Cis-trans isomerization of the Xaa-Pro bond is an important step in protein folding and a critical determinant of protein structure (27). Peptidyl-prolyl cis-trans isomerases (PPIases or rotamases) belong to three unrelated families: FK506-binding proteins (FKBPs), cyclophilins, and parvulins. The first two families are collectively known as immunophilins because of their ability to tightly bind immunosuppressive drugs: cyclophilins bind cyclosporine A, while FKBPs bind the molecules rapamycin and FK506. In lymphocytes, the complexes formed by the drugs and their receptors ultimately cause the immunosuppression effect, either by inhibiting the calcineurin-dependent activation of gene expression or by interfering with interleukin-dependent signal transduction. Surprisingly, vascular plants harbor a vast array of immunophilins (52 in Arabidopsis [19]), some of which combine an FKBP or a cyclophilin domain with other protein domains (complex immunophilins). Complex immunophilins are also found in mammalian cells. This suggests that immunophilins play an important role in signal transduction in the plant cell as well (3). Indeed, Arabidopsis, like animals, contains a TOR (target of rapamycin) protein kinase that is expressed in proliferating tissue and binds rapamycin in the presence of FKBP (28). Thus, plants could use immunophilins to regulate protein translation according to the energy status of the cell. Little is known about the individual functions of most immunophilins, in particular whether they act in the form of receptor-ligand complexes. Cyclosporine binding has been demonstrated for two cyanobacterium-like Plasmodium cyclophilins (14). In Arabidopsis, mutations in three FKBPs have been found to cause developmental defects (15, 22, 35) or dominant male sterility (22). Finally, a role in auxin signaling has been attributed to a parvulin-type PPIase acting on specific substrate proteins (5).
The unicellular green alga Chlamydomonas reinhardtii appears as a model of choice for the study of plant FKBPs and cyclophilins. Its single-cell type can be readily exposed to controlled concentrations of immunosuppressive drugs, and a powerful genetic system has been developed by decades of work on photosynthesis, organelle biogenesis, flagellar function, and other basic cellular processes (18). A Chlamydomonas cytosolic cyclophilin has been identified and shown to be induced by low-CO2 conditions (32). This gene is repressed during sulfur starvation in a SAC1-independent manner, together with two chloroplast cyclophilins (37), suggesting a link with cell growth. And a homologue of the FKBP12-interacting protein AtFIP37 (8) is also encoded in the Chlamydomonas genome. A comprehensive description of Chlamydomonas immunophilins thus appears desirable.
With the recent release by the Joint Genome Institute (JGI) of a draft nuclear genome sequence, Chlamydomonas has fully entered the genomics era (16). Among the primary goals of genomics, and one of its toughest challenges, is the comprehensive description of gene content. To delineate transcripts for protein-coding genes along the genome, the Joint Genome Institute has used a variety of algorithms relying either mostly on homology (Genewise [2]) or on coding capacity (greenGenie [24]) or expression signals (FgeneSH [31]). For each locus, the preferred model is chosen and refined, and its untranslated regions (UTR) are determined, making use of expressed sequence tag (EST) data.
In the context of a draft sequence, such as version 2.0 of the Chlamydomonas genome, it is expected that the accuracy of gene prediction will be limited by a variety of factors, including but not limited to the following. (i) Incomplete coverage of the genome: 2 to 5% of ESTs, depending on libraries, do not map onto the genome (O. Vallon and C. Hauser, unpublished results), an indication of the proportion of genes that have not yet been hit by genome sequencing. (ii) Sequence gaps within or at the ends of genes, hiding some of the information necessary to predict the gene correctly. Wisely, the programs have been allowed to build models across sequence gaps, even to incorporate them within an exon. While allowing a better coverage, this will inevitably result in ill-predicted gene structures, fusion of neighboring genes, and other problems. (iii) Assembly artifacts, which are difficult to avoid in a whole-genome shotgun sequencing approach. Repeated sequences are an obvious source of such artifacts, as are chimeric DNA clones. This can result in various fragments of a gene being found in different scaffolds. (iv) Limitations in the algorithms themselves: the programs use regular properties of transcribed sequences, of transcription, splicing and termination signals, etc., which, although established statistically and tested rigorously, may not always apply. A case in point is alternative splicing, whereby the molecular machinery of splicing interprets in multiple ways the sequence information in the pre-mRNA, whereas gene prediction programs will only choose the most likely intron/exon structure.
Thus, the Chlamydomonas genomics project, just like any other, must at some point face the question of the reliability and completeness of its gene model data set. This is crucial, since this data set is to serve as a basis for most of the postgenomic analysis. In Drosophila, a large-scale experiment has been devised to confront gene prediction programs and experimental approaches (1), based on high-resolution gene mapping in a well-known region of the genome. In Chlamydomonas, the early sequencing of a large stretch of genomic DNA has allowed benchmarking of greenGenie and a preliminary assessment of gene content (24).
It is not the scope of this paper to provide a complete analysis of a particular fragment of the genome. Rather, I will try to describe the Chlamydomonas instantiation of two well-known medium-size gene families, cyclophilins and FK506-binding proteins. Both show a high degree of sequence conservation across phyla, so that simple BLAST searches are expected to provide an exhaustive identification of all family members in Chlamydomonas. By comparing Chlamydomonas immunophilins with those of vascular plants, we can hope to identify which isoforms could be involved in specific aspects of signal transduction and development, inasmuch as they will differ between a multicellular organism and a unicellular organism. We can also shed light on the evolution of gene families with isoforms directed to many intracellular compartments.
The aim of this paper is therefore threefold: to describe Chlamydomonas immunophilins and parvulins, an important class of proteins that can become the subject of experimentation with this microbe; to analyze phylogenetic relationships between family members, in particular identifying early and late gene duplication events that have given rise to the present-day diversity; and to examine the validity of Chlamydomonas gene models whenever possible by comparison either with Chlamydomonas ESTs or with sequences of orthologues in other organisms. This last perspective, although in no way a quantitative assessment of gene prediction in Chlamydomonas, can help identify common artifacts in the current genomic data set. It can thus serve as a guide for those who want to use this information in the study of their favorite genes. Our hope is that it can also help improve gene models in future versions of the Chlamydomonas genome.
METHODOLOGY
Chlamydomonas genes were identified on the v2.0 draft genome sequence available at http://genome.jgi-psf.org/chlre2/chlre2.home.html, with the search interface or with TBlastN, using Arabidopsis proteins as the query. This information was compared with that derived from the various EST assemblies (available at http://www.chlamy.org/search.html), and gene models were corrected when discrepant with reliable EST data (excluding genomic contaminants). Other immunophilin sequences were retrieved from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=protein) and MIPS (Munich Information Center for Protein Sequences [http://mips.gsf.de/proj/thal/db/index.html]) databases. Alignment of the Chlamydomonas proteins with the homologues from Arabidopsis or other organisms was done with CLUSTALW, using the Blosum62 matrix. The alignment was optimized manually in BIOEDIT, using protein domain boundaries determined by SMART (http://smart.embl-heidelberg.de/). The Chlamydomonas models which did not align satisfactorily with their homologues over their entire length were reexamined; 5′ ends and intron/exon boundaries were changed when necessary so as to optimize protein alignment, unless they had experimental support. The modified protein sequences were entered in the JGI genome database, in the model notes of the corresponding gene model. The whole sets of sequences used and their final alignments are available as supplemental material in FASTAformat.
Phylogenetic trees were built using the optimized alignments after trimming to the conserved domain, i.e., excluding the N-terminal targeting peptide and the unique domains. This was judged preferable, since the part of the alignment covering the N-terminal transit peptides (TPs) and additional domains was not meaningful. The neighbor-joining method, run at http://bioweb.pasteur.fr/seqanal/interfaces/clustalw.html#trees,was used with Kimura's correction and bootstrapping (n = 1,000). Prediction of intracellular localization made use of TargetP (http://www.cbs.dtu.dk/services/TargetP/). Note that Chlamydomonas chloroplast TPs are different from those of higher plants (11). Thus, the indication of a chloroplast or mitochondrial location was only taken as indicative of targeting to either of these organelles.
