Abstract
Differential scanning fluorimetry (DSF) is a fluorescence-based assay to evaluate protein stability by determining protein melting temperatures. Here, we describe the application of DSF to investigate aminoacyl-tRNA synthetase (AARS) stability and interaction with ligands. Employing three bacterial AARS enzymes as model systems, methods are presented here for the use of DSF to measure the apparent temperatures at which AARSs undergo melting transitions, and the effect of AARS substrates and inhibitors. One important observation is that the extent of temperature stability realized by an AARS in response to a particular bound ligand cannot be predicted a priori. The DSF method thus serves as a rapid and highly quantitative approach to measure AARS stability, and the ability of ligands to influence the temperature at which unfolding transitions occur.
Keywords: Differential scanning fluorimetry, Aminoacyl-tRNA synthetase, Histidyl-tRNA synthetase, SYPRO Orange, tRNA, Amino acid
1. Introduction: Development of differential scanning fluorimetry assay
Aminoacyl-tRNA synthetases (AARS) synthesize aminoacylated tRNA in one of the earliest steps of protein synthesis. The aminoacylation reaction involves binding of the three substrates amino acid, tRNA, and ATP to each AARS to produce the final aminoacylated tRNA product. Traditionally, biophysical techniques to measure AARS enzyme stability and unfolding have included spectroscopic methods such as intrinsic tryptophan fluorescence in the presence of a denaturant like urea or guanidine-HCl, monitoring fluorescence with 1-anilino-8-naphthalene-sulfonic (ANS) dye, and circular dichroism [1–4]. An example of a classic method for determining a protein melting point without the use of exterior dyes is differential scanning calorimetry (DSC) [5,6]. Differential scanning calorimetry calculates the amount of heat required to raise the temperature of a sample by a given number of degrees, computing the difference between the sample and a buffer reference cell. This approach has been employed to study protein stability in several AARS systems [7,8]. Isothermal titration calorimetry (ITC) has also proven useful for studying the interactions of small molecule ligands with AARSs is [9–11]. In addition to providing binding constants and stoichiometry, ITC is a direct process that does not involve chemical alteration of either the enzyme or the substrate, while providing detailed thermodynamic parameters for a given interaction. The chief disadvantage of ITC is that it requires expensive instrumentation and, depending on the specific heat of a particular interaction, may require relatively high concentrations of the target protein in order derive adequate signal. Accordingly, both calorimetric approaches can provide valuable information, but typically require large quantities of protein, extensive analysis, and are low throughput.
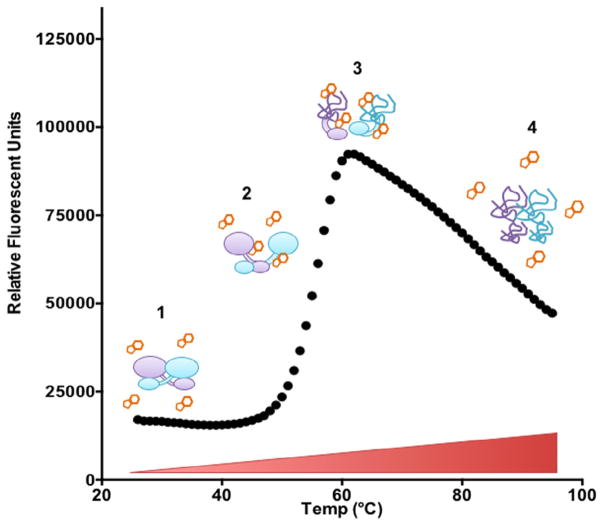
Differential scanning fluorimetry (DSF) is a method that enables researchers to monitor thermal denaturation of purified proteins with a fluorescent reporter. The DSF assay relies on the interactions of the fluorophore dye SYPRO Orange with the exposed regions of partially unfolded proteins rich in hydrophobic amino acids (Fig. 1). The folded structure of proteins sequesters these hydrophobic amino acid residues from water, burying them in the highly folded regions of the protein’s core domains. As the solution temperature is increased incrementally, denaturation of the protein exposes increasing numbers of hydrophobic residues. This creates an opportunity for SYPRO Orange dye binding, which gives rise to an increase in fluorescence (excitation and emission of 490 and 575 nm, respectively).
Fig. 1.
Molecular basis of differential scanning fluorimetry. (1) Folded protein is incubated in solution with SYPRO Orange dye. (2) Temperature increases, causing protein to unfold and SYPRO Orange dye to bind exposed hydrophobic residues. This results in an increase in fluorescence signal. (3) Protein is denatured and maximum SYPRO Orange dye binding is achieved. (4) Denatured protein begins to aggregate and SYPRO Orange dye binding decreases. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
DSF was first developed as a drug screen for small molecule studies, but is also applicable as a screen to optimize stabilizing buffer conditions for aaRS crystallization [12–14] and to obtain ligand binding parameters [15]. Many examples of DSF have been reported for proteins such as Carbonic Anhydrase-II [16], cytoplasmic sulfotransferase 1C1 [17], cAMP-dependent protein kinase (PKA) [18], MAPK13 [19], and pregnane xenobiotic receptor (PXR) [20] to name a few [21]. A particular advantage of the DSF method is that the experimental set up can be implemented in a high-throughput mode by use of a 96-well microtiter plate [22]. The measurements can be performed in a real-time PCR instrument that is standard equipment for most molecular biology core facilities [23]. Fluorescence output is plotted against temperature to determine the melting temperature (Tm), by fitting the Boltzmann equation to fluorescent data [24].
