Abstract
Triple-negative breast cancer (TNBC) cells are modulated in reaction to tumor-infiltrating lymphocytes. However, their specific responses to this immune pressure are unknown. In order to address this question, we first used mRNA sequencing to compare the immunophenotype of the TNBC cell line MDA-MB-231 and the luminal breast cancer cell line MCF7 after both were cocultured with activated human T cells. Despite similarities in the cytokine-induced immune signatures of the two cell lines, MDA-MD-231 cells were able to transcribe more IDO1 than MCF7 cells. The two cell lines had similar upstream JAK/STAT1 signaling and IDO1 mRNA stability. However, using a series of BC cell lines, IFNγ stimulated IDO1 protein expression and enzymatic activity only in ER−, not ER+, cell lines. Treatment with 5-aza-deoxycytidine reversed the suppression of IDO1 expression in MCF7 cells, suggesting that DNA methylation was potentially involved in IDO1 induction. By analyzing several breast cancer datasets, we discovered subtype-specific mRNA and promoter methylation differences in IDO1, with TNBC/basal subtypes exhibiting lower methylation/higher expression and ER+/luminal subtypes exhibiting higher methylation/lower expression. We confirmed this trend of IDO1 methylation by bisulfite pyrosequencing breast cancer cell lines and an independent cohort of primary breast tumors. Taken together, these findings suggest that IDO1 methylation regulates anti-immune responses in breast cancer subtypes and could be used as a predictive biomarker for IDO1 inhibitor–based immunotherapy.
Keywords: DNA methylation, IDO1, breast cancer, activated T cells, RNAseq
Introduction
The cancer immunoediting paradigm suggests that tumor-infiltrating lymphocytes (TIL), along with other immune cells, play a major role in modulating immune surveillance at the tumor site (1). In breast cancer (BC), infiltration of CD8+ T cells into the tumor site is a positive prognostic factor (2–4). The leukocyte composition of TILs in BC consists predominantly of activated CD8+ cytotoxic T lymphocytes (CTLs) with high expression of activation marker CD69 and costimulatory receptor CD28, compared to peripheral T cells (5). Interferon γ (IFNγ), a cytokine secreted by CD8+ T cells, can prevent both carcinogen-induced and spontaneous epithelial tumors, and type 1 CD8+ T cells (Tc1) are the main source of IFNγ-mediated tumor growth suppression in a murine TCR transgenic and breast tumor model (6). Moreover, patients with tumors that elicit potent CTL responses tend to respond better to chemotherapy and have a more favorable prognosis (7). Triple-negative breast cancer (TNBC) is intrinsically more immunogenic than other subtypes (8). Gene expression profiling studies have identified interferon-regulated gene clusters, called immune response (IR) modules, in estrogen receptor–negative (ER−) and basal-like breast tumors (9–12). Together these studies indicate the presence of active immune surveillance mediated by TILs within the tumor microenvironment in BC.
Tumor cells respond to the presence of TILs by developing counteracting mechanisms to escape immune surveillance. Spranger et al. demonstrated, using a melanoma mouse model, that immunosuppressive factors like IDO1, PD-L1, and T-regulatory cell recruitment into the tumor microenvironment required the presence of CTLs, suggesting that immunosuppressive pathways are intrinsically driven by the active immune system (13). Coculture of MDA-MB-231 cells with activated T cells revealed that IFNγ is primarily responsible for IDO1 expression; however TH2 cytokines negatively regulated IFNγ-induced IDO1 expression, suggesting the importance of cytokine balance at tumor sites (14). IDO1 expression and kynurenine, a metabolite of the functional IDO1 enzyme, correlate in basal-like BC (15). Moreover, PD-L1 is highly expressed in TNBC (16). Poschke et al., reported that even at an early stage, breast tumors induced differentiation and immunosenescence of CD8+ T cells with loss of CD28 and upregulation of PD-1 in BC patients compared to normal individuals (17), indicating that tumor cells can efficiently counteract the immune system.
Although studies thus far have provided insights into tumor suppressor functions of TILs, it remains poorly understood how tumor cells intrinsically counteract immune surveillance at the gene expression level. Towards addressing this knowledge gap, we performed RNA sequencing (RNAseq) analysis on ER+ and TNBC cell lines that were cocultured with activated human T cells. Our results revealed activation of the IR module in BC cells along with upregulation of immunosuppressive genes IDO1 and PD-L1, providing in vitro evidence that expression of immune responsive genes in BC are primarily due to recruitment of TILs to tumor sites. Subsequent analysis, in a panel of BC cell lines and primary BC tissue samples, of one of the most upregulated genes by activated human T cells, IDO1, revealed a subtype-specific regulation of IDO1 expression by DNA methylation, suggesting that IDO1 promoter methylation may be a predictive biomarker for BC. Based on our analysis, we predict that the unique epigenetic background of TNBC makes it a T cell–inflamed tumor type that may preferentially benefit from IDO1 inhibitor–based immunotherapy.
Materials and Methods
Primary breast tumor samples and cell culture
Primary normal and BC tissues were obtained from the University of Texas Health Center at San Antonio and the Department of Pathology at Augusta University in compliance with the Institutional Review Boards at the respective institutions. BT474, MCF7, T47D, ZR-751, MDA-MB-231, SUM159, and BT549 cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS). MDA-MB-231 and MCF7 cell lines were purchased from ATCC in July 2009. All other cell lines were obtained from Dr. Muthusamy Thangaraju’s laboratory in 2013 and have been thoroughly tested and authenticated. Morphology, karyotyping, and PCR-based approaches were used to confirm the identity of the cell lines as indicated previously (18). The analysis of gene expression profiles that we previously reported (19) also confirmed that these cell lines belong to their anticipated molecular subtypes such as basal, luminal A and B subtypes, respectively. The BT474-PTEN-LTT line was established by Hasan Korkaya and cultured as described previously (20). The parental BT474 cell line was authenticated by short tandem repeat (STR) analysis. Most of the cell lines used in this study were cultured for less than three months for the experiments described. MDA-MB-231 and MCF7 cells were used in the experiments for about 12 months, however the cell cultures were re-started from frozen stocks at least twice during the duration of this study.
Coculture of breast cancer cells with PBMCs
Blood samples from healthy donors were purchased from a local blood bank. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Paque (GE Healthcare) density gradient separation. CD8+ T cells were negatively selected using an EasySepTM Human CD8+ T cell Isolation kit (STEMCELL Technologies). To activate T cells, a total of 3 million PBMCs or CD8+ T cells were treated with immobilized monoclonal antibody (mAb) to CD3 (5μg/mL, Cat# 317304, Biolegend) and soluble anti-CD28 (2μg/mL, Cat# 555725, BD Biosciences) in 1ml of RPMI-1640 media supplemented with 10% fetal bovine serum (FBS). The isotype control antibody used was mouse IgG2a,κ (Cat# 554645) from BD Biosciences. After 48 hrs, PBMCs and the conditioned-media were harvested. Activated PBMCs or CD8+ T cells were transferred into 0.4μm Transwell Insert (Corning) and placed into 6-well plates with pre-seeded MDA-MB-231 and MCF7 cells. MDA-MB-231 or MCF7 cells were cocultured with PBMCs alone or the conditioned-media or a combination of both for 24 hrs, and then harvested for further analysis. Control experiments were performed in the same manner except that isotype control antibodies were used.
Cytokine array analyses
Cytokines secreted by activated T cells were analyzed using RayBiotech Human cytokine array C1000 (RayBiotech) following manufacturer’s instructions. Briefly, cytokine array membranes were blocked with blocking buffer for 30 minutes at room temperature, followed by incubation with 1mL of supernatant from T-cell activation culture on each membrane for overnight at 4°C. After two washes followed by incubation with biotinylated antibody cocktail and streptavidin-HRP concentrate for 2 hours at room temperature, the signal was developed as instructed onto film and spot intensities were determined using ImageJ software (21).
RNAseq library construction and data analyses
Libraries for mRNA sequencing were prepared using KAPA Stranded mRNAseq Kit (Kapa Biosystems) following manufacturer’s instructions (Version 2.14). Briefly, total RNA (1.5 μg) was depleted of ribosomal RNA using supplied mRNA capture magnetic beads. Eluted mRNA was fragmented to a size of 300–400bp by incubating at 85°C for 6 minutes followed by 1st strand and 2nd strand cDNA synthesis. From here on, AMPure XP beads (Beckman Coulter) were used for subsequent cleanup wherever required. Double-strand cDNA was marked by A-tailing followed by ligation with Illumina barcoded adapters (5 μL). The libraries of six different indexes were pooled for multiplexing and analyzed by Bioanalyzer. A final 10 nM of pooled libraries were sequenced for 30 to 60 million 50-bp paired-end reads on Illumina Hi-seq 2500 platform (Illumina). TopHat (22) was used for mapping sequence reads to the human genome (hg19). BedTools (23) and bedGraphToBigWig generated stranded binary wiggle files after in-house scripts split reads mapped to each strand. FeatureCounts (24) computed the number of reads mapped to each gene and transcript, followed by DESeq (25) for differential expression analysis. In-house scripts and R Bioconductor 2.15.1 (26) were used to compare TCGA data and generate heatmaps and Venn diagrams.