DIVERSITY OF IMMUNOPHILINS
The multiplicity of PPIases in eukaryotes and in particular in plants is an evolutionary and functional puzzle. Most cellular compartments possess not only PPIases of the different types but also multiple members of each. For example, the thylakoid lumen of Arabidopsis is proposed to harbor no fewer than 10 FKBPs and 5 cyclophilins, while the cytosol has 4 and 12, respectively, of each type, plus two parvulins. Since physiological functions are dependent upon the environment and interactions of the protein, it is of interest to examine whether phylogenetic trees are congruent with subcellular localization. An effort was made to include in the analysis isoforms from the red alga Cyanidioschyzon merolae (25) and a cyanobacterium (Synechocystis sp. strain PCC6803) in hopes of stressing ancestral relationships between orthologues and paralogues. The diverse functions of immunophilins must have been acquired at different times in their phylogeny, and different plant lineages may have different complements of immunophilins. The Cyanidioschyzon genome, however, with its seven FKBPs and four cyclophilins, is unusually reduced in size and may not be representative of the red algal lineage. The diatom Thalasisosira pseudonana, a complex alga resulting from secondary endosymbiosis of a red alga, has at least 16 FKBPs and 8 cyclophilins (http://genome.jgi-psf.org/thaps1/thaps1.home.html).
FK506-BINDING PROTEINS
There are 26 gene models in Chlamydomonas with homology to FKBP-type PPIases (Table 1). Altogether, they define 23 genes, compared to 23 in Arabidopsis, 7 in C. merolae, and 2 in Synechocystis. The Chlamydomonas proteins, sometimes after slight corrections of the sequence, were aligned to those from other photosynthetic organisms, plus human FKBP1 (see supplemental Fig. S1). From the deduced phylogenetic tree, (Fig. 1), a clear relation of orthology could be deduced in many cases between Chlamydomonas and Arabidopsis proteins. This indicates that the diversity of FKBPs was already established in their last common ancestor, probably close to the root of the “green” lineage of plants. Whenever possible, Chlamydomonas genes were given names based on the Arabidopsis nomenclature (19), except that the root FKB was used instead of FKBP (to conform with the three-letter preference for Chlamydomonas). When no Arabidopsis orthologue was obvious, additional numbers were coined, with no attempt to make them coincide with the molecular weight. Some of the Chlamydomonas proteins are probably inactive as PPIases. Table 2 shows the conservation pattern for those residues which have been implicated in drug-binding and rotamase activity in human FKBP12 (21). While positions 55, 88, and 92 appear nondiscriminatory, the others may be used to distinguish active from inactive isoforms, in particular positions 56 and 57, which form a β-sheet with the substrate. Not surprisingly, the Chlamydomonas and Arabidopsis orthologues generally show the same pattern of conservation of these residues.
TABLE 1.
Immunophilin and parvulin genes in C. reinhardtii
| Protein group and gene name | Gene model(s) | Subcellular localizationa | Notes, additional domains | Note(s) on gene model | EST support (Chlre2 ACEG) |
|---|---|---|---|---|---|
| FK506-binding proteins | |||||
| FKB12 | C_230098 | Cytosol | No issue | Complete, >63 EST (23.3.2.11) | |
| FKB15-1 | C_480074 | Secreted (23) | Linked to FKBP15-2 and 15-4 | No issue | Complete, >45 EST (48.4.3.12) |
| FKB15-2 | C_480037 | Secreted (22) | In tandem with FKB15-4 | Sequence gap; sequence corrected based on EST: modify exon 1, remove exon 4 | Partial (48.37.1.5) |
| FKB15-4 | C_480038 | Secreted (22) | In tandem with FKB15-2 | No issue | Complete (48.36.2.31) |
| FKB16-1 | C_440012 | TL (56 + RR) | No issue | Complete (44.12.1.0) | |
| FKB16-2 | C_580070 | TL (26 + RR) + others? | In tandem with FKB16-5 | Three splicing variants: -A (main, lumen), -B (stroma), -C (mitochondrion?) | Complete (A: 58.1.3.12; B:AW720777; C: 58.1.2.11) |
| FKB16-3 | C_380115 + C_18290001 | TL? | Represented on 2 scaffolds due to misassembly; sequence incomplete | Complete (38.11.1.0) | |
| FKB16-4 | C_1080039 | TL (27 + RR) | 3′ end is also in left arm of scaff_3031 | Complete (108.1.18.21) | |
| FKB16-5 | C_580071 | M?(23) | In tandem with FKB16-2 | No issue | Complete (58.6.1.0) |
| FKB16-6 | C_1850018 | TL (51 + RR) | Sequence gap | Partial (185.5.2.51; 185.5.1.31) | |
| FKB16-7 | C_3330006 | ? | Overlaps with upstream gene in tandem | Main isoform has shorter exon 7; C_3330006 describes a splicing variant with 20 AA insertion between 5th and 6th beta-strand | Complete (333.1.7.12) |
| FKB16-8 | C_140146 | TL (? + RR) | Covers 2 other gene models, both unsupported by EST | Complete (14.13.1.0) | |
| FKB16-9 | C_320049 | TL (36 + RR) | Sequence corrected: remove intron 2 | Partial (32.69.2.51) | |
| FKB17-1 | C_220072 | TL (? + RR) | No issue | Complete (22.105.1.0) | |
| FKB17-2 | C_140132 | TL (membrane-anchored?) | No issue | Complete (14.17.1.0) | |
| FKB18 | C_1630014 | TL (29 + RR) | No issue | Complete (163.1.5.12) | |
| FKB19 | C_910042 | TL (? + RR) | No issue | Complete (91.10.1.0) | |
| FKB20-2 | C_210108 | TL (31 + RR) | 3′ overlaps largely with convergent gene C_210018 | No issue | Complete (21.2.2.11) |
| FKB42 | C_680056 | Cytosol | FKBP; TPR; calmodulin-binding site; transmembrane region; unknown central domain; 2 PQQ (WD40-like); leucine repeat | Sequence gap | Partial (68.20.3.11; 68.18.1.0) |
| FKB53 | C_240139 | Cytosol | N-terminal domain rich in acidic residues; FKBP | Sequence corrected: lengthen exon7 3′; additional His-rich loop before helix 1 due to insertion of CCAC(C/A)A repeats | No |
| FKB62 | C_2440003 + C_20610001 | Cytosol | 3 FKBP; TPR | Split between 2 scaffolds due to misassembly; sequence incomplete | Partial (244.3.1.5) |
| FKB99 | C_280158 | Nucleus | 3 trans-membrane spans; 2 or 3 FKBP; TPR; calmodulin-binding motif; nuclear localization signal; possibly a fibrinoin C-terminal domain | Sequence gaps impact model; possibly a fusion of several genes | Partial (28.9.1.5, 28.52.1.5, 28.117.1.0) |
| TIG1 | C_4210001 + C_4210002 | Stroma | Trigger factor; FKBP; N- and C-terminal domains involved in ribosome binding | Sequence gap; corrected based on EST | Partial (421.1.3.52; 421.1.1.51; 421.1.4.32) |
| Cyclophilins | |||||
| CYN16 | C_240021 | Cytosol | Sequence corrected: 4th exon overlooked due to noncanonical splice site | No | |
| CYN17 | C_20323 | Cytosol | Alternative splicing: removes intron 1 | Complete (2.14.3.12) | |
| CYN18-1 | C_20046 | Cytosol | Sequence lengthened at 5′ end | No | |
| CYN18-2 | C_1010007 | Cytosol | No issue | Complete (101.31.1.0) | |
| CYN19-1 | C_250142 | Cytosol | Possibly also a C-terminal GCIP domain (cyclin D-interacting protein) | Sequence corrected: extend 5′ end, probable sequence error in genome; could be a fusion with downstream gene (sequence gap) | Partial:CF557930 |
| CYN19-2 | C_3230001 | Cytosol | Described as CYP1 (AF052206), induced in low-CO2 conditions (32) and sulfur starvation (37) | No issue | Complete (323.5.1.0) |
| CYN19-3 | C_790042 | Organelle? | Sequence gap shortens 3′ UTR | Complete (79.26.3.11) | |
| CYN20-1 | C_290072 | Secreted | Sequence corrected: extend 5′, wrong computation of exon 7. | Complete (29.14.1.0) | |
| CYN20-2 | C_70215 | TL? | No issue | Complete (7.35.2.11) | |
| CYN20-3 | C_740054 | Stroma | Induced during sulfur starvation (37) | No issue | Complete (74.9.3.11) |
| CYN20-4 | C_990048 | ? | An expressed pseudogene | Neither genome nor EST code for full-length protein | Partial (99.4.1.0) |
| CYN20-5 | C_660007 | Organelle? | Possible alternative splicing (5.132.1.31) | Complete (66.56.2.11) | |
| CYN22 | C_910055 | Cytosolic | Intron 1 is unusually long (1,741 nt) | Complete (91.12.1.0) | |
| CYN23 | C_2260002 | Secreted | 3′ UTR needs shortening; 2 splicing variants with mutually exclusive exon 4 positions (226.9.2.11) | Complete (226.9.3.12) | |
| CYN26-2 | C_460089 | TL | No issue | Complete (46.6.1.0) | |
| CYN28 | C_1220009 | TL | Sequence corrected: exons 1 and 2 missed | Complete (122.27.1.0) | |
| CYN37 | C_90033 | TL? | No issue | Partial (9.112.1.0) | |
| CYN38 | C_30248 + C_30247 | TL | N-terminal domain of unknown function; induced during sulfur starvation (37) | Sequence gap impacts models (5′ truncation, fusion with downstream gene); corrected based on EST | Complete (20021010.2563.1) |
| CYN40 | C_180001 | Cytosol | Cyclophilin, TPR domain | No issue | Partial (18.87.1.5, 18.56.1.5) |
| CYN51 | C_200140 | Organelle? | N-terminal domain of unknown function, cyclophilin | Sequence corrected (gap) | Complete (20.23.1.0) |
| CYN52 | C_1560006 | Secreted | 2 cyclophilin domains; next to CYN53 | Sequence corrected (gap), alternative splice removing exon 2 causes frameshift | Complete (156.16.2.11) |
| CYN53 | C_1560025 | Organelle? | N-terminal domain + 2 cyclophilin domains; next to CYN52 | Sequence corrected (gap, noncanonical splice site) | No |
| CYN57 | C_1280001 | Cytosolic? | C-terminal S/K-R/E RNA-binding domain | No issue | No |
| CYN59 | C_480065 | Nucleus? | cyclophilin, RNP1-RRM RNA-binding, RDG-rich domains | Sequence partially corrected (gap, exon 4 missed) | No |
| CYN65 | C_800078 | Cytosolic | N-terminal with U-box (modified RING Zn finger) | Sequence corrected (5th intron missed) | Partial (80.7.1.0) |
| CYN71 | C_19080001 + C_320104 | Cytosolic? | 3 WD40 repeats, cyclophilin domain | Sequence partially corrected (gaps split gene between 2 scaffolds) | Partial (1112019F10, 1112127B05) |
| Parvulins | |||||
| PIN3 | C_100048 | Organelle? | Similar to AtPIN3: parvulin, rhodanese domain | No issue | Partial (10.79.1.5) |
| PIN4 | C_530020 | Cytosol? | N-terminal FHA (forkhead) domain, binding phosphoproteins | Sequence corrected (5th exon ill predicted) | Complete (20021010.2553.1) |
Parenthetical numbers are lengths of predicted targeting peptides.