Here, we present a set of protocols for determining the apparent melting transitions of aminoacyl-tRNA synthetases by use of DSF. After a presentation of the specific methodology for a single experiment, additional examples are discussed in which the melting transitions of three bacterial Class II AARS enzymes are determined. In addition, we also show how DSF can be used to study the binding of cognate amino acids and inhibitors to AARSs, and how DSF can be used to evaluate tRNA binding to certain AARS enzymes.
2. Differential scanning fluorimetry method
2.1. Protein (aaRS) preparation and purification
Purified E. coli enzymes Alanyl-tRNA synthetase (AlaRS), Histidyl-tRNA synthetase (HisRS) and Threonyl-tRNA synthetase (ThrRS) were isolated by Ni-column chromatography as previously described [25–27]. This represents the minimum degree of recommended purification; some enzymes may require additional steps for best results. We typically employ an additional step, such as ion exchange chromatography. The effect of affinity tags on protein unfolding has not been investigated systematically. Given that affinity tags have limited structure, there is little rationale to expect major deviations from the melting temperatures determined in the absence of the 6X-His tag. The results might also be affected by non-specifically bound residual RNA or DNA, which can sometimes co-purify with AARS. Extra purification steps were not required for the specific enzymes describe in this report, but may be required for other AARSs. The final preparations are typically dialyzed into a storage buffer consisting of 50 mM HEPES pH 7.5, 150 mM KCl, 1 mM DTT and 40% glycerol, and then concentrated before storage at −20 °C.
2.2. tRNA purification and preparation
2.3. Reaction set-up for differential scanning fluorimetry
All reagents used in the DSF assays are listed in Table 1. All buffers were pre-made in sterile 0.22 μM filtered deionized water.
The dilution buffer used in our thermal shift experiments is identical to the standard reaction buffer typically used for aminoacylation assays: 20 mM HEPES pH 7.5, and 150 mM KCl. However, salt concentrations, pH, and buffers can alter AARS Tm and therefore should be evaluated. Final glycerol concentrations in our preparations varied between 1% to 10%, as this is a stabilizing agent for HisRS [30]. Glycerol can be excluded from the enzyme preparation to examine its effects on Tm.
Prepare a 10X stock of enzyme at a concentration of 100 μM. A useful starting point with respect to final aaRS concentration in the individual DSF reaction wells is 10 μM. Protein concentration can contribute to DSF profiles and therefore a range of AARS concentrations should be evaluated for optimal solubility and stability. In our assay to determine Tm and the effect of bound ligand on protein Tm, approximately 0.5 mg of protein distributed over 48 wells was used. Based on the requirement of 10 μg of sample for each well, approximately 1 mg of enzyme would be needed to fill a 96 well microtiter plate.
Prepare substrates to be evaluated for binding and stability as 10X concentrated stocks (ie. amino acid, nucleotide (ATP or AMP), and tRNA. In our experiments, typical 10X concentrated substrate stocks were 50 mM amino acid and 100 μM for tRNA. These were diluted to final concentrations of 5 mM and 10 μM, respectively.
Dilute SYPRO Orange (Molecular Probes #S6651) (5000X concentrated solution) to a 20X stock in a dark Eppendorf tube (Sigma SIAL311AA2C). If using a transparent Eppendorf tube, protect from light by wrapping tube in foil.
Typical reactions for determination of melting temperature and the effect of ligands on AARS stability featured 25 μL reaction volumes. (The issue of the number or replicates to ensure robust statistics is treated below.) The final enzyme concentration that we routinely use is 10 μM.
The sample mix is prepared by combining enzyme, ligand, buffer, and SYPRO Orange in an Eppendorf tube. Refer to Table 2 (samples without ligands) and Table 3 (samples containing ligands) for typical examples of initial test reagent concentrations.
Once samples are prepared, immediately transfer 20 μL of the sample mix to the wells of a 96 well plate either individually, or using a multichannel pipette. Avoid generating air bubbles in the sample well(s) during this process. The reactions in Table 2 were prepared at a final volume of 25 μL to ensure that each well gets a full 20 μL, accounting for some loss in the transfer process. Following transfer to the microtiter plate, reactions containing SYPRO Orange should not be allowed to stand for longer than 15–20 min before reading in PCR instrument.
Cover plate with ThermalSeal as listed in Table 1.
Place the 96-well plate in a swinging bucket centrifuge configured with adapters for 96-well plates and centrifuge at 700–1000×g to ensure that the entire sample volume resides the bottom of the well before placing the plate in a PCR instrument.
Table 1.
Reagents necessary for DSF experiment.
| Reagent or supply | Supplier | Catalogue number |
|---|---|---|
| Ultrol grade HEPES buffer | CalBioChem | 391338 |
| KCl | Fisher | 138149 |
| SYPRO Orange Dye 5000 X | Molecular probes | S6651 |
| MillexGV 0.22 uM filter | MerkMillipore | 309603 |
| Sterile syringe 5 mL | BD Biosciences | 309603 |
| Siliconized low retention micocentrifuge tubes | Fisherbrand | 02-681-311 |
| 96 well PCR Plate non-warping | Phenix Research | MPS-3580-NW |
| ThermalSeal RT2 Fi | Phenix Research | LMT-RT2-RR |
Table 2.
Suggested mix for screening new aaRS for Tm.
| Sypro Orange + AARS in buffer alone | |||
|---|---|---|---|
|
| |||
| 6X SyprO for 0.5 mg/mL WT HisRS in 20 uL well | |||
|
| |||
| Final [concentration] | vol.uL/ sample | Sample for 3 wells | |
| 20X SyproOrange | 6X | 2.5 | 10.5 |
| Diluted 10X Enzyme Stock (100 uM) | 10 uM | 2.5 | 10.5 |
| Dilution Buffer | na | 10.5 | 42 |
| Final volume | – | 25 | 75 |
Table 3.
