Abstract
Gynecological cancer is difficult to be diagnosed at early stages. The relatively high mortality rate has been a serious issue accordingly. We herein reported a diagnosis method by using circulating tumor cells (CTCs) which have been extensively explored as a potential tool for diagnostics and prognostics of ovarian cancers. Nonetheless, the detection of CTCs still remains a challenge because of the difficulty in isolating them from whole blood samples since they are shed into the vasculature from primary tumors and circulate irregularly in the bloodstream in extremely low concentrations. In this work, we reported a new, integrated microfluidic system capable of (1) red blood cells lysis, (2) white blood cell (WBC) depletion via a negative selection process, and (3) capture of target cancer cells from whole blood samples using aptamer-binding technology. Furthermore, this is the first time that an aptamer was used to capture ovarian cancer cells owing to its high affinity. The new microfluidic chip could efficiently perform the entire process in one hour without human intervention at a high recovery rate and a low false positive detection rate when compared with antibody-based systems. A high recovery rate for the isolation of CTCs within a short period of time has been reported when compared to the traditional negative or positive selection approach by using traditional antibody biomarkers. More importantly, “false positive” results from WBCs could be significantly alleviated due to the high specificity of the cancer cell-specific aptamers. The developed integrated microfluidic system could be promising for the isolation and detection of CTCs, which could be used for early diagnosis and prognosis of cancers.
ABBREVIATIONS AND NOMENCLATURE
- BSA
Bovine serum albumin
- CaAM
Calcein Green AM
- CTCs
Circulating tumor cells
- DMEM
Dulbcco's Minimum Essential Medium
- EDTA
Ethylenediaminetetraacetic acid
- EGFR
Epidermal growth factor receptor
- EMVs
Electromagnetic valves
- EpCAM
Epithelial cell adhesion molecule
- FDA
Food and Drug Administration
- FBS
Fetal bovine serum
- MPC
Magnetic particle separator
- OCCs
Ovarian cancer cells
- PBS
Phosphate buffer saline
- PDMS
Polydimethylsiloxane
- PMMA
Polymethylmethacrylate
- RBCs
Red blood cells
- SELEX
Systematic evolution of ligands and exponential enrichment
- WBCs
White blood cells
I. INTRODUCTION
Gynecological cancer is still challenging to be diagnosed at early stages. Bio-imaging approaches such as computerized tomography scan and ultrasound examination are still the main gold standards in hospitals. Unfortunately, these approaches could not detect cancer carcinoma at early stages due to their resolution issues. Especially, ovarian cancer carcinoma often occurs deep inside women's body without any symptom at early stages. Recently, circulating tumor cells (CTCs) have shown great promise as biomarkers for cancer diagnostics and prognostics, as their presence in the bloodstream can suggest metastatic relapse. CTCs have been reported to originate from metastatic lesions, indicating that these cancer cells can detach from the primary tumor and enter the vasculature.1 Previous studies have demonstrated that cancer patients can be classified based on the severity of tumor metastasis, and the enumeration of CTCs can predict the stage of cancer carcinoma progression/development.2,3 Although the quantification of CTC levels in human blood has consequently drawn considerable attention in recent years, efforts to do so have generally been unsuccessful due to the exceptionally low concentrations of CTCs in blood samples. To date, various CTC isolation/quantification approaches have been explored extensively,4 though only a few, such as the CellSearch system, are Food and Drug Administration (FDA)-approved.5 The major challenge for CTC detection is to isolate rare cancer cells from whole blood and reduce the number of blood cells (red and white blood cells) such that one can accurately identify the target cancer cells without false positive or false negative results. Previous studies have detected CTCs from whole blood samples with either microstructures6 or cancer-specific biomarkers (e.g., epithelial cell adhesion molecule, EpCAM).7,8 Unfortunately, microstructures may often damage cells. On the other hand, antibody-based cancer biomarkers are generally of low specificity and can generate false positive results. Furthermore, such cancer biomarkers may also yield a false negative issue since their affinity to cancer cells may be low.7,8 Recently, other approaches using inertial forces,9 microfilters,10 dielectrophoretic forces,11,12 or other technologies13–16 have been extensively explored. However, they all are characterized by the significant generation of either false positive or false negative results.
Typical processes involved in the isolation of CTCs from whole blood include red blood cell (RBC) lysis, white blood cell (WBC) depletion (negative selection), and CTC capture (positive selection).12 Recently, our group developed a microfluidic system for WBC depletion via anti-CD45-coated beads.17 However, it did not lyse RBCs, nor did it feature a positive selection process to directly capture CTCs. Note that the lysis of RBCs could affect the subsequent capture of CTCs and therefore play an important role in sample pretreatment. Recently, cancer cell-specific aptamers have been used in place of the EpCAM antibody used for capturing CTCs in the CellSearch device. For instance, an aptamer (CX-BG1-10-A) was previously screened from an on-chip cell-SELEX (systematic evolution of ligands by the exponential enrichment) process;18,19 CX-BG1-10-A was demonstrated to be highly specific to ovarian cancer cells (OCCs) and could bind them with high affinity,20 thereby allowing for their isolation from mixed cell lines.21 Note that it has not yet been used for human blood samples. Furthermore, an aptamer-based strategy against an epidermal growth factor receptor (EGFR) was reported to capture glioblastoma.22 These aptamers were coated on the substrate and have captured cancer cells successfully. However, the capture rate was less than the one with the EGFR-antibody. Likewise, 1 ml of whole blood spiking with “colon” cancer cell lines was tested by using aptamers on a microfluidic device.23 While the sample fluid flew through microchannels which were equipped with micropillars, the CTCs could be captured since the microchannel/micropillars was immobilized with specific aptamers and micropillars were used to increase the contact surface to achieve a high recovery rate of cancer cells. However, after the isolation process and counting by fluorescent-labelling, they could not collect cancer cells for further use. Similarly, aptamers with multiple fluorescent labeling were used to detect CTCs from whole blood for “breast” cancer and “prostate” cancer.24 However, it did not perform isolation of CTCs and did not use any microfluidic device. In our work, we integrated several functional microfluidic devices (micropumps, microvalves, and micromixers) in a single chip such that the entire process including RBC lysis, WBC depletion, and CTC isolation could be automated for “ovarian cancer.” With this approach, it may reduce human intervention and reagent consumption. The entire process could be completed within 1 h. Furthermore, the commonly observed false-positive issue could be greatly alleviated.
