Abstract
The rat tyrosine hydroxylase gene promoter contains an E-box/dyad motif and an octameric and heptameric element that may be recognized by classes of transcription factors highly expressed during nervous system development. In a one-hybrid genetic screen, we used these sites as targets to isolate cDNAs encoding new transcription factors present in the brain. We identified ZENON, a novel rat POZ protein that contains two clusters of Kruppel-like zinc fingers and that presents several features of a transcription factor. ZENON is found in nuclei following transient transfection with the cDNA. The N-terminal zinc finger cluster contains a DNA binding domain that interacts with the E box. Cotranfection experiments revealed that ZENON induces tyrosine hydroxylase promoter activity. Unlike other POZ proteins, the ZENON POZ domain is not required for either activation of transcription or self-association. In the embryonic neural tube, ZENON expression is restricted to neurons that have already achieved mitosis and are engaged in late stages of neuronal differentiation (late postmitotic neurons). ZENON neuronal expression persists in the adult brain; therefore, ZENON can be considered a marker of mature neurons. We propose that ZENON is involved in the maintenance of panneuronal features and/or in the survival of mature neurons.
The two major cell types in the nervous system, neurons and glia, both comprise a large number of subtypes. The generation and maintenance of this phenotypic diversity require extracellular signals that are converted into tightly regulated transcriptional cascades. Many transcription factors involved in these cascades—often basic helix-loop-helix (bHLH), homeodomain, or zinc finger-containing proteins—remain to be identified. Description of these factors would help elucidate the molecular events leading to the differentiation of the various cell types in the nervous system.
One way to identify these factors is to isolate novel proteins that interact with sequences promoting the neural expression of a highly developmentally regulated gene. A good candidate is the gene encoding tyrosine hydroxylase (TH), the rate-limiting enzyme in catecholamine biosynthesis (44, 56). The expression of the TH gene is a common feature of all neurons and neuroendocrine cells that synthesize and release catecholamines (dopamine, norepinephrine, and epinephrine). These cells are extremely diverse in their functional, morphological, and anatomical properties and have different embryonic origins. Depending on their locations in the adult central nervous system (CNS) and peripheral nervous system (PNS), catecholaminergic cells originate from different parts of the neural tube and neural crest (6, 73, 74). In the embryonic neural tube, catecholaminergic cell groups arise from various neuromeres (64, 78), suggesting that a large number of extracellular signals and transcription factors govern the specification and maintenance of catecholaminergic identity.
The production and functional analysis of knockout mice revealed the essential roles of several transcription factors in the specification and differentiation of catecholaminergic cells. Mash1, Phox2a, and Phox2b are involved in noradrenergic specification (25, 28, 54, 61). Nurr1 and Lmx1b are fundamental for establishing dopaminergic identity in the substantia nigra (13, 68, 70, 71, 88). GATA-3, dHAND, and eHAND are implicated in sympathetic differentiation (29, 46). However, there is no in vivo evidence for the direct regulation of TH gene expression by these factors, suggesting that they act early in the transcriptional cascades leading to catecholaminergic identity. Presumably, other as-yet-unidentified factors regulate transcription by interacting directly with TH regulatory sequences. The rat TH proximal promoter contains within the first 220 bp upstream from the transcription start site many elements that are potential transcription factor binding sites. With the exception of the AP1/TRE and CRE sites (76), the roles of these proximal elements have not been documented in vivo. The production and analysis of transgenic mice have demonstrated that large parts of the rat TH promoter region are necessary to direct expression in the appropriate cells, but the elements responsible for cell-specific expression have not been mapped precisely (8, 47, 53, 69, 72). In contrast, in vitro studies have revealed the roles of several proximal elements, including the E-box/dyad, AP1/TRE, and CRE elements (37, 41, 42, 75, 86), octameric and heptameric (oct/hept) elements (18, 86), SP1 (84), and the NBRE-like 1 motif (36).
The E-box/dyad and oct/hept elements appear to be the most promising for selecting novel transcription factors present in the nervous system. They are potentially bound by a wide variety of transcription factors thought to play fundamental roles in the mechanisms underlying cell type diversity in the nervous system. The E box may be recognized by bHLH and some zinc finger transcription factors (34, 55) and is surrounded by a dyad symmetry element that could interact with other families of transcription factors. The juxtaposed oct/hept elements can bind several classes of homeodomain proteins, including POU and paired-like proteins (18, 87). Two E-box/dyad motif-interacting proteins, rITF2 and CDP2, were previously identified (85). None of these proteins was found to be a novel transcription factor specifically expressed in the nervous system, perhaps because the cDNA library used was built from a neuroendocrine cell line and not from freshly dissected neural tissue and also because the screening strategy was based on in vitro interactions between DNA and proteins.
