Abstract
While there are myriad mechanisms of primary and acquired resistance to conventional and next-generation hormonal therapies in prostate cancer, the potential role of androgen receptor splice variants (AR-Vs) has recently gained momentum. AR-Vs are abnormally truncated isoforms of the androgen receptor (AR) protein that lack the COOH-terminal domain but retain the NH2-terminal domain and DNA-binding domain and are thus constitutively active even in the absence of ligands. Although multiple preclinical studies have previously implicated AR-Vs in the development of castration resistance as well as resistance to abiraterone and enzalutamide, recent technological advances have made it possible to reliably detect and quantify AR-Vs from human clinical tumor specimens including blood samples. Initial clinical studies have now shown that certain AR-Vs, in particular AR-V7, may be associated with resistance to abiraterone and enzalutamide but not taxane chemotherapies when detected in circulating tumor cells. Efforts are now underway to clinically validate AR-V7 as a relevant treatment-selection biomarker in the context of other key genomic aberrations in men with metastatic castration-resistant prostate cancer. Additional efforts are underway to therapeutically target both AR and AR-Vs either directly or indirectly. Whether AR-Vs represent drivers of castration-resistant prostate cancer, or whether they are simply passenger events associated with aggressive disease or clonal heterogeneity, will ultimately be answered only through these types of clinical trials.
Introduction
In 1941, Huggins and Hodges1 first demonstrated the clinical efficacy of hormonal manipulation for the treatment of metastatic prostate cancer. Androgen deprivation therapy (ADT), involving surgical or chemical castration, remains the standard first-line option for men with metastatic prostate cancer, and suppression of androgen receptor (AR) signaling has been the therapeutic goal in prostate cancer drug development for seven decades. It is well known, however, that ADT only provides temporary clinical benefit and progression to castration-resistant prostate cancer (CRPC) almost always occurs after a variable period of time. In general, prostate cancer progression upon first-line ADT continues to rely on AR signaling sustained by adrenal and intratumoral androgens as well as upregulation of AR protein expression in tumor cells. The established concept that sustained AR signaling is a key molecular determinant of CRPC has directly contributed to the successful clinical development of abiraterone and enzalutamide,2,3 both of which have been approved by the US Food and Drug Administration (FDA) to treat metastatic CRPC on the basis of survival improvements.4–7 However, a significant subset of CRPC patients demonstrates primary resistance to the two agents, and nearly all patients that are treated eventually develop acquired resistance during the course of treatment. Therefore, understanding and managing primary and acquired resistance to abiraterone and enzalutamide has become a critical unmet need.8 One potential explanation for this resistance is the generation of AR splice variants (AR-Vs). In this review, we will discuss evolving insights into AR-V expression in prostate cancers and their implications in contemporary prostate cancer clinical care, as well as current efforts in therapeutic targeting of AR-Vs aiming to overcome resistance to novel hormonal therapies.
Mechanisms Of Androgen/Ar Resistance
A significant fraction of prostate tumors treated with androgen/AR-directed therapies, including abiraterone and enzalutamide, will demonstrate a molecular signature consistent with continued ‘addiction’ to AR signaling. General mechanisms of androgen/AR resistance focusing on the AR pathway have been covered in several recent reviews.8–11 Tumors treated by therapies designed to suppress AR signaling are expected to acquire molecular alterations in this axis to maintain their addiction. Indeed, the AR gene is frequently amplified or mutated (less common than AR amplification) in CRPC.12,13 In a recent study involving 150 metastatic CRPC cases, AR amplifications or mutations were found in ∼ 62% of these cases.13 In contrast, focal amplification of the AR gene was detected in < 1% of hormone-naive prostate cancers (n = 596).12 In addition, overexpression of both the canonical full-length AR (AR-FL) and AR-Vs are frequently observed in CRPC. However, a wider spectrum of molecular aberrations may be responsible for sustained AR signaling, requiring carefully designed studies to dissect key drivers and determinants of resistance.11,14 For example, characterization of the relative frequency of previously reported molecular aberrations (including CYP17A1, AKR1C3, HSD3B1, GR and PR) in the context of aforementioned AR aberrations may help to further clarify their importance and clinical relevance. As prostate cancer is now being managed by increasingly more potent androgen/AR-directed therapies, it is reasonable to anticipate a rise in tumors in which AR expression may be low or even absent. These tumors may demonstrate histological and molecular features of neuroendocrine differentiation and/or small cell carcinoma, in which loss and/or mutations of the RB1, TP53 and PTEN genes are often observed. A recent report suggested that up to a quarter of prostate tumors resistant to abiraterone or enzalutamide may demonstrate distinct morphological and molecular features intermediate between typical acinar adenocarcinoma and neuroendocrine differentiation/small cell carcinoma.15 It is currently unknown whether AR-Vs may be present in some of these tumors, and indeed in many of these cases serum PSA can be quite elevated suggesting ongoing AR activation. The various resistance mechanisms also portend increasingly complex patterns of intra-and inter-tumor heterogeneity that will need to be accounted for in the clinical setting and relevant study designs.
AR Splice Variants
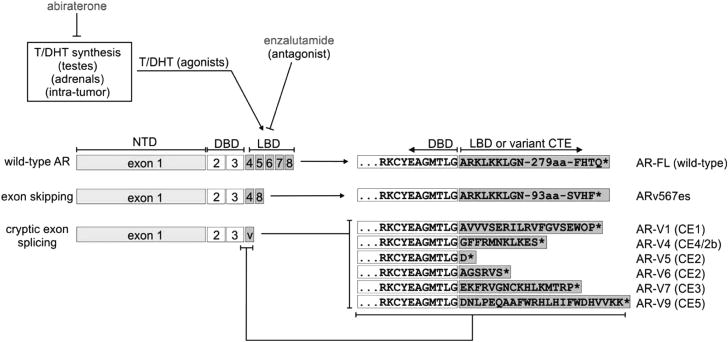
The availability of enzalutamide and abiraterone has facilitated studies aimed at understanding the role of AR-Vs in the presence of potent inhibitors of AR signaling. AR-Vs10,16 are alternatively spliced isoforms of the AR mRNA usually resulting in premature termination of the AR protein product. Most AR-Vs retain the NH2-terminal transactivating domain (NTD) but are missing variable portions of the COOH-terminal domain including the ligand-binding domain (LBD) (Figure 1). In particular, AR-V7, the most frequently expressed AR-V,16 has been implicated in resistance to enzalutamide and abiraterone in both preclinical experiments and a few recent clinical studies (see later sections). Conceptually, AR-V7 along with a number of other AR-Vs, is a biologically plausible mechanism of resistance to abiraterone and enzalutamide. AR-Vs lack the LBD, which is the intended therapeutic target of all existing androgen/AR-directed therapies (Figure 1). Preclinical studies have confirmed that AR-Vs are capable of mediating constitutively active AR signaling (that is, in the absence of androgens or AR-FL),17–21 that expression levels of AR-Vs are typically elevated in CRPC and in response to inhibition of AR-FL signaling,19–24 and that AR-V expression is associated with disease progression in retrospective studies.18,19,25 In clinical CRPC specimens, individual AR-Vs are often co-expressed with AR-FL and are usually less abundant than the wild-type transcripts.13,19,21,26 For example, quantitative PCR with reverse transcription (RT-PCR) analysis of circulating tumor cells from men with CRPC has revealed the median mRNA ratio of AR-V7/AR-FL is 21% (range, 1.8–208%).27 RNA-seq analysis of CRPC autopsy and biopsy tissue has similarly revealed a broad range of AR-V7 expression ratios,13,27 with the median level of AR-V7 expression as a function of overall AR expression being ∼ 5% in metastatic biopsies.13 However, because AR-FL is a very abundant transcript (increased by ∼ 10-fold in CRPC compared with hormone-sensitive prostate cancer),19 a ratio of AR-V7/AR-FL at 5–20% would bring the levels of AR-V7 in CRPC specimens on par with the levels of AR-FL in an untreated hormone-naive tumor. Presently the levels of nuclear AR-Vs required to drive an androgen-independent transcriptome remains unclear. However, even a low level of AR-V expression may be sufficient in the setting of castration or potent full-length AR blockade. Another AR-V, designated ARv567es, is also expressed at levels equivalent to AR-FL in certain contexts.28 These quantitative data provide the context to fathom a clinically relevant role of AR-Vs as ligand-independent transcription factors that may or may not require AR-FL.
Figure 1.