Quantitative RT-PCR analyses
Total RNA extracted from the breast cancer cell lines with Qiazol (Qiagen) was subjected to reverse transcription using High Capacity cDNA Reverse Transcription kit (Applied Biosystems) with random hexamers and oligo dT primers. Quantitative PCR (qPCR) was set up using Power SYBR Green Master Mix (Life Technologies) and qPCR was performed on a CFX96 C1000 Real-Time thermal cycler (Bio-Rad). β-Actin or 18S were used as housekeeping genes wherever required. Primer sequences are provided in Supplementary Materials.
Flow cytometry analyses
MDA-MB-231 and MCF7 cells were incubated overnight with conditioned medium from T-cell activation assays. The harvested cells were stained with PE-labeled anti-CD274 (PD-L1) antibody (eBioscience) or Mouse IgG2b, K Isotype control PE (eBioscience), respectively, and analyzed on LSRII flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software.
IDO enzymatic assays
MDA-MB-231 and MCF7 cells were cultured in RPMI-1640 complete medium with or without IFNγ treatment (100ng/mL) for 12 hours. The culture media was replaced with synthetic RPMI-1640 medium and aliquots were collected at indicated time points. The IDO1 enzymatic assay was performed as described previously (27). The same concentration of IFNγ (100ng/mL) was used in all experiments unless otherwise stated.
IDO1 mRNA stability studies
MDA-MB-231 and MCF7 breast cancer cells were cultured in 6-well plates. The cells were pretreated with IFNγ for 4 hours before replacing with media containing actinomycin-D (1 μg/mL). Cells were harvested using Qiazol for total RNA and analyzed by qRT-PCR at indicated time points. By normalizing IDO1 mRNA at each point to 18S ribosomal RNA, mRNA stability was assessed. The stability of IRF1 mRNA was determined similarly.
3′-UTR reporter assays
The 3′-UTR sequence of IDO1 was amplified from HCT116 genomic DNA. The 669 bp 3′-UTR was cloned into pmirGLO luciferase plasmid using XhoI and XbaI cloning sites. The 3′UTR reporter plasmid (1 μg) was transiently transfected into MDA-MB-231 and MCF7 cells. Normalized luciferase activity was determined using Dual-Luciferase Reporter Assay System (Promega, Cat# E1910). Luminescence was measured as relative light units (RLUs) using Sirius luminometer SIRIUS luminometer (Berthold Detection System GmbH).
IDO protein stability studies
MDA-MB-231 cells were pre-stimulated with IFNγ for 12 hrs to induce IDO1 protein expression. IFNγ was withdrawn and replaced with media containing of cycloheximide (50μg/mL; CHX), 5μM proteosome inhibitor (MG132), or both agents, respectively. Cells were harvested at the indicated time points and IDO1 was analyzed by immunoblot analysis. MCF7 cells were treated in a similar way with MG132 only and analyzed for IDO1 expression.
Bisulfite pyrosequencing analysis
Genomic DNA isolated from BC cell lines using AllPrep DNA/RNA kit (Qiagen) was bisulfite-converted using EZ DNA Methylation Kit (Zymo Research). Pyrosequencing analysis was performed as described previously (28). The heatmap representing percentage methylation status of CpG sites was generated using Partek Genomic Suite 6.6 (Partek).
Chromatin immunoprecipitation (ChIP) assay
MDA-MB-231 and MCF7 cells were treated with IFNγ for the indicated times or left untreated. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature and quenched with 0.125 M glycine for 5 min to stop cross-linking. Cells were lysed by freezing at −80°C and thawing on ice in hypotonic cell lysis buffer containing protease inhibitors for 15 min followed by centrifugation to pellet the nuclei. SDS-Nuclear lysis buffer was added to the nuclei pellet along with protease inhibitor and incubated on ice for 30 minutes with frequent vortexing to lyse the nuclei. The cross-linked chromatin was sonicated using the Bioruptor (Diagenode) to obtain 200–500 bp DNA fragments. Diluted chromatin was incubated with 5μg of RNA polymerase II (Pol II) antibody (Abcam) with rotation at 4°C overnight. Protein A/G magnetic beads (Invitrogen) were blocked with 2% BSA and incubated with chromatin-antibody complexes for 2 hrs at 4°C. Subsequent washes were performed for 5 min each at 4°C with low salt buffer, high salt buffer, LiCl wash buffers and a finally wash with TE buffer. After eluting the DNA from magnetic beads, crosslinking reversal and phenol-chloroform extractions were performed. qPCR analysis was performed to determine fold enrichment compared to 1% of input DNA. Primers used for qPCR analysis are available in the supplementary materials.
5-aza-2′-deoxycytidine (5-azadC) treatment
MCF7 cells were plated at 30% confluency in a 6-well plate. Cells were treated with 2 μM 5-azadC every the other day for 4 days. After day 4, MCF7 cells were stimulated with IFNγ overnight. On day 5, cells were harvested for total RNA extraction followed by qRT-PCR to assess IDO1 mRNA expression.
Luciferase reporter assays
A 1.3 kb DNA fragment from the IDO1 promoter was cloned into the pGL2 basic luciferase reporter plasmid (Promega). To obtain an unmethylated promoter, the reporter construct was transformed into the dam−/dcm− E. coli strain (New England Biolabs). To obtain a fully methylated reporter, the construct was methylated with SssI methylase (New England Biolabs) in vitro. Breast cancer cell lines were transfected with 1 μg of methylated or unmethylated reporter plasmid DNA and 50 ng of renilla luciferase plasmid DNA using Lipofectamine 2000 (Life Technologies). Following transfection, cells were treated with or without IFNγ (100 ng/mL) overnight. Reporter assays were performed using Dual-Luciferase Reporter Assay System (Promega).
Human breast tumor datasets
Matched gene expression, methylation, metabolomics, and immunohistochemistry staining data along with clinical information from BC patients were obtained from Terunuma et al. (29). As described in (29), gene expression data was generated using Gene Chip Human Gene 1.0 ST Arrays (Affymetrix), global DNA methylation profiling was performed using Human Methylation 450 BeadChips (Illumina), and the concentrations of 296 named metabolites (including kynurenine) in 67 human breast tumors and 65 tumor-adjacent noncancerous tissues were measured using mass spectrometry based-metabolomics (29). TCGA Methylation 450K array and RNAseq datasets were downloaded from UCSC Cancer Genome Browser (https://genome-cancer.ucsc.edu).
Statistical analysis
To test the association of kynurenine levels with methylation changes, BC samples were divided into high and low kynurenine subgroups based on the relative abundance of kynurenine within each tumor (normalized by per kg frozen tissue weight). Breast tumors that had measured kynurenine levels in the top 15th percentile of all breast tumors were assigned to the high kynurenine subgroup. Similarly, breast tumors with kynurenine levels in the lowest 15th percentile of all breast tumors were placed in the low kynurenine subgroup. Methylation sample outliers were detected using the R package “lumi” based on 1.5 times median distance from the center of all tumor samples and excluded from statistical analysis (30). BC samples were also divided into high and low IDO1 groups based on median expression. Two-sided student t-tests were used to determine statistically significant differences between breast tumor subgroups at P < 0.05. Survival analysis was performed using the R package “survival” to determine differences in survival outcome between patients with breast tumors having the highest and lowest 20-percentile of kynurenine concentrations. Statistically significant differences in survival between breast tumor subgroups were determined using the log-rank test at P < 0.05.