As a first step toward identifying novel transcription factors in the nervous system, we used the E-box/dyad and oct/hept elements as targets in a functional in vivo genetic assay. An adult rat brain cDNA library and a rat embryo cDNA library built from mesencephalic and rhombencephalic catecholaminergic nuclei were screened, leading to the isolation of several transcription factors. We describe here the discovery of a gene encoding a novel POZ zinc finger protein designated ZENON, for “zinc finger gene expressed in neurons.” The most striking feature of ZENON is that its expression is confined mainly to neurons that have already achieved mitosis and are differentiated or engaged in late stages of neuronal differentiation (mature neurons).
MATERIALS AND METHODS
One-hybrid screening.
The target sites used in this assay were the E-box/dyad motif (5′-TTCAGAGGCAGGTGCCTGTGAC-3′) and the oct/hept element and surrounding region (5′-CAGTGGATGCAATTAGATCTAATGGGACGGA-3′) (underlining indicates the E-box, octameric element, and heptameric element). Three copies of these sites were independently inserted upstream from the HIS3 reporter gene into the single blunt-ended XhoI site of yeast expression vector pYi2267OHIS (12). The orientation of each E-box and oct/hept element in the resulting plasmids was confirmed by sequence analysis to be the same as that in the rat native TH promoter.
The two reporter plasmids were linearized with StuI and introduced into yeast strain YM954 (83) by the polyethylene glycol-lithium acetate method (22). Recombinants were selected by growth on plates lacking uracil. Genomic DNA was prepared from five colonies for each reporter plasmid and digested with HindIII. Southern blot analysis was used to confirm integration at the URA3 locus, with the ampicillin resistance gene as the probe. Two strains in which only one copy of the reporter plasmid had been integrated were selected for the screening and were named YM-E and YM-OH.
Two libraries of rat cDNAs fused to the Gal4 activation domain (pGAD10; Clontech) were screened with YM-E and YM-OH: an adult rat brain library (Clontech) and an embryonic day 14 (E14) rat library built from brain regions containing the mesencephalon and the pons by using a two-hybrid cDNA library construction kit (Clontech). After transformation, the reporter strains were used to seed selective medium lacking leucine (library marker) and histidine to select for transcription of the HIS3 reporter gene. We added 20 mM 3-aminotriazole to the medium to prevent the growth of colonies due to leaky HIS3 expression. Several colonies were isolated over a 10-day period. The corresponding cDNA clones were extracted and amplified in bacteria. The specificity of HIS3-positive phenotype activation was checked by transforming YM-E and YM-OH in parallel with each purified plasmid. cDNAs conferring reproducible growth on selective medium for only one of the two strains were integrally sequenced and used for BLAST queries (5).
Isolation of the complete ZENON coding sequence.
The 5′ coding sequence was obtained by single-stranded ligation of cDNA (SLIC) (20). Single-stranded cDNA was synthesized from rat brain poly(A)+ RNA with an antisense primer contained in the 3′ extended cDNA clone: 5′-AGAATCCTGGGTTTGGGTCTTTGG-3′. After ligation with oligonucleotide A5′NV, nested PCR was performed with the first SLIC primer (A5′-1) and specific primer Z3′-1 (5′-GTCATGTTCCCTCAGAGGCTGACTC-3′) and then with the second SLIC primer (A5′-2) and specific primer Z3′-2 (5′-CTCAGTGATGGAAGAAACGGCATA-3′). The resulting PCR product was inserted into pGEM-T Easy (Promega) and sequenced.
The 3′ coding sequence was obtained by 3′ anchored PCR (20). Single-stranded cDNA was synthesized from rat brain poly(A)+ RNA with oligonucleotide RA3′NV. Several rounds of nested PCR were successively performed to obtain the complete 3′ coding sequence: the first step with A3′-1 and Z5′-1a, Z5′-1b, Z5′-1c, Z5′-1d, Z5′-1e, and Z5′-1f and the second step with A3′-2 and Z5′-2a, Z5′-2b, Z5′-2c, Z5′-2d, Z5′-2e, and Z5′-2f. The resulting PCR products were inserted into pGEM-T Easy and sequenced.