Domain structures of full-length androgen receptor and androgen receptor splice variants. Full-length androgen receptor and androgen receptor splice variants share a core structure composed of the transcriptionally active AR NH2-terminal domain encoded by exon 1 and the DNA-binding domain encoded by exons 2 and 3. Androgen receptor splice variants (AR-Vs) lack the AR ligand-binding domain (LBD), which is the binding site for agonists testosterone (T) and DHT and competitive antagonists such as enzalutamide. Instead of a ligand-binding domain, androgen receptor splice variants contain CTE of variable length and sequence, which arise from exon skipping or splicing of various CE. Amino-acid sequences of CTEs from selected androgen receptor splice variants are shown. AR, androgen receptor; CE, cryptic exons; CTE, COOH-terminal extensions; DHT, dihydrotestosterone.
Structure and Function of AR Variants
Detailed structures of various AR-Vs have been described in recent reviews.10,16 The key domains shared among wild-type AR-FL and all AR-Vs are the NTD and DNA-binding domain (DBD). AR-Vs lack the LBD and instead have divergent COOH-terminal extensions encoded by mRNAs derived from exon-skipping events or incorporation of alternative 3′-terminal cryptic exons (Figure 1). The AR-NTD is an intrinsically disordered protein domain, and is responsible for the majority of AR transcriptional activity. The AR-DBD is composed of two zinc fingers that mediate AR/DNA interactions and also AR/AR dimerization. There have been multiple review articles written on various aspects of molecular structure and function of AR-FL.29,30 Importantly, several lines of evidence support the notion that the function and regulation of the AR-NTD and DBD may proceed similarly between AR-FL and AR-Vs. For example, the AR transactivation unit-5 (TAU5) domain in the AR-NTD has been shown to function as a key transactivation domain for AR-FL under conditions of no/low androgen.31 The transcription factor FOXO1 has been shown to bind the AR TAU5 domain and thereby repress transcriptional activity of AR-FL and AR-Vs.32 This indicates that TAU5 has similar roles in mediating ligand-independent transcriptional activation in the context of AR-FL and AR-Vs. In addition, the highest-affinity genome-wide binding sites for both AR-FL and AR-FL vs AR-Vs are canonical inverted repeat androgen response elements (AREs), which indicates that AR-FL and AR-Vs display similar regulation of the DBD.33 In line with this, dimerization between AR-V monomers requires the D-box dimerization interface in the second zinc finger of the AR-DBD,33,34 which is the same interface required for dimerization of AR-FL.35
An outstanding question regarding AR-V structure/function is the identity of the precise domains(s) that promote nuclear localization of AR-Vs. The hinge region located between the AR DBD and LBD harbors the canonical AR nuclear localization signal, which is required for nuclear localization of AR-FL following ligand-binding.36 However, only certain AR-Vs encoded by mRNAs that retain AR exon 4 (such as ARv567es) retain this hinge region. Some AR-Vs encoded by mRNAs lacking exon 4 (such as AR-V7) do not harbor this hinge region, yet are still able to localize efficiently to the nucleus in a constitutive manner, possibly due to a NLS-like signal in their unique COOH-terminal extensions,19,26 although mutation of these residues has only a modest impact on the ability of AR-V7 to localize to the nucleus and does not impact on transcriptional activity.37 One factor contributing to this efficient nuclear localization may be that most AR-Vs lack a nuclear export signal (NES) encoded by AR exon 6, which is required for nuclear exclusion of AR-FL in the absence of ligand.38 In other studies however, some AR-Vs (including AR-V1 and AR-V9) did not demonstrate efficient nuclear localization although they lack NES, suggesting that some of the differential nuclear localization activities could be mediated by the short variant-specific COOH-terminal extensions.26
Dimerization and Constitutive Activation
Because AR-Vs often co-exist with AR-FL in CRPC, the role of AR-FL in mediating the function of AR-Vs remains unclear and warrants further studies. It is possible that AR-Vs may form heterodimers with AR-FL.20,21 If so, therapeutic targeting of AR-FL alone may theoretically disrupt AR-V-mediated functions.21 Xu et al.34 characterized protein–protein interactions between AR-Vs (AR-V7 and ARv567es) and AR-FL using bimolecular fluorescence complementation and bioluminescence resonance energy transfer assays. In this study, AR-Vs were found to form heterodimers with AR-FL and also to form homodimers with themselves in the absence of androgens, possibly through NTD-to-CTD interactions (NTD of AR-V and CTD of AR-FL) as well as DBD-to-DBD interactions (present in both AR-Vs and AR-FL). These data suggest potential interdependency between AR-FL and AR-Vs. However, the focus on overexpression models (as opposed to cells expressing endogenous AR-FL and AR-Vs) may limit the generalizability and clinical relevance of the findings.39 Indeed, overexpression experiments using diverse readouts (co-immunoprecipitation, bimolecular fluorescence complementation) have consistently been successful in detecting AR-FL/AR-V heterodimers.20,34,40 However, it remains plausible that the detected interactions may represent transient, DNA-dependent interactions given that AR-FL and AR-Vs have overlapping DNA-binding sites.33 In line with this, reports testing heterodimerization between endogenously expressed AR-FL and AR-Vs have had mixed results. For example, an endogenous interaction between AR-FL and AR-Vs was not detectable in the 22Rv1 or CWR-R1 cell lines18,41,42 and in only one instance were interactions between AR-FL and ARv567es detected in lysates from patient-derived xenografts.20 Because of these mixed findings, it remains unclear whether AR-FL and AR-Vs directly interact or in what contexts, particularly in patients. Studies designed to detect dimer formation in situ may help to shed additional light on this important topic.
Molecular Origins of AR Variants
The mRNA and protein expression levels of AR-Vs relative to AR-FL varies within normal and malignant hormone-naive and castration-resistant prostate tissues,13,19,22,26,43 circulating tumor cells27,44 and prostate cancer cell lines.17–19 This suggests that AR-V synthesis is not simply a byproduct of AR splicing. One mechanism that can dramatically alter the ratio of AR-V expression to AR-FL is rearrangement of the AR gene. For example, the 22Rv1 and CWR-R1 cell lines, which express high levels of AR-V7 and display AR-V-driven resistance to AR-targeted therapies, harbor large intragenic structural rearrangements in AR.45–47 Interestingly, the CWR-R1 cell line is heterogeneous, and the cell sub-population harboring a rearranged AR allele was shown to be the cell sub-population with AR-V7-driven antiandrogen resistance. Conversely, the CWR-R1 cell sub-population without this rearranged AR gene expressed very low levels of AR-V7 and displayed sensitivity to antiandrogens.46 In addition, intragenic AR rearrangements have also been shown to dramatically impact expression of ARv567es. For example, the LuCaP86.2 and LuCaP136 patient-derived xenografts harbor intragenic deletions and inversions, respectively, of a segment of the AR gene containing AR exons 5, 6 and 7.28,47 In the case of these alleles, AR-FL expression is abolished, and ARv567es is the only AR species synthesized.28
Additional mechanisms have been demonstrated to impact AR-V mRNA and protein levels in prostate cancer cells, which may function independently from or synergistically with structural alterations in the AR gene. An early observation in several prostate cancer cell lines was that specific inhibition of the AR-FL protein (via castration, antiandrogen treatment or siRNA) led to increased expression of AR-V7.22,23 However, concomitant increases in AR-FL expression have also been observed with these manipulations, which is likely the result of negative feedback inhibition through AR transcriptional autoregulation.48 Similarly, overexpression of components of the canonical and non-canonical NF-κB signaling pathways in LNCaP cells (including IKK2, p65/RelA and p52) led to increased expression of AR-FL and AR-Vs, and inhibition of components of these pathways via siRNA or chemical inhibitors led to decreased expression of AR-FL and AR-Vs.49–51 Further, knockdown of hnRNPA1 (encoded by the HNRNPA1 gene) and hnRNPA2 (encoded by the HNRNPA2B1 gene), which are both regulated by NF-κB, decreased AR-FL and AR-V7 protein expression in 22Rv1 and VCaP prostate cancer cells.50 Additional hnRNPs, including hnRNP1 (encoded by the PTBP1 gene) and hnRNPH1 (encoded by the HNRNPH1 gene), have also been shown to physically associate with the AR gene locus and positively affect expression of AR-FL and AR-Vs in various cell lines.52 Interestingly, splicing factors SF2 (encoded by the SRSF1 gene) and U2AF65 (encoded by the U2AF2 gene) have been shown to associate with the AR pre-RNA near the exon CE3/3b (encoding the 3′-terminal exon of AR-V7) splice acceptor. Further, knockdown of these factors impaired splicing of exon 3 to CE3 but not exons 3 to 4 in LNCaP95, a cell line derived from the parental LNCaP line with high levels of AR-V7.53 Overall, these studies have provided a diverse set of factors that can affect AR-V synthesis, but it should be noted that most of these factors also have similar effects on AR-FL synthesis, indicating that in these cell line models, production of AR-Vs may be influenced by aberrant AR transcription and perhaps a consequential change in splicing dynamics, rather than aberrant AR splicing that specifically affect the expression of AR-Vs.