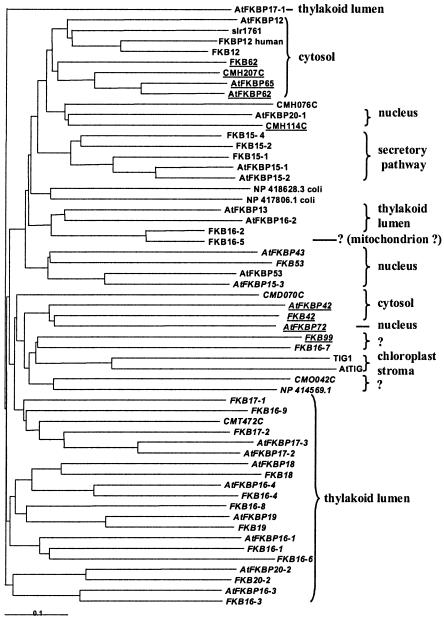
FIG. 1.
Phylogenetic tree of FKBPs. Multidomain FKBPs are underlined. Isoforms that are presumably inactive (Table 2) are in italics. The presumed localization of the mature protein is indicated on the right. Note that nuclear and thylakoid lumen FKBPs arise in various branches of the tree.
TABLE 2.
Conservation of key residues in FKBPsa
| Protein | Residue at aa position:
|
% Conservedb | TargetP prediction | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27 | 37 | 38 | 43 | 47 | 55 | 56 | 57 | 60 | 82 | 83 | 88 | 92 | 100 | |||
| FKBP12_human | Y | F | D | R | F | E | V | I | W | A | Y | H | I | F | 100 | — (2) |
| FKB12 C_230098 | : | : | : | : | : | : | : | : | : | : | : | I | — | : | 86 | — (2) |
| FKB15-1 C_480074 | : | : | : | : | : | S | : | : | : | G | : | G | K | : | 71 | S (2): 23 |
| FKB15-2 C_480037 | : | : | : | : | I | Q | : | : | : | : | : | G | — | : | 71 | S (1): 22 |
| FKB15-4 C_480038 | : | : | : | : | I | Q | : | : | : | G | : | G | — | : | 64 | S (1): 22 |
| FKB16-1 C_440012 | : | I | Y | F | V | : | L | : | L | G | : | G | P | : | 36 | C (4): 56 |
| FKB16-2A C_580070 | : | : | : | : | L | Q | : | : | : | G | : | G | V | : | 64 | M (3): 26 |
| FKB16-3 C_380115 | : | : | : | K | Y | Q | : | : | L | : | F | A | R | : | 50 | M (5): 46 |
| FKB16-4 C_1080039 | : | M | T | G | L | G | T | L | L | G | : | L | E | I | 14 | C (5): 27 |
| FKB16-5 C_580071 | : | : | : | : | L | Q | : | : | : | G | : | G | V | : | 64 | M (3): 23 |
| FKB16-6 | : | X | X | X | X | S | L | P | V | G | W | K | A | : | >14 | C (3): 51 |
| FKB16-7 C_3330006 | F | L | Q | L | L | R | L | F | F | G | L | V | A | I | 0 | C (3): 82 |
| FKB16-8 C_140146 | : | M | T | G | Y | N | : | L | L | : | : | K | E | L | 29 | C (1): 78 |
| FKB16-9 C_320049 | V | : | L | E | L | Y | : | T | L | G | : | K | R | Y | 21 | C (2): 36 |
| FKB17-1 C_220072 | : | : | : | Y | A | — | P | : | I | G | F | A | A | Y | 29 | M (3): 70 |
| FKB17-2 C_140132 | : | : | E | : | I | P | F | T | N | G | F | G | R | Y | 21 | M (4): 113 |
| FKB18 C 1630014 | : | V | S | G | Y | K | P | P | L | G | : | G | — | L | 14 | C (4): 29 |
| FKB19 C_910042 | W | : | E | K | Y | K | : | : | F | G | : | N | L | : | 36 | M (4): 12 |
| FKB20-2 C_210108 | : | I | : | Q | A | G | M | : | F | G | P | G | F | : | 29 | C (4): 31 |
| FKB42 C_680056 | : | M | : | T | V | A | Q | E | L | G | : | F | C | Y | 21 | — (2) |
| FKB53 | : | : | : | G | : | N | H | L | L | : | : | X | X | X | >43 | — (4) |
| FKB62 | : | : | : | : | : | F | E | A | : | : | : | V | — | : | 64 | — (2) |
| FKB99 C_280158 | : | Y | S | G | Y | R | : | P | : | G | : | M | V | — | 29 | — (4) |
| TIG1 C_4210001+2 | G | E | I | G | : | — | — | V | I | D | V | S | K | : | 14 | M (2): 63 |
| AtFKBP12 AT5G64350 | C | : | W | E | : | A | : | : | : | : | : | F | G | : | 57 | — (3) |
| AtFKBP13 AT5G45680 | : | : | : | : | L | : | : | : | : | : | : | K | C | : | 79 | C (3): 14 |
| AtFKBP15-1 AT3G25220 | : | : | : | : | I | Q | : | : | : | G | : | G | K | : | 64 | S (3): 18 |
| AtFKBP15-2 AT5G48580 | : | : | : | : | : | Q | : | : | : | G | : | G | T | : | 71 | S (1): 25 |
| AtFKBP15-3 AT5G05420 | : | : | : | K | Y | K | : | : | L | G | : | G | S | : | 50 | — (3) |
| AtFKBP16-1 AT4G26555 | : | H | S | S | V | D | : | : | L | G | : | T | : | : | 43 | C (1): 71 |
| AtFKBP16-2 AT4G39710 | : | : | : | : | L | K | : | : | L | : | : | S | N | Y | 57 | C (3): 34 |
| AtFKBP16-3 AT2G43560 | : | : | : | K | Y | Q | : | : | L | : | F | S | R | : | 50 | C (2): 36 |
| AtFKBP16-4 AT3G10060 | W | I | : | : | Y | Q | A | : | F | : | : | D | R | : | 43 | C (2): 56 |
| AtFKBP17-1 AT4G19830 | : | : | : | H | : | K | : | : | I | G | : | S | L | : | 57 | C (3): 63 |
| AtFKBP17-2 AT1G18170 | L | V | : | K | L | P | Y | S | L | G | F | G | E | Y | 7 | C (3): 79 |
| AtFKBP17-3 AT1G73655 | V | V | : | K | L | P | Y | S | L | G | F | G | E | Y | 7 | C (3): 28 |
| AtFKBP18 AT1G20810 | F | I | S | A | Y | K | P | P | M | G | : | G | — | L | 7 | C (4): 67 |
| AtFKBP19 AT5G13410 | W | : | E | K | : | : | : | : | F | G | : | D | S | : | 50 | C (1): 29 |
| AtFKBP20-1 AT3G55520 | : | : | : | D | : | S | : | : | : | : | : | G | D | : | 71 | — (2) |
| AtFKBP20-2 AT3G60370 | : | I | : | Q | A | A | L | V | F | G | P | G | F | : | 21 | C (2): 31 |
| AtFKBP42 AT3G21640 | : | : | E | E | I | K | E | L | L | : | : | F | N | Y | 29 | — (2) |
| AtFKBP43 At3g12330 | : | : | : | E | L | N | : | : | L | G | : | G | K | Y | 43 | — (4) |
| AtFKBP53 AT4G25340 | : | : | : | K | : | S | : | : | : | G | : | G | Q | : | 64 | |
| AtFKBP62 AT3G25230 | : | : | : | : | : | Q | : | : | : | : | : | S | — | : | 79 | — (1) |
| AtFKBP65 AT5G48570 | : | : | : | : | : | H | : | : | : | : | : | S | — | : | 79 | — (2) |
| AtFKBP72 AT3G54010 | : | Y | : | N | L | L | : | P | F | : | : | P | G | W | 36 | |
| AtTIG AT5G55220 | E | S | A | G | : | R | L | L | F | K | Q | G | Q | : | 14 | C (3): 26 |
| CMD070C | F | : | : | S | : | N | L | V | V | : | F | S | — | Y | 29 | C (3): 38 |
| CMH076C | : | : | : | : | : | Q | : | : | : | : | : | I | — | : | 79 | M (4): 7 |
| CMH114C | : | : | : | : | : | S | : | : | : | : | : | V | — | : | 79 | C (1): 41 |
| CMH207C | : | : | : | : | : | S | : | : | : | : | : | A | — | : | 79 | M (3): 117 |
| CMO042C | : | I | E | G | V | T | L | P | V | N | F | R | — | Y | 7 | M (4): 11 |
| CMT472C | : | : | : | K | L | Q | M | V | V | G | F | I | — | Y | 21 | C (2): 51 |
| NP_414569.1 coli | F | A | E | N | A | S | L | S | L | : | F | Y | R | : | 14 | — (3) |
| NP_417806.1_coli | : | : | : | : | L | — | : | : | : | : | : | A | G | : | 71 | S (1): 25 |
| NP_418628.3_coli | : | : | : | : | A | — | : | : | : | : | : | R | A | : | 71 | S (4): 27 |
| slr1761 Syn 6803 | : | : | : | : | : | Q | : | : | : | : | : | R | G | : | 79 | M (3): 107 |
| % Conserved | 77 | 65 | 67 | 39 | 35 | 9 | 60 | 60 | 40 | 44 | 72 | 2 | 4 | 65 | ||
IsoformspresumedtobeinactiveasPPlaseareinitalics.IntracellulartargetingpredictedbyTargetPisindicated(C,chloroplast;M,mitochondrion; S, secretory pathway; —, none),togetherwiththelevelofconfidence(1,highest;4,lowest)andthepredictedlengthofthetransitpeptide.“:”indicatesidentitytohumanFKBP12.