In order to detect rare CTCs in whole blood samples herein, an automatic, rapid, and sensitive system capable of performing all experimental processes without any human intervention is necessary. Therefore, we surface-coated the aforementioned OCC-specific aptamer onto magnetic beads to serve as an affinity reagent for capturing CTCs. The aptamer could capture cancer cells in the human whole blood. This is the first time that this kind of aptamer has been used in the human whole blood, not simple cell lines. Furthermore, the high specificity of this aptamer toward WBCs was further demonstrated, which play an important role for CTCs to exclude false-positive results. Furthermore, a new integrated microfluidic system that could perform three critical processes for CTC isolation on a single chip via cell selection and enrichment methods was reported. The advantages of integrating three important steps to isolate CTCs from human whole blood are described as follows. First, the RBC lysis process could enhance the chance of capturing target cancer cells. Depletion of the WBCs could lower the false positive ratio efficiently. After these two processes, aptamer-coated beads could capture cancer cells from whole blood with a high recovery rate. In this work, we further optimized the RBC lysis, WBC depletion, and CTC capture rates. We also demonstrated that CTCs could be isolated successfully with a relatively low false positive rate.
The novelty of this work has been summarized as follows.
-
1.
We optimized the RBC lysis process to reduce reagent consumption such that it could be performed within a short period of time. The RBC lysis process could improve the recovery rates of cancer cells in the following CTC isolation processes.
-
2.
Negative and positive selection processes were performed to increase the recovery rates of cancer cells and alleviate the false-positive issue. Furthermore, the entire process for CTC isolation from human whole blood was automated on a single chip without any human intervention.
-
3.
The aptamer could capture cancer cells specifically in the human whole blood. This is the first time that the CX-BG1-10-A aptamer for “ovarian cancer” was used in the human whole blood. Furthermore, the high specificity of this aptamer toward WBCs has never been reported, which play an important role in alleviating the false-positive issue.
II. MATERIALS AND METHODS
A. Working process of on-chip CTC isolation
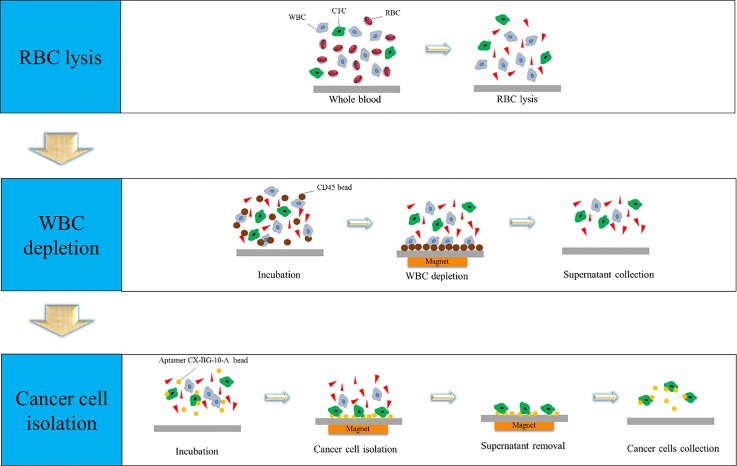
The whole cell selection and enrichment process included a RBC lysis step, a WBC depletion step, and a cancer cell capture step (Fig. 1). RBC lysis buffer (Sigma-Aldrich, USA) was first used for the chemical lysis of RBCs from whole blood (provided from the National Cheng Kung University Hospital, Tainan, Taiwan). Next, a negative selection process, whereby WBCs were depleted from the blood sample, was performed to deplete WBCs by using anti-CD45 coated magnetic beads. Briefly, the anti-CD45-coated beads were shown to bind to WBCs with high affinity, allowing for their removal via an externally supplied magnetic force. After WBC depletion, a positive selection process featuring CX-BG1-10-A aptamer-coated magnetic beads was used to capture CTCs in human blood. Specifically, after this cancer cell-specific aptamer bound to the CTCs, the bead-cell complexes could be removed from the remaining blood sample with a magnet (Fig. 1).
FIG. 1.
Schematic diagram of the CTC isolation process including (1) RBC lysis, (2) WBC depletion, and (3) CTC capture. RBC lysis buffer was first used to lyse RBCs. Then, anti-CD45 magnetic beads were used to deplete WBCs, and the supernatant containing the cancer cells was collected. Finally, aptamer-coated magnetic beads were used to isolate the CTCs in the supernatant from the previous step.