Contemporary prostate cancer genome-sequencing studies have not been able to confirm an early report of a Q640X stop point-mutation in prostate cancer that can give rise to an AR-V-like truncated AR protein species.13,43,54 This raises the question: if truncation of the AR LBD is an effective resistance mechanism in prostate cancer, why are not truncating mutations observed? One possibility may be that mRNAs harboring premature termination codons in exons 4–7 of the AR gene would have subsequent exons splice downstream of this mutant termination codon, which is the classical signal for degradation by nonsense-mediated mRNA decay.55 Therefore, rearrangement of the AR gene and/or changes in splicing dynamics may be the only mechanism(s) available to achieve expression of AR-V proteins in prostate cancer cells.
AR and AR-V Transcriptomes
AR-Vs were initially found to function as constitutively active transcription factors that can activate transcription of AR-regulated target genes, such as KLK3 (PSA), KLK2 (HK2), TMPRSS2 and NKX3-1.17 Subsequent studies indicated that AR-Vs can not only support the broad androgen/AR transcriptional program 22,46 but may also uniquely transcriptionally activate alternative targets such as AKT1,18 genes associated with M-phase regulation of the cell cycle including UBE2C and CCNA2 [ref. 22] and the FOXA1-repressed target genes EDN2 and ETS2.56 In contrast, a ChIP-seq study found that ARv567es displayed the same genome-wide binding preference as AR-FL, but engaged chromatin sites with weaker affinity.33 Chromatin sites found to be engaged by both ARv567es and AR-FL included UBE2C, CCNA2, EDN2 and ETS2, which would not be expected if these genes were unique transcriptional targets of AR-Vs.33 One explanation for the apparent discrepancy, among many others,57 may be the known biphasic nature of androgen signaling.46 This biphasic signaling profile, wherein prostate cancer cells proliferate maximally when exposed to androgens in the 0.1–1.0 nM range, but display paradoxical proliferative inhibition when androgens are 10 nM and above, is one of the justifications for trials of high-dose testosterone therapy for men with castration-resistant prostate cancer.58 Accordingly, many of the gene targets that have been proposed to be unique to AR-Vs, including UBE2C and CCNA2, may be induced by AR-FL in the low androgen environment but repressed under high suppressive androgen levels.46 Similarly, AR-V7 has been shown to activate (while AR-FL has been shown to repress) the tricarboxylic acid cycle-related genes MDH1 and OGDH. This differential gene expression has been linked to differences in metabolism noted for LNCaP cells expressing AR-V7 versus LNCaP cells treated with androgen.59 Overall, these studies have documented several differences when AR-FL and AR-V transcriptional targets have been evaluated head-to-head. Further investigation is required to determine whether these and other AR-V gene targets are truly unique, or instead gene targets common to AR-FL and AR-Vs that display threshold and/or biphasic responses to varying levels of AR transcriptional output.
Preclinical Data On AR-V Inhibition
Despite many studies documenting expression of AR-Vs in prostate cancer cell lines, patient-derived xenografts, patient tissues and circulating tumor cells, relatively few studies have evaluated the potential therapeutic efficacy of directly inhibiting AR-Vs. This is because many of the manipulations or treatments that have been applied to AR-V-expressing prostate cancer cells can inhibit both AR-FL and AR-Vs. Therefore, it has been difficult to discern the relative contributions of these two AR species to key biological parameters. For example, development of enzalutamide resistance in the LNCaP cell line model has been shown to be associated with increased expression of AR-FL as well as AR-V7. Antisense oligonucleotides (ASOs) that inhibited AR-FL expression inhibited growth and induced apoptosis in these enzalutamide-resistant cells. However, these effects were equivalent for various AR-targeted ASOs, even those that blocked AR-FL but not AR-V7. Alternatively, in 22Rv1 cells, ASOs that blocked expression of AR-FL and AR-V7 were more effective in inhibiting growth and inducing apoptosis than ASOs that only blocked AR-FL.60 Similarly, a separate study showed that LNCaP cells stably expressing AR-Vs remained sensitive to inhibition of AR-FL via siRNA-mediated knockdown or treatment with enzalutamide,21 indicating that mere expression of AR-Vs in this cell line model may not be sufficient to drive all parameters of resistance. In contrast, in cases where AR-V synthesis is due to underlying rearrangements in the AR gene, selective ablation of AR-V expression can impair proliferation of prostate cancer cells and restore responses to androgens and antiandrogens.28,46
AR Variants and Epithelial Plasticity
Emerging data suggest a complex relationship between AR biology and epithelial plasticity, defined as the ability of cells to reversibly undergo phenotypic changes.61,62 These phenotypic changes range from alterations in gene expression to protein translation, changes in invasion, proliferation and metastasis, and changes in morphology, such as the mesenchymal or neuroendocrine transition. In transgenic mice engineered to overexpress either AR-V7 or AR-V567es, increases in invasion and stemness/plasticity biomarkers were observed during castration-resistant outgrowth.63,64 This was accompanied by a promotion of paracrine signaling in the tumor microenvironment, which then contributed further into treatment resistance and growth/invasion. It has long been appreciated that castration itself can induce mesenchymal biomarker expression in cell lines, xenografts and patient samples, including induction of N- and OB-cadherin expression, SNAIL and ZEB1, and loss of cytokeratin and PSA expression.65,66 Loss of cytokeratin expression has been associated with activation of stemness pathways, such as NOTCH or GLI, as well as chemotherapy resistance in prostate cancer.67 Growth of prostate cancer cells in charcoal-stripped media can also induce higher levels of AR-FL and AR-Vs.22,23 Finally, circulating tumor cells from men with metastatic CRPC frequently express both epithelial and mesenchymal markers, suggesting that this plasticity is important in lethal disease.68 A key question that emerges from these observations is: what is the relationship between such plasticity and AR biology?
Overexpression of AR-V7 has been demonstrated to result in higher levels of SNAIL, TWIST, N-cadherin and ZEB1, despite a lack of impact on cytokeratin or E-cadherin downregulation.69,70 These data suggest that overexpression of AR-Vs may induce a partial EMT in some contexts. These data have also been observed with androgens and overexpression of AR-FL, which may also drive an EMT program in some contexts. Induced cellular plasticity and invasion has also been observed with exogenous androgens, which may also drive an EMT program in some contexts of low AR activity and TGF-β signaling dependence.71 These authors observed suppression of epithelial plasticity by overexpression of AR-FL, but did not measure AR-Vs in their system. A relationship between SNAIL overexpression and induction of both AR-FL and AR-V7 has recently been demonstrated, which promotes resistance to enzalutamide.72 SNAIL has been demonstrated to be overexpressed in patients with metastatic prostate cancer and high-grade disease, and is commonly found in neuroendocrine prostate cancer metastases.73–76 Overexpression of SNAIL was accompanied by increased migration and invasion in the context of these alterations in AR biology. Although in some contexts, SNAIL may lead to loss of AR activity and plasticity or neuroendocrine prostate cancer transformation, in other contexts it appears that SNAIL may lead to alternative splicing of AR and/or increased AR expression, which may promote enzalutamide resistance. Further work will clarify the mechanisms of this relationship, including the impact of epithelial plasticity proteins on alternative splicing and epigenetic regulation of AR. However, these data suggest that AR biology may be directly impacted by cellular differentiation programs normally operative during embryology, and which are re-awakened during metastasis. In addition, AR inhibition may lead to generation of AR-Vs which can then activate these latent programs as part of a reciprocal relationship.
Clinical Significance of AR-V7
Although at least 22 AR-Vs have been discovered to date in tumor samples from patients with metastatic CRPC,13 AR-V7 is the most clinically relevant variant. This is because it is the most frequently observed and the most abundant AR-V in clinical specimens, and is the only variant that can be detected reproducibly at both the mRNA and protein levels. In addition, a number of retrospective studies (assessing AR-V7 using a variety of methods) have suggested that this variant is associated with more rapid disease progression and shorter survival in men with metastatic CRPC.18,19,25,77,78
The first prospective study to evaluate the prognostic impact of AR-V7 was conducted by Efstathiou et al.79 In that study, 60 men with bone-metastatic CRPC were treated with enzalutamide and underwent bone marrow biopsies at baseline and after 8 weeks. The presence of AR-V7 (on >1% of tumor cells, detected at the protein level using immunohistochemistry on formalin-fixed paraffin-embedded specimens) was associated with primary resistance to enzalutamide. More specifically, AR-V7 protein was detected in 57% of men who developed disease progression within 4 months of starting enzalutamide, but was not detected in any patient who responded to enzalutamide for longer than 6 months. These finding were inline with an earlier study by the same investigators80 evaluating the combination of abiraterone and enzalutamide in 60 men with bone-metastatic CRPC. In that study, AR-V7 protein was detected in bone marrow biopsies from 66% of patients who developed progression within 4 months, but in none of the men who responded to therapy for more than 6 months.