With respect to human FKBP12.
The shortest Chlamydomonas FKBP is FKB12. It is the one represented by the largest number of ESTs and also the only single-domain FKBP unambiguously targeted to the cytosol. It is similar to the well-characterized cytosolic FKBP12 that interacts with the mTOR protein kinase required for cell cycle progression (7) and in Arabidopsis with AtFIP37, a phosphatidyl-inositol kinase essential for development (34). In the phylogenetic tree (Fig. 1), it groups with the Arabidopsis and human FKBP12 and the unique Synechocystis FKBP. Interestingly, the Chlamydomonas protein sequence is closer to that of human protein, and shows a better conservation of critical residues, than that of Arabidopsis (Table 2). Branching near this clade are a series of complex FKBPs with TPR repeats, including FKB62, further described below. AtFKBP20-1, like two related red algal proteins, is predicted to be targeted to the nucleus: no Chlamydomonas orthologue can be found in either the genome or the EST database. Chlamydomonas FBK16-7 appears unrelated and has no obvious orthologue in Arabidopsis.
Other single-domain FKBPs include the three isoforms putatively directed to the secretory pathway, FKB15-1, -15-2, and -15-4. They are closely related in sequence, and all linked on scaffold 48; thus, they probably arose from recent gene duplications, obviously distinct from the duplication that gave rise to AtFKBP15-1 and -15-2 in Arabidopsis (19). As a group, secretory pathway FKBPs are characterized by the presence of two conserved Cys residues, already noted in human and yeast FKBP13 (21): they form a disulfide bridge stabilizing the loop crossing region in this particular environment. Interestingly, while the Arabidopsis proteins have C-terminal signals (KNEL and NDEL) that may retain them in the endoplasmic reticulum (ER), as is the case for human FKBP13, the Chlamydomonas proteins lack such signals or the recently proposed CVLF signal (36). They may be secreted, and operate in the cell wall compartment. This is also true of the Chlamydomonas cyclophilins, which raises the question of which protein, if any, is responsible for PPIase activity in the ER lumen.
Eleven FKBPs in Arabidopsis have been shown or predicted to be targeted to the thylakoid lumen and suggested to have common ancestry (19). Based on high sequence conservation with the Arabidopsis orthologue and on the presence of a putative bipartite transit sequence, 11 Chlamydomonas FKBPs can be predicted to localize to the thylakoid lumen as well: they have been called FKB16-1, -16-2, -16-3, -16-4, -16-6, -16-8, -16-9, -17-1, -18, -19, and -20-2. Thus, diversity of lumen-targeted FKBPs is probably an ancient trait in the green lineage. This is in marked contrast with the red algae: only one C. merolae FKBP (CMT472C) branches together with the thylakoid lumen FKBPs of green organisms. I note, though, that the diversification of lumen-targeted FKBPs must have continued after the separation of algae and plants: FKB16-5 is found in tandem with, and is extremely similar to, FKB16-2, while FKB16-1 has FKB16-6 as its closest relative, not AtFKBP16-1. Symmetrically, AtFKBP17-2 and -17-3 are also more closely related to one another than to Chlamydomonas FKB17-2. Strictly speaking, unambiguous orthology can be claimed only between FKB16-3, -16-4, -18, -19, and -20-2 and the Arabidopsis genes of same numbering.
Like their Arabidopsis counterparts (19), all these proteins show a twin-arginine motif typical of proteins translocated via the TAT pathway (two Arg residues followed by a hydrophobic stretch; see supplemental Fig. S1). In general, it is followed by a transit peptidase cleavage site in the form Ala-Xaa-Ala, indicating that the proteins are soluble in the lumen. Why are all the lumenal FKBPs transported by the TAT pathway, which is believed to transport proteins in the folded state? It could be because folding of small FKBPs is a rapid process, occurring before they can be presented to the translocation apparatus. Alternatively, it could be related to the binding of a specific effector, similar to FK506, in the chloroplast stroma, so that the binary complex would be the transported entity. In any event, the question remains of why so many different, sometimes extremely well-conserved FKBP-type PPIases localize to a compartment that harbors only a small fraction of the proteome. I note that among these lumenal FKBPs, only FKB16-2, like the cognate AtFKBP16-2 and AtFKBP13, shows a good conservation of the residues involved in PPIase activity (Table 2). The suggestion that AtFKBP20-2 (with only two critical residues conserved) is involved in isomerization of a critical Pro residue in LHCII (29) may need to be reexamined.
An interesting case is that of FKB17-2, which also appears to be targeted to an organelle and shows a twin-arginine signal, but where the AXA signal peptidase cleavage site is absent. Interestingly, the entire sequence following the two Arg residues is extremely well conserved between FKB17-2 and its two orthologues, AtFKBP17-2 and AtFKBP17-3 (see supplemental Fig. S1), which are predicted to localize to the thylakoid lumen but which also lack a cleavage site. Since sequence conservation in signal peptides is in general very low, this leads us to propose that this region is part of the mature protein. It may constitute a transmembrane helix spanning the thylakoid membrane, similar to that observed in another membrane-anchored TAT pathway substrate, the Rieske protein (10).
In terms of localization, FKB16-2 presents an interesting puzzle. Three distinct splicing variants are documented in the EST data. The main isoform, FKB16-2A, like the Arabidopsis orthologue AtFKBP16-2, has an organellar TP and an RR motif with a cleavage site and hence is probably directed to the thylakoid lumen. But alternative splicing generates another isoform, FKB16-2C, with a deletion of the RR motif. This protein would thus be predicted to reside in the chloroplast stroma. And yet another one, FKB16-2B, has a slightly different N-terminal sequence that could direct it to another location, possibly the mitochondrion.