Suggested mix for screening new aaRS for substrate binding.
| SyproOrange + AARS + aminoacid in buffer | |||
|---|---|---|---|
|
| |||
| 6X SyprO for 0.5 mg/mL WT HisRS in 20 uL well | |||
|
| |||
| Final [concentration] | vol.uL/ sample | Sample for 3 wells | |
| 20X SyproOrange | 6X | 2.5 | 10.5 |
| Diluted 10X Enzyme Stock (100 uM) | 10 uM | 2.5 | 10.5 |
| 10X amino acid stock (50 mM) | 5 mM | 2.5 | 10.5 |
| Dilution Buffer | NA | 15 | 35 |
| Final volume | – | 25 | 75 |
2.4. Melting temperature determination by use of qPCR device
These studies used the Advanced Genome Technologies Core (AGTC) at University of Vermont qPCR instrument Applied Biosystems 7500 fast real-time qPCR. The basic program for dye-binding temperature scans is designated in the instructions for the ABI instrument as the “Protein Thermal Shift Solution” (http://www.slideshare.net/ThermoFisher/protein-thermal-shift-solution-using-applied-biosystems-realtime-pcr-systems), and has been described previously [13,24]. Briefly, the ROX (carboxy-X-rhodamine) detector for the ABI 7500 fast instrument was selected with no passive dye reference to monitor SYPRO Orange fluorescence emission. In a typical run, the scan is programmed to initiate at 25 °C, followed by a temperature gradient in which the samples are heated at a scan rate of 1 °C per minute until a final temperature of 95 °C is reached. During the heating process, fluorescence intensity is measured every 1 °C. Under these conditions, a complete scan of a 96 well plate can typically be completed in 1.5 h.
2.5. Data analysis
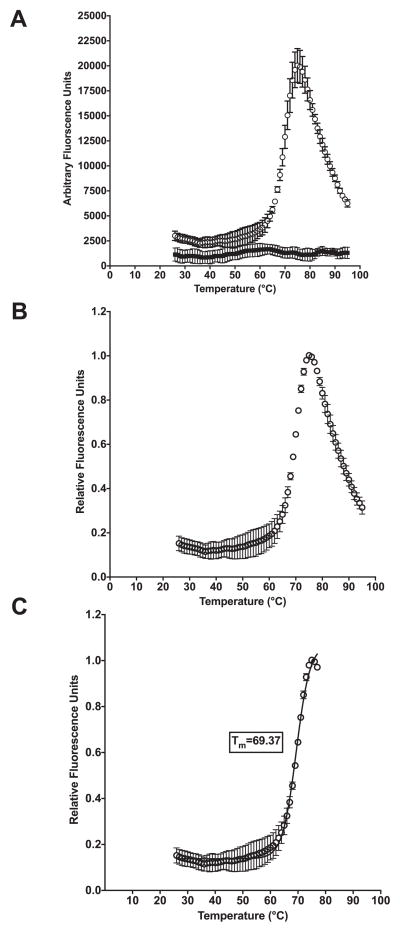
Once the PCR run is complete, export fluorescent data (component data from ABI 7500 Fast instrument, in .xls format) into an Excel sheet. In this format, the corresponding wells should be annotated with the requisite sample information (i.e. well number, enzyme concentration, substrate concentration). The fluorescent data can then be exported from the annotated Excel sheet into Graphpad Prism 6 for Tm analysis. Fluorescent values are typically plotted on the Y-axis, while temperature is plotted on the X-axis (Fig. 2A). Alternatively, relative fluorescence can be calculated by dividing all fluorescence data in a given well by the maximum fluorescence value achieved during the run within the same well and plotted on the Y-axis (Fig. 2B). The thermal shift data are truncated by excluding fluorescent values after maximum fluorescence intensity is achieved (before fluorescent values begin to decrease as a result of protein aggregation) (Fig. 2C). To determine the melting temperature (Tm) the Boltzmann equation is fit to fluorescent data:
where F(T) is the fluorescence at a particular temperature, F(pre) is the fluorescence before the transition or melting at the start of the region of analysis (ROA), F(post) is the fluorescence after the transition or melting at the end of the ROA, Tm is the melting temperature, and C is a slope factor that corresponds to the enthalpy of the reaction (applied biosystems Protein Thermal Shift Studies: https://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_095141.pdf). If the data is a monophasic curve Boltzmann will be an appropriate fit to determine Tm (Fig. 2C).
Fig. 2.
Representative data analysis with lysozyme. (A) Arbitrary fluorescence units for 6 well run with Lysozyme (empty circles) and buffer control incubated with SYPRO Orange (filled squares) obtained from ABI real-Fast PCR instrument plotted against temperature. (B) Data shown as relative fluorescence units plotted against temperature. (C) Relative fluorescence data truncated and fit with the Boltzmann equation to determine melting temperature (Tm).
ΔTm is substrate dependent and can be determined by the equation shown below:
Here Tm represents the protein in the presence of ligand while T0 is the unbound protein. This will yield a positive ΔTm if binding ligand increases in stability (Tm > T0). A negative ΔTm value would imply a decrease in stability as a function of ligand binding. For samples that have more then one transition of unfolding or multi-phasic melt curves, the Boltzmann equation cannot be used to reliably fit the data to determine Tm (Fig. 2C). However, the derivative method can be used to determine the Tm. By plotting the first derivative of fluorescence emission as a function of temperature (−dF/dT) each unfolding transition can be easily evaluated as the peak values (TmD) [13].
While additional software is available to aid DSF data analysis, it is not completely necessary, as Excel and Graphpad Prism 6 were employed for the analysis reported here. As a useful resource, Frank Niesen and colleagues have prepared and made available Microsoft Excel spreadsheets in which to analyze DSF data (ftp://ftp.sgc.ox.ac.uk/pub/biophysics).