B. Chip design and fabrication process
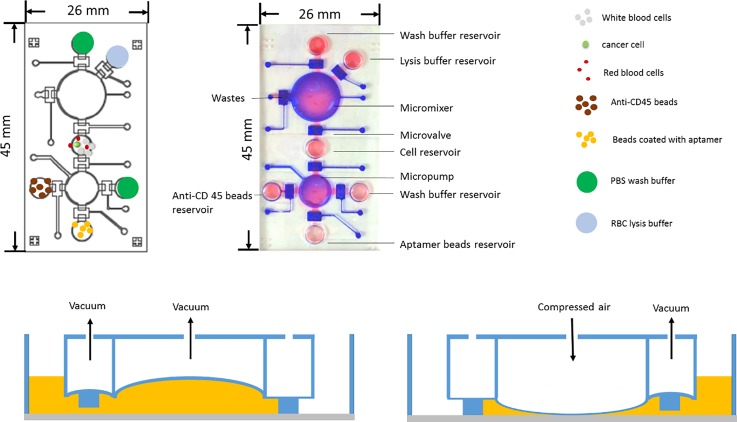
An integrated microfluidic chip that could perform the entire CTC isolation process was designed and fabricated. The 26 mm × 45 mm microchip was equipped with micropumps, micromixers, microvalves, and several chambers [Figs. 2(a) and 2(b)]. The operation and characterization of the micropumps and micromixers can be found in a previous work.25 The chip [Fig. 2(b)] features a RBC lysis buffer reservoir, an anti-CD45 bead reservoir, and an aptamer bead reservoir. Micropumps, micromixers, and microvalves were designed and programmed for transportation and mixing of samples and reagents.25,26 Note that electromagnetic valves (EMVs) were used to control the positive (compressed air) and negative gauge pressure (suction force) such that the polydimethylsiloxane (PDMS) membranes in these pneumatically driven microdevices (micropumps, micromixers, and microvalves) could be deformed to control the transportation and mixing process [Figs. 2(c) and 2(d)]. Note that the mixing process was performed, while the sample was moving in and out between the cell reservoir and the micropump. After 7 times of mixing and transportation processes [Figs. 2(c) and 2(d)], the entire process was finished and all samples were well mixed. The accommodated volumes for the micropump and cell reservoir were 0.157 ml [0.5 cm × 0.5 cm × 0.2 cm (depth)] and 1 ml, respectively.
FIG. 2.
(a) Schematic illustration of the microfluidic chip. (b) A photograph of the fabricated chip. Red and blue colors indicate the liquid transport and air control channels, respectively. (c) and (d) The deformation of the PDMS membranes drives the transport and mixing of samples and reagents.
The microfluidic chip was made of two PDMS layers, including a thick PDMS layer and a thin PDMS membrane layer. A glass substrate was then used to seal the microchannel and chambers. Detailed information on the fabrication of the microfluidic chip can be found in a previous work.17 Some brief information about the fabrication process was described in supplementary material Fig. 1. Briefly, two master molds were first devised by a computer-numerical-control machine (EGX-400, Roland Inc., Japan) equipped with a 0.5-mm drill bit on polymethylmethacrylate (PMMA) plates to form the master molds (a rotational speed of 27 000 rpm and a feed rate of 15 mm min−1). The inverse microstructures of the microdevices were then formed via a PDMS replication process. The PDMS that was poured into the PMMA molds was placed in a vacuum chamber and cured at 80 °C for 1 h. The two PDMS layers replicated from the two PMMA master molds were formed subsequently. Finally, the microfluidic device was completed by using an oxygen plasma treatment to bind the two PDMS layers onto the glass substrate.17,27 It is worth noting that this chip integrated several functional microfluidic devices, including micropumps, micromixers, and microvalves such that they can automate three major operating processes (including red blood cell lysis, white blood cell depletion, and cancer cell isolation), which has never been demonstrated in the literature.
C. Preparation of the spiked blood samples and reagents
The human ovarian serous-type carcinoma cell line BG-128 was chosen as the target rare cancer cells since the aptamer was screened to target this cell line.20 Dulbecco's minimum essential medium (DMEM, Life Technologies, USA) containing 10% fetal bovine serum (FBS, Life Technologies, USA) was used to culture the BG-1 cells at 37 °C with 5% CO2 in air. Human blood samples were provided from the National Cheng Kung University Hospital, Tainan, Taiwan and maintained at 4 °C prior to use. One milliliter of blood sample was inoculated with BG-1 cells at a final concentration of 104 cells ml−1. Cells were stained with 0.1 M CellTrace™ Calcein Green AM (CaAM, Life Technologies, USA) prior to inoculation. Ten microliter aliquots of blood spiked with stained BG-1 cells were quantified under a fluorescence microscope (Bx43, Olympus, Japan) to ensure that the BG-1 cell concentration was approximately 104 cells ml−1 (∼100 cells counted within a 10-μl aliquot). Note that we used this cancer cell concentration to characterize the performance of the developed device. Later, spiked cancer cells (100 cells in 1 ml blood) were also used to simulate the real samples.
