Abstract
CRISPR/Cas9 systems have been advanced as promising tools in the HIV eradication armamentarium for sequence-specific disruption or latency reversal. Enthusiasm is balanced by concerns about off-target host genome modification and effects on HIV evolution. In the chronically HIV-1-infected U1 promonocytic latency model, we have confirmed stimulation of HIV-1 production by a mutant Cas9-transcriptional activator and guide RNAs with two guide RNAs apparently more potent than one. However, significant increases were also observed in the absence of guide RNAs. We encourage continued careful evaluation of non-sequence-specific and off-target effects of Cas9-mediated approaches.
Introduction
The first use of the CRISPR/Cas9 system to disrupt HIV-1 proviral DNA was reported in 2013[1]. These results have since been validated in vitro[2] and in a rodent model[3]. Enthusiasm for CRISPR/Cas9-based HIV-1 eradication approaches arises in part from sequence-specificity and, in contrast with so-called “shock-and-kill” treatments, potential avoidance of cell activation and toxicity. However, off-target activity of Cas9 has been reported. There are varying estimates of the prevalence of off-target effects, and attempts have been made minimize these effects through genetic engineering of Cas proteins[4][5][6]. Nevertheless, in the HIV-1 patient, the gene editing system would have to be introduced into a large number of cells to reduce the size of known HIV-1 reservoirs; even a vanishingly small percentage of off-target activity could result in potentially dangerous genomic alterations. Viral evolution in response to Cas9 has also raised some concerns[7][8].
An alternative application of CRISPR/Cas9 in HIV-1 studies seeks to achieve the opposite goal: upregulation of HIV expression instead of disruption or excision. In this approach, a nuclease-mutated Cas9 (dead Cas9 or dCas9) fused to a transcriptional activator domain[9][10] reverses HIV-1 latency. Instead of cutting genomic DNA, the dCas9 transcriptional activator stimulates HIV-1 transcription, resulting in increased display of viral products and possible elimination of HIV-1-harboring cells by the immune system. The feasibility of this approach has been demonstrated in vitro by several groups[11][12][13].
Objective
We evaluated the CRISPR/Cas9-activator HIV-1 transcriptional activation approach in the U1 HIV-1 latency model. Our goals were to determine the degree of reactivation as measured by virus release as well as the degree of non-specific activation in the absence of guide RNAs.
Results & Discussion
Cas9-transcriptional activator-associated HIV-1 transcriptional activation was observed in the U1 promonocytic latency model (U937 derivative with two integrated copies of HIV-1 provirus[14]) even in the absence of added guide RNAs (Figure 1). 48 hours after a Cas9-VP160 fusion protein/gRNA dual expression plasmid (Addgene #48240) was electroporated into the U1 promonocytic line, two HIV-1 gRNAs, previously described and designated T5 and T6[1], were associated separately with a significant increase in HIV-1 p24 release (by a mean 4.7- and 4.8-fold, respectively). When plasmids expressing the two gRNAs were introduced together, there was a relatively higher 5.8-fold increase (p <0.001). However, even the parent plasmid without a guide RNA sequence effected a significant increase of, on average, 3.5-fold (p <0.01). There was no difference between baseline of no treatment and mock electroporation (no plasmid). Treatment with a positive control, nonspecific activator (Tumor necrosis factor-alpha, TNFalpha), resulted in approximately 9-fold activation compared with Cas9-Vp160 alone. In contrast, electroporation of a GFP-containing DNA vector had no effect on HIV production.
Figure 1.
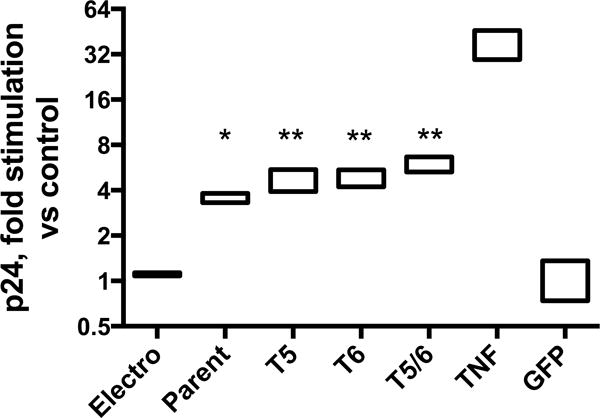
Conclusions
We confirm that HIV-1 LTR-specific guide RNAs and Cas9-VP160 can stimulate HIV-1 production, here, in the U1 promonocytic latency model. Significant but lesser upregulation was observed with Cas9-VP160 in the absence of guide RNAs, but not mock electroporation, suggesting that mutant Cas9-transcriptional activator can achieve independent upregulation of HIV-1 production.