2.6. Replicates and controls
Technical Replicates for each condition (sample mix) should minimally be done in triplicate within one 96-well plate run. The reactions shown in this protocol (Tables 2 and 3) can be prepared in a single Eppendorf tube and are sufficient for a triplicate analysis based on three wells of a microtiter plate. A single set of technical replicates provides an indication of the variability between wells for the PCR instrumentation being used. We suggest repeating this step an additional one to two times for a total of 6–9 wells of three technical replicates. When dealing with a new system, each substrate or ligand tested in the assay should be evaluated for SYPRO Orange dye binding by incubation in buffer with dye in the absence of enzyme. We have determined that SYPRO Orange does not bind tRNA and therefore tRNA substrates can be tested for binding with aminoacyl-tRNA synthetases.
3. Results: Sample applications employing lysoyzyme and aminoacyl-tRNA synthetases
3.1. Lysozyme test experiment
Hen egg white lysozyme (Sigma) serves as a useful test protein to acquire familiarity with the method. A 60 μL reaction containing 0.1 mg/mL lysozyme in 50 mM HEPES pH 7.0, 100 mM KCl serves as a simple test experiment. In this standard DSF reaction, lysozyme exhibits an apparent published melting temperature transition (Tm) of 70.9 ± 0.7 °C [31]. In our apparatus and using these conditions, we determined a Tm of 69.4 ± 0.1 °C for lysozyme (Fig. 2C).
3.2. Measurements of AARS protein stability
We applied the DSF method to three Class IIa AARS bacterial enzymes purified by Ni-column chromatography with no additional purification necessary (Table 4). While all AARSs catalyze the same basic reaction, the Class IIa enzymes also share several structural and biochemical characteristics [32]. The Class IIa AARS proteins have conserved catalytic domain consists of a seven-stranded β-sheet flanked by α-helices, and nearly all (PheRS is the exception) catalyze amino acid transfer to the 3′ hydroxyl of the tRNA A76. With limited exceptions [33], the Class II AARSs are dimers or multimers of dimers [34–36], and recognize the major groove of the acceptor stem of their cognate tRNA. Here, we focused on three typical Class IIa enzymes (AlaRS, HisRS, and ThrRS), and their interactions with canonical substrates: amino acid, ATP, and tRNA.
Table 4.
Melting point (Tm) values of ThrRS, HisRS and AlaRS in presence of various substrates.
| Enzyme | Apo Tm (°C) | Amino Acid 5 mM Tm (°C) | ATP 10 mM Tm (°C) | In vitro tRNA 10 uM Tm (°C) |
|---|---|---|---|---|
| ThrRS | 53.9 ± 0.5 | § | 53.8 ± 0.4 | N.D. |
| HisRS | 53.4 ± 1.1 | 57.2 ± 1.1 | 55.8 ± 0.5 | 55.6 ± 0.1 |
| AlaRS | 55.5 ± 0.2 | 55.6 ± 0.1 | 55.0 ± 0.1 | 55.9 ± 0.04 |
Tm ± standard error of the mean.
Multi-phase melting transitions observed. N.D. not determined.
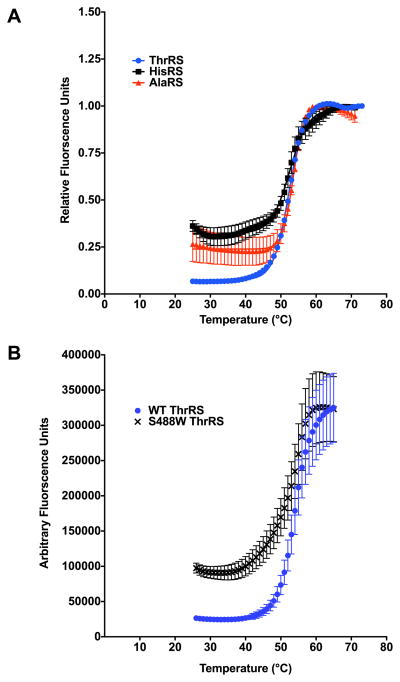
The application of the DSF method to E. coli HisRS, AlaRS, and ThrRS demonstrated that all three have very comparable temperature stabilities, falling in the range of 53 °C–55 °C (Fig. 3A) and (Table 4). In addition, DSF was shown to reliably and reproducibly determine Tm values for two different HisRS preparations only varying by approximately 1 °C. We also evaluated a ThrRS active site substitution [37], S488W, by DSF to determine if changes in temperature stability accompanied the mutant substitution (Fig. 3B). While a significant difference in Tm relative to wild type was not detected for the S488W substitution, there was a 4-fold increase in pre-melt fluorescence at lower temperatures (Fig. 3B). This may be a result of adding an additional hydrophobic residue for SYPRO Orange binding but is likely indicative of an altered conformation of the S488W ThrRS protein that is different from WT folded structure, as this residue is found in the active site pocket. Similar studies examining the effects of disease related AARS substitutions have been investigated by DSF for mitochondrial AspRS [38].
Fig. 3.
Protein melting curves for representative bacterial Class II aminoacyl-tRNA synthetases. (A) WT ThrRS (blue circles), HisRS (black diamond) and AlaRS (red squares) in the absence of substrates. (B) WT ThrRS (blue circles) and active site substitution S488W (black x). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The experimental conditions described here to determine Tm represent useful initial conditions to begin analysis of an AARS with an unknown apparent melting temperature. These conditions have been successfully used in the characterization of purified human AARS enzymes (data not shown), and represent a convenient experimental starting point. Other buffer conditions (salt concentration, pH, addition of MgCl2) should be explored to maximize AARS stability when the enzymes exhibit less consistent solubility behavior. Additionally, a negative ΔTm can be indicative of a positive hit for binding [24]. One possible explanation for a lower ΔTm value relative to Tm is that ligands or substrates bind the unfolded protein much more tightly [39].