RBC lysis buffer (catalog No. 420301, Biolegend, USA) was used to chemically lyse RBCs. It was diluted 10 fold prior to use with deionized water per the manufacturer's recommendations. It was thawed at room temperature and then gently mixed in the micromixer with 1 μl of human blood at pH values ranging from 7.2 to 7.4. Various concentrations of buffer were used at a variety of mixing times to optimize RBC lysis efficiency on-chip. Lysed RBCs were further quantified under a microscope. As a comparison, lysis was also conducted on the benchtop using a product description protocol.
WBCs were captured and depleted with Dynabeads® anti-human CD45-coated magnetic beads (4.5 μm in diameter, Life Technologies, USA). WBC depletion is a critical step in the CTC enrichment process due to the abundance of WBCs in human blood. A buffer containing Ca2+- and Mg2+-free phosphate buffer saline (PBS) supplemented with 0.1% bovine serum albumin (BSA) and 2 mM ethylenediaminetetraacetic acid (EDTA, pH 7.4) was used as the isolation buffer during the experiment following the manufacturer's instruction, and the concentration of anti-CD45 beads was adjusted to 3.5 × 108 beads per ml prior to use. Approximately 106–107 WBCs and 7 × 107 magnetic beads were mixed for initial WBC depletion efficiency experiments, and a magnetic particle separator (MPC, Dynabeads MPC®-1, Life Technologies, USA) was used to separate bead-WBC complexes after a 30-min incubation. The residual WBCs in the supernatant that did not bond with anti-CD45 beads were quantified with a cell counter (TC10, Bio-Rad, USA) to estimate depletion efficiency.
Dynabeads MyOne™ carboxylic acid magnetic beads (1 μm diameter, 4 × 106 beads ml−1, Invitrogen, USA) coated with BG1-specific aptamers were used to capture BG-1 cells. The CX-BG1-10-A aptamer (total 72 base-pairs, 5′-GGCAGGAAGACAAACACCCGGAAAAATCCAGCAAAAACAACTAAAAAAAAACCAATGGTCTGTGGTGCTGTA-3′) was modified with an amine group at the 5′ end and conjugated to the fluorescent dye (FAM) so that cell-aptamer-bead complexes could be visualized under a fluorescence microscope. The process for binding the aptamer onto the carboxylic acid magnetic beads can be found in a previous work.27 The recovery rates of the BG-1 cells from CTC-spiked blood samples (described above) were quantified for both on-chip and benchtop approaches, and the optimal magnetic bead/CTC ratio was determined empirically.
III. RESULTS AND DISCUSSION
A. RBC lysis efficiency
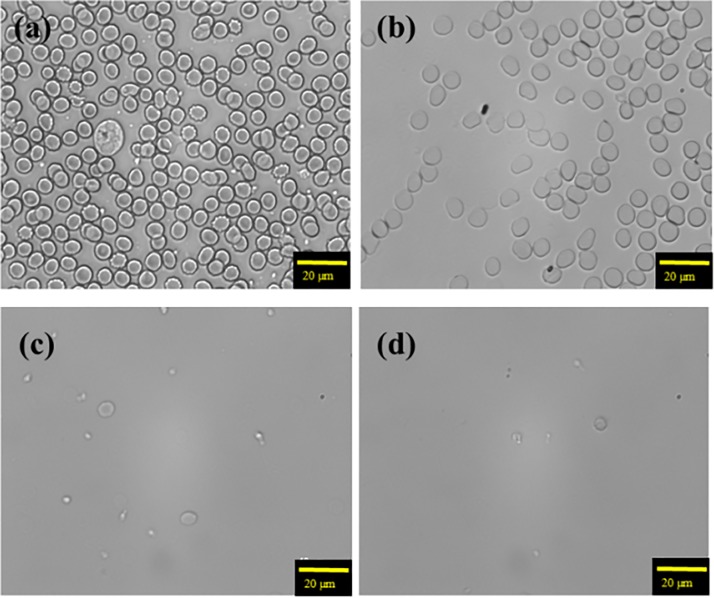
RBCs were first lysed within the microfluidic chip after 0, 5, 10, or 15 min of incubation in the lysis buffer (Fig. 3). Experimental data showed that RBCs could be lysed at a high rate after 10 min at an applied frequency and a gauge pressure of 0.5 Hz and 30 kPa, respectively. As a comparison, a benchtop protocol required 20 min for complete RBC lysis, suggesting a time-saving advantage of the developed microfluidic chip.
FIG. 3.
The RBC lysis buffer was mixed with RBC lysis buffer for (a) 0 min, (b) 5 min, (c) 10 min, and (d) 15 min. It indicates that RBCs could be completely lyzed in just 10 min for the on-chip process.
B. WBC depletion efficiency
After RBC lysis, anti-CD45 magnetic beads were used to deplete WBCs. Such a WBC depletion step is necessary to reduce the inadvertent binding of WBCs to the EpCAM antibody for the CellSearch system, which is also crucial for the aptamer-based assay. In order to optimize the operating parameters for the depletion of WBCs, the optimal ratio between the anti-CD45 beads and WBCs was first explored. Note that the micromixer was operated at 0.5 Hz for 5 min at a gauge pressure of 30 kPa, and the depletion process was typically conducted for 5 min. After each round of WBC depletion (up to three rounds were conducted), the WBC-bead complexes were collected by an external magnet that was placed underneath the micromixer. A single round of WBC depletion was characterized with an efficiency of only 40.71 ± 6.18% at a low beads-to-cell ratio (2:1). The depletion efficiency increased to be 75.13 ± 4.42% and 79.70 ± 2.07% at bead-to-cell ratios of 6:1 and 7:1, respectively (supplementary material Fig. 2). A 99.9% WBC depletion efficiency was achieved after three rounds of depletion. Note that even though the WBC depletion efficiency could be as high as 99.9%, there are about 104 WBCs remaining in whole blood (1 ml). Therefore, non-specific cancer biomarkers (e.g., EpCAM) could still be a concern, even after having reduced the WBC concentration by 99.9%. In this work, we used a cancer cell specific aptamer to reduce the false positive results.