Results
Activated human T cells induced a cytokine-mediated gene expression signature in BC cells
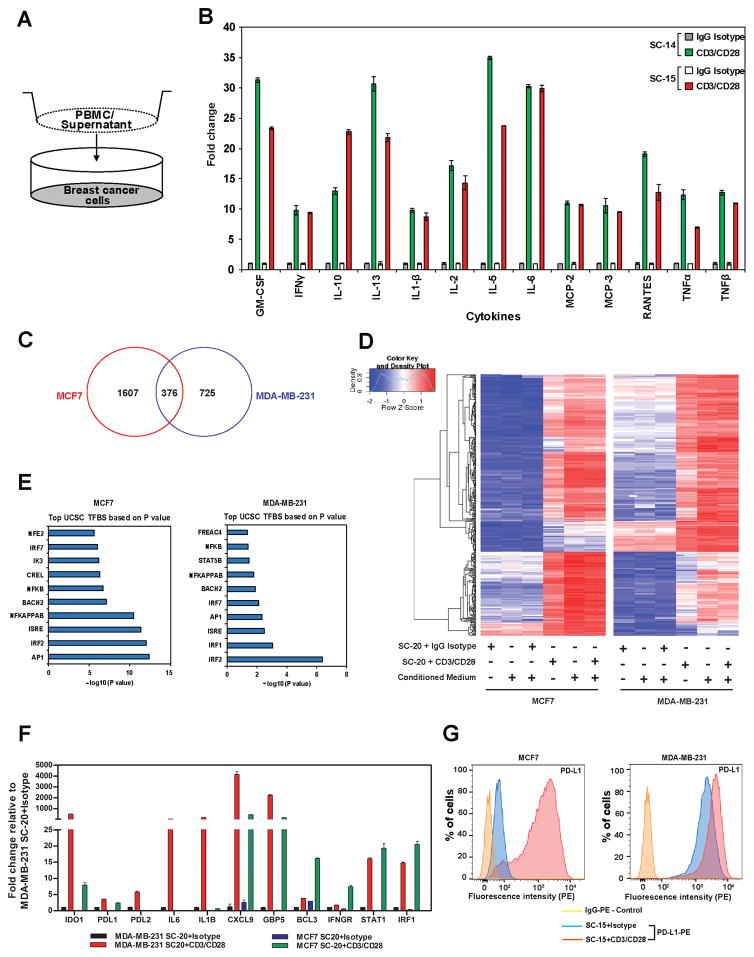
To determine the immunomodulatory effects of cytokines secreted by activated human T cells on BC cells, we performed RNAseq analysis on MCF7 and MDA-MB-231 cells, after coculturing them with normal PBMCs activated with CD3/CD28 mAbs in a contact-independent manner (Fig. 1A). Using cytokine-array analysis (Fig. 1B and Supplementary Fig. S1), we first confirmed that a similar cytokine profile with high levels of IFNγ, TNFα, TNFβ, IL2, GM-CSF, and IL10 was produced in the culture media of PBMCs activated with anti-CD3/CD28 antibodies as previously reported (31). Since anti-CD3/CD28 treatment mainly activates the T-cell fraction in PBMCs, activated PBMCs will be referred as activated T cells hereafter. RNAseq analysis revealed that 1607 genes in MCF7 and 725 genes in MDA-MB-231 cells were upregulated by coculturing with activated T cells, with 376 genes upregulated in both cell lines (Fig. 1C). Cluster analysis of the 376 genes revealed that similar gene expression patterns were observed for MDA-MB-231 or MCF7 cells when they were cocultured with activated T cells, conditioned media from T-cell activation assays, or a combination of both (Fig. 1D). Analyses using DAVID and IPA tools demonstrated that a majority of genes upregulated in both cell lines were involved in immune signaling and enriched with IRF binding sites in their promoters (Fig. 1E). The top ranked genes upregulated in both cell lines include many known IFNγ-inducible genes such as CXCL9, CXCL10, CXCL11, IDO1, IRF1, GBP5, PDL1, PDL2, and STAT1. qRT-PCR analysis validated the RNAseq results, demonstrating that many of these genes were expressed at basal levels in MDA-MB-231 and significantly upregulated when exposed to activated T cells (Fig. 1F). Flow cytometric analysis of PD-L1 revealed strong basal expression in MDA-MB-231 cells and enhanced cell surface expression in both MDA-MB-231 and MCF7 cells after coculture with activated T cells compared to control T cells (Fig. 1G).
Figure 1.
Coculture of activated human T cells induced immune response gene signature in BC cells.
A, Schematic representation of coculture experiment. B, Cytokines secreted by PBMCs that were activated with anti-CD3/CD28 antibodies. PBMCs from two donors (SC-14, SC-15) were analyzed using cytokine array. C, Venn diagram showing the number of genes upregulated in MCF7 and MDA-MB-231 cells when cocultured with activated PBMCs. D, Heat map resulted from a hierarchical cluster analysis of 376 genes upregulated in both MDA-MB-231 and MCF7 cells after they were cocultured with PBMCs. E, List of top transcription factor binding sites enriched in the promoter of upregulated genes after cocultured with PBMCs in MCF7 and MDA-MB-231, respectively. F, Validation of RNAseq results by qRT-PCR. G, PD-L1 cell surface expression measured by flow cytometry after being cocultured with control or anti-CD3/CD28 activated PBMCs (SC-15). Data shown in B, F, and G are from representative experiments using at least 2 independent donor PBMC samples. The RNA-seq experiment in C was carried out using a single donor PBMC sample, but under three different experimental settings as described in the Materials and Methods.
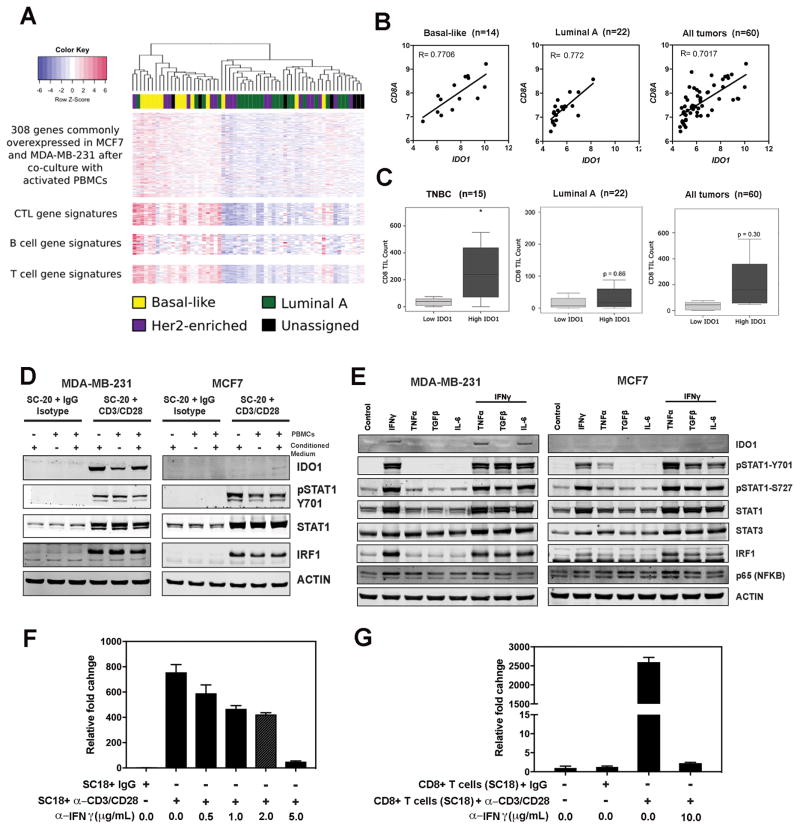
To further understand the gene signatures upregulated by coculturing with activated T cells, we compared the expression profiles of 308 (out of 376) genes with previously defined CTL, B-cell, and T-cell expression signatures genes (32), using the TCGA RNAseq dataset. Cluster analysis of these 308 genes in 274 BC patients suggested a positive association with CTL, B-cell, and T-cell gene signatures (Supplementary Fig. S2). Previously, Terunuma et al. have carried out an integrated metabolomics, gene expression and DNA methylation analysis in a set of matched primary breast tumor specimens (29). By analyzing this published data set, we found that expression profiles of these 308 genes displayed even stronger association with B-cell, T-cell, and CTL gene signatures in basal-like subtype BC patients (Fig 2A). Further analysis of the Terunuma dataset revealed a significant positive correlation between IDO1 and CD8A expression values across molecular subtypes of BC patients (Fig. 2B). However, immunohistochemical staining of TILs in BC tumors seemed to suggest that tumors expressing higher amounts of IDO1 mRNA generally had a significantly higher number of CD8+ TILs in TNBC samples, whereas IDO1 expression outside of the TNBC subgroup did not associate significantly with CD8+ TILs (Fig 2C).
Figure 2.
Activated T cells induce differential IDO1 expression in BC cells. A, The expression profiles of basal subtype tumors display a stronger correlation with immune gene signatures. The heat map demonstrated a positive correlation between the expression profiles of these 308 genes and CTL, B-cell and T-cell gene signatures in BC patient samples. B, Positive correlation between IDO1 and CD8A mRNA expression in basal-like, luminal A, and all BC patients. C, Immunohistochemistry staining of TILs in BC samples revealed significant association of IDO1 mRNA expression with the number of TILs in TNBC samples. BC samples were also divided into high and low IDO1 groups based on median expression levels. Two-sided student t-tests were used to determine statistically significant differences between breast tumor subgroups at P < 0.05. D, Immunoblot analysis of MDA-MB-231 and MCF7 cells cocultured with control and activated human PBMCs for 24 hrs, respectively. E, Immunoblot analysis of MDA-MB-231 and MCF7 cells after treatment with IFNγ (100ng/mL), TNFα (1ng/mL), TGFβ (1ng/mL), and IL6 (5ng/mL), alone or in combination with IFNγ overnight and probing with the indicated antibodies. F, qRT-PCR analysis of IDO1 expression in MDA-MB-231 cells treated with conditional media from activated PBMC cultures. The PBMC sample (SC18) was treated with CD3/CD28 antibodies or isotype control antibodies as described in the Materials and Method section. G, qRT-PCR analysis of IDO1 expression in MDA-MB-231 cells cocultured with activated and non-activated CD8+ T cells (SC18). The IFNγ neutralizing antibody was added to the activated PBMC or CD8+ T-cell cultures in the amount as indicated and incubated for 30 minutes before adding MDA-MB-231 cells to the cocultures. Results in D and E are representative blots from 2–3 independent experiments. Data shown in F and G are from representative experiments using at least 2 independent donor PBMC samples.