The primer sequences were as follows: Z5′-1a, 5′-GCAGCCAGTTCTCGTCGGTG-3′; Z5′-1b, 5′-AACCGTTCCATGTAACAGGGGAG-3′; Z5′-1c, 5′-GAGAGTGACCCCGAAACCCG-3′; Z5′-1d, 5′-AAGGAAAGTCAAGTGGAGGAAGGAA-3′; Z5′-1e, 5′-TCAGTATTGTAACAAGGCTTTCACGC-3′; Z5′-1f, 5′-CGGGAGCATGACGGGAAAG-3′; Z5′-2a, 5′-AGCAACCACCAGTCCCCTTCAC-3′; Z5′-2b, 5′-ATCTTCGGGCAAAACCACAAATG-3′; Z5′-2c, 5′-CTGTGTGGAGATGAGTGAAGCATTTG-3′; Z5′-2d, 5′-GAGGAAGGAACGAGGGAGCAGA-3′; Z5′-2e, 5′-ACACAGGGGAGAAGCGTTACCACT-3′; and Z5′-2f, 5′-CACCAAAAGAAGCACTTATTCAAGACCT-3′.
Eukaryotic ZENON expression.
ZENON sequences encoding residues 1 to 1202 and residues 196 to 1202 were inserted into vector pFLAG-CMV5b (Sigma) to obtain pFLAG-ZENON and pFLAG-ΔPOZ, respectively. These plasmids encode ZENON in frame with a C-terminal FLAG peptide. 293T kidney epithelial cells were grown in Dulbecco's modified Eagle medium supplemented with 10% heat-inactivated fetal calf serum and 1% penicillin-streptomycin.
For whole-cell extracts, 3 × 106 cells were used to seed 10-cm culture dishes on the day before transfection. Cells were transfected by the calcium phosphate precipitation method with 25 μg of pFLAG-ZENON or pFLAG-ΔPOZ per dish. Cells were harvested 48 h after transfection and lysed in lysis buffer, containing 5 mM HEPES (pH 7.9), 25% glycerol, 1.5 mM MgCl2, 0.5 mM dithiothreitol (DTT), 1 μM ZnSO4, 0.5 mM phenylmethylsulfonyl fluoride, and protease inhibitors without EDTA (Roche). NaCl then was added to a final concentration of 300 mM to release the nuclear extracts. Cell debris was pelleted by ultracentrifugation, and supernatants, corresponding to whole-cell extracts, were divided into aliquots and frozen at −80°C.
For Western blot analysis, 20 μg of whole-cell extracts was subjected to electrophoresis in a 9% polyacrylamide gel and blotted onto a nitrocellulose membrane. The ZENON-FLAG fusion proteins were detected by using anti-FLAG M2 antibody (Sigma) according to the manufacturer's instructions.
For subcellular localization, 105 cells per well were plated in 24-well culture dishes 24 h before transfection. Cells were transfected with 0.8 μg of pFLAG-ZENON per well or with the same amount of insert-free pFLAG-CMV5b as a control. Transfected cells were incubated for 48 h, and nuclei then were stained with Hoechst reagent diluted in culture medium (7 μg/ml). Cells were rinsed and fixed by incubation for 1 min in 50% ethanol-50% acetone. Immunocytochemical analysis was performed with the anti-FLAG antibody according to the manufacturer's instructions. Goat anti-mouse antibody conjugated to the Cy3 fluorophore was used as a secondary antibody (1:400; Jackson Laboratories). No staining was observed when the cells were transfected with insert-free pFLAG-CMV5b, demonstrating the specificity of the anti-FLAG antibody.
Electrophoretic mobility shift assays (EMSAs).
Sequences encoding L-ZENON (amino acids 177 to 752) or R-ZENON (amino acids 751 to 1203) were introduced into pGEX-4T1 (Amersham Pharmacia Biotech) in frame with the glutathione S-transferase (GST)-encoding sequence. The production of recombinant proteins in BL21 cells (Stratagene) was induced by incubation with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 37°C. Purification was performed with a glutathione-Sepharose resin (Amersham Pharmacia Biotech) according to the manufacturer's recommendations. ZENON proteins were cleaved from GST by thrombin digestion.
Primers containing the BssHII and Bst98I restriction sites were designed to amplify a PCR fragment spanning the rat TH proximal promoter from positions −259 to −59. The pTH probe was obtained by BssHII and Bst98I digestion of the PCR product. The E probe was generated from a double-stranded oligonucleotide with the same sequence as the motif used for one-hybrid screening, except that a GGG sequence was added at the 5′ end. The sequences of the ΔE and PAN oligonucleotides used for competition were 5′-GGGTTCAGAGGTCCGTGCCTGTGAC-3′ and 5′-GGGGAAAAGGACAGGTGCCCTGATT-3′ (underlining indicates mutated and intact E boxes), respectively. For radiolabeling of the probes, the Klenow fragment of DNA polymerase I was used to fill the 5′ overhangs with 32P-labeled nucleotides.