C. Recovery rate of ovarian cancer cells
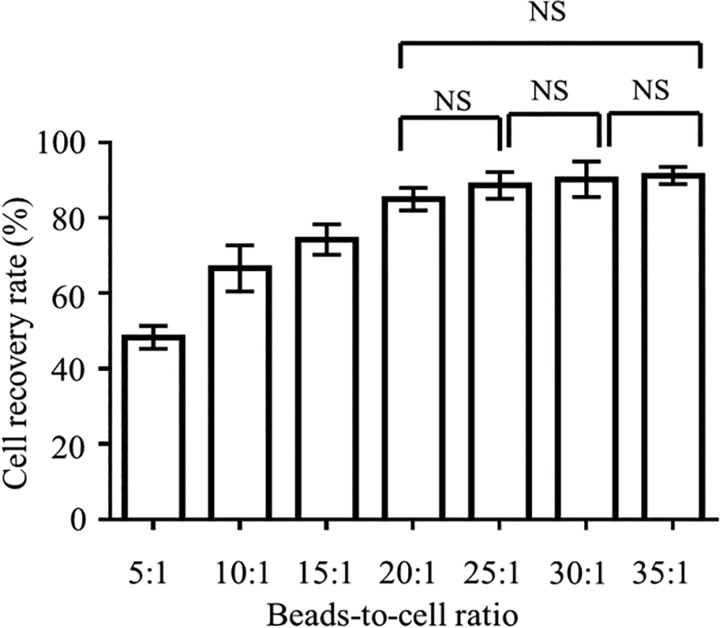
To explore the CTC recovery rate from whole blood samples after the RBC lysis and WBC depletion processes, about 100 fluorescently stained BG-1 cells were spiked into 1 ml of human blood (n = 3 replicates), and the CTCs captured by aptamer-coated magnetic beads were quantified after a 5-min incubation at bead:cell ratios ranging from 5 to 35 (Fig. 4). Experimental results showed that the CTC recovery rate increased with increasing bead-to-cell ratio until reaching a plateau at 20:1. Therefore, this ratio was chosen for subsequent experiments. At this ratio, BG-1 cancer cells could be successfully captured by the aptamer-coated magnetic beads on-chip (Fig. 5), revealing that the aptamer exhibits high affinity toward OCCs, even when their cellular concentrations were low (104 ml−1).
FIG. 4.
OCC recovery rate (%) in a range of bead:cell ratios. All incubations were performed for 5 min. Error bars represent standard deviation (n = 3). The effect of the bead:cell ratio on the recovery rate was statistically significant (one-way ANOVA effect of ratio, p < 0.01). NS = No significant differences.
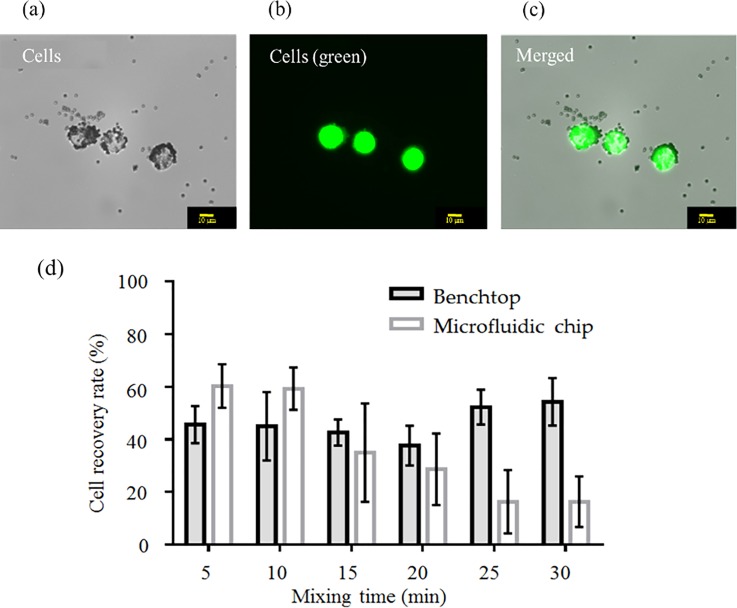
FIG. 5.
Recovery of OCCs with a microfluidic chip and a benchtop protocol. After elimination of WBCs from the media, BG-1 OCCs (BG-1) were successfully captured with aptamer (CX-BG1-10)-coated magnetic beads. The OCCs were stained with a cancer cell-specific fluorescent probe (CaAM) to aid in visualization. (a) Cancer cells under bright field illumination. (b) OCCs under fluorescence microscopy; (c) Merged photograph to confirm that the fluorescence was emitted from the OCCs. (d) After the negative selection process, OCCs were successfully captured with CX-BG1-10-A-coated magnetic beads using either a benchtop protocol or the microfluidic chip (n = 3 samples for each method. Error bars represent standard deviation (n = 3 samples for each method). Two-way ANOVA found significant effects of both mixing time and method (chip vs. traditional) on the cell recovery rate (p < 0.01 for both factors).