Activated T cells induced differential IDO expression in BC cells
RNAseq analysis demonstrated that both MCF7 and MDA-MB231 cells expressed full-length IDO1 mRNA, albeit at different levels, when cocultured with activated T cells. Neither alternative spliced transcripts of IDO1 nor alternative promoter usage was detected after examining the RNAseq results at the IDO1 locus (Supplemental Fig. S3). However, coculture with activated T cells induced IDO1 protein expression in MDA-MB-231 cells, but not in MCF7 cells, despite the hyperphosphorylation of STAT1 at Y701 and significant upregulation of STAT1 and IRF1 expression, as detected by immunoblot, suggesting the activation of JAK/STAT signaling in both cell lines (Fig. 2D). Because the conditioned media from the activated T-cell cultures is a mixture of cytokines, we treated MDA-MB-231 and MCF7 cells with TNFα, TGFβ, and IL6, alone and in combination with IFNγ, to determine how cytokines modulate IDO1 expression (Fig. 2E). Interferons, particularly IFNγ, induce IDO1 expression in various types of cells (33). We indeed observed that IFNγ-induced IDO1 expression in MDA-MB-231 cells, but not in MCF7 cells (Fig. 2E). The combination of IFNγ and TNFα slightly increased IDO1 expression in MDA-MB-231 cells, whereas TGFβ abrogated IFNγ-induced IDO expression (Fig. 2E). IL6 did not have any significant effect on IFNγ-induced IDO expression. TNFα, TGFβ, and IL6 alone did not induce IDO1 expression in either cell line. This observation is in line with previous studies where IFNγ and TNFα synergistically induced IDO1 expression (34,35), whereas TGFβ suppressed IFNγ-induced IDO1 expression (36). In addition, we found that IFNγ-neutralizing antibody can inhibit the upregulation of IDO1 mRNA in MDA-MB-231 cells by conditioned media from the activated PBMC cultures in a dose-dependent manner, suggesting that IFNγ is one of the most important soluble factors in the condition media that induces IDO1 expression (Fig. 2F). Furthermore, we demonstrated that coculture with activated CD8+ T cells also induced IDO1 expression in MDA-MB-231 cells and this induction could be blocked by the IFNγ-neutralizing antibody (Fig. 2G).
IDO1 mRNA expression and IDO1 enzyme activity higher in basal-like BCs and TNBC cell lines
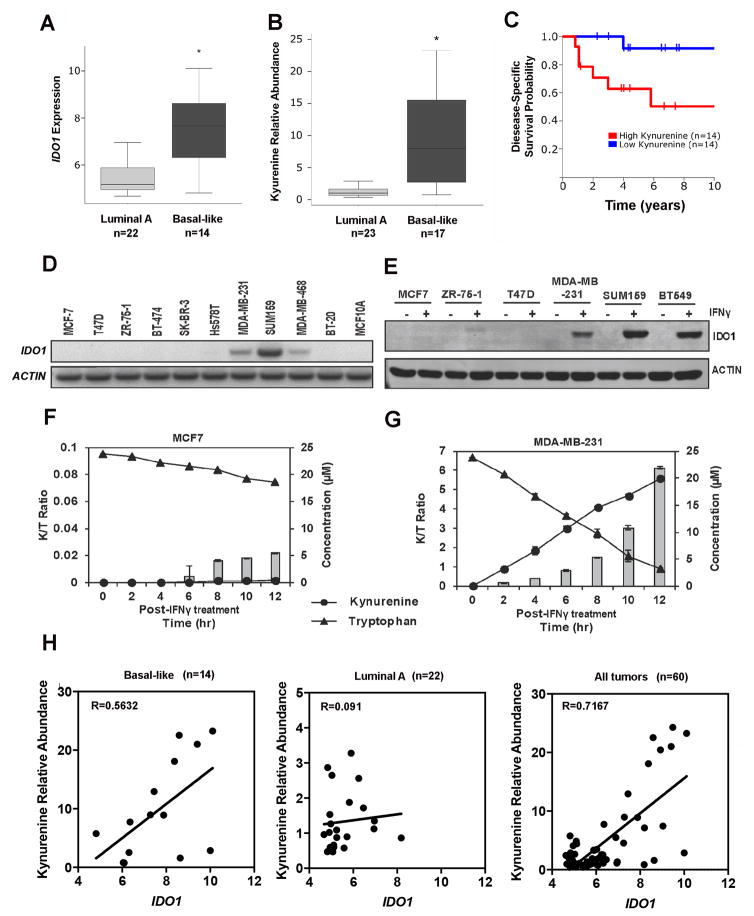
From the comprehensive Terunuma data set (29), we found that IDO1 mRNA was upregulated significantly in basal-like tumors when compared to the luminal subtype of breast tumors (Fig. 3A). Basal-like BC tissues also had higher concentrations of the tryptophan metabolite, kynurenine (Fig. 3B). These results corresponded with previous reports based on gene expression and metabolomics data (37). We also found that higher kynurenine was associated with poor disease-specific survival of a subset of BC patients (Fig. 3C).
Figure 3.
IDO1 expression in basal-like BC cell lines and patient samples. A. IDO1 mRNA expression was upregulated in basal-like breast tumors. Student t test was used to compare basal-like and luminal subgroups of BC based on a large microarray data set. B, Kynureinine was elevated in basal-type tumors based on metabolic profiling data. C, Kynurenine concentrations in BC tissues were associated with survival based on Kaplan-Meier analysis. D, Semi-quantitative RT-PCR analysis of basal IDO1 mRNA expression in 10 BC cell lines. E, Overnight treatment of IFNγ (100 ng/mL) induced IDO1 protein expression in ER−/TNBC cell lines, but not in ER+ cell lines. F and G, IDO1 enzymatic assays in MDA-MB-231 and MCF7, respectively. IFNγ (100 ng/mL) induced enzymatically active IDO1 protein expression causing tryptophan catabolism and kynurenine accumulation over time in MDA-MB-231, but not, MCF7 cells. H, Positive correlation between IDO1 mRNA expression and kynurenine relative abundance within tumor was observed in basal-like and all BC patient samples, but not in luminal A samples, based on the results of metabolite and mRNA analysis. Results in D and E are representative gel images from 2–3 independent experiments. Data in F and G are shown by mean ± SEM from three independent measurements.
Semi-quantitative PCR analysis of IDO1 mRNA in 10 BC cell lines consisting of three ER+ (MCF7, T-47D, ZR-75-1), two HER2+ (BT-474, SK-BR-3), five TNBC cell lines (HS578T, MDA-MB-231, SUM159, MDA-MB-468, BT20), and one immortalized breast epithelial cell line (MCF10A) revealed that IDO1 mRNA was not detectable in any ER+ and HER2+ cell lines, but was detected in three out of five TNBC cell lines (Fig. 3D). Despite detectable IDO1 mRNA in three TNBC cell lines, immunoblot analysis failed to detect IDO1 protein in any of these cell lines. However, IDO protein was readily induced by IFNγ in TNBC cell lines (MDA-MB-231, SUM159, and BT549), but not in ER+ cell lines (MCF7, T47D, and ZR-75-1) (Fig. 3E). To determine if IFNγ-induced IDO1 protein was enzymatically active, we performed HPLC analysis for tryptophan degradation and kynurenine accumulation in the culture media, which is represented as the kynurenine to tryptophan (K/T) ratio. MDA-MB-231 cells expressed enzymatically active IDO1 protein after overnight IFNγ treatment, as demonstrated by a time-dependent increase in K/T ratio (Fig. 3F). Although tryptophan concentration slightly decreased in MCF7 cells, kynurenine did not accumulate. We speculate that tryptophan was consumed for normal cell metabolism rather than catabolized by IDO1 in MCF7 cells (Fig. 3G). We observed a positive correlation between kynurenine concentrations and IDO1 mRNA expression within the basal-like BC subtype and for all BC samples in the Terunuma data set (29), but not with the luminal A subtype (Fig. 3H). Therefore, the in vivo metabolite data seem to support our in vitro experimental results.
IFNγ-induced IDO1 expression mediated by JAK/STAT1/IRF1 signaling pathway
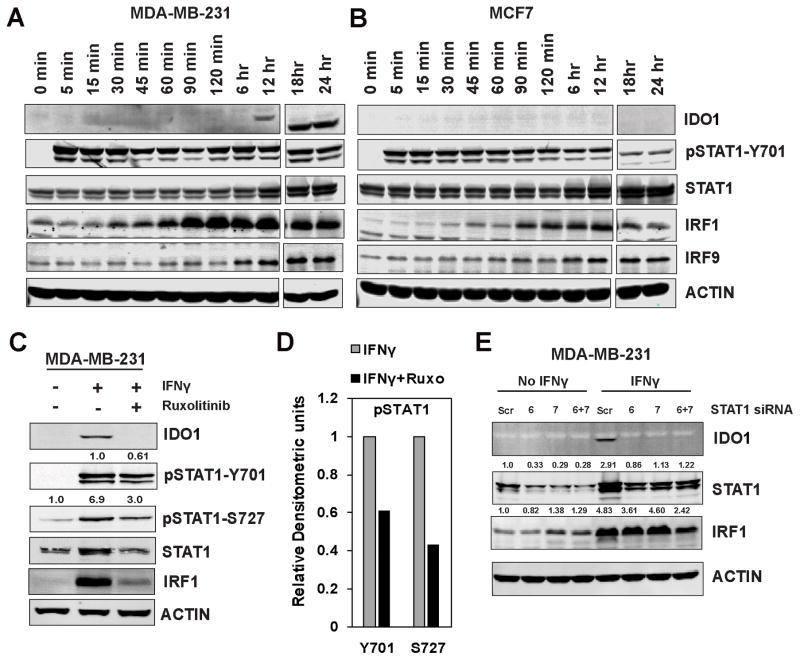
IFNγ-induced IDO1 expression is mediated through activation of the IFNGR1/JAK/STAT pathway (38). We investigated the STAT1/IRF1 pathway activity in MDA-MB-231 and MCF7 cells (Fig. 4A and B, respectively). By conducting a time-course analysis, we observed that STAT1 was phosphorylated at Y701 within minutes of addition of IFNγ, followed by significant upregulation of IRF1 expression by 90 minutes. Although IRF1 protein was expressed early and remained constant, IDO1 protein expression was not observed until 12 hours after IFNγ treatment in MDA-MB-231 cells (Fig. 4A). IRF1 protein expression in MCF7 cells was similar to that of MDA-MB-231 cells, but no IDO1 protein expression was observed in MCF7 cells (Fig. 4B).