More recently, Antonarakis et al.27 have developed an assay to serially evaluate AR-V7 at the mRNA level from circulating tumor cells (CTCs), using a RT-PCR detection method (positivity is defined as detection of AR-V7 cDNA at ≤36 PCR cycles). These authors conducted a prospective study assessing the prognostic role of AR-V7 in 31 CRPC patients receiving abiraterone and 31 patients receiving enzalutamide. In the abiraterone-treated cohort, AR-V7 was detected at baseline in 19% of patients. AR-V7–positive men had lower PSA response rates (0 vs 68%), shorter progression-free survival (hazard ratio (HR) 16.5) and shorter overall survival (HR 9.9) to abiraterone than AR-V7–negative men. The baseline prevalence of AR-V7 in the enzalutamide-treated cohort was 39%. Again, AR-V7–positive men had lower PSA response rates (0 vs 53%), shorter progression-free survival (HR 8.5) and shorter overall survival (HR 4.3) to enzalutamide than AR-V7-negative men. Notably, the prevalence of AR-V7 was higher in enzalutamide-treated men who had previously received abirater-one and in abiraterone-treated men who had previously received enzalutamide; AR-V7 prevalence was lowest in men who did not receive either agent. In addition, when assessing serial CTC samples over time, the authors reported that all men with baseline detection of AR-V7 remained AR-V7-positive during the course of therapy with abiraterone and enzalutamide, while 14% of men with negative AR-V7 status at baseline converted to AR-V7-positive during treatment; these patients had intermediate clinical outcomes.
These findings were supported by an independent study recently published by Steinestel et al.81 In this prospective study, the authors used a CTC-based RT-PCR assay to detect AR-V7 in the context of various therapies for CRPC, including 22 patients receiving abiraterone (n = 10) or enzalutamide (n = 12). To this end, the PSA response rate to abiraterone or enzalutamide was 7% among AR-V7-positive patients and 63% among AR-V7-negative patients. Notably, one AR-V7-positive patient did have a PSA response to abiraterone, suggesting that a small proportion of AR-V7-positive men may derive some benefit from abiraterone or enzalutamide. In addition, this study confirmed that the prevalence of AR-V7 was higher in patients who had previously received abiraterone or enzalutamide compared with those who had not.
Another important question is whether the presence of AR-V7 is relevant in the setting of taxane chemotherapy, especially because two preclinical studies had previously produced conflicting results in this regard.82,83 To answer this question, Antonarakis et al.84 performed a second prospective study using their CTC-based AR-V7 RT-PCR assay on 37 patients beginning treatment with docetaxel (n = 30) or cabazitaxel (n = 7). The prevalence of AR-V7 in these patients, most of which had previously received abiraterone and/or enzalutamide, was 46%. Encouragingly, PSA responses were observed in both AR-V7–positive and AR-V7–negative men (41 vs 65%). Similarly, progression-free survival was not statistically different in AR-V7-positive and negative patients. As a hypothesis-generating exercise, the authors then incorporated data from their prior study of 62 abiraterone- and enzalutamide-treated patients, and showed that clinical outcomes appeared to be better with taxanes compared with enzalutamide or abiraterone in AR-V7-positive men, while outcomes did not appear to differ by treatment type in AR-V7-negative men. More specifically, in AR-V7-positive patients, PSA responses were higher in taxane-treated vs enzalutamide- or abiraterone-treated men (41 vs 0%), and progression-free survival was longer in taxane-treated men (HR 0.21). A very interesting observation from this study was that a significant proportion of patients (58%) with baseline positive AR-V7 converted to AR-V7-negative during treatment with docetaxel or cabazitaxel. Whether or not such transitions in AR-V7 status may re-sensitize such patients to further AR-directed therapies is unknown.85
In an independent prospective clinical trial, Onstenk et al.86 developed a CTC assay to evaluate AR-V7 mRNA in 29 CRPC patients starting therapy with cabazitaxel. The prevalence of AR-V7 at baseline in these patients, who had all received prior docetaxel as well as abiraterone in most cases, was 55%. In support of the previous study, the authors showed no significant differences between AR-V7-negative and AR-V7-positive patients with respect to PSA responses (18 vs 8%), progression-free survival (HR 0.8) or overall survival (HR 1.6). Therefore, taken together with the findings from the previous study, the preliminary evidence to date suggests that presence of AR-V7 may not be a marker of resistance to taxane chemotherapy and may therefore represent a treatment-selection biomarker in CRPC.
Clinical qualification of AR-V7: A Treatment-Selection Biomarker?
Despite the intriguing clinical correlations discussed above, these findings remain preliminary and will require systematic prospective validation and clinical qualification. As such, it remains premature at this time to use CTC-based AR-V7 testing in routing clinical practice to inform treatment decisions. AR-V detection and presence in men with metastatic CRPC must be understood in the context of a number of additional known resistance and progression-related genomic and epigenomic alterations (Figure 2). These include both AR-dependent and AR-independent mechanisms, immune tolerance, DNA repair defects and aberrations in key oncogenes or tumor suppressors implicated in CRPC progression. Although there are a number of AR-V7 validation studies currently being conducted, this review will highlight two examples. Other studies aiming to confirm (or refute) the clinical relevance of AR-V7 in CRPC patients are summarized in Table 1.
Figure 2.
Mechanisms of castration-resistant progression, reflecting various molecular disease states. AR, androgen receptor; LBD, ligand-binding domain; mCRPC, metastatic castration-resistant prostate cancer.
Table 1. Selected biomarker trials evaluating the clinical utility of AR-V7 in CRPC patients.
| Therapeutic agents | Trial phase | Description | Key outcomes | Biomarker platform | NCT number |
|---|---|---|---|---|---|
| Cabazitaxel vs abiraterone/enzalutamide [PRIMCAB] | Phase 2 | Randomized open-label trial of cabazitaxel vs abiraterone or enzalutamide in mCRPC patients refractory to enzalutamide or abiraterone, with prospective validation of AR-V7 biomarker | rPFS | From CTCs; mRNA-based, AdnaTest (Qiagen) | NCT02379390 |
| Cabazitaxel vs abiraterone/enzalutamide [OZM-054] | Phase 2 | Randomized open-label trial of cabazitaxel vs abiraterone or enzalutamide in mCRPC patients with poor-prognosis features who have not previously received abiraterone or enzalutamide | PSA response rate and/or radiographic response | From whole-blood RNA; mRNA-based, PAXgene (PreAnalytiX, Hombrechtikon, Switzerland) | NCT02254785 |
| Abiraterone, enzalutamide, taxanes [PCF 00056936] | Phase 2 | Prospective observational study in mCRPC patients starting standard-of-care abiraterone or enzalutamide, with potential switch to taxane chemotherapy upon progression, evaluating mechanisms of resistance related to AR-V7 and other biomarkers. Three AR-V7 assays will be compared. | PFS, OS | (1) From CTCs; mRNA-based, AdnaTest (Qiagen) (2) From CTCs; mRNA-based, RosetteSep (StemCell Technologies, Vancouver, BC, Canada) (3) From CTCs; protein-based, Epic Sciences | NCT02269982 |
| Abiraterone, enzalutamide [GUTG-001] | Phase 2 | Randomized open-label sequencing study of abiraterone→ enzalutamide vs enzalutamide→ abiraterone in mCRPC | PSA response rate | From whole-blood RNA; mRNA-based, PAXgene (PreAnalytiX) | NCT02125357 |
| Abiraterone, enzalutamide [BARRIER-P] | Phase 2 | Open-label trial of standard-of-care enzalutamide or abiraterone for mCRPC, evaluating biomarkers of response and resistance including AR-V7 | PSA response rate, PSA progression | From whole-blood RNA; mRNA-based, PAXgene (PreAnalytiX) | NCT02429193 |
Abbreviations: AR-V7, androgen receptor splice variant 7; CTC, circulating tumor cell; mCRPC, metastatic castration-resistant prostate cancer; OS, overall survival; rPFS, radiographic progression-free survival.
The first study is the Sanofi-sponsored PRIMCAB trial (NCT02379390), whose target population is men with metastatic CRPC who have developed clinical disease progression within 6 months of starting abiraterone or enzalutamide. Such patients (n = 274) will be randomized equally to receive either cabazitaxel chemotherapy or the alternative AR-directed therapy. The primary end point of this trial is radiographic progression-free survival. As a secondary end point, the trial will prospectively evaluate baseline AR-V7 mRNA status from CTCs as a putative predictive biomarker in this setting, where the prevalence of AR-V7 mRNA is expected to be ∼ 33%. Johns Hopkins will serve as the central laboratory for AR-V7 testing in this trial. Exploratory analyses will also evaluate transitions in AR-V7 status at the time of progression.