Limitations
This report focuses solely on HIV-1 release from U1 cells in vitro as measured by HIV-1 protein release. We did not assess HIV-1 transcription directly, nor do we report here the effects of the transcriptional activator on host transcripts. This report involves a single HIV-1 cell culture latency model, the U937 derivative known as U1. We do not address T-cells, thought to be the major reservoir of latent HIV in the body. Possibly, our results are applicable only to the U1 model.
Conjectures
These encouraging, if modest, results should be viewed as confirmatory of other findings, but with a potentially cautionary element. The Cas9-transcriptional activator system has strong potential for improvement as well as low but not null likelihood of off-target effects. Improvements could be made by use of higher fidelity Cas9 to reduce off-target effects and more potent Cas9-associated transcriptional activator domain(s) to increase activation[15]; use of more than two HIV-1 specific gRNAs; further optimization of gRNA sequences by performing careful screens (as for example in[12]); simultaneous gRNA-mediated upregulation of host genes (e.g., TRBP[16]) to enhance HIV-1 transcription, protein production, or viral release; and with an eye towards clinical translation, simultaneous upregulation of retroviral restriction factor production for added protection of uninfected cells against any released virus.
Methods
Cas9-Vp160-encoding plasmid pAC154-160Dual (Jaenisch lab, purchased from Addgene, #48240) with or without HIV-1-specific gRNAs T5 and T6 (sequences from[1], or a GFP-containing DNA vector were electroporated into U1 cells (AIDS Reagent) using a Bio-Rad Gene Pulser II. HIV-1 p24 production was measured by ELISA (PerkinElmer). Results were normalized by viable cell count (Muse, Merck Millipore), although introduction of plasmid did not have a significant effect on viability compared with mock electroporation. Human recombinant tumor necrosis factor-alpha (TNFalpha, Sigma Aldrich) was introduced at 10 ng/mL.
Statistical analysis was done using Prism version 7 for Mac OS X (GraphPad Software, Inc.). One-way ANOVA was performed followed by Dunnett’s multiple comparisons test for individual comparisons (Supplementary data).
Supplementary Material
Acknowledgments
Funding statement
The authors acknowledge support from T32GM008752 (Cellular and Molecular Medicine training grant supporting BM) and seed funds from the Department of Molecular and Comparative Pathobiology (to KWW).
Footnotes
Ethics statement
Not applicable.
No fraudulence is committed in performing these experiments or during processing of the data. We understand that in the case of fraudulence, the study can be retracted by Matters.
Contributor Information
Veronica Kim, Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine.
Brian M Mears, Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine.
Bonita H Powell, Molecular and Comparative Pathobiology, The Johns Hopkins University School of Medicine.
References
- 1.Ebina H, et al. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3 doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu W, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proceedings of the National Academy of Sciences. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaminski R, et al. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Therapy. 2016;23:690–695. doi: 10.1038/gt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, et al. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular Therapy-Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yee JK. Off-target effects of engineered nucleases. The FEBS Journal. 2016;283:3239–3248. doi: 10.1111/febs.13760. [DOI] [PubMed] [Google Scholar]
- 6.O’Geen H, et al. How specific is CRISPR/Cas9 really? Current Opinion in Chemical Biology. 2015;29:72–78. doi: 10.1016/j.cbpa.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang C, et al. CRISPR/Cas9: a double-edged sword when used to combat HIV infection. Retrovirology. 2016;13 doi: 10.1186/s12977-016-0270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueda S, et al. Anti-HIV-1 potency of the CRISPR/Cas9 system insufficient to otency of the CRISPR/Cas9 system insufficient to fully inhibit viral replication. Microbiology and Immunology. 2016;60:483–496. doi: 10.1111/1348-0421.12395. [DOI] [PubMed] [Google Scholar]
- 9.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature Biotechnology. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng A, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci Rep. 2015;5:16277. doi: 10.1038/srep16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saayman S, et al. Potent and Targeted Activation of Latent HIV-1 Using the CRISPR/dCas9 Activator Complex. Mol Ther. 2015;24:488–498. doi: 10.1038/mt.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bialek J, et al. Targeted HIV-1 Latency Reversal Using CRISPR/Cas9-Derived Transcriptional Activator Systems. PLOS ONE. 2016;11:e0158294. doi: 10.1371/journal.pone.0158294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folks T, et al. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238:800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- 15.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nature Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanghvi VR, Steel LF. The Cellular TAR RNA Binding Protein, TRBP, Promotes HIV-1 Replication Primarily by Inhibiting the Activation of Double-Stranded RNA-Dependent Kinase PKR. Journal of Virology. 2011;85:12614–12621. doi: 10.1128/jvi.05240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