We also tested the recovery rate of BG-1 cells in PBS buffer using a traditional benchtop protocol [supplementary material Fig. 3(a)]. Since the affinity of the aptamer towards the target cancer cells was already proven to be high20 when using a PBS buffering system, PBS was also used herein. An OCC recovery rate as high as 80% could be achieved for mixing of 5–20 min. However, the high standard deviation suggests a marked degree of human manipulation error. When aptamer-based OCC capture was instead performed on the integrated microfluidic chip, the standard deviation was much lower (supplementary material Fig. 3(b)]. Note that the cell recovery rate was measured to be around 80% when mixing for 5 min and up to 90% after mixing for 20 min. The developed microfluidic system may then be preferable to the benchtop protocol given its similar capture efficiency and lower variance in the results generated.
A comparison of recovery rates for OCCs between the microfluidic chip and the benchtop protocol was also made after having depleted the WBCs. Note that the culture media were placed with PBS in this case. After performing the negative selection process to deplete WBCs, the benchtop method recovered around 60% of the cells within 10 min [supplementary material Fig. 4(a)]. In this case, OCCs could theoretically be captured by the aptamers in the absence of potentially interfering WBCs. When the mixing was applied for 15–30 min, the recovery rate was close to 80%. The microfluidic chip was more efficient at OCC recovery in the absence of WBCs; 81.5% of the OCCs could be captured in just 10 min (supplementary material Fig. 4(b)]. As the microfluidic chip was adept at capturing CTCs resuspended in PBS buffer, as well as in WBC-depleted media, we next used BG-1 cell-inoculated blood to determine its capture efficacy against traditional benchtop methods. RBC lysis and WBC depletion were both performed prior to cell capture, and the on-chip and benchtop protocols captured ∼50%–60% of the OCCs with aptamer-coated beads within 5 min [Fig. 5(d)]. The ability of the aptamer-coated beads to capture OCCs was therefore lower in whole blood in comparison to BG-1 cells resuspended in PBS. There are a myriad of reasons why this may be the case, although it is likely because some molecules in the blood reduce the likelihood of an aptamer-coated bead coming into contact with OCCs. Although the on-chip and benchtop methods were similarly effective at capturing OCCs at shorter mixing durations (5–10 min), the high shearing forces resultant from longer mixing times (15 min or more) were associated with lower cell recovery rates on the microfluidic chip.
Supplementary material Fig. 5 shows the recovery rate for spiking 105 cancer cells into human whole blood directly without RBC lysis (even though we used CX-BG1-10-A aptamer coated-beads to isolate cancer cells). As shown in this figure, the recovery rates of the cancer cells were very low (35% and 10% for on-chip and on-bench, respectively), indicating that RBC lysis plays an important role in CTC isolation. It is noteworthy that the recovery rates could be improved to about 80% and 60% for on-chip and on-bench processes, respectively, if RBC lysis was performed.
To determine the detection limits of the microfluidic chip, we inoculated 1 ml of human blood (n = 3 aliquots) with only 100 cells (Table I). The spiked OCCs could be captured from whole blood at an average recovery rate of about 62% (ranging from 40% to 82%). When compared to the commercially available CellSearch system, (80%–82% recovery at best), our developed system exhibited lower recovery rates. However, our system is characterized by a lower yield of false positive results due to the highly specific nature of the aptamer, in contrast to the CellSearch system, which may recognize that WBCs are cancerous in 20% of instances on average.30,31
TABLE I.
One hundred ovarian cancer cells inoculated into 1 ml of human blood and isolated and detected on-chip.
| CTC recovery rate (n = 3) | ||||
|---|---|---|---|---|
| Original CTCs | Test 1 | Test 2 | Test 3 | Mean ± std. dev. |
| Sample 1 | 35 | 66 | 48 | 50 ± 5 |
| Sample 2 | 89 | 105 | 64 | 86 ± 20 |
| Sample 3 | 42 | 51 | 27 | 40 ± 12 |
D. False positive and false negative issues
An important issue that needs to be addressed properly is the occurrence of false positive results, which are commonly generated when using traditional antibody biomarkers (e.g., EpCAM). There exists the possibility that our aptamer may bind to WBCs that were not eliminated in the deletion step. It is also possible that the presumably cancer-cell specific anti-CD45 probes bound to OCCs, thereby causing the target cells to be removed from the media prior to their positive selection via aptamer-coated beads. Therefore, we also tested whether anti-CD45 magnetic beads could bind OCCs. Regarding the former concern (i.e., false positive results), the capture rate of WBCs by the aptamer-coated beads was only 0.026% (supplementary material Fig. 6 and supplementary material Tables I and II).
Furthermore, if we performed the negative selection process in which the majority of the WBCs were eliminated from the blood and then consequently performed CTC capture, the number of inadvertently captured WBCs was markedly low (<0.001%; Table II). Of the captured cells, 40-fold more were the target OCCs. In contrast, 10%–25% of the CellSearch-isolated cells were off-target cell types.30,31 Moreover, anti-CD45 magnetic beads did not capture any OCCs in whole human blood samples (supplementary material Fig. 7), whereas the other systems inadvertently captured at least 15% of the cell populations for other cancer cells when using anti-CD45 beads.29 Therefore, it is concluded that BG-1 cells may not possess the CD45 antigen on their cell membranes.