3.3. AARS amino acid binding
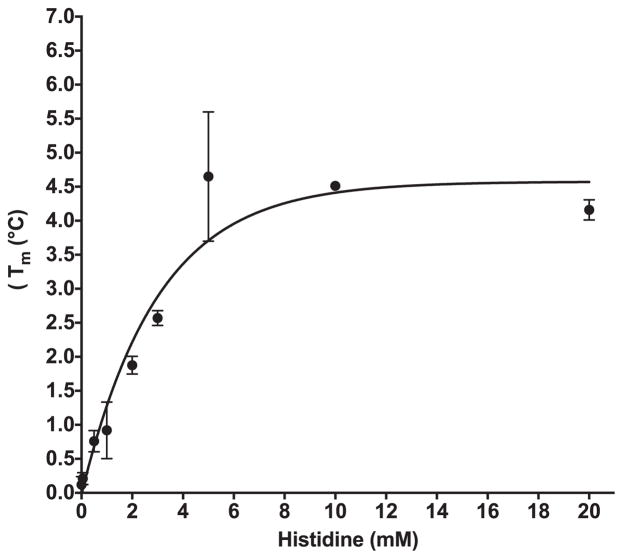
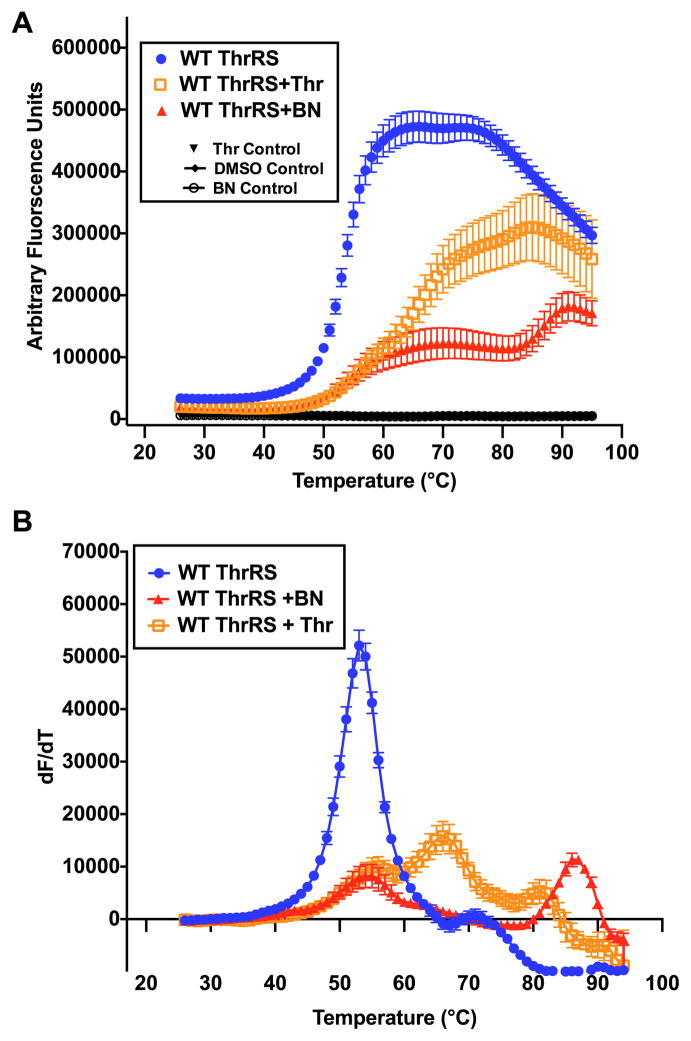
As DSF was originally designed for small molecule studies, it can easily be applied to evaluate amino acid binding by AARS enzymes, assuming a significant shift in Tm occurs upon binding [40]. By titrating increasing amounts of histidine while holding HisRS enzyme concentration constant, the change in Tm (ΔTm) can be recorded as a function of amino acid concentration (Fig. 4). A maximum increase in Tm of 4.4 °C was achieved with 10 mM histidine (Fig. 4). In some cases, ligand dependent changes in Tm values determined by DSF have been shown to correlate with activity and calorimetrically determined binding measurements [41]. However, it appears that the concentration of histidine required to produce 50% of the ligand dependent increase in melting temperature is higher than the Kd determined by spectrofluorometric methods. In other systems, Tm shifts greater than 4 °C have been shown to correlate to IC50 values less then 1 μM [17]. In our experiments, a maximal apparent Tm increase of 4.4 °C was observed upon titration of histidine to HisRS; this required several orders of magnitude higher concentrations than the 35.4 μM KM determined kinetically [42]. Notably, we observed that not all Class II enzymes respond identically to the presence of amino acid. As shown in Table 4, AlaRS exhibited essentially no shift in Tm in the presence of saturating amounts of alanine, in contrast to the results obtained when HisRS was titrated with corresponding amino acid. ThrRS binding to threonine resulted in a three phase melting curve. This multi-phase melting transition is best shown as a first derivative plot (Fig. 6A).
Fig. 4.
Stabilization of Histidyl-tRNA synthetase as a function of histidine concentration.
Fig. 6.