Figure 4.
STAT1-mediated IFNγ signaling and IDO1 expression in BC cells. A and B, Immunoblot analysis of IFNγ-treated MDA-MB-231 and MCF7 cell lysates in time-course experiments and probing for indicated proteins. C, JAK inhibitor ruxolitinib effectively blocked IFNγ-induced STAT1-mediated IDO1 and IRF1 protein expression in MDA-MB-231 cells. D, Densitometric analysis of pSTAT1 bands. E, Knockdown of STAT1 with two different siRNAs effectively blocked IFNγ-mediated IDO1 protein expression. Results in A and B are blots from a single time course experiment. Results in C, D and E are representative of at least two independent experiments.
In addition, pretreatment of MDA-MB-231 with ruxolitinib, a JAK kinase inhibitor, followed by overnight exposure to IFNγ, significantly decreased IRF1 expression and blocked IDO1 induction in MDA-MB-231 cells (Fig. 4C) by effectively blocking STAT1 phosphorylation at Y701 and S727 (Fig. 4D). Similar results were observed in two other TNBC cell lines, SUM159 and BT549 (Supplementary Fig. S4). Knockdown of STAT1 by siRNA resulted in a loss of IFNγ-induced IDO1 protein expression in MDA-MB-231 cells (Fig. 4E), confirming that STAT1 was the primary transcription factor that regulated IDO1 transcription in response to IFNγ.
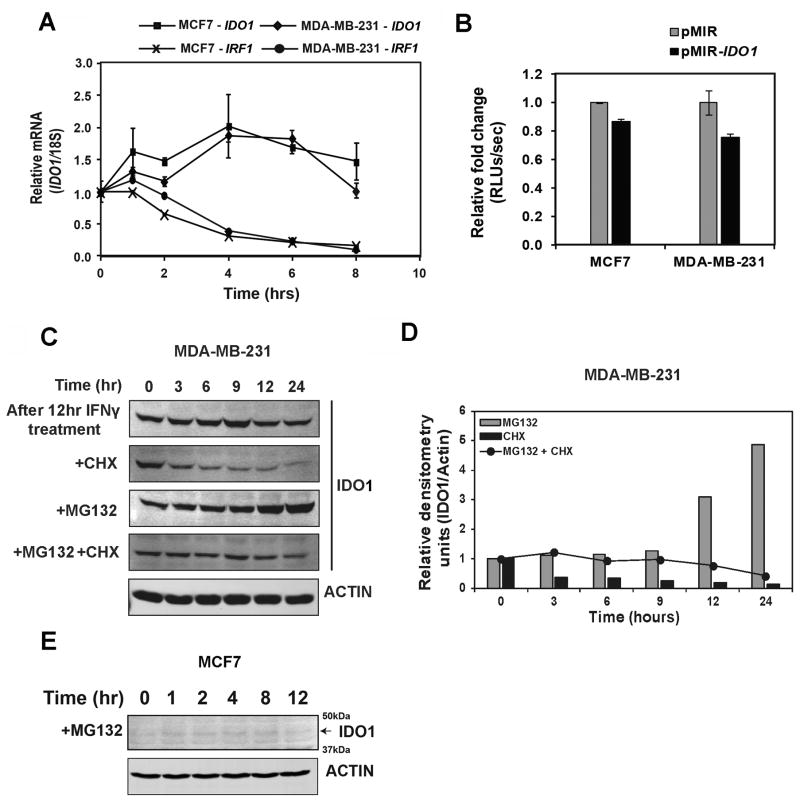
IDO1 mRNA and protein stability do not contribute to differential induction of IDO1 expression
Because IDO1 expression can also be regulated post-transcriptionally, we investigated IDO1 mRNA stability and proteasome-mediated degradation of the IDO1 protein. IDO1 mRNA was quite stable with no obvious difference between MDA-MB-231 and MCF7 cells when pretreated with IFNγ followed by actinomycin-D, an inhibitor of RNA transcription (Fig. 5A). Using an IDO1 3′-UTR reporter construct, we also found no difference in terms of 3′-UTR regulation of IDO1 mRNA stability between MCF7 and MDA-MB-231 cells (Fig. 5B). To assess the role of proteasome-mediated degradation of IDO1 and protein stability in contributing to differential expression of IDO1 protein between MCF7 and MDA-MB-231 cells, we treated MDA-MB-231 cells with the protein translation inhibitor CHX after 12 hours of IFNγ exposure and subsequent withdrawal. We observed a steady decrease in the detectable IDO1 protein with time by immunoblot analysis, suggesting protein degradation (Fig. 5C and D). On the other hand, treatment with MG132, a proteasome inhibitor, resulted in a gradual accumulation of IDO1 protein over time, suggesting proteasome-mediated degradation. Treatment with both CHX and MG132 resulted in steady levels of lDO1 protein. Similarly, in the absence of CHX or MG132 treatment and after IFNγ withdrawal, we observed steady state levels of IDO1 protein, as opposed to decreased IDO1 protein with time, which may have been due to sustained activity of STAT1/IRF1 transcription factors after cytokine activation. This also explains why IDO1 protein accumulated with MG132 treatment. However, under similar treatment of MCF7 cells with MG132, we did not observe any expression of IDO1 protein (Fig. 5E).
Figure 5.
Post-transcriptional regulation of IDO1 expression in BC cells. A, Analysis of IDO1 mRNA stability in MDA-MB-231 and MCF7 cells after both cell lines were stimulated with IFNγ (100ng/mL) for 4 hrs followed by treatments with actinomycin-D (1 μg/mL), a transcription inhibitor. B, Analysis of 3′-UTR based post-transcriptional regulation of IDO1 expression with a reporter in which the 3-UTR region of IDO1 was cloned into pmirGLO 3′-UTR reporter plasmid. C, Immunoblot analysis of the protein stability of IFNγ-induced IDO1 after treatment with CHX (50 μg/mL) and MG132 (5 μM) in MDA-MB-231 cells. D, Densitometric analysis of panel C for IDO1 expression in MDA-MB-231 cells. E, Immunoblot analysis of the protein stability of IFNγ-induced IDO1 after treatment with MG132 (5μM) in MCF7 cells. Data in A and B are shown by mean ± SEM from three independent measurements. Results in C, D and E are representative of at least two independent experiments.
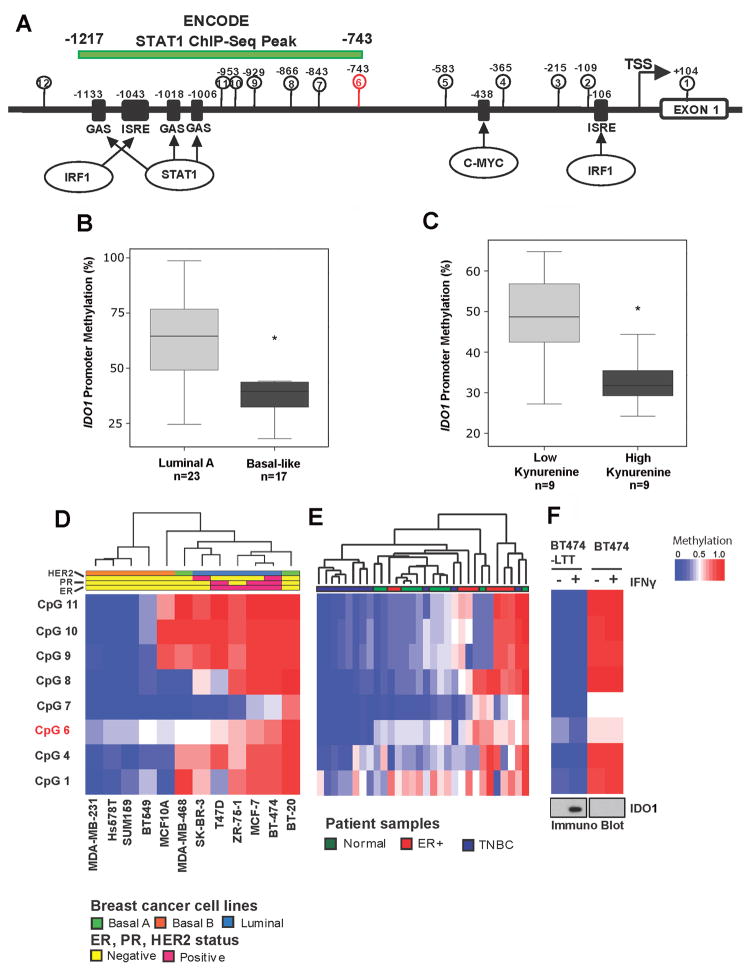
Hypomethylation of IDO1 promoter in basal-like subtype compared to other molecular subtypes
It has been suggested that epigenetic regulation plays a role in regulating IFNγ-induced IDO1 mRNA expression (39), so we investigated the promoter methylation status of IDO1 in BC cells. The IDO1 promoter was relatively CpG poor with just 11 CpG sites between +104 to −1200bp (Fig. 6a). CpG sites 2, 9, 10 and 11 were in close proximity to the palindromic gamma-activated sequences (GAS) and IFN-stimulated response elements (ISREs) in the IDO1 promoter. The STAT1 binding peak (−743 to −1217bp) identified by ENCODE ChIP-seq data overlapped with CpG sites 6–11, providing confidence that methylation of these critical CpG sites may regulate the recruitment of transcription factors and eventually regulate IDO1 gene transcription.
