter we used is an enhanced version of the CTR4 promoter described previously as pCTR4-2 (13). This promoter results in 78-fold-induced expression in the absence of copper in the H99 background (13). Our promoter replacement construct consists of 858 bp of genomic sequence upstream of the TRR1 promoter, the G418 resistance marker, the enhanced copper promoter, and 994 bp of TRR1 genomic sequence. Homologous recombination of this promoter replacement construct results in the deletion of 280 bp upstream of the TRR1 gene and the insertion of the 2.4-kb resistance marker and inducible promoter. During the transformation of this construct and the passaging of the transformants, all media contained 200 μM BCS (bathocuproninedisulfonic acid) to keep the promoter induced.
After obtaining stable transformants, we screened for homologous recombination of the promoter replacement construct by PCR. The resulting strain, PTRR1::PCTR4-2, was then grown in the presence of copper to reduce the expression of the TRR1 gene. By use of real-time PCR, we detected a more than 23-fold reduction in TRR1 expression after a 2-h treatment with 25 μM CuSO4 (data not shown). As shown in Fig. 3, PTRR1::PCTR4-2 grows on yeast extract-peptone-dextrose (YPD) with 200 μM BCS just slightly slower than wild-type H99 whereas it is unable to survive on YPD with 25 μM CuSO4. This suggests that thioredoxin reductase is necessary for the viability of C. neoformans. This result is not surprising, since thioredoxin reductase has been shown to be essential for erythrocytic stages in Plasmodium falciparum (8) and appears to be essential for growth in S. aureus (19). But this is an important finding, since thioredoxin reductase has not been shown to be essential in any fungal organism or in any eukaryote harboring the low-molecular-weight isoform.
FIG. 3.
Growth of H99 and strain with promoter replacement in the absence and presence of copper. Five 10-fold dilutions were plated on YPD plates with the copper chelater, BCS, or copper sulfate.
The S. cerevisiae genome encodes a full mitochondrial thioredoxin system in addition to its cytoplasmic system (15), which could help explain why Trr1 is not essential in yeast. In addition to its extreme growth defects, the S. cerevisiae trr1Δ mutant is very sensitive to H2O2 and high temperatures and has a methionine auxotrophy (10, 14). The thioredoxin reductase from P. falciparum is of the high-molecular-weight form but is still thought to be a good drug target on the basis of slight differences in the active site for which mechanisms of specific inactivation have been shown (2). Since its structure is highly conserved across species and since most pathogenic microorganisms, including other fungi (9), encode the low-molecular-weight isoform, thioredoxin reductase would make an excellent drug target when its importance to viability is also conserved.
Acknowledgments
We thank Tamara Doering and John Perfect for providing plasmids. We also thank the C. neoformans H99 sequencing project, Duke Center for Genome Technology (http://cgt.genetics.duke.edu), the Broad Institute (www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans), and the Genome Sequence Centre, British Columbia Cancer Research Centre (http://www.bcgsc.bc.ca/) as well as the C. neoformans serotype D Genome Project, Stanford Genome Technology Center, funded by the National Institute of Allergy and Infectious Diseases-National Institutes of Health under cooperative agreement U01 AI47087, and The Institute for Genomic Research, funded by the National Institute of Allergy and Infectious Diseases-National Institutes of Health under cooperative agreement U01 AI48594.
This work was supported by an American Heart Association fellowship to T.A.M. and by National Institute of Allergy and Infectious Diseases-National Institutes of Health grants RO1-AI051209 and RO1-AI50184 to J.K.L.