Evaluation of threonine and borrelidin binding by ThrRS (A) ThrRS (blue circles) fluorescence thermal melting data as a function of temperature shown with threonine binding (empty orange squares) and BN binding (filled red triangles) (B) Fluorescence data converted to first derivative plot. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. DSF analysis of tRNA binding
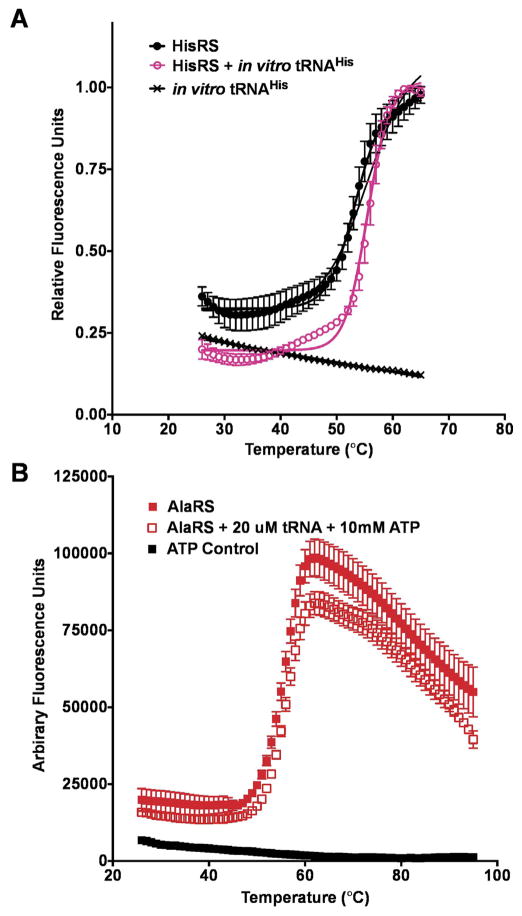
We used DSF to monitor the effect of tRNA binding on AARS stability as a function of Tm. Notably, we observed that in vitro transcribed tRNA did not exhibit appreciable SYPRO Orange binding. The addition of tRNAHis was accompanied by a 2.3 °C shift in Tm for HisRS (Fig. 5A). In addition to the temperature shift, tRNAHis binding to the HisRS enzyme was also associated with a 20% reduction in baseline fluorescence at pre-melting temperatures. There may be several different possible explanations for the observed reduction in fluorescence at 25 °C as a function of tRNA binding. First, tRNA binding might act to protect HisRS hydrophobic residues from SYPRO Orange dye binding. Second, the HisRS enzyme might adopt a different conformation upon tRNA binding such that hydrophobic residues typically available for SYPRO Orange binding in the Apo state are buried. In contrast to the effect of tRNA binding on the HisRS Tm, adding tRNAAla to AlaRS only produced a 0.5 °C shift in Tm for AlaRS. While the mechanism by which AlaRS recognizes the specificity determining G3:U70 base pair has been exhaustively investigated [43,36,44], much less is known about structural changes in the enzyme that accompany tRNA binding. The absence of a distinctive shift in the Tm for AlaRS in the presence of saturating ATP and in vitro transcribed tRNAAla is in striking contrast to HisRS (Fig. 5B). This may indicate that the enthalpic and hydrophobic contributions of AlaRS tRNAAla binding compete and mask changes in Tm [24]. Generally, high ΔTm values are associated with hydrophobic binding [24].
Fig. 5.
Aminoacyl-tRNA synthetase stabilization upon tRNA binding. (A) Thermal melting curve of HisRS in the absence (black diamonds) and presence of tRNAHis pink empty circles) (B) Thermal melting curve of AlaRS (filled red squares) and in the presence of ATP and tRNAAla (empty red squares). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Inhibitor binding studies
Previously, DSF has been used to examine histidine, AMP, and amino acid inhibitor analog binding to the Trypanosoma cruzi HisRS histidine-binding pocket [45]. Here, we examined the response of Tm for E. coli ThrRS to the presence of the natural product inhibitor Borrelidin (BN) [46]. ThrRS is a potent angiogenic factor that induces growth of blood vessels [47], and BN is a potent inhibitor of this activity [47]. The binding of borrelidin to ThrRS is best described by slow tight binding kinetics [48], and leads to inhibition of aminoacylation function and angiogenesis. Interestingly, binding of BN binding to the ThrRS enzyme produced a multiphase melting curve similar to that seen for threonine (Fig. 6A). To determine the apparent Tm values for this data, we used a first derivative plot (Fig. 6B). By use of this analysis, a tri-phasic curve was derived for threonine binding with Tm values of 55.8 °C, 66.5 °C and 81.1 °C, and a biphasic-melting curve was obtained for BN with Tm values of 54.6 °C and 86.7 °C. Interpretation of the biphasic curves observed when E. coli ThrRS is incubated with amino acid and inhibitor might be facilitated by application of DSF to individual domains of ThrRS. Ideally, one could determine which Tm corresponds to unfolding of a specific domain(s). While a complete physical description of these complex binding events awaits further study, these observations underscore the remarkable variability in the response of different AARSs to their cognate ligands.
4. Concluding remarks
Here, we have described detailed experimental conditions for application of the DSF protocol to three different bacterial AARS enzymes. For these enzymes, representative experiments demonstrate that this approach can be useful to monitor amino acid and inhibitor binding. Given that DSF provides a rapid “first pass” approach to examine ligand binding, Tm information can be complimented by other biophysical experiments, such as crystallography [45], provided the necessary protein regents are in abundance. Supporting experiments can also include dynamic light scattering (DLS), synchrontron radiation circular dichroism (SRCD), and size exclusion chromatography (SEC) [38]. These techniques could be useful under conditions where DSF gives complex results. Isothermal titration calorimetry experiments can also aid in validation of an apparent Kd for amino acid and inhibitor binding studies while also providing much higher accuracy. Circular dichroism experiments may be helpful in determining what structural elements are changing as a consequence of substrate binding. Future applications of SYPRO Orange and DSF for AARS studies not described here include protein-protein interactions and cellular thermal shift experiments [49].