The second study is a Prostate Cancer Foundation (PCF)-sponsored prospective biomarker trial (NCT02269982) evaluating three different CTC-based AR-V7 assays in 120 men with metastatic CRPC who have not received taxane chemotherapy for CRPC. In an attempt to enrich for patients with evaluable CTCs,1 eligible subjects must have at least two of the following high-risk features: radiographic progression, hemoglobin <10 g dl−1, alkaline phosphatase above normal, lactate dehydrogenase above normal, PSA doubling time <3 months, prior abiraterone or enzalutamide use, presence of visceral metastases, presence of pain requiring narcotics or detectable CTC using the CellSearch platform. In this non-interventional trial, patients will receive standard-of-care abiraterone or enzalutamide and then may also chose to receive standard-of-care taxane at progression. AR-V7 testing will be performed before AR-directed therapy, at progression on AR-directed therapy, and also at progression on chemotherapy (for those patients subsequently receiving taxane treatment). Each patient will undergo AR-V7 testing with three clinical assays at each time point: the Johns Hopkins mRNA-based assay, the Weill-Cornell mRNA-based assay (which also evaluates other AR-Vs), and the EPIC Sciences protein-based AR assay (San Diego, CA, USA). The coordinating center for this trial is the Duke Cancer Institute. In this study, the relationship of AR-Vs with outcome will be analyzed in the context of CTC enumeration, clinical phenotypes and other genomic aberrations detected in CTCs and cell-free DNA through copy number analysis and whole-exome sequencing, including AR amplification and other pathways implicated in CRPC (Figure 2).
Therapeutic Targeting of AR-V7
While there are currently no agents in clinical use that can specifically target AR-V7 or other AR-Vs in prostate cancer, a number of interesting compounds are now in clinical development that may have AR-V-directed activities. Here we will highlight three of these agents, while others are summarized in Table 2.
Table 2. Selected therapeutic trials using agents with potential activity against AR-V7-expressing CRPC.
| Investigational agents | Trial phase | Description | Key outcomes | Biomarker platform | NCT number |
|---|---|---|---|---|---|
| Galeterone vs enzalutamide [ARM0R3-SV] | Phase 3 | Randomized open-label trial of enzalutamide vs galeterone in treatment-naive AR-V7-positive mCRPC patients | rPFS, OS | From CTCs; mRNA-based, AdnaTest (Qiagen) | NCT02438007 |
| EPI-506 (AR-NTD inhibitor) [EPI-506-CS-000I] | Phase 1/2 | Single-arm trial in men with mCRPC after progression on enzalutamide or abiraterone (one prior taxane also permitted) | Safety, PSA response rate | From CTCs; protein-based, Epic Sciences | NCT02606123 |
| Niclosamide+enzalutamide [NCI-2015-01246] | Phase 1 | Open-label trial of niclosamide plus enzalutamide in AR-V7-positive abiraterone-refractory mCRPC | Safety, PSA response rate | From CTCs; mRNA-based, AdnaTest (Qiagen) | NCT02532114 |
| High-dose testosterone [RESTORE] | Phase 2 | Single-arm trial of high-dose testosterone for abiraterone- or enzalutamide-refractory mCRPC | PSA response rate, safety | From CTCs; mRNA-based, AdnaTest (Qiagen) | NCT02090114 |
| High-dose testosterone vs enzalutamide [TRANSFORMER] | Phase 3 | Randomized study of high-dose testosterone vs enzalutamide for abiraterone-refractory mCRPC | rPFS | From CTCs; mRNA-based, AdnaTest (Qiagen) | NCT02286921 |
| Cabazitaxel [CARVE] | Phase 2 | Single-arm open-label trial of cabazitaxel in mCRPC patients with AR-V7-positive CTCs who have previously received docetaxel | PSA response rate | From CTCs; mRNA-based, CellSearch (Janssen, Horsham, PA, USA) | NCT02621190 |
| GSK525762 (BET inhibitor) [GSK-115521] | Phase 1 | Open-label trial of GSK525762 in solid tumors, including CRPC | Safety, response rate | No AR-V7 testing | NCT01587703 |
| GS-5829 (BET inhibitor) [GS-US-350-1604] | Phase 1/2 | Open-label trial of GS-5829 specifically for mCRPC, used alone (phase 1) and in combination with enzalutamide (phase 2) | Safety (phase 1); PFS at 24 weeks (phase 2) | From CTCs; protein-based, Epic Sciences | NCT02607228 |
| Ipilimumab+nivolumab [STARVE-PC] | Phase 2 | Single-arm trial of ipilimumab plus nivolumab in AR-V7-positive mCRPC | PSA response rate, safety | From CTCs; mRNA-based, AdnaTest (Qiagen) | NCT02601014 |
Abbreviations: AR-NTD, androgen receptor N-terminal domain; BET, bromodomain extra-terminal motif; CTC, circulating tumor cell; mCRPC, metastatic castration-resistant prostate cancer; OS, overall survival; rPFS, radiographic progression-free survival.
The first drug is galeterone, manufactured by Tokai Pharmaceuticals, Boston, MA, USA. Galeterone is an oral AR signaling inhibitor that possesses three mechanisms of AR-directed action: it inhibits CYP17 lyase, it antagonizes the AR ligand-binding domain, and it destabilizes AR protein via an unknown, proteasome-dependent mechanism.87 Interestingly, treatment of AR-V-expressing prostate cancer cells with galeterone also leads to reduced AR-V expression (including AR-V7), presumably through this same proteasome-dependent mechanism.88 In a post hoc analysis of the phase 2 ARMOR2 trial, six out of seven men with reduced/lost expression of the AR COOH-terminal domain (as determined by immunohistochemistry on CTCs using an AR COOH-terminal domain-specific antibody) achieved a >50% PSA reduction with galeterone. Based on these preliminary data, a registrational phase-3 trial, ARMOR3-SV, was launched in Q4 2015 (NCT02438007). Eligible patients are those with AR-V7-positive metastatic CRPC without prior treatment with abiraterone, enzalutamide, or taxane chemotherapies. AR-V7 testing will be conducted using a CLIA-certified assay developed by Qiagen (Hilden, Germany). Patients with CTCs positive for AR-V7 mRNA (n = 148) will be equally randomized to receive either galeterone 2550 mg daily or enzalutamide 160 mg daily. The primary end point is radiographic progression-free survival, with the key secondary end point being overall survival. Notably, ARMOR3-SV is the first registrational trial in prostate cancer to use a biomarker-selection precision-medicine trial design, and will test the efficacy of a multi-targeted AR-directed agent in men with detectable CTCs who would not be predicted to benefit substantially from a pure AR-FL inhibitor.
The second agent is EPI-506 (an oral prodrug of EPI-002, manufactured by ESSA), which is the first drug capable of targeting the AR-NTD.89 Specifically, EPI-002 is one out of four stereoisomers of racemic EPI-001 which is a chlorinated bisphenol compound that can bind covalently to the AR-NTD. Because the NTD is common to both in the AR-FL and in all of the AR-Vs (including AR-V7), treatment with EPI-506 would be expected to extinguish all forms of AR signaling. Indeed, preclinical studies with EPI-002 have shown that this compound has activity in several AR-V expressing cell lines and xenograft models, including LNCaP95 and VCaP.90,91 However, EPI-001 has also been shown to have anti-proliferative activity at higher doses in AR-null prostate cancer cells, and demonstrates effects that are independent of the AR-NTD, including inhibition of AR expression, selective PPARγ agonist activity, and a general pH-dependent alkylating activity.92 Based on these encouraging preclinical data, a phase 1 trial (including a subsequent phase 2 expansion) was initiated in Q1 2016 (NCT02606123). Eligible patients will be those with meta-static CRPC who have previously received either abiraterone or enzalutamide; one prior taxane therapy is also permitted but not required.93 Exploratory analyses of AR-V7 and AR mutations will also be conducted in this trial, but this information with not be used for patient selection or stratification.