Figure 6.
IDO1 promoter is hypomethylated in basal-like BC subtype. A, Schematic representation of IDO1 gene promoter. B, Methylation status of probe cg10262052 at IDO1 promoter obtained from 450K methylation array data. Student’s t-test was used to compare basal-like vs. lumina A subtypes of breast tumors. C, Inverse correlation between IDO1 promoter methylation and kynurenine levels was observed in the tumors with matched DNA methylation and metabolite profiles. D and E, Heat maps illustrating CpG methylation levels in the IDO1 promoter as determined by pyrosequencing in BC cell lines and primary tumors. F, Intensity plot of CpG methylation levels in BT474-PTEN−LTT and its parental cell lines before and after IFNγ treatment. Immunoblot of IDO1 protein in the same cell line is shown at the bottom. Pyrosequencing data in D, E, and F were acquired from a single experiment. Only low quality data marked by the analysis software were repeated until satisfactory results were obtained.
We first analyzed IDO1 promoter methylation status in primary BC samples using the 450K methylation array data (29). Among the three probes for the IDO1 gene in the 450K array, only probe cg10262052 was present within the IDO1 promoter, and it was designated as CpG site 6 on the IDO1 promoter map (Fig. 6A). When we analyzed the methylation status of probe cg10262052, we found that basal-like tumors were hypomethylated when compared to luminal subtype breast tumor tissues (Fig. 6B). A survey of the TCGA dataset resulted in very similar observations; IDO1 promoter methylation was significantly lower in ER− tumors compared to ER+ tumors (Supplementary Fig. S5A) and in basal-like tumors compared to other subtypes and normal breast tissue (Supplementary Fig. S5B). IDO1 methylation and mRNA expression appeared to be inversely correlated in the TCGA datasets (Supplementary Fig. S5C). We also found that the hypomethylation of the IDO1 promoter was also associated with elevated kynurenine levels (Fig. 6C) suggesting that promoter hypomethylation is indicative of IDO1 enzymatic activities.
To extend methylation analysis to other CpG sites in the IDO1 promoter, we designed quantitative bisulfite pyrosequencing assays. A heat map representing the methylation levels of critical CpG sites in the IDO1 promoter was generated using bisulfite pyrosequencing data in a panel of BC cell lines (Fig. 6D). The pyrosequencing results show that the three TNBC cell lines (MDA-MB-231, HS578T and SUM159) were hypomethylated at all 8 CpG sites analyzed. On the other hand, ER+ (MCF7, T-47D and ZR-75-1) and HER2+ (SK-BR-3 and BT-474) cell lines are relatively hypermethylated. Normal breast epithelial cell line MCF10A was hypomethylated at 5 out of 8 CpG sites analyzed. MCF10A was readily inducible to express IDO1 protein with IFNγ treatment (data not shown). We extended our promoter methylation analysis to primary BC samples to confirm that the above observation is not specific to cell lines. The IDO1 promoter was significantly hypomethylated in TNBC tumors when compared to ER+ breast tumors, suggesting the role of promoter DNA methylation in IDO1 transcriptional regulation (Fig. 6E). The methylation levels of the IDO1 promoter in normal breast tissue samples were quite variable (Fig. 6E), but generally lie in between ER+ and TNBC tumors, suggesting that IDO1 promoter methylation levels in normal breast cancer tissue samples may depend on their histological composition. A brief survey of whole genome bisulfite sequencing data in luminal epithelial and myoepithelial cells from the Epigenomics Roadmap Project indeed revealed higher IDO1 promoter methylation in luminal epithelial cells and very low/absent methylation in myoepithelial cells (Supplementary Fig. 6).
DNA methylation regulates IDO1 promoter activity
Korkaya et al. reported that the PTEN-depleted BT474 cell line (BT474-PTEN−LTT) exhibits loss of epithelial markers and expresses mesenchymal markers compared to its parental line upon long-term treatment with Trastuzumab (20). By analyzing IDO1 promoter methylation of BT474-PTEN−LTT and the parental BT474 cell lines, we show that BT474-PTEN−LTT was significantly hypomethylated compared to its parental line (Fig. 6F) and that this hypomethylation corresponded with IFNγ-inducible IDO1 protein expression, as shown by immunoblot analysis. Data from this set of experiments suggests that IDO1 promoter methylation dictated IDO1 gene expression, and loss of IDO1 promoter methylation was a potential hallmark of epithelial-mesenchymal transitioning (EMT) in BC.
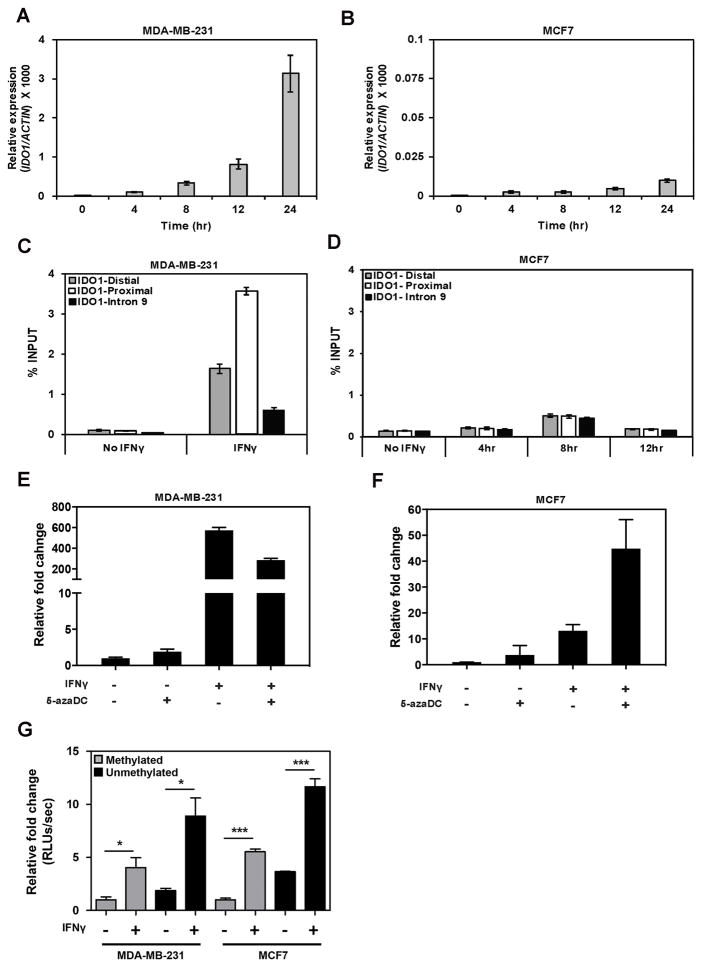
When treated with IFNγ, IDO1 mRNA level increased over time in both MCF7 and MDA-MB-231 cells. However, IDO1 mRNA expression was 430-fold higher in MDA-MB-231 than in MCF7 cells at 24hrs (Fig. 7A and B). To determine whether the differential expression observed was mainly due to a difference in transcriptional activity at the IDO1 promoter in these two cell lines, we performed ChIP assays to measure the RNA Pol II loading to the IDO1 promoter upon IFNγ treatment (Fig. 7C and D). After 4 hrs of IFNγ treatment, RNA Pol II was significantly more enriched on the IDO1 promoter in MDA-MB-231 cells than in MCF7 cells, even when compared to the highest Pol II enrichment at 8 hrs in MCF7 cells after IFNγ treatment, suggesting that differential IDO1 expression between MDA-MB-231 and MCF7 cells is due to a discrepancy in recruitment of RNA Pol II to the IDO1 promoter.
Figure 7.
IDO1 promoter methylation regulates IDO1 gene expression. A and B, Analysis of IDO1 mRNA expression by qRT-PCR in time course treatment of MCF7 and MDA-MB-231 cells with IFNγ (50ng/mL). C, ChIP analysis of RNA Poll II at IDO1 gene promoter in MDA-MB-231 cells after treatment with IFNγ for 4hr. D, ChIP analysis of RNA Pol II at IDO1 gene promoter in MCF7 cells after time course treatment with IFNγ for up to 8hr. E and F, qRT-PCR analysis of IDO1 expression in MDA-MB-231 (E) and MCF7 (F) cells after treatment with 5-azadC (2μM) for three days followed by IFNγ stimulation overnight. G, Luciferase reporter activity measured in MDA-MB-231 and MCF7 cells transfected with methylated or unmethylated IDO1 promoter reporter constructs after treatment with or without IFNγ. Data in this figure are shown by mean ± SEM from three independent measurements. Results are representative of at least two independent experiments.