I note that the N-terminal targeting sequence of FKB16-5 differs markedly from that of its closely related paralogue FKB16-2: it is predicted to be an organellar TP but does not contain a hydrophobic stretch after the two arginines, so that the protein would be predicted to be retained in the chloroplast stroma or in the mitochondrial matrix. In Arabidopsis, no FKBP is predicted to localize to the mitochondrion, where rotamase activity is carried out by two cyclophilins. Since Chlamydomonas has no orthologue for these two mitochondrial cyclophilins (see below), it is tempting to speculate that rotamase activity in the Chlamydomonas mitochondrion is carried out by FKBPs. It could be carried out by FKB16-5 and/or FKB16-2B, which both show a decent conservation of the residues important for rotamase activity (Table 2).
In addition to these simple FKBPs, a series of complex FKBPs can be found in the plant genomes, which combine an FKBP and a TPR domain formed of three tetratricopeptide (TPR) repeats. The latter domain is generally involved in protein-protein interactions, in particular as a binding domain for HSP90 chaperones (26). These proteins are predicted to reside either in the cytosol or in the nucleus. Overall, their function is poorly understood, but they may play an important role in signal transduction: mutation of AtFKBP72, also known as Pasticcino 1, leads to ectopic cell proliferation (35), while that of AtFKBP42 causes a twisted dwarf phenotype (20). In Chlamydomonas, three proteins are found to combine FKBP and TPR domains. C_680056 (1,785 residues) has been named FKB42 on the basis of the similarity of its N-terminal 350 residues to the sequence of ATFKBP42: a single FKBP domain, 3 TPR repeats, a calmodulin binding site, and a C-terminal membrane-anchoring domain (20). This combination is also present in human (FKBP38) and C. merolae (CMH207C) and may thus be an ancient eukaryotic trait. In addition, C_680056 comprises an unknown domain, two PQQ domains (WD40-like repeats; COG1520), and a C-terminal Leu-rich repeat, making this protein arguably one of the most complex encoded by the Chlamydomonas genome (note that we cannot rule out that the model fuses two neighboring genes). Chlamydomonas FKB62 (split between two scaffolds) contains at least two, probably three, FKBP domains in tandem, followed by a TRP domain. This is similar to the closely related Arabidopsis proteins AtFKBP62 and AtFKBP65 (which probably arose recently from the same large duplication that generated AtFKBP15-1 and 15-2). Finally, C_280158 (2,437 residues) shows a hydrophobic domain with three probable transmembrane helices, followed by one (possibly two) FKBP domain and a TRP domain, plus a calmodulin-binding motif and a nuclear localization signal. This is in part similar to AtFKBP62, -65 and -72. Unfortunately, C_280158 suffers from sequence gaps and possible gene fusion, so that its relationship to Arabidopsis FKBPs is not clear. I give this gene the provisory name FKB99.
Another type of complex FKBP is represented by FKB53, with its negatively charged N-terminal domain (45% E/D; theoretical pI = 3.45 over the first 131 amino acids). It is very close to AtFKBP53 (17), in which the N-terminal domain contains both acidic and basic residues (23.3% E/D, 15% R/K; pI = 4.45). AtFKBP53 and the related AtFKBP43 have been proposed to interact with DNA via their Arg/Lys-rich domain, but it is unclear how this could fit with the negative charge on the Chlamydomonas orthologue.
Finally, the most divergent FKBP is trigger factor, a PPIase and chaperone associated with the ribosome and involved in the early steps of protein folding (9). In Chlamydomonas, EST data are consistent with a single gene, which I call TIG1, represented by two overlapping gene models. Phylogenetic analysis (data not shown) indicates that the trigger factors of Chlamydomonas and Arabidopsis are related to that of Synechocystis rather than to that of Rickettsia and other Proteobacteria, believed to be close to the ancestor of mitochondria. It is probably directed to the chloroplast and clearly descends directly from the trigger factor gene of the cyanobacterial endosymbiont. Similarly, none of the Chlamydomonas or Arabidopsis FKBPs appeared to be related to that of Rickettsia, suggesting a complete loss of any FKBP that could have been present in the early mitochondrial endosymbiont.
CYCLOPHILINS
In the Chlamydomonas genome, I found 28 gene models that contain similarity to Arabidopsis cyclophilin genes (Table 1). They are believed to represent 25 genes and one pseudogene. This compares well with the 29 described for Arabidopsis (19, 30) and is much more than the 4 described for the red alga and the 3 described for Synechocystis, indicating that a vigorous diversification occurred specifically in the green lineage of plants. CYN20-4 probably is a pseudogene, in spite of being supported by cDNA data: the EST and genomic sequences, concordant, are incapable of coding for a full-length cyclophilin.
Chlamydomonas proteins were aligned with those of Arabidopsis, Synechocystis, and C. merolae, plus human cyclophilin A for structural comparison (see supplemental Fig. S2), and the alignment was used to generate a phylogenetic tree (Fig. 2). Here again, unambiguous one-to-one orthology could often be observed between Arabidopsis and Chlamydomonas proteins. Chlamydomonas genes were named based on the closest Arabidopsis homologue except that the root CYN was used (CYP being reserved for cytochrome P450). The residues implicated in PPIase activity (38) are fully conserved in only a fraction of the cyclophilins analyzed (Table 3). This does not necessarily mean that these proteins are not enzymatically active, since only a few substitutions have been tested. Only nine of the Chlamydomonas proteins show conservation of the W121 residue in helix II that is crucial for cyclosporine binding, independently of PPIase activity, and orthologues are generally consistent at that position (except AtCyp18-2/CYN18-2).
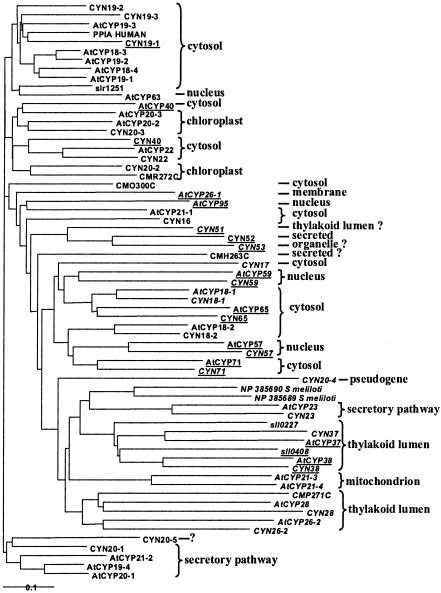
FIG. 2.
Phylogenetic tree of cyclophilins. Multidomain cyclophilins are underlined. Isoforms that are presumably inactive (Table 3) are in italics. The proposed localization of the mature protein is indicated on the right. Note that subcellular localization and phylogeny in general do not coincide.
TABLE 3.