The third agent with potential activity against AR-V-expressing prostate cancer is the anti-helminthic drug niclosamide. In a drug library screen aimed at identifying FDA-approved drugs capable of targeting AR-V7, niclosamide emerged as an unexpected hit.94,95 Further mechanistic studies suggested that this agent functioned by promoting degradation of AR-FL and AR-V7 through a proteasome-dependent pathway. Interestingly, this study noted that AR-V7 degradation occurred more rapidly than AR-FL degradation in niclosamide-treated cells. Remarkably, niclosamide demonstrated significant antitumor activity in a number of AR-V-expressing CRPC cell lines (C4-2 and CWR22Rv1), as well as in a CWR22Rv1 xenograft model. Notably, 22Rv1 was found to be completely resistant to enzalutamide, but niclosamide resulted in tumor growth restriction in this model, while the combination of niclosamide and enzalutamide produced maximal tumor inhibition. Based on these preclinical data, a phase 1 clinical trial was launched in Q4 2015 (NCT02532114) for men with abiraterone-pretreated CRPC who test positive for AR-V7 using a CTC-based AR-V7 assay developed at the University of Washington, Seattle. In this trial, patients will receive enzalutamide 160 mg plus escalating doses of oral niclosamide (500 mg three times daily, 1000 mg three times daily and 1500 mg three times daily). Exploratory analyses will evaluate changes in AR-V7 status during the course of niclosamide treatment and at the time of progression.
Unresolved Questions and Future Research
The principal question that arises from the data presented is whether AR-Vs, particularly AR-V7, are drivers of malignant progression and treatment resistance in the clinic, or whether AR-Vs are passenger markers of aggressive disease. For example, high levels of AR copy number amplification may be associated with altered splicing which may lead to the detection of these variants in CTCs or tissues, but it may be that AR-FL gains rather than AR-V expression is important given their relative abundance. Recent data suggest a strong relationship between AR copy gains detectable in plasma cell-free DNA and poor outcomes with abiraterone in men with metastatic CRPC.96 The relationship between AR copy gains and altered splicing of AR in patients is unclear, and which biomarker is most associated with poor response remains undetermined. In addition, heterogeneity of CTCs clearly exists in men with mCRPC, and while AR-Vs may be detectable, they may co-exist with other aggressive disease genotypes and phenotypes, such as neuroendocrine transformation, de-differentiation and stem-like phenotypes, as well as AR-null CTCs (Figure 2,13,97). The dissemination of CTCs, which allows AR-Vs to be detectable, may itself be a marker of highly aggressive/invasive disease and epithelial plasticity which may or may not be causally related to AR-Vs.98 In addition, targeting of AR-Vs in preclinical models appears to be context-dependent. In some contexts, reduction of AR-Vs may restore sensitivity to AR antagonists, while in other contexts AR-V action appears expendable while AR-FL activity appears dominant. Some of this model dependence may be related to the presence of agonistic mutations that prevent AR-Vs from emerging in the presence of drugs such as enzalutamide, while in other contexts, AR-Vs may be found at low levels or unlinked from tumor cell growth.60
Men with metastatic CRPC present with a large number of genomic alterations that impact on DNA repair pathways, PI3K pathway signaling, cell cycle pathways, stemness/differentiation pathways (WNT and NOTCH signaling), epigenetic signaling and p53 loss among many others.13 In addition, epigenetic divergent evolution toward a neuroendocrine phenotype may lead to a loss of AR dependence,99 which may co-exist and eventually overtake AR-dependent clones under the selection pressures of hormonal therapy. These genomic lesions may take on a more important role when AR-FL is suppressed, and while AR-Vs may become detectable due to AR-FL suppression, their persistence may be transient, and may be expendable in the context of these additional mutations. Furthermore, the metastatic process in prostate cancer is accompanied by widespread alterations in splicing decisions, which may impact many genes other than AR. Silencing of AR and a movement away from AR dependency is clearly operative in several cases after progression on enzalutamide/abiraterone, and in this context it is unlikely that further AR inhibition (even with inhibitors that target the AR-NTD or DBD, or anti-AR immunotherapy) would have therapeutic efficacy.
To discern the driver vs. passenger role of AR-Vs in the clinical setting, trials in which agents that selectively inhibit AR-Vs (or both AR-FL and AR-Vs) are needed. Such trials are listed in Table 2, as agents such as galeterone, EPI-506, and perhaps others may have activity in AR degradation or inhibition of AR more broadly. Demonstrating reversal of resistance with these agents would provide proof-of-concept that at least some of the resistance to AR-LBD inhibitors is mediated by AR-Vs. However, if efficacy is modest or short-lived, it would imply that AR-Vs are merely markers of aggressive disease, and that other approaches beyond AR targeting are needed, such as immunotherapy or combinations with poly (ADP-ribose) polymerase inhibitors or chemotherapy.100 However, the fact that AR-V protein expression appears to increase during hormonal therapy indicates strong clonal selection or plasticity induced by drug treatment, and suggests that further targeting of the AR is likely to provide clinical benefit. The key question is whether the root/trunk cells in CRPC remain AR-negative and de-differentiated, even while spawning more differentiated progeny. If this is the case, potent AR inhibition with NTD inhibitors may only select for more aggressive AR-null clones such as the neuroendocrine prostate cancer transformation over time.
Given this, ongoing and future trials of novel AR inhibitors should measure and account for this heterogeneity in both genotype and phenotype, and track it longitudinally. One such ongoing study (NCT02269982) is doing exactly this, using cell-free RNA/DNA, CTC-derived RNA/DNA, and measures of tumor heterogeneity in the context of AR-V detection. Given that data suggest that AR-Vs may promote plasticity itself in a feed-forward loop, which may eventually lead to escape from AR dependency, measures of the relationship between plasticity, stemness, neuroendocrine prostate cancer transformation and AR biology in CRPC patients receiving standard-of-care therapies are needed over time. Only through such pharmacodynamic and mechanistic studies can we develop broader therapeutic approaches, such as immunotherapies or combinations of targeted agents, to address this heterogeneity within patients and between patients. Finally, such predictive biomarker-driven studies may permit the optimization of care delivery to those men with CRPC who are most likely to benefit, saving patients and society from the burdens of cost and excess toxicity from ineffective agents.
Conclusions
We have reviewed the origin, structure, and biology of AR variants, and have demonstrated the strong clinical associations of measurements of AR-Vs in CTCs with clinical outcomes in CRPC patients receiving novel AR-targeted therapies and taxane chemotherapies. AR variants are likely an important contributor to CRPC progression and AR therapy resistance, and emerging biomarkers of AR variant expression in patients should help to select men with prostate cancer most likely to benefit from AR-targeted therapies or to select men who are appropriate for other systemic approaches. Much of these data suggest that in some patients, targeting of the AR-NTD or DBD may provide greater therapeutic benefit than targeting the AR LBD alone, or that combination approaches with AR LBD inhibitors may be beneficial. A number of biomarker-based predictive trials are ongoing to examine AR variants in the contexts of standard enzalutamide or abiraterone therapy. In addition, AR variant expression is being utilized in several trials of novel hormonal or immunologic agents in order to demonstrate clinical benefit in AR variant-driven tumors. As most of oncology moves toward using precision biomarkers, we anticipate that measures of AR biology, including AR-Vs, in the context of a broad genomic characterization of patients, will help to select patients for AR-directed therapies vs chemotherapies, to monitor more closely those men who appear to have only a modest AR dependence due to tumor heterogeneity or plasticity, and to direct patients who appear to have AR-independent disease to other therapies (including radium-223, 239 immunotherapy approaches or clinical trials). The ultimate goal driving these trials is a personalized medicine approach to optimizing care based on the underlying and treatment-induced genotype and phenotype of men with mCRPC.
Acknowledgments
ESA has received funding from the Prostate Cancer Foundation, the Patrick C. Walsh Fund, and NIH grants R01 CA185297 and P30 CA006973. AJA has received funding from a Prostate Cancer Foundation and Movember Global Treatment Sciences Challenge Award. SMD is currently funded by a Movember/Prostate Cancer Foundation Challenge Award, American Cancer Society Research Scholar Grant RSG-12-031-01-TBE, NIH grant R01 CA174777, US Department of Defense Prostate Cancer Research Program grants W81XWH-12-2-0093, W81XWH-13-1-0518, W81XWH-15-1-0633, and W81XWH-15-1-0501, and a grant from the Minnesota Partnership for Biotechnology and Medical Genomics. JL is currently funded by a Prostate Cancer Foundation grant, NIH grant R01 CA185297, and US Department of Defense Prostate Cancer Research Program grants W81XWH-13-2-0093 and W81XWH-15-2-0050.
Footnotes
Conflict of Interest: ESA has served as a paid consultant/advisor for Janssen, Astellas, Sanofi, Dendreon, Essa, and Medivation; has received research funding to his institution from Janssen, Johnson & Johnson, Sanofi, Dendreon, Exelixis, Genentech, Novartis and Tokai; and is a co-inventor of a technology that has been licensed to Tokai. AJA has served as a paid consultant for Sanofi-aventis, Dendreon, Janssen, Eisai, Bayer and Medivation/Astellas; is on the speaker's bureau for Sanofi-aventis and Dendreon; and receives research funding to his institution from Janssen, Medivation/Astellas, Sanofi-aventis, Active Biotech, Bayer, Dendreon, Novartis and Pfizer. SMD has served as a paid consultant/advisor for Medivation/Astellas. JL has served as a paid consultant/advisor for Astellas, Gilead and Sanofi; has received research funding to his institution from Orion, Mirati, Astellas, Sanofi and Gilead; and is a co-inventor of a technology that has been licensed to A&G and Tokai.