TABLE II.
Capture tests of aptamer-coated beads after WBC depletion. In general, <0.001% of the total WBC population were inadvertently captured by the microfluidic chip.
| False positive cells (n = 3) | |||||
|---|---|---|---|---|---|
| Original WBC cells | Test 1 | Test 2 | Test 3 | Mean ± std. dev. | |
| Sample 1 | 7.30 × 106 | 0 | 2 | 0 | 1 |
| Sample 2 | 6.12 × 106 | 1 | 13 | 0 | 4 |
| Sample 3 | 3.45 × 106 | 0 | 0 | 1 | 1 |
IV. CONCLUSIONS
A newly designed, integrated microfluidic chip capable of RBC lysis, WBC depletion, and OCC capture via aptamer-coated magnetic beads was demonstrated in the current study, which was able to detect OCCs inoculated at concentrations of only 100 cells ml−1 in human blood samples. A recovery rate of 62 ± 3% was achieved when larger numbers of cells (10 000) were spiked into the blood samples. Moreover, significantly fewer WBCs were captured, and the false positive detection rate was at least two orders of magnitude lower than that of a commercially available OCC detection assay (CellSearch). Higher cell recovery rates (i.e., improved sensitivity) are likely to be documented upon performing multiple rounds of deletion and enrichment. As such, the developed microfluidic chip represents a promising means of detecting low-abundance CTCs, specifically OCCs, from human blood samples and therefore could be a useful tool for early diagnosis of ovarian cancer or other types of cancers if proper aptamers have been used.
V. SUPPLEMENTARY MATERIAL
See supplementary material for the chip fabrication process, optimization of the beads-to-cell ratio for on-chip white blood cell depletion, the recovery rate of human ovarian serous-type carcinoma cell line BG-1 cells captured with aptamer (CX-BG1-10)-coated magnetic beads and bench-top assays, and images of captured ovarian cancer cells.
ACKNOWLEDGMENTS
The authors would like to acknowledge financial support from the National Health Research Institute of Taiwan (NHREI-EX104-10428EI to GBL) and Taiwan's Ministry of Science and Technology (MOST 105-2119-M-007-009 to GBL).
Note: Preliminary results in this manuscript were presented at the 19th International Conference on Miniaturized Systems for Chemistry and Life Sciences, MicroTAS 2015 Conference, Gyeongju, Korea, Oct 25–29, 2015.
References
- 1. Shao C., Liao C. P., Hu P., Chu C. Y., Zhang L., Bui M. H., Ng C. S., Josephson D. Y., Knudsen B., Tighiouart M., Kim H. L., Zhau H. E., Chung L. W., Wang R., and Posadas E. M., PLoS One 9, e88967 (2011). 10.1371/journal.pone.0088967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sleijfer S., Gratama J. W., Sieuwerts A. M., Kraan J., Martens J. W., and Foekens J. A., Eur. J. Cancer 43, 2645–2650 (2007). 10.1016/j.ejca.2007.09.016 [DOI] [PubMed] [Google Scholar]
- 3. Pepe M. S., Etzioni R., Feng Z., Potter J. D., Thompson M. L., Thornquist M., Winget M., and Yasui Y., J. Natl. Cancer Inst. 93, 1054–1061 (2001). 10.1093/jnci/93.14.1054 [DOI] [PubMed] [Google Scholar]
- 4. Mocellin S., Keilholz U., Rossi C. R., and Nitti D., Trends Mol. Med. 12, 130–139 (2006). 10.1016/j.molmed.2006.01.006 [DOI] [PubMed] [Google Scholar]
- 5. Lowes L. E., Hedley B. D., Keeney M., and Allan A. L., Cytometry, Part A: J. Int. Soc. Anal. Cytol. 81, 983–995 (2012). 10.1002/cyto.a.22158 [DOI] [PubMed] [Google Scholar]
- 6. Kang J. H., Krause S., Tobin H., Mammoto A., Kanapathipillai M., and Ingber D. E., Lab Chip 12, 2175–2181 (2012). 10.1039/c2lc40072c [DOI] [PubMed] [Google Scholar]
- 7. Nagrath S., Sequist L. V., Maheswaran S., Bell D. W., Irimia D., Ulkus L., Smith M. R., Kwak E. L., Digumarthy S., Muzikansky A., Ryan P., Balis U. J., Tompkins R. G., Haber D. A., and Toner M., Nature 450, 1235–1239 (2007). 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mauk M. G., Ziober B. L., Chen Z., Thompson J. A., and Bau H. H., Ann. N. Y. Acad. Sci. 1098, 467–475 (2007). 10.1196/annals.1384.025 [DOI] [PubMed] [Google Scholar]