Conservation of key residues in cyclophilinsa
| Protein | Length | Position
|
% Conserved | TargetP prediction | |||
|---|---|---|---|---|---|---|---|
| 55 | 60 | 121 | 126 | ||||
| PPIA_HUMAN | 165 | R | F | W | H | 100 | — (2) |
| CYN16 C_240021 modified | 190 | * | * | Q | * | 75 | — (3) |
| CYN17 C_20323 | 180 | * | G | F | R | 25 | — (2) |
| CYN18-1 C_20046 modified | 162 | * | * | H | Y | 50 | — (3) |
| CYN18-2 C_1010007 | 157 | * | * | * | * | 100 | — (2) |
| CYN19-1 C_250142 | >338 | * | * | * | * | 100 | — (2) |
| CYN19-2 C_3230001 | 172 | * | * | * | * | 100 | — (2) |
| CYN19-3 C_790042 | 201 | * | * | * | * | 100 | — (3) |
| CYN20-1 C_290072 | 222 | * | * | * | * | 100 | S (4): 36 |
| CYN20-2 C_70215 | 243 | * | * | * | * | 100 | C (4): 39 |
| CYN20-3 C_740054 | 200 | * | * | * | * | 100 | M (2): 30 |
| CYN20-4 C_990048 | 229 | X | X | — | — | <50 | — (4) |
| CYN20-5 C_660007 | 340 | * | * | H | * | 75 | M (4): 48 |
| CYN22 C_910055 | 187 | * | * | * | * | 100 | — (3) |
| CYN23 C_2260002 | 235 | * | * | H | Y | 50 | S (2): 19 |
| CYN26-2 C_460089 modified | 310 | G | Y | D | N | 0 | C (4): 40 |
| CYN28 C_1220009 modified | 350 | * | E | R | N | 25 | C (3): 45 |
| CYN37 C_90033 | 288 | A | V | A | F | 0 | — (3) |
| CYN38 C_30248 modified | 413 | * | * | N | F | 50 | M (4): 27 |
| CYN40 C_180001 | 369 | * | * | H | * | 75 | — (4) |
| CYN51 C_200140 modified | 465 | * | * | H | Y | 50 | M (4): 9 |
| CYN52 C_1560006 modified | 482 | * | * | H | * | 75 | S (1): 28 |
| CYN53 C_1560025 modified | 583 | * | * | H | Y | 50 | M (3): 12 |
| CYN57 C_1280001 | 585 | * | Y | * | N | 50 | S (3): 24 |
| CYN59 C_480065 modified | 486 | N | * | S | * | 50 | — (3) |
| CYN65 C_800078 modified | 586 | * | * | H | * | 100 | — (4) |
| CYN71 C_320104/C_19080001 | ? | * | * | — | — | <50 | ? |
| AtCYP18-1 At1g01940 | 160 | * | * | H | Y | 50 | — (4) |
| AtCYP18-2 At2g36130 | 164 | * | * | S | * | 100 | — (3) |
| AtCYP18-3 At4G38740 | 172 | * | * | * | * | 100 | — (2) |
| AtCYP18-4 At4G34870 | 172 | * | * | * | * | 100 | — (3) |
| AtCYP19-1 At2G16600 | 173 | * | * | * | * | 100 | — (2) |
| AtCYP19-2 At2g21130 | 174 | * | * | * | * | 100 | — (2) |
| AtCYP19-3 At3g56070 | 176 | * | * | * | * | 100 | — (5) |
| AtCYP19-4 At2G29960 | 201 | * | * | * | * | 100 | S (1): 23 |
| AtCYP20-1 At5G58710 | 204 | * | * | * | * | 100 | S (1): 23 |
| AtCYP20-2 At5g13120 | 259 | * | * | * | * | 100 | C (1): 70 |
| AtCYP20-3 At3g62030 | 260 | * | * | * | * | 100 | C (1): 77 |
| AtCYP21-1 At4g34960 | 224 | * | * | * | * | 100 | S (2): 27 |
| AtCYP21-2 At3g55920 | 228 | * | * | * | * | 100 | S (1): 31 |
| AtCYP21-3 At2G47320 | 230 | * | * | D | L | 50 | M (4): 47 |
| AtCYP21-4 At3G66654 | 236 | * | Y | D | L | 25 | C (4): 61 |
| AtCYP22 At2g38730 | 199 | * | * | * | * | 100 | — (5) |
| AtCYP23 At1g26940 | 226 | * | * | H | Y | 50 | S (1): 22 |
| AtCYP26-1 At3g22920 | 232 | H | L | Q | * | 25 | — (3) |
| AtCYP26-2 At1g74070 | 314 | K | Y | E | V | 0 | C (1): 45 |
| AtCYP28 At5g35100 | 289 | K | Q | Q | N | 0 | C (2): 70 |
| AtCYP37 At3g15520 | 464 | T | A | S | F | 0 | C (2): 65 |
| AtCYP38 At3g01480 | 437 | * | * | N | Y | 50 | C (1): 36 |
| AtCYP40 At2g15790 | 361 | * | * | H | * | 75 | — (2) |
| AtCYP57 At4g33060 | 504 | * | * | * | * | 100 | — (1) |
| AtCYP59 At1g53720 | 506 | T | * | Y | * | 50 | — (4) |
| AtCYP63 At3g63400 | 570 | * | * | H | * | 75 | — (4) |
| AtCYP65 At5g67530 | 595 | * | * | H | * | 100 | — (2) |
| AtCYP71 At3g44600 | 631 | * | * | * | * | 100 | — (3) |
| AtCYP95 At4g32420 | 837 | * | S | Q | N | 25 | — (2) |
| CMH263C | 238 | * | * | P | * | 75 | S (2): 22 |
| CMO300C | 168 | * | * | * | * | 100 | — (2) |
| CMP271C | 310 | Y | A | E | N | 0 | C (5): 47 |
| CMR272C | 251 | * | * | Q | * | 75 | C (5): 78 |
| NP_385689_S_meliloti | 190 | * | * | F | Y | 50 | S (1): 23 |
| NP_385690_S_meliloti | 169 | * | * | * | Y | 75 | — (2) |
| sll0227 Syn 6803 | 246 | * | * | G | Y | 50 | S (2): 28 |
| sll0408 Syn 6803 | 403 | * | * | N | Y | 50 | — (2) |
| slr1251 Syn 6803 | 170 | * | * | * | * | 100 | — (2) |
IsoformspresumedtobeinactiveasPPlaseareinitalics.IntracellulartargetingpredictedbyTargetPisindicated(C,chloroplast; M, mitochondrion; S, secretory pathway; —,none),togetherwiththelevelofconfidence(1,highest;4,lowest)andthepredictedlengthofthetransitpeptide.*,identitytoPPIA_HUMAN.
Most of the Chlamydomonas cyclophilins lack an N-terminal extension and thus are predicted to be cytosolic. The closest relatives of human cyclophilin A are encoded by a group of three genes (CYN19-1, CYN19-2, and CYN19-3). They are closely related to four Arabidopsis homologues (AtCyp18-3, -18-4, -19-1, -19-2, and -19-3), but there is no clear gene-to gene orthology. As outlined before (30), the similarity and close linkage of AtCYP18-4 and -18-3 on chromosome 4 and of AtCYP19-1 and -19-2 on chromosome 2 suggests two successive gene duplications, the latter involving an entire chromosomal fragment. Gene diversification probably occurred through a different route in Chlamydomonas: CYN19-1 and -19-2 are also found on chromosome 1, but far apart. Interestingly, CYN19-1 has, following its cyclophilin domain, a domain similar to GCIP-interacting protein, whose best hit in the Arabidopsis genome is At2g16860, closely linked to AtCYP19-1. Whether this reflects an ancient functional relationship remains uncertain. One of the cyanobacterial cyclophilins branches near this clade, together with the nucleus-located AtCyp63. Both lack the conserved Cys-62 and -115 residues probably involved in glutathionylation of cyclophilin A (12). There is no orthologue for this entire group in the red algal genome.
Another group of cyclophilins shows complex orthology relationships. CYN20-1 is related to AtCYP20-1, -19-4, and -21-2, all clearly directed to the secretory pathway. CYN20-5 is similar to these proteins, but it has a long N-terminal extension that could direct it to an organelle. Note that this branch is separate from that which harbors CYN23 and AtCYP23, also unambiguously directed to the secretory pathway but characterized by an insertion after helix II. This confirms the hypothesis that plant ER cyclophilins are polyphyletic (4). None show an ER retention signal, suggesting that they are secreted to the periplasm. Note that no Cyanidioschyzon cyclophilin branches in either of these clades. The only PPIase in this genome with anything approaching a potential ER-targeting signal is the cyclophilin CMH263C.
In several clades, univocal orthology and concordant N-terminal sequences leave no doubt as to the final location of the protein. Thus, CYN26-2 and CYN28, like their respective orthologues and the red algal CMP271C, appear targeted to the thylakoid lumen. They have insertions between β-strands 5 and 6 and after helix II. Extended loops (this time between strand 2 and helix 1 and after strand 4) are also found in the group formed by CYN37, CYN38, and the related Arabidopsis proteins. Two Synechocystis cyclophilins are found at the root of each branch, indicating an ancient diversification inherited from the cyanobacterial endosymbiont. AtCYP38 (TLP40) is one of the most extensively studied cyclophilins of higher plants (13, 33) and has been shown to be a lumenal protein. This is also probably true of CYN38 and of the two related cyanobacterial cyclophilins. The localization of the related CYN37 remains uncertain, since it does not show a convincing organelle targeting sequence, in contrast to its orthologue AtCYP37, and its N-terminal domain is truncated. The putative leucine zipper in the N-terminal domain of AtCYP38 is not conserved in this group of related sequences, and the role of this entire domain is unknown.
Branching close to this clade are the two mitochondrion-targeted proteins AtCyp21-3 and -21-4. They do not have orthologues either in the red alga or in Chlamydomonas, which suggests a recent origin. As mentioned above, the question of which protein carries out PPIase activity in this organelle in algae remains open. Several Chlamydomonas cyclophilins, like CYN16 and CYN17, have no orthologue in Arabidopsis, but they lack an N-terminal extension that could direct them to an organelle.