References
- 1.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941 J Urol. 2002;167:948–951. [PubMed] [Google Scholar]
- 2.Attard G, Cooper CS, de Bono JS. Steroid hormone receptors in prostate cancer: a hard habit to break? Cancer Cell. 2009;16:458–462. doi: 10.1016/j.ccr.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8:440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.deBono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 8.Nakazawa M, Antonarakis ES, Luo J. Androgen receptor splice variants in the era of enzalutamide and abiraterone. Horm Cancer. 2014;5:265–273. doi: 10.1007/s12672-014-0190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferraldeschi R, Welti J, Luo J, Attard G, de Bono JS. Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene. 2014 doi: 10.1038/onc.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ware KE, Garcia-Blanco MA, Armstrong AJ, Dehm SM. Biologic and clinical significance of androgen receptor variants in castration resistant prostate cancer. Endocr Relat Cancer. 2014;21:T87–T103. doi: 10.1530/ERC-13-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merson S, Yang ZH, Brewer D, Olmos D, Eichholz A, McCarthy F, et al. Focal amplification of the androgen receptor gene in hormone-naive human prostate cancer. Br J Cancer. 2014;110:1655–1662. doi: 10.1038/bjc.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karantanos T, Evans CP, Tombal B, Thompson TC, Montironi R, Isaacs WB. Understanding the mechanisms of androgen deprivation resistance in prostate cancer at the molecular level. Eur Urol. 2015;67:470–479. doi: 10.1016/j.eururo.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small EJ, Huang J, Youngren J, Sokolov A, Aggarwal RR, Thomas G, et al. Characterization of neuroendocrine prostate cancer (NEPC) in patients with meta-static castration resistant prostate cancer (mCRPC) resistant to abiraterone (Abi) or enzalutamide (Enz): preliminary results from the SU2C/PCF/AACR West Coast Prostate Cancer Dream Team (WCDT) ASCO Annual Meeting Proceedings: abstr 5003. 2015 [Google Scholar]
- 16.Lu C, Luo J. Decoding the androgen receptor splice variants. Transl Androl Urol. 2013;2:178–186. doi: 10.3978/j.issn.2223-4683.2013.09.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci USA. 2010;107:16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Z, Chen S, Sowalsky AG, Voznesensky OS, Mostaghel EA, Nelson PS, et al. Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin Cancer Res. 2014;20:1590–1600. doi: 10.1158/1078-0432.CCR-13-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mostaghel EA, Marck BT, Plymate SR, Vessella RL, Balk S, Matsumoto AM, et al. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clin Cancer Res. 2011;17:5913–5925. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate. 2011;71:1656–1667. doi: 10.1002/pros.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA, et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci USA. 2013;110:17492–17497. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan SC, Dehm SM. Constitutive activity of the androgen receptor. Adv Pharmacol. 2014;70:327–366. doi: 10.1016/B978-0-12-417197-8.00011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan X, Cai C, Chen S, Chen S, Yu Z, Balk SP. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33:2815–2825. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dehm SM, Regan KM, Schmidt LJ, Tindall DJ. Selective role of an NH2-terminal WxxLF motif for aberrant androgen receptor activation in androgen depletion independent prostate cancer cells. Cancer Res. 2007;67:10067–10077. doi: 10.1158/0008-5472.CAN-07-1267. [DOI] [PubMed] [Google Scholar]
- 32.Bohrer LR, Liu P, Zhong J, Pan Y, Angstman J, Brand LJ, et al. FOXO1 binds to the TAU5 motif and inhibits constitutively active androgen receptor splice variants. Prostate. 2013;73:1017–1027. doi: 10.1002/pros.22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE, et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015;43:5880–5897. doi: 10.1093/nar/gkv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W, et al. Androgen receptor splice variants dimerize to transactivate target genes. Cancer Res. 2015;75:3663–3671. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Royen ME, van Cappellen WA, de Vos C, Houtsmuller AB, Trapman J. Step-wise androgen receptor dimerization. J Cell Sci. 2012;125:1970–1979. doi: 10.1242/jcs.096792. [DOI] [PubMed] [Google Scholar]
- 36.Cutress ML, Whitaker HC, Mills IG, Stewart M, Neal DE. Structural basis for the nuclear import of the human androgen receptor. J Cell Sci. 2008;121:957–968. doi: 10.1242/jcs.022103. [DOI] [PubMed] [Google Scholar]
- 37.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. 2012;287:19736–19749. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saporita AJ, Zhang Q, Navai N, Dincer Z, Hahn J, Cai X, et al. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem. 2003;278:41998–42005. doi: 10.1074/jbc.M302460200. [DOI] [PubMed] [Google Scholar]
- 39.Antonarakis ES, Luo J. Prostate cancer: AR splice variant dimerization-clinical implications. Nat Rev Urol. 2015;12:431–433. doi: 10.1038/nrurol.2015.184. [DOI] [PubMed] [Google Scholar]
- 40.Liang M, Adisetiyo H, Liu X, Liu R, Gill P, Roy-Burman P, et al. Identification of androgen receptor splice variants in the Pten deficient murine prostate cancer model. PLoS One. 2015;10:e0131232. doi: 10.1371/journal.pone.0131232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tepper CG, Boucher DL, Ryan PE, Ma AH, Xia L, Lee LF, et al. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res. 2002;62:6606–6614. [PubMed] [Google Scholar]
- 42.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research Network. Electronic address scmo, Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71:2108–2117. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31:4759–4767. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20:457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin R, Yamashita H, Yu X, Wang J, Franco OE, Wang Y, et al. Inhibition of NF-kappa B signaling restores responsiveness of castrate-resistant prostate cancer cells to anti-androgen treatment by decreasing androgen receptor-variant expression. Oncogene. 2015;34:3700–3710. doi: 10.1038/onc.2014.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nadiminty N, Tummala R, Liu C, Lou W, Evans CP, Gao AC. NF-kappaB2/p52:c-Myc:hnRNPA1 pathway regulates expression of androgen receptor splice variants and enzalutamide sensitivity in prostate cancer. Mol Cancer Ther. 2015;14:1884–1895. doi: 10.1158/1535-7163.MCT-14-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nadiminty N, Tummala R, Liu C, Yang J, Lou W, Evans CP, et al. NF-kappaB2/p52 induces resistance to enzalutamide in prostate cancer: role of androgen receptor and its variants. Mol Cancer Ther. 2013;12:1629–1637. doi: 10.1158/1535-7163.MCT-13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y, Jia D, Kim H, Abd Elmageed ZY, Datta A, Davis R, et al. Dysregulation of miR-212 promotes castration resistance through hnRNPH1-mediated regulation of AR and AR-V7: implications for racial disparity of prostate cancer. Clin Cancer Res. 2015;22:1744–1756. doi: 10.1158/1078-0432.CCR-15-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–3150. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ceraline J, Cruchant MD, Erdmann E, Erbs P, Kurtz JE, Duclos B, et al. Constitutive activation of the androgen receptor by a point mutation in the hinge region: a new mechanism for androgen-independent growth in prostate cancer. Int J Cancer. 2004;108:152–157. doi: 10.1002/ijc.11404. [DOI] [PubMed] [Google Scholar]
- 55.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 56.Krause WC, Shafi AA, Nakka M, Weigel NL. Androgen receptor and its splice variant, AR-V7, differentially regulate FOXA1 sensitive genes in LNCaP prostate cancer cells. Int J Biochem Cell Biol. 2014;54:49–59. doi: 10.1016/j.biocel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo J, Pienta KJ. Words of wisdom: re: androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Eur Urol. 2013;64:339–340. doi: 10.1016/j.eururo.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Schweizer MT, Antonarakis ES, Wang H, Ajiboye AS, Spitz A, Cao H, et al. Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: results from a pilot clinical study. Sci Transl Med. 2015;7:269ra2. doi: 10.1126/scitranslmed.3010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shafi AA, Putluri V, Arnold JM, Tsouko E, Maity S, Roberts JM, et al. Differential regulation of metabolic pathways by androgen receptor (AR) and its con-stitutively active splice variant, AR-V7, in prostate cancer cells. Oncotarget. 2015;6:31997–32012. doi: 10.18632/oncotarget.5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto Y, Loriot Y, Beraldi E, Zhang F, Wyatt AW, Nakouzi NA, et al. Generation 2.5 antisense oligonucleotides targeting the androgen receptor and its splice variants suppress enzalutamide-resistant prostate cancer cell growth. Clin Cancer Res. 2015;21:1675–1687. doi: 10.1158/1078-0432.CCR-14-1108. [DOI] [PubMed] [Google Scholar]