To determine whether the IDO1 promoter activity is directly regulated by DNA methylation, we first investigated if 5-azadC, a demethylating agent, can reverse the epigenetic silencing of IDO1. When MCF7 cells were treated with 2 μM 5-azadC for 4 days, IDO1 mRNA expression increased by 7-fold (Fig. 7F). With combinatorial treatments of IFNγ and 5-azadC, IDO1 mRNA expression increased to 45-fold, indicating the synergism between demethylation treatment and IFNγ-induction. As expected, we did not observe similar synergy in MDA-MB-231 cells that were treated in the same way, because the IDO1 promoter is already hypomethylated in MDA-MB-231 cells; pretreatment with 5-azadC decreased the IFNγ-induced upregulation of IDO1 by nearly half (Fig. 7E). Next, we transfected either unmethylated or fully methylated pGL2-IDO1 promoter reporter constructs into MCF7 and MDA-MB-231 cells, then treated them with IFNγ overnight (Fig. 7G). Luciferase activity of the unmethylated promoter reporter was two-fold higher than the fully methylated promoter reporter under both basal and IFNγ stimulated conditions in MCF7 and MDA-MB-231 cells, confirming the role of CpG methylation in regulating IDO1 promoter activity.
Discussion
TILs serve as indicators of immune response and are a possible key to controlling tumor progression. Among the TILs, CD8+ cytotoxic T cells are responsible for killing cancer cells and other damaged cells. Mahmoud, S. M. et al. demonstrated the positive impact of CD8+ TILs on BC prognosis (2). Liu et al. revealed that high numbers of TILs in basal-like breast tumors proved to be a favorable prognostic indicator (40). In this study, using an in vitro coculture system, we showed that cytokines secreted by activated T cells induced significant changes in gene expression profiles of MDA-MB-231 and MCF7 cells. Analysis of TCGA data for 308 genes upregulated in both cell lines after coculture with activated T cells suggested that the expression profiles of these genes generally correlated with expression profiles of T cells, B cells, and CD8+ CTLs in BC samples. A subset of 96 genes whose expression positively correlated with CD8A expression displayed an even stronger positive correlation with CTL signature genes in TNBC. Some of these genes, such as IDO1, CXCL9, CXCL10, CXCL11, GBP5, PSMB9, and STAT1, are also identified as signature genes for the basal immune-activated (BILA) subgroup of TNBC patients (41). Immunosuppressive factors such as IDO1 and PD-L1 are intrinsic features of the TNBC subtype, as they are already expressed at basal level in the TNBC MDA-MB-231 cell line under unstimulated conditions. Increased CXCL9 and CXCL10 secretion by breast tumor cells may further lead to the recruitment of natural killer cells and CTLs as seen in murine models (42). Re-analysis of the Terunuma data set (29) also revealed a positive correlation between CD8 TILs and IDO1 mRNA expression within the basal-like BC subtype, but not with the luminal A subtype. Overall, results from our in vitro study support the model proposed by Gajewski et al. (43), in which it was suggested that induction of immunosuppressive pathways in cancer is intrinsically driven by the active immune system.
Evading antitumor immunity is a hallmark for the development and progression of cancer (44). One such mechanism used by tumors to escape from the host immune system is the activation of the IDO1 enzyme. Previous studies have demonstrated that IDO1 mRNA expression is higher in basal-like BC (37). Our analysis of several large gene expression data sets confirmed that IDO1 is indeed increasingly expressed more in ER− tumors than in ER+ tumors, notably in basal-like subtype compared to luminal subtype breast tumors. Although immunohistochemistry studies of IDO1 expression in BC patients have yielded somewhat contradicting results with regard to the expression of IDO1 protein and ER status (45), the metabolic profiling data analyzed in our study revealed more kynurenine in basal-like subtype compared to luminal subtype breast tumors. Higher kynurenine is associated with poor survival in BC patients. Using matched 450K-methylation array data in the same patient cohort, we found that differential IDO1 expression was associated with its promoter methylation status. We also demonstrated an inverse correlation between IDO1 promoter methylation and gene expression using a panel of BC cell lines. Only cells with a hypomethylated promoter such as MDA-MB-231 express enzymatically active IDO1 enzyme upon IFNγ stimulation. Since the methylation patterns observed in BC cell lines was recapitulated in primary BC samples, it is likely that IDO1 promoter methylation can predict inducibility of IDO1 by IFNγ in vivo. The analysis of kynurenine levels within tumor samples seems to support this notion, because kynurenine relative abundances are significantly correlated with IDO1 expression and promoter methylation in primary BC samples.
Activation of ERα signaling was reported to attenuate IFNγ-induced expression of HLA class II genes such as CIITA and HLA-DR in ER+ BC cell lines (46). However, careful examination of the IDO1 promoter sequence failed to identify valid estrogen response elements (EREs) and analysis of the ERα ChIP-seq data set in the ENCODE database also did not reveal any ERα binding event in IDO1 locus in estradiol-treated MCF7 and T47D cells. Therefore, we do not believe that ERα signaling is directly involved in the regulation of IDO1 expression. Epigenome Roadmap data suggests that IDO1 promoter methylation is higher in normal luminal epithelial cells and very low/absent methylation in myoepithelial cells. Thus, IDO1 methylation in breast cancer tissue may be indicative of the cell type at the origin of the tumor, or reflect cellular differentiation processes associated with tumor progression. Basal-like breast cancer cells including MDA-MB-231 cells were shown to display myoepithelial features (47). Nevertheless, the hypomethylation of IDO1 promoter may be an intrinsic marker of basal-like or TNBC patients. We found that promoter hypermethylation observed in BT474 cells was completely lost and IFNγ inducibility of IDO1 was restored in its isogenic BT474-PTEN−LTT cell line, which has undergone extensive morphological changes associated with EMT compared to its parental line. Even though the molecular mechanism that drives this phenotype is currently unclear, these results indicate that IDO1 promoter methylation is dynamic and it can undergo demethylation under appropriate conditions. PTEN loss has also been reported to contribute to upregulation of PD-L1 expression in TNBC tumors (16), which could also play a similar role here.
IDO1 is a classic IFNγ-inducible gene. IRF1, like IDO1, is also an IFNγ-responsive gene activated through STAT1. With IFNγ treatments, a second wave of IDO1 transcription is mediated by the IRF1 transcription factor that binds to the ISRE in the IDO1 promoter. Using time-course experiments we demonstrated that the IFNGR1/JAK/STAT1/IRF1 signaling cascade was functional in both ER+ and TNBC cell lines. However, IDO1 protein synthesis was delayed compared to the rapid expression of IRF1. Thus, STAT1 together with IRF1 may be required for driving efficient transcription of IDO1 gene in response to IFNγ. However, siRNA knockdown of STAT1 and treatment with JAK inhibitor ruxolitinib blocked IFNγ-mediated IDO1 induction, suggesting that STAT1 is the primary transcription factor that drives IDO1 expression. IDO1 protein is also regulated by mRNA (36) and protein stability (48) upon IFNγ stimulation. We found that the stability of IDO1 mRNA was similar between MCF7 and MDA-MB-231 cell lines. In fact, IDO1 mRNA was more stable than IRF1. A previous report demonstrated that SOCS3 was responsible for proteasomal degradation of the IDO1 protein in dendritic cells (48). However, treatment of MCF7 cells with proteasome inhibitor MG132 failed to induce IDO1 expression with IFNγ, suggesting that protein stability may not be the primary reason for lack of IDO1 expression in MCF7 cells. Although we cannot completely rule out other mechanisms, promoter methylation status seemed the most distinct difference between MCF7 and MDA-MB-231 cells, representing ER+ and TNBC subtypes, respectively. Several CpG sites in the IDO1 promoter are located in close proximity to GAS binding sites and overlap with a STAT1 ChIP-seq peak observed in IFNγ-treated Hela-S3 cells. We speculate that these methylated CpG sites may recruit methyl-binding proteins or other repressor complexes to obstruct STAT1 from binding to GAS sequences, and thus control its promoter activity.
In summary, our data reveal an important distinguishing attribute of TNBC, which has significant implications for therapy. TNBC often consists of aggressive tumors with high mutational burden, and may thus be highly immunogenic. Consistent with this, the more T cells that were recruited into the tumor, the better the survival outcome. However, we found that TNBC cells also tended to have highly inducible IDO1 expression, with demethylated promoters and production of functional IDO1 in response to T cell–derived IFNγ. Therefore, TNBC patients may represent ideal candidates for IDO1 inhibitor therapy. Many TNBC patients already have a robust immune response, if the counter-regulatory inhibition by IDO1 could be removed, their overall prognosis would be further improved. This might be true particularly for patients with a pre-existing T cell–inflamed tumor microenvironment (13). We further predict that IDO1 promoter hypomethylation is a molecular feature of TNBCs that represents a T cell–inflamed tumor phenotype, and this this subclass of BC could greatly benefit from IDO1 inhibitor–based immunotherapy.