Complex cyclophilins appear in several distinct branches of the phylogenetic tree. CYN59, like AtCYP59, has an RRM domain involved in RNA binding but lacks the Zn finger. Its C terminus is rich in Arg and Gly residues and may be homologous to the Arg/Lys-rich domain of the Arabidopsis protein. CYN65 is entirely orthologous to the cytosolic AtCYP65, with its N-terminal U box (modified RING Zn finger). CYN57 is orthologous to AtCYP57 and probably also nucleus located. The sequence of CYN71 is incomplete, but it shares with AtCYP71 an N-terminal domain of unknown function. As a group, these complex cyclophilins form a clade with CYN18-1 and -18-2 and their Arabidopsis orthologues, with which they share a compact structure of the cyclophilin domain with short loops. The common ancestor of green algae and land plants probably showed a variety of complex cyclophilins. No cyanobacterial or red algal cyclophilins are found in this group, suggesting that it appeared after the green and red lineage separated.
Of particular interest are three complex Chlamydomonas cyclophilins with no orthologues in Arabidopsis. CYN52 and -53 have two cyclophilin domains in tandem, a feature not hitherto found in any other organism. Phylogenetic analysis (data not shown) shows that internal duplication predated gene duplication, since the N- and C-terminal cyclophilin domains are more similar from one gene to the other than to each other. As is often found in Chlamydomonas, these closely related genes are found next to one another on the genome. The related CYN51 has only one cyclophilin domain and thus appears closer to the ancestor. It shares with CYN53 a new type of domain, also found in higher plants (for example, AT4g33380 and At4g17070). This domain has apparently been lost in CYN52. Interestingly, while CYN53 appears directed to the chloroplast stroma or mitochondrial matrix, the N-terminal sequence of CYN51 has typical features of a dual targeting sequence, suggesting that the protein could end up in the thylakoid lumen. CYN52, in contrast, is unambiguously directed to the secretory pathway (TargetP score of 0.951). Clearly, this subfamily of cyclophilins deserves further study.
CYN40 is another type of complex cyclophilin, with a C-terminal TPR domain. It is probably cytosolic, like its Arabidopsis orthologue, AtCYP40, and so are the related simple cyclophilins CYN22 and AtCYP22. Also in this group are the organelle-targeted CYN20-2 and CYN20-3. They received their names from AtCYP20-2 and -20-3, but this is based more on their putative localization than on sequence similarities. Based on the presence of a potential thylakoid transfer sequence in CYN20-2, I propose that it is directed to the thylakoid lumen, whereas CYN20-3 would be a stromal protein.
Several Arabidopsis cyclophilins have no orthologue in Chlamydomonas: AtCyp21-1, AtCyp26-1, and AtCyp95. While the last is presumably nucleus located, AtCyp26-1 is predicted to be membrane anchored. It is expressed only in flowers (19), so it may function in a development pathway specific to spermatophytes. I note that no cDNA sequence is available for this gene and that no other plant has the N-terminal hydrophobic stretch predicted at the C terminus of the Arabidopsis protein, so that its membrane anchoring may need to be checked.
PARVULINS
Parvulins constitute a third type of PPIases, for which no ligand binding has been described. They act specifically on [Thr(P)/Ser(P)]-Pro peptide bonds and are inhibited by juglone. While there are three parvulins in Arabidopsis (19, 23), only one gene model in Chlamydomonas has homology to parvulins (PIN3; C_100048). Its closest relative is the chloroplast AtPIN3, with which it shares a C-terminal rhodanese domain. Both are probably chloroplast located (Table 1). In addition, searches in the EST database revealed another gene, PIN4, whose sequence had been mispredicted in the gene model. It encodes a parvulin domain highly similar to that of AtPIN1 and AtPIN2 and an N-terminal Forkhead domain usually found in proteins involved in nuclear signaling (6). Interestingly, this type of domain binds phosphopeptides, including peptides phosphorylated on Tyr residues. It has never been found before in a parvulin. Parvulins generally contain an N-terminal WW domain specialized in binding Pro-rich peptides. AtPin1 is unusual in that it shows no substrate-binding domain. Obviously, substrate recognition in parvulin-mediated signaling involves a variety of mechanisms and protein domains.
ASSESSMENT OF GENE MODELS
Of the 50 genes described in this report, the protein product of 26 (at least one of them, for those with alternative splicing) was correctly predicted by a gene model. By correctly, I mean that I was unable to find any flaw in them based on criteria of consistency with EST data and likelihood of generating a functional protein. The expressed pseudogene CYN20-4 cannot be assessed by these criteria, but it provides an interesting example of evolution caught in the act: mutations can scramble the information content of the coding sequence long before they abolish the ability of the gene to be transcribed.
Alternative splicing was found in four genes. For FKB16-2 and CYN17, the isoform described by the model was the one most represented in the EST database, but this was not true for FKB16-7 and CYN23. For 10 genes, the sequence was corrected based on EST data. For example, I found several cases where the 3′ or 5′ UTR was incorrectly predicted due to faulty interpretation of EST (in general because of overlapping genes). For several genes, internal exons were ill predicted, and I always verified that the EST data gave a protein with a better alignment to the other family members. Thus, gene modeling could be improved by placing more emphasis on concordance with EST contigs. In other cases, sequence correction was possible because the EST data bridged a gap in the nucleotide sequence. The missing sequence was sometimes found by BLAST in the unplaced reads, not used in the assembly, suggesting a possible use of EST contigs to guide gap closure.
Sometimes, even when the genomic sequence was complete and no EST data were available, I proposed to change the gene models in order to restore good alignment of the protein products. This implied extending the 5′ end (CYN18-1 and CYN19-1) or changing the intron exon boundaries. For example, I could add a fourth exon to CYN16 simply by using a noncanonical splice site. In the absence of experimental data, I cannot ascertain that my propositions are valid: the gene models could be right, and the genes could either be divergent at that position or be pseudogenes in the making. Still, I feel that there is a window of improvement for the computation of gene models, and my bias would be to make heavier use of homology-based modeling.
Several cases were found where the genome sequence is probably erroneously assembled. This was usually evidenced as one arm of a small scaffold being repeated in another scaffold, next to a gap (possibly due to a chimeric DNA clone). Thus, three genes were split between two gene models on different scaffolds. For FKB16-4, I found that the gene sequence was partly repeated on another, small scaffold, but this did not affect the model. For CYN19-1, adding a C nucleotide at position 543109 of scaffold_25 changed the reading frame in such a way that use of the next canonical 5′ intron splice site was possible and full conservation with the Arabidopsis orthologue was achieved. Sequencing errors are predicted to appear at fewer than 1/10,000 positions in the sequence; this could be one of them. Finally, two strange cases were found of a “bug” in the prediction. In C_290072 (CYN20-1), the sixth exon is presented as starting at position +2 with respect to the exon that can be deduced from EST data or predicted using the canonical 3′ splice site. This introduces a frameshift that throws off the alignment. In C_530020 (PIN4), the 4-nucleotide-long fifth exon obeys no consensus and probably also results from a computation error.
CONCLUSIONS
Chlamydomonas FKBPs and cyclophilins enjoy the same level of diversity that characterizes vascular plants. This appears to result both from an early diversification of the two gene families in the common ancestor of land plants and green algae and from a complex interplay of gene duplication, gene extinction, and mutation of N-terminal sequences thereafter. Alternative splicing also contributes to this diversity, in one case by changing localization of the mature protein. In the two gene families, chloroplast isoforms have evolved within diverse evolutionary lineages, and the same is true for ER and nucleus-targeted cyclophilins. This emphasizes how easily proteins are redirected to another compartment during evolution. Some genes found in vascular plants are absent in the alga, for example, AtCyp26-1, which is expressed only in flowers. Symmetrically, Chlamydomonas harbors novel domains or combination of domains, for instance, cyclophilins with two cyclophilin domains or a parvulin with a Forkhead domain. While only a fraction of the immunophilins appear to be enzymatically active, PPIase activity has probably been maintained in most compartments of the cell. It remains unclear which protein, if any, carries out peptidyl-prolyl cis-trans isomerization in the Chlamydomonas mitochondrion. Probably the most tantalizing of the remaining questions is that of which endogenous or exogenous ligands, if any, combine with plant immunophilins to carry out joint signaling functions.