- 61.Bitting RL, Schaeffer D, Somarelli JA, Garcia-Blanco MA, Armstrong AJ. The role of epithelial plasticity in prostate cancer dissemination and treatment resistance. Cancer Metastasis Rev. 2014;33:441–468. doi: 10.1007/s10555-013-9483-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das R, Gregory PA, Hollier BG, Tilley WD, Selth LA. Epithelial plasticity in prostate cancer: principles and clinical perspectives. Trends Mol Med. 2014;20:643–651. doi: 10.1016/j.molmed.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 63.Liu G, Sprenger C, Sun S, Epilepsia KS, Haugk K, Zhang X, et al. AR variant ARv567es induces carcinogenesis in a novel transgenic mouse model of prostate cancer. Neoplasia. 2013;15:1009. doi: 10.1593/neo.13784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun F, Chen HG, Li W, Yang X, Wang X, Jiang R, et al. Androgen receptor splice variant AR3 promotes prostate cancer via modulating expression of autocrine/paracrine factors. J Biol Chem. 2014;289:1529–1539. doi: 10.1074/jbc.M113.492140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin J, Liu X, Laffin B, Chen X, Choy G, Jeter CR, et al. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10:556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Y, Wang BE, Leong KG, Yue P, Li L, Jhunjhunwala S, et al. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 2012;72:527–536. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]
- 67.Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, Castillo-Martin M, Quinn SA, Rodriguez-Barrueco R, et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell. 2012;22:373–388. doi: 10.1016/j.ccr.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cottard F, Asmane I, Erdmann E, Bergerat JP, Kurtz JE, Ceraline J. Constitutively active androgen receptor variants upregulate expression of mesenchymal markers in prostate cancer cells. PLoS One. 2013;8:e63466. doi: 10.1371/journal.pone.0063466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kong D, Sethi S, Li Y, Chen W, Sakr WA, Heath E, et al. Androgen receptor splice variants contribute to prostate cancer aggressiveness through induction of EMT and expression of stem cell marker genes. Prostate. 2015;75:161–174. doi: 10.1002/pros.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu ML, Kyprianou N. Role of androgens and the androgen receptor in epithelial-mesenchymal transition and invasion of prostate cancer cells. FASEB J. 2010;24:769–777. doi: 10.1096/fj.09-136994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ware KE, Schaeffer D, Zhang T, Garcia-Blanco MA, Armstrong AJ. AR-V7 regulation during epithelial plasticity. Cancer Res. 2015;75:1847. [Google Scholar]
- 73.Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 2015;12:922–936. doi: 10.1016/j.celrep.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 74.Heebøll S, Borre M, Ottosen PD, Dyrskjøt L, Ørntoft TF, Tørring N. Snail1 is over-expressed in prostate cancer. APMIS. 2009;117:196–204. doi: 10.1111/j.1600-0463.2008.00007.x. [DOI] [PubMed] [Google Scholar]
- 75.Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, et al. ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: implications for Gleason grading. Cancer Cell. 2010;17:319–332. doi: 10.1016/j.ccr.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McKeithen D, Graham T, Chung LW, Odero-Marah V. Snail transcription factor regulates neuroendocrine differentiation in LNCaP prostate cancer cells. Prostate. 2010;70:982–992. doi: 10.1002/pros.21132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qu Y, Dai B, Ye D, Kong Y, Chang K, Jia Z, et al. Constitutively active AR-V7 plays an essential role in the development and progression of castration-resistant prostate cancer. Sci Rep. 2015;5:7654. doi: 10.1038/srep07654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X, Morrissey C, Sun S, Ketchandji M, Nelson PS, True LD, et al. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PLoS One. 2011;6:e27970. doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Efstathiou E, Titus M, Wen S, Hoang A, Karlou M, Ashe R, et al. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur Urol. 2015;67:53–60. doi: 10.1016/j.eururo.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Efstathiou E, Titus MA, Wen S, SanMiguel A, Hoang A, De Haas-Amatsaleh A, et al. Enzalutamide (ENZA) in combination with abiraterone acetate (AA) in bone metastatic castration resistant prostate cancer (mCRPC) ASCO Annual Meeting Proceedings; abstr 5000. 2014 [Google Scholar]
- 81.Steinestel J, Luedeke M, Arndt A, Schnoeller TJ, Lennerz JK, Wurm C, et al. Detecting predictive androgen receptor modifications in circulating prostate cancer cells. Oncotarget. 2015 doi: 10.18632/onco-target.3925. e-pub ahead pf print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thadani-Mulero M, Portella L, Sun S, Sung M, Matov A, Vessella RL, et al. Androgen receptor splice variants determine taxane sensitivity in prostate cancer. Cancer Res. 2014;74:2270–2282. doi: 10.1158/0008-5472.CAN-13-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang G, Liu X, Li J, Ledet E, Alvarez X, Qi Y, et al. Androgen receptor splice variants circumvent AR blockade by microtubule-targeting agents. Oncotarget. 2015;6:23358–23371. doi: 10.18632/oncotarget.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Nakazawa M, et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 2015;1:582–591. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakazawa M, Lu C, Chen Y, Paller CJ, Carducci MA, Eisenberger MA, et al. Serial blood-based analysis of AR-V7 in men with advanced prostate cancer. Ann Oncol. 2015;26:1859–1865. doi: 10.1093/annonc/mdv282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Onstenk W, Sieuwerts AM, Kraan J, Van M, Nieuweboer AJ, Mathijssen RH, et al. Efficacy of cabazitaxel in castration-resistant prostate cancer is independent of the presence of AR-V7 in circulating tumor cells. Eur Urol. 2015;68:939–945. doi: 10.1016/j.eururo.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 87.Purushottamachar P, Godbole AM, Gediya LK, Martin MS, Vasaitis TS, Kwegyir-Afful AK, et al. Systematic structure modifications of multitarget prostate cancer drug candidate galeterone to produce novel androgen receptor down-regulating agents as an approach to treatment of advanced prostate cancer. J Med Chem. 2013;56:4880–4898. doi: 10.1021/jm400048v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kwegyir-Afful AK, Ramalingam S, Purushottamachar P, Ramamurthy VP, Njar VC. Galeterone and VNPT55 induce proteasomal degradation of AR/AR-V7, induce significant apoptosis via cytochrome c release and suppress growth of castration resistant prostate cancer xenografts in vivo. Oncotarget. 2015;6:27440–27460. doi: 10.18632/oncotarget.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer cell. 2010;17:535–546. doi: 10.1016/j.ccr.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 90.Kato M, Banuelos CA, Imamura Y, Leung JK, Caley DP, Wang J, et al. Co-targeting androgen receptor splice variants and mTOR signaling pathway for the treatment of castration-resistant prostate cancer. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-15-2119. e-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Myung JK, Banuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH, et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest. 2013;123:2948–2960. doi: 10.1172/JCI66398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brand LJ, Olson ME, Ravindranathan P, Guo H, Kempema AM, Andrews TE, et al. EPI-001 is a selective peroxisome proliferator-activated receptor-gamma modulator with inhibitory effects on androgen receptor expression and activity in prostate cancer. Oncotarget. 2015;6:3811–3824. doi: 10.18632/oncotarget.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Montgomery R, Antonarakis E, Hussain M, Fizazi K, Joshua A, Attard G, et al. A phase 1/2 open-label study of safety and antitumor activity of EPI-506, a novel AR N-terminal domain inhibitor, in men with metastatic castration-resistant prostate cancer (mCRPC) with progression after enzalutamide or abiraterone. J Clin Oncol. 2015;33 abstr TPS5072. [Google Scholar]
- 94.Liu C, Lou W, Armstrong C, Zhu Y, Evans CP, Gao AC. Niclosamide suppresses cell migration and invasion in enzalutamide resistant prostate cancer cells via Stat3-AR axis inhibition. Prostate. 2015;75:1341–1353. doi: 10.1002/pros.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, et al. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res. 2014;20:3198–3210. doi: 10.1158/1078-0432.CCR-13-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Romanel A, Tandefelt DG, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7:312re10. doi: 10.1126/scitranslmed.aac9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bitting RL, Healy P, Halabi S, George DJ, Goodin M, Armstrong AJ. Clinical phenotypes associated with circulating tumor cell enumeration in metastatic castration-resistant prostate cancer. Urol Oncol. 2015;33:110.e1–110.e9. doi: 10.1016/j.urolonc.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 99.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]