5. Helpful links
Excel Sheets and other useful DSF data Analysis information by Frank Niesen: ftp://ftp.sgc.ox.ac.uk/pub/biophysics
Using DSF for KD Analysis video: http://www.jove.com/video/51809/determination-protein-ligand-interactions-using-differential-scanning
ABI Protein Thermal Shift Studies: https://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_095141.pdf)
Trial download for ABI PCR instrumentation protein melt data analysis: www.appliedbiosystems.com/proteinmelt
Acknowledgments
The authors are grateful to Tim Hunter for his assistance in setting up the ABI instrumentation and valuable conversations. Also, authors would like to thank the UVM’s Advanced Genome Technology Core (AGTC) for use of the RT-PCR instrumentation for these studies. Dr. Susan Robey-Bond is acknowledged for advice on experimental design and comments on the manuscript.
Funding for this work was provided by National Institutes of Health [GM54899 to C.S.F. and T32 HL007594-30 to J.A.A.
References
- 1.Banerjee B, Banerjee R. Urea unfolding study of E. coli Alanyl-tRNA synthetase and its monomeric variants proves the role of C-terminal domain in stability. J Amino Acids. 2015;2015:805681. doi: 10.1155/2015/805681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park YC, Bedouelle H. Dimeric tyrosyl-tRNA synthetase from Bacillus stearothermophilus unfolds through a monomeric intermediate. A quantitative analysis under equilibrium conditions. J Biol Chem. 1998;273(29):18052–18059. doi: 10.1074/jbc.273.29.18052. [DOI] [PubMed] [Google Scholar]
- 3.Reed CJ, Bushnell S, Evilia C. Circular dichroism and fluorescence spectroscopy of cysteinyl-tRNA synthetase from Halobacterium salinarum ssp. NRC-1 demonstrates that group I cations are particularly effective in providing structure and stability to this halophilic protein. PLoS One. 2014;9(3):e89452. doi: 10.1371/journal.pone.0089452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dignam JD, Qu X, Chaires JB. Equilibrium unfolding of Bombyx mori glycyl-tRNA synthetase. J Biol Chem. 2001;276(6):4028–4037. doi: 10.1074/jbc.M006840200. [DOI] [PubMed] [Google Scholar]
- 5.Sturtevant JM. Heat capacity and entropy changes in processes involving proteins. Proc Natl Acad Sci USA. 1977;74(6):2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sturtevant JM. Biochemical applications of differential scanning calorimetry. Annu Rev Phys Chem. 1987;38:463–488. [Google Scholar]
- 7.Ray S, et al. Fusion with anticodon binding domain of GluRS is not sufficient to alter the substrate specificity of a chimeric Glu-Q-RS. Protein J. 2014;33(1):48–60. doi: 10.1007/s10930-013-9537-7. [DOI] [PubMed] [Google Scholar]
- 8.Dignam JD, Nada S, Chaires JB. Thermodynamic characterization of the binding of nucleotides to glycyl-tRNA synthetase. Biochemistry. 2003;42(18):5333–5340. doi: 10.1021/bi030031h. [DOI] [PubMed] [Google Scholar]
- 9.Neuenfeldt A, et al. Thermodynamic properties distinguish human mitochondrial aspartyl-tRNA synthetase from bacterial homolog with same 3D architecture. Nucleic Acids Res. 2013;41(4):2698–2708. doi: 10.1093/nar/gks1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes SJ, et al. Functional asymmetry in the lysyl-tRNA synthetase explored by molecular dynamics, free energy calculations and experiment. BMC Struct Biol. 2003;3:5. doi: 10.1186/1472-6807-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blais SP, et al. TRNAGlu increases the affinity of glutamyl-tRNA synthetase for its inhibitor glutamyl-sulfamoyl-adenosine, an analogue of the aminoacylation reaction intermediate glutamyl-AMP: mechanistic and evolutionary implications. PLoS One. 2015;10(4):e0121043. doi: 10.1371/journal.pone.0121043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poklar N, et al. PH and temperature-induced molten globule-like denatured states of equinatoxin II: a study by UV-melting, DSC, far- and near-UV CD spectroscopy, and ANS fluorescence. Biochemistry. 1997;36(47):14345–14352. doi: 10.1021/bi971719v. [DOI] [PubMed] [Google Scholar]
- 13.Huynh K, Partch CL. Analysis of protein stability and ligand interactions by thermal shift assay. Curr Protoc Protein Sci. 2015;79:28.9.1–14. doi: 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simeonov A. Recent developments in the use of differential scanning fluorometry in protein and small molecule discovery and characterization. Expert Opin Drug Discovery. 2013;8(9):1071–1082. doi: 10.1517/17460441.2013.806479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matulis D, et al. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry. 2005;44(13):5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 16.Klinger AL, et al. Inhibition of carbonic anhydrase-II by sulfamate and sulfamide groups: an investigation involving direct thermodynamic binding measurements. J Med Chem. 2006;49(12):3496–3500. doi: 10.1021/jm058279n. [DOI] [PubMed] [Google Scholar]
- 17.Vedadi M, et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci USA. 2006;103(43):15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrne DP, et al. CAMP-dependent protein kinase (PKA) complexes probed by complementary differential scanning fluorimetry and ion mobility-mass spectrometry. Biochem J. 2016 doi: 10.1042/BCJ20160648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yurtsever Z, et al. First comprehensive structural and biophysical analysis of MAPK13 inhibitors targeting DFG-in and DFG-out binding modes. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbagen.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekiguchi M, et al. High-throughput evaluation method for drug association with pregnane X receptor (PXR) using differential scanning fluorometry. J Biomol Screening. 2013;18(9):1084–1091. doi: 10.1177/1087057113491826. [DOI] [PubMed] [Google Scholar]