REFERENCES
- 1.Arner, E.S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267:6102-6109. [DOI] [PubMed] [Google Scholar]
- 2.Davioud-Charvet, E., M. J. McLeish, D. M. Veine, D. Giegel, L. D. Arscott, A. D. Andricopulo, K. Becker, S. Muller, R. H. Schirmer, C. H. Williams, and G. L. Kenyon. 2003. Mechanism-based inactivation of thioredoxin reductase from Plasmodium falciparum by Mannich bases. Implication for cytotoxicity. Biochemistry 42:13319-13330. [DOI] [PubMed] [Google Scholar]
- 3.Enjalbert, B., A. Nantel, and M. Whiteway. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fradin, C., M. Kretschmar, T. Nichterlein, C. Gaillardin, C. D'Enfert, and B. Hube. 2003. Stage-specific gene expression of Candida albicans in human blood. Mol. Microbiol. 47:1523-1543. [DOI] [PubMed] [Google Scholar]
- 5.Godon, C., G. Lagniel, J. Lee, J. M. Buhler, S. Kieffer, M. Perrot, H. Boucherie, M. B. Toledano, and J. Labarre. 1998. The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem. 273:22480-22489. [DOI] [PubMed] [Google Scholar]
- 6.Hirt, R. P., S. Muller, T. M. Embley, and G. H. Coombs. 2002. The diversity and evolution of thioredoxin reductase: new perspectives. Trends Parasitol. 18:302-308. [DOI] [PubMed] [Google Scholar]
- 7.Hong, S. M., H. W. Lim, I. H. Kim, K. Kim, E. H. Park, and C. J. Lim. 2004. Stress-dependent regulation of the gene encoding thioredoxin reductase from the fission yeast. FEMS Microbiol. Lett. 234:379-385. [DOI] [PubMed] [Google Scholar]
- 8.Krnajski, Z., T. W. Gilberger, R. D. Walter, A. F. Cowman, and S. Muller. 2002. Thioredoxin reductase is essential for the survival of Plasmodium falciparum erythrocytic stages. J. Biol. Chem. 277:25970-25975. [DOI] [PubMed] [Google Scholar]
- 9.Kutty, G., S. N. Huang, and J. A. Kovacs. 2003. Characterization of thioredoxin reductase genes (trr1) from Pneumocystis carinii and Pneumocystis jiroveci. Gene 310:175-183. [DOI] [PubMed] [Google Scholar]
- 10.Machado, A. K., B. A. Morgan, and G. F. Merrill. 1997. Thioredoxin reductase-dependent inhibition of MCB cell cycle box activity in Saccharomyces cerevisiae. J. Biol. Chem. 272:17045-17054. [DOI] [PubMed] [Google Scholar]
- 11.Missall, T. A., M. E. Pusateri, and J. K. Lodge. 2004. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 51:1447-1458. [DOI] [PubMed] [Google Scholar]
- 12.Mustacich, D., and G. Powis. 2000. Thioredoxin reductase. Biochem. J. 346:1-8. [PMC free article] [PubMed] [Google Scholar]
- 13.Ory, J. J., C. L. Griffith, and T. L. Doering. 2004. An efficiently regulated promoter system for Cryptococcus neoformans utilizing the CTR4 promoter. Yeast 21:919-926. [DOI] [PubMed] [Google Scholar]
- 14.Pearson, G. D., and G. F. Merrill. 1998. Deletion of the Saccharomyces cerevisiae TRR1 gene encoding thioredoxin reductase inhibits p53-dependent reporter gene expression. J. Biol. Chem. 273:5431-5434. [DOI] [PubMed] [Google Scholar]
- 15.Pedrajas, J. R., E. Kosmidou, A. Miranda-Vizuette, J. A. Gustafsson, A. P. H. Wright, and G. Spyrou. 1999. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J. Biol. Chem. 274:6366-6373. [DOI] [PubMed] [Google Scholar]
- 16.Salmon, T. B., B. A. Evert, B. Song, and P. W. Doetsch. 2004. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 32:3712-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandalova, T., L. Zhong, Y. Lindquist, A. Holmgren, and G. Schneider. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc. Natl. Acad. Sci. USA 98:9533-9538. [DOI] [PMC free article] [PubMed]
- 18.Tarrio, N., S. D. Prado, M. E. Cerdan, and M. I. G. Siso. 2004. Isolation and characterization of two nuclear genes encoding glutathione and thioredoxin reductases from the yeast Kluyveromyces lactis. Biochim. Biophys. Acta 1678:170-175. [DOI] [PubMed] [Google Scholar]
- 19.Uziel, O., I. Borovok, R. Schreiber, G. Cohen, and Y. Aharonowitz. 2004. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]