Supplementary Material
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique (UPR 1261).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Ashburner, M. 2000. A biologist's view of the Drosophila genome annotation assessment project. Genome Res. 10:391-393. [DOI] [PubMed] [Google Scholar]
- 2.Birney, E., M. Clamp, and R. Durbin. 2004. GeneWise and Genomewise. Genome Res. 14:988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiman, A., and I. Camus. 2002. The involvement of mammalian and plant FK506-binding proteins (FKBPs) in development. Transgenic Res. 11:321-335. [DOI] [PubMed] [Google Scholar]
- 4.Chou, I. T., and C. S. Gasser. 1997. Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol. Biol. 35:873-892. [DOI] [PubMed] [Google Scholar]
- 5.Dharmasiri, N., S. Dharmasiri, A. M. Jones, and M. Estelle. 2003. Auxin action in a cell-free system. Curr. Biol. 13:1418-1422. [DOI] [PubMed] [Google Scholar]
- 6.Durocher, D., and S. P. Jackson. 2002. The FHA domain. FEBS Lett. 513:58-66. [DOI] [PubMed] [Google Scholar]
- 7.Edinger, A. L., C. M. Linardic, G. G. Chiang, C. B. Thompson, and R. T. Abraham. 2003. Differential effects of rapamycin on mammalian target of rapamycin signaling functions in mammalian cells. Cancer Res. 63:8451-8460. [PubMed] [Google Scholar]
- 8.Faure, J. D., D. Gingerich, and S. H. Howell. 1998. An Arabidopsis immunophilin, AtFKBP12, binds to AtFIP37 (FKBP interacting protein) in an interaction that is disrupted by FK506. Plant J. 15:783-789. [DOI] [PubMed] [Google Scholar]
- 9.Ferbitz, L., T. Maier, H. Patzelt, B. Bukau, E. Deuerling, and N. Ban. 2004. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431:590-596. [DOI] [PubMed] [Google Scholar]
- 10.Finazzi, G., C. Chasen, F. A. Wollman, and C. de Vitry. 2003. Thylakoid targeting of Tat passenger proteins shows no delta pH dependence in vivo. EMBO J. 22:807-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzen, L. G., J. D. Rochaix, and G. von Heijne. 1990. Chloroplast transit peptides from the green alga Chlamydomonas reinhardtii share features with both mitochondrial and higher plant chloroplast presequences. FEBS Lett. 260:165-168. [DOI] [PubMed] [Google Scholar]
- 12.Fratelli, M., H. Demol, M. Puype, S. Casagrande, P. Villa, I. Eberini, J. Vandekerckhove, E. Gianazza, and P. Ghezzi. 2003. Identification of proteins undergoing glutathionylation in oxidatively stressed hepatocytes and hepatoma cells. Proteomics 3:1154-1161. [DOI] [PubMed] [Google Scholar]
- 13.Fulgosi, H., A. V. Vener, L. Altschmied, R. G. Herrmann, and B. Andersson. 1998. A novel multi-functional chloroplast protein: identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J. 17:1577-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavigan, C. S., S. P. Kiely, J. Hirtzlin, and A. Bell. 2003. Cyclosporin-binding proteins of Plasmodium falciparum. Int. J. Parasitol. 33:987-996. [DOI] [PubMed] [Google Scholar]
- 15.Geisler, M., H. U. Kolukisaoglu, R. Bouchard, K. Billion, J. Berger, B. Saal, N. Frangne, Z. Koncz-Kalman, C. Koncz, R. Dudler, J. J. Blakeslee, A. S. Murphy, E. Martinoia, and B. Schulz. 2003. TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 14:4238-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossman, A. R., E. E. Harris, C. Hauser, P. A. Lefebvre, D. Martinez, D. Rokhsar, J. Shrager, C. D. Silflow, D. Stern, O. Vallon, and Z. Zhang. 2003. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2:1137-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrar, Y., C. Bellini, and J. D. Faure. 2001. FKBPs: at the crossroads of folding and transduction. Trends Plant Sci. 6:426-431. [DOI] [PubMed] [Google Scholar]
- 18.Harris, E. H. 1989. The Chlamydomonas source book: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 19.He, Z., L. Li, and S. Luan. 2004. Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 134:1248-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamphausen, T., J. Fanghanel, D. Neumann, B. Schulz, and J. U. Rahfeld. 2002. Characterization of Arabidopsis thaliana AtFKBP42 that is membrane-bound and interacts with Hsp90. Plant J. 32:263-276. [DOI] [PubMed] [Google Scholar]
- 21.Kay, J. E. 1996. Structure-function relationships in the FK506-binding protein (FKBP) family of peptidylprolyl cis-trans isomerases. Biochem. J. 314:361-385. [PMC free article] [PubMed] [Google Scholar]
- 22.Kurek, I., R. Dulberger, A. Azem, B. B. Tzvi, D. Sudhakar, P. Christou, and A. Breiman. 2002. Deletion of the C-terminal 138 amino acids of the wheat FKBP73 abrogates calmodulin binding, dimerization and male fertility in transgenic rice. Plant Mol. Biol. 48:369-381. [DOI] [PubMed] [Google Scholar]
- 23.Landrieu, I., L. De Veylder, J. S. Fruchart, B. Odaert, P. Casteels, D. Portetelle, M. Van Montagu, D. Inze, and G. Lippens. 2000. The Arabidopsis thaliana PIN1At gene encodes a single-domain phosphorylation-dependent peptidyl prolyl cis/trans isomerase. J. Biol. Chem. 275:10577-10581. [DOI] [PubMed] [Google Scholar]
- 24.Li, J. B., S. Lin, H. Jia, H. Wu, B. A. Roe, D. Kulp, G. D. Stormo, and S. K. Dutcher. 2003. Analysis of Chlamydomonas reinhardtii genome structure using large-scale sequencing of regions on linkage groups I and III. J. Eukaryot. Microbiol. 50:145-155. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki, M., O. Misumi, I. T. Shin, S. Maruyama, M. Takahara, S. Y. Miyagishima, T. Mori, K. Nishida, F. Yagisawa, Y. Yoshida, Y. Nishimura, S. Nakao, T. Kobayashi, Y. Momoyama, T. Higashiyama, A. Minoda, M. Sano, H. Nomoto, K. Oishi, H. Hayashi, F. Ohta, S. Nishizaka, S. Haga, S. Miura, T. Morishita, Y. Kabeya, K. Terasawa, Y. Suzuki, Y. Ishii, S. Asakawa, H. Takano, N. Ohta, H. Kuroiwa, K. Tanaka, N. Shimizu, S. Sugano, N. Sato, H. Nozaki, N. Ogasawara, Y. Kohara, and T. Kuroiwa. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653-657. [DOI] [PubMed] [Google Scholar]
- 26.Pratt, W. B., P. Krishna, and L. J. Olsen. 2001. Hsp90-binding immunophilins in plants: the protein movers. Trends Plant Sci. 6:54-58. [DOI] [PubMed] [Google Scholar]
- 27.Reimer, U., and G. Fischer. 2002. Local structural changes caused by peptidyl-prolyl cis/trans isomerization in the native state of proteins. Biophys. Chem. 96:203-212. [DOI] [PubMed] [Google Scholar]
- 28.Robaglia, C., B. Menand, Y. Lei, R. Sormani, M. Nicolai, C. Gery, E. Teoule, D. Deprost, and C. Meyer. 2004. Plant growth: the translational connection. Biochem. Soc. Trans. 32:581-584. [DOI] [PubMed] [Google Scholar]
- 29.Romano, P. G., A. Edvardsson, A. V. Ruban, B. Andersson, A. V. Vener, J. E. Gray, and P. Horton. 2004. Arabidopsis AtCYP20-2 is a light-regulated cyclophilin-type peptidyl-prolyl cis-trans isomerase associated with the photosynthetic membranes. Plant Physiol. 134:1244-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romano, P. G., P. Horton, and J. E. Gray. 2004. The Arabidopsis cyclophilin gene family. Plant Physiol. 134:1268-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salamov, A. A., and V. V. Solovyev. 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10:516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somanchi, A., and J. V. Moroney. 1999. As Chlamydomonas reinhardtii acclimates to low-CO2 conditions there is an increase in cyclophilin expression. Plant Mol. Biol. 40:1055-1062. [DOI] [PubMed] [Google Scholar]
- 33.Vener, A. V., A. Rokka, H. Fulgosi, B. Andersson, and R. G. Herrmann. 1999. A cyclophilin-regulated PP2A-like protein phosphatase in thylakoid membranes of plant chloroplasts. Biochemistry 38:14955-14965. [DOI] [PubMed] [Google Scholar]
- 34.Vespa, L., G. Vachon, F. Berger, D. Perazza, J. D. Faure, and M. Herzog. 2004. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 134:1283-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vittorioso, P., R. Cowling, J.-D. Faure, M. Caboche, and C. Bellini. 1998. Mutation in the Arabidopsis PASTICCINO1 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Mol. Cell. Biol. 18:3034-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarei, M. M., M. Eghbali, A. Alioua, M. Song, H. G. Knaus, E. Stefani, and L. Toro. 2004. An endoplasmic reticulum trafficking signal prevents surface expression of a voltage- and Ca2+-activated K+ channel splice variant. Proc. Natl. Acad. Sci. USA 101:10072-10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Z., J. Shrager, M. Jain, C. W. Chang, O. Vallon, and A. R. Grossman. 2004. Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot. Cell 3:1331-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zydowsky, L. D., F. A. Etzkorn, H. Y. Chang, S. B. Ferguson, L. A. Stolz, S. I. Ho, and C. T. Walsh. 1992. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1:1092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.