- 9. Hou H. W., Warkiani M. E., Khoo B. L., Li Z. R., Soo R. A., Tan D. S., Lim W. T., Han J., Bhagat A. A., and Lim C. T., Sci. Rep. 3, 1259 (2013). 10.1038/srep01259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adams D. L., Zhu P., Makarova O. V., Martin S. S., Charpentier M., Chumsri S., Li S., Amstutz P., and Tang C. M., RSC Adv. 4, 4334–4342 (2014). 10.1039/C3RA46839A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gascoyne P. R. C. and Shim S., Cancers 6, 545–579 (2014). 10.3390/cancers6010545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang S. B., Wu M. H., Lin Y. H., Hsieh C. H., Yang C. L., Lin H. C., Tseng C. P., and Lee G. B., Lab Chip 13, 1371–1383 (2013). 10.1039/c3lc41256c [DOI] [PubMed] [Google Scholar]
- 13. Hong B. and Zu Y., Theranostics 3, 377–394 (2013). 10.7150/thno.5195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu Y., Phillips J. A., Yan J., Li Q., Fan Z. H., and Tan W., Anal. Chem. 81, 7436–7442 (2009). 10.1021/ac9012072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shah A. M., Yu M., Nakamura Z., Ciciliano J., Ulman M., Kotz K., Stott S. L., Maheswaran S., Haber D. A., and Toner M., Anal. Chem. 84, 3682–3688 (2012). 10.1021/ac300190j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lien K. Y., Chuang Y. H., Hung L. Y., Hsu K. F., Lai W. W., Ho C. L., Chou C. Y., and Lee G. B., Lab Chip 10, 2875–2886 (2010). 10.1039/c005178k [DOI] [PubMed] [Google Scholar]
- 17. Luo W. Y., Tsai S. C., Hsieh K., and Lee G. B., J. Micromech. Microeng. 25, 084007 (2015). 10.1088/0960-1317/25/8/084007 [DOI] [Google Scholar]
- 18. Sefah K., Shangguan D., Xiong X., O'Donoghue M. B., and Tan W., Nat. Protocols 5, 1169–1185 (2010). 10.1038/nprot.2010.66 [DOI] [PubMed] [Google Scholar]
- 19. Hung L. Y., Wang C. H., Che Y. J., Fu C. Y., Chang H. Y., Wang K., and Lee G. B., Sci. Rep. 5, 10326 (2015). 10.1038/srep10326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hung L. Y., Wang C. H., Hsu K. F., Chou C. Y., and Lee G. B., Lab Chip 14, 4017–4028 (2014). 10.1039/C4LC00587B [DOI] [PubMed] [Google Scholar]
- 21. Wan Y., Kim Y. T., Li N., Cho S. K., Bachoo R., Ellington A. D., and Iqbal S. M., Cancer Res. 70, 9371–9380 (2010). 10.1158/0008-5472.CAN-10-0568 [DOI] [PubMed] [Google Scholar]
- 22. Simaeys D. V., Lopez-Colon D., Sefah K., Sutphen R., Jimenez E., and Tan W., PLoS One 5, e13770 (2010). 10.1371/journal.pone.0013770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sheng W., Chen T., Kamath R., Xiong X., Tan W., and Fan Z. H., Anal. Chem. 84, 4199–4206 (2012). 10.1021/ac3005633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zeng Z., Parekh P., Zhao N., and Zu Y., FASEB J. 27(1), 4017–4026 (2013) 10.1096/fj.13-228668. [DOI] [PubMed] [Google Scholar]
- 25. Huang C. W., Huang S. B., and Lee G. B., J. Micromech. Microeng. 16, 2265 (2006). 10.1088/0960-1317/16/11/003 [DOI] [Google Scholar]
- 26. Yang Y. N., Hsiung S. K., and Lee G. B., Microfluid. Nanofluid. 6, 823–833 (2009). 10.1007/s10404-008-0356-7 [DOI] [Google Scholar]
- 27. Lin C. H., Lee G. B., Lin Y. H., and Chang G. L., J. Micromech. Microeng. 11, 726 (2001). 10.1088/0960-1317/11/6/316 [DOI] [Google Scholar]
- 28. Zhang Z., Bast R. C., Yu J. Y., Li J., Sokoll L. J., Rai A. J., Rosenzweig J. M., Cameron B., Wang Y. Y., Meng X. Y., Berchuck A., Haaften-Day C. V., Hacker N. F., de Bruijn H. W., van der Zee A. G., Jacobs I. J., Fung E. T., and Chan D. W., Cancer Res. 64, 5882–5890 (2004). 10.1158/0008-5472.CAN-04-0746 [DOI] [PubMed] [Google Scholar]
- 29. Wang C. H., Chang C. J., Wu J. J., and Lee G. B., Nanomed.: Nanotechnol., Biol., Med. 10, 809–818 (2014). 10.1016/j.nano.2013.10.013 [DOI] [Google Scholar]
- 30. Müller V., Riethdorf S., Rack B., Janni W., Fasching P. A., Solomayer E., Aktas B., Sabine K. B., Pantel K., and Fehm T., Breast Cancer Res. 14, R118 (2012). 10.1186/bcr3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eifler R. L., Lind J., Falkenhagen D., Weber V., Fischer M. B., and Zeillinger R., Cytometry, Part B, Clin. Cytometry 80, 100–111 (2011). 10.1002/cyto.b.20560 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the chip fabrication process, optimization of the beads-to-cell ratio for on-chip white blood cell depletion, the recovery rate of human ovarian serous-type carcinoma cell line BG-1 cells captured with aptamer (CX-BG1-10)-coated magnetic beads and bench-top assays, and images of captured ovarian cancer cells.