Supplementary Material
Acknowledgments
Grant Support
This work was partially supported by grants from the NIH R21CA185833, R01GM113242 and Georgia Cancer Center Startup Fund (to HS). This research was also supported by the following grants to Arun Sreekumar: NIH R21CA185516, UO1CA179674 NSF DMS-1161759, Funds from the Alkek Center for Molecular Discovery, Agilent Technologies Center of Excellence in Mass Spectrometry at Baylor College of Medicine and P30CA125123 awarded to the Dan L. Duncan Cancer Center. Dr. Huidong Shi is a Georgia Cancer Coalition Distinguished Scientist.
The authors thank Mr. Rahul Shinde for helping with HPLC analysis.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest. 2007;117:1137–46. doi: 10.1172/JCI31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29:1949–55. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 3.Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, et al. Tumor-Associated Lymphocytes As an Independent Predictor of Response to Neoadjuvant Chemotherapy in Breast Cancer. J Clin Oncol. 2010;28:105–13. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 4.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31:860–7. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 5.Ruffell B, Au A, Rugo HS, Esserman LJ, Hwang ES, Coussens LM. Leukocyte composition of human breast cancer. Proc Natl Acad Sci U S A. 2012;109:2796–801. doi: 10.1073/pnas.1104303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobrzanski MJ, Reome JB, Hylind JC, Rewers-Felkins KA. CD8-mediated type 1 antitumor responses selectively modulate endogenous differentiated and nondifferentiated T cell localization, activation, and function in progressive breast cancer. J Immunol. 2006;177:8191–201. doi: 10.4049/jimmunol.177.11.8191. [DOI] [PubMed] [Google Scholar]
- 7.Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, et al. Association between CD8+T-cell infiltration and breast cancer survival in 12 439 patients. Ann Oncol. 2014;25:1536–43. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 8.Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, et al. Tumor-Infiltrating Lymphocytes and Response to Neoadjuvant Chemotherapy With or Without Carboplatin in Human Epidermal Growth Factor Receptor 2-Positive and Triple-Negative Primary Breast Cancers. J Clin Oncol. 2015;33:983–91. doi: 10.1200/JCO.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 9.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–65. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godin-Ethier J, Pelletier S, Hanafi LA, Gannon PO, Forget MA, Routy JP, et al. Human activated T lymphocytes modulate IDO expression in tumors through Th1/Th2 balance. J Immunol. 2009;183:7752–60. doi: 10.4049/jimmunol.0901004. [DOI] [PubMed] [Google Scholar]
- 15.Tang X, Lin CC, Spasojevic I, Iversen ES, Chi JT, Marks JR. A joint analysis of metabolomics and genetics of breast cancer. Breast Cancer Res. 2014;16:415. doi: 10.1186/s13058-014-0415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res. 2014;2:361–70. doi: 10.1158/2326-6066.CIR-13-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poschke I, De Boniface J, Mao Y, Kiessling R. Tumor-induced changes in the phenotype of blood-derived and tumor-associated T cells of early stage breast cancer patients. Int J Cancer. 2012;131:1611–20. doi: 10.1002/ijc.27410. [DOI] [PubMed] [Google Scholar]
- 18.Pathania R, Ramachandran S, Mariappan G, Thakur P, Shi H, Choi JH, et al. Combined Inhibition of DNMT and HDAC Blocks the Tumorigenicity of Cancer Stem-like Cells and Attenuates Mammary Tumor Growth. Cancer Res. 2016;76:3224–35. doi: 10.1158/0008-5472.CAN-15-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo D, Wilson JM, Harvel N, Liu J, Pei L, Huang S, et al. A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of breast cancer cells. J Transl Med. 2013;11:57. doi: 10.1186/1479-5876-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korkaya H, Kim GI, Davis A, Malik F, Henry NL, Ithimakin S, et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell. 2012;47:570–84. doi: 10.1016/j.molcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 25.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laich A, Neurauter G, Widner B, Fuchs D. More Rapid Method for Simultaneous Measurement of Tryptophan and Kynurenine by HPLC. Clin Chem. 2002;48:579–81. [PubMed] [Google Scholar]
- 28.Pei L, Choi JH, Liu J, Lee EJ, McCarthy B, Wilson JM, et al. Genome-wide DNA methylation analysis reveals novel epigenetic changes in chronic lymphocytic leukemia. Epigenetics. 2012;7:567–78. doi: 10.4161/epi.20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terunuma A, Putluri N, Mishra P, Mathe EA, Dorsey TH, Yi M, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124:398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 31.Thompson CB, Lindsten T, Ledbetter JA, Kunkel SL, Young HA, Emerson SG, et al. CD28 activation pathway regulates the production of multiple T-cell-derived lymphokines/cytokines. Proc Natl Acad Sci U S A. 1989;86:1333–7. doi: 10.1073/pnas.86.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iglesia MD, Vincent BG, Parker JS, Hoadley KA, Carey LA, Perou CM, et al. Prognostic B-cell Signatures Using mRNA-Seq in Patients with Subtype-Specific Breast and Ovarian Cancer. Clin Cancer Res. 2014;20:3818–29. doi: 10.1158/1078-0432.CCR-13-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werner-Felmayer G, Werner ER, Fuchs D, Hausen A, Reibnegger G, Wachter H. Characteristics of interferon induced tryptophan metabolism in human cells in vitro. Biochim Biophys Acta. 1989;1012:140–7. doi: 10.1016/0167-4889(89)90087-6. [DOI] [PubMed] [Google Scholar]
- 34.Babcock TA, Carlin JM. Transcriptional activation of indoleamine dioxygenase by interleukin 1 and tumor necrosis factor alpha in interferon-treated epithelial cells. Cytokine. 2000;12:588–94. doi: 10.1006/cyto.1999.0661. [DOI] [PubMed] [Google Scholar]
- 35.Robinson CM, Hale PT, Carlin JM. The role of IFN-gamma and TNF-alpha-responsive regulatory elements in the synergistic induction of indoleamine dioxygenase. J Interferon Cytokine Res. 2005;25:20–30. doi: 10.1089/jir.2005.25.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan W, Collado-Hidalgo A, Yufit T, Taylor M, Varga J. Modulation of cellular tryptophan metabolism in human fibroblasts by transforming growth factor-beta: selective inhibition of indoleamine 2,3-dioxygenase and tryptophanyl-tRNA synthetase gene expression. J Cell Physiol. 1998;177:174–86. doi: 10.1002/(SICI)1097-4652(199810)177:1<174::AID-JCP18>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 37.Jacquemier J, Bertucci F, Finetti P, Esterni B, Charafe-Jauffret E, Thibult M-L, et al. High expression of indoleamine 2,3-dioxygenase in the tumour is associated with medullary features and favourable outcome in basal-like breast carcinoma. Int J Cancer. 2012;130:96–104. doi: 10.1002/ijc.25979. [DOI] [PubMed] [Google Scholar]
- 38.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–74. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 39.Xue Z-T, Sjögren H-O, Salford LG, Widegren B. An epigenetic mechanism for high, synergistic expression of indoleamine 2,3-dioxygenase 1 (IDO1) by combined treatment with zebularine and IFN-γ: Potential therapeutic use in autoimmune diseases. Mol Immunol. 2012;51:101–11. doi: 10.1016/j.molimm.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Lachapelle J, Leung S, Gao D, Foulkes WD, Nielsen TO. CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 2012;14:R48. doi: 10.1186/bcr3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua S, et al. Comprehensive Genomic Analysis Identifies Novel Subtypes and Targets of Triple-negative Breast Cancer. Clin Cancer Res. 2015;21:1688–98. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bronger H, Kraeft S, Schwarz-Boeger U, Cerny C, Stockel A, Avril S, et al. Modulation of CXCR3 ligand secretion by prostaglandin E2 and cyclooxygenase inhibitors in human breast cancer. Breast Cancer Res. 2012;14:R30. doi: 10.1186/bcr3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–22. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Soliman H, Rawal B, Fulp J, Lee J-H, Lopez A, Bui M, et al. Analysis of indoleamine 2–3 dioxygenase (IDO1) expression in breast cancer tissue by immunohistochemistry. Cancer Immunol Immunother. 2013;62:829–37. doi: 10.1007/s00262-013-1393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mostafa AA, Codner D, Hirasawa K, Komatsu Y, Young MN, Steimle V, et al. Activation of ERalpha signaling differentially modulates IFN-gamma induced HLA-class II expression in breast cancer cells. PLoS One. 2014;9:e87377. doi: 10.1371/journal.pone.0087377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon LA, Mulligan KT, Maxwell-Jones H, Adams M, Walker RA, Jones JL. Breast cell invasive potential relates to the myoepithelial phenotype. Int J Cancer. 2003;106:8–16. doi: 10.1002/ijc.11172. [DOI] [PubMed] [Google Scholar]
- 48.Orabona C, Pallotta MT, Volpi C, Fallarino F, Vacca C, Bianchi R, et al. SOCS3 drives proteasomal degradation of indoleamine 2,3-dioxygenase (IDO) and antagonizes IDO-dependent tolerogenesis. Proc Natl Acad Sci U S A. 2008;105:20828–33. doi: 10.1073/pnas.0810278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.