- 21.Boivin S, Kozak S, Meijers R. Optimization of protein purification and characterization using Thermofluor screens. Protein Expr Purif. 2013;91(2):192–206. doi: 10.1016/j.pep.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Pantoliano MW, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screening. 2001;6(6):429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 23.Lo MC, et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332(1):153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 25.Pasman Z, et al. Substrate specificity and catalysis by the editing active site of Alanyl-tRNA synthetase from Escherichia coli. Biochemistry. 2011;50(9):1474–1482. doi: 10.1021/bi1013535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan W, Augustine J, Francklyn C. A tRNA identity switch mediated by the binding interaction between a tRNA anticodon and the accessory domain of a class II aminoacyl-tRNA synthetase. Biochemistry. 1996;35(21):6559–6568. doi: 10.1021/bi952889f. [DOI] [PubMed] [Google Scholar]
- 27.Minajigi A, Francklyn CS. RNA-assisted catalysis in a protein enzyme: the 2′-hydroxyl of tRNA(Thr) A76 promotes aminoacylation by threonyl-tRNA synthetase. Proc Natl Acad Sci USA. 2008;105(46):17748–17753. doi: 10.1073/pnas.0804247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milligan JF, et al. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA template. Nucl Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francklyn CS, et al. Methods for kinetic and thermodynamic analysis of aminoacyl-tRNA synthetases. Methods. 2008;44(2):100–118. doi: 10.1016/j.ymeth.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francklyn C, Harris D, Moras D. Crystallization of histidyl-tRNA synthetase from Escherichia coli. J Mol Biol. 1994;241(2):275–277. doi: 10.1006/jmbi.1994.1498. [DOI] [PubMed] [Google Scholar]
- 31.Rosa N, et al. Meltdown: a tool to help in the interpretation of thermal melt curves acquired by differential scanning fluorimetry. J Biomol Screening. 2015;20(7):898–905. doi: 10.1177/1087057115584059. [DOI] [PubMed] [Google Scholar]
- 32.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 33.Nishio K, Kawakami M. Purification and properties of alanyl-tRNA synthetase from Bombyx mori: a monomeric enzyme. J Biochem. 1984;96(6):1867–1874. doi: 10.1093/oxfordjournals.jbchem.a135021. [DOI] [PubMed] [Google Scholar]
- 34.Naganuma M, et al. Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization. Proc Natl Acad Sci USA. 2009;106(21):8489–8494. doi: 10.1073/pnas.0901572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dignam JD, et al. Allosteric interaction of nucleotides and tRNA(ala) with E. coli alanyl-tRNA synthetase. Biochemistry. 2011;50(45):9886–9900. doi: 10.1021/bi2012004. [DOI] [PubMed] [Google Scholar]
- 36.Naganuma M, et al. The selective tRNA aminoacylation mechanism based on a single G*U pair. Nature. 2014;510(7506):507–511. doi: 10.1038/nature13440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egri SB. UVM College of Arts and Sciences College Honors Theses. University of Vermont; 2015. The Role of Protein Structure Conformational Change in Secondary Functions of Threonyl-tRNA Synthetase; p. 42. [Google Scholar]
- 38.Sauter C, et al. Neurodegenerative disease-associated mutants of a human mitochondrial aminoacyl-tRNA synthetase present individual molecular signatures. Sci Rep. 2015;5:17332. doi: 10.1038/srep17332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cimmperman P, et al. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys J. 2008;95(7):3222–3231. doi: 10.1529/biophysj.108.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vivoli M, et al. Determination of protein-ligand interactions using differential scanning fluorimetry. J Vision Exp. 2014;91:51809. doi: 10.3791/51809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bullock AN, et al. Structural basis of inhibitor specificity of the human protooncogene proviral insertion site in moloney murine leukemia virus (PIM-1) kinase. J Med Chem. 2005;48(24):7604–7614. doi: 10.1021/jm0504858. [DOI] [PubMed] [Google Scholar]
- 42.Hawko SA, Francklyn CS. Covariation of a specificity-determining structural motif in an aminoacyl-tRNA synthetase and a tRNA identity element. Biochemistry. 2001;40(7):1930–1936. doi: 10.1021/bi0025316. [DOI] [PubMed] [Google Scholar]
- 43.Hou YM, Francklyn C, Schimmel P. Molecular dissection of a transfer RNA and the basis for its identity. TIBS. 1989;14:233–237. doi: 10.1016/0968-0004(89)90033-9. [DOI] [PubMed] [Google Scholar]
- 44.Sokabe M, et al. The structure of alanyl-tRNA synthetase with editing domain. Proc Natl Acad Sci USA. 2009;106(27):11028–11033. doi: 10.1073/pnas.0904645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koh CY, et al. A binding hotspot in Trypanosoma cruzi histidyl-tRNA synthetase revealed by fragment-based crystallographic cocktail screens. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 8):1684–1698. doi: 10.1107/S1399004715007683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang P, et al. Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase. Nat Commun. 2015;6:6402. doi: 10.1038/ncomms7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirando AC, et al. Aminoacyl-tRNA synthetase dependent angiogenesis revealed by a bioengineered macrolide inhibitor. Sci Rep. 2015;5:13160. doi: 10.1038/srep13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruan B, et al. A unique hydrophobic cluster near the active site contributes to differences in borrelidin inhibition among threonyl-tRNA synthetases. J Biol Chem. 2005;280(1):571–577. doi: 10.1074/jbc.M411039200. [DOI] [PubMed] [Google Scholar]
- 49.Jafari R, et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9(9):2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]