Binding reaction mixtures contained 1 μg of recombinant protein, 0.02 pmol of radiolabeled probe, 10 ng of poly(dI-dC), 10% glycerol, 50 mM KCl, 20 mM HEPES (pH 7.9), 1 mM DTT, and 1 μM ZnSO4 in a total volume of 10 μl. After incubation for 15 min, DNA-protein complexes were resolved by nondenaturing electrophoresis (5% polyacrylamide, 0.25× Tris-borate-EDTA electrophoresis buffer). The gel was dried for autoradiography.
RT-PCR.
Total RNA was extracted from 2 × 106 cells (PC12, 293T, and HeLa) and treated with DNase I by using the RNeasy system (Qiagen). Reverse transcription (RT) was performed from 1 μg of RNA with random primers and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. ZENON and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were amplified from 10% of the RT reaction mixture under identical PCR conditions (35 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 1 min with standard polymerase from Promega). Identical primers chosen within conserved regions were used to amplify ZENON and GAPDH from human cells (293T and HeLa) and rat cells (PC12). The sequences of the ZENON primers used were as follows: 5′-CTCAAGGACGACTTTCACAGTGA-3′ and 5′-GGGGAATTTGAGAAGTTGCG-3′. The sequences of the GAPDH primers used were as follows: 5′-GACCCCTTCATTGACCTCAACTA-3′ and 5′-CAGTGTAGCCCAGGATGCC-3′.
Transient transfections and luciferase assays.
HeLa and 293T cells were grown in Dulbecco's modified Eagle medium supplemented with 10% heat-inactivated fetal calf serum, and PC12 cells (rat pheochromocytoma) were grown in RPMI 1640 supplemented with 10% horse serum and 5% fetal calf serum. Cells were electroporated as previously described (51) with 1 pmol of 5kbTH-Luc or 5kbTH(ΔE)-Luc (65) and 1.5 pmol of effector pcDNA3 (Invitrogen), pcDNA3-Z, pcDNA3-ΔP, or pcDNA3-ΔC. The latter plasmids contain various fragments of ZENON cDNA subcloned into pcDNA3. To normalize for transfection efficiency, 2 μg of Renilla luciferase expression plasmid (pRL-TK; Promega) was also added. In a control experiment, we checked that ZENON effectors did not induce Renilla luciferase transcription. For each set of conditions, the amount of DNA was adjusted to 17.5 μg with pBluescript in a final volume of 15 μl, which was added to the cells. Firefly and Renilla luciferase activities were measured 48 h after transfection as previously described (4) by using a dual-luciferase reporter assay system (Promega).
GST pull-down assays.
GST fusion proteins and whole-cell extracts containing complete or partial ZENON-FLAG polypeptides were obtained as described above. After immobilization on glutathione-Sepharose resin, GST fusion proteins were incubated for 30 min at room temperature with cell extracts in binding buffer (10% glycerol, 100 mM KCl, 20 mM HEPES [pH 7.9], 1 mM DTT, 0.4% NP-40) supplemented with 10% bovine serum albumin (BSA). Successive washes with binding buffer containing 10%, 5%, and no BSA were performed prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with the anti-FLAG antibody.
Northern blotting.
A fragment of ZENON cDNA corresponding to amino acids 210 to 582 was subcloned into the EcoRI site of pcDNA3. We used 1 μg of this plasmid, linearized by XhoI digestion, to synthesize a 32P-labeled riboprobe with RNA polymerase T7 (riboprobe combination system Sp6/T7; Promega). RNA was prepared from fresh tissues by the acidic phenol-chloroform-guanidine thiocyanate method. Poly(A)+ RNA was extracted with Dynabeads oligonucleotide (dT)25 (Dynal). After denaturation in formamide and formaldehyde for 5 min at 65°C, 0.5 to 2 μg of poly(A)+ RNA was resolved by electrophoresis in 1% agarose denaturing gels and transferred by capillary blotting to nitrocellulose membranes (Protran; Schleicher & Schuell). The blots were hybridized with 107 cpm of 32P-labeled riboprobe as previously described (4).
In situ hybridizations.
Riboprobes were synthesized from various cDNAs inserted into pcDNA3 or pGEM-T Easy. After linearization of the plasmids, nonradioactively labeled riboprobes were synthesized in the presence of 3.5 nmol of digoxigenin (Dig)-11-UTP or fluorescein (Fluo)-12-UTP (Roche) by using riboprobe combination system Sp6/T7. The ZENON riboprobe spans the whole coding sequence except for the POZ domain (amino acids 188 to 1203). Rat microtubule-associated protein 2 (MAP2) cDNA (1.3 kb) was obtained from rat brain by RT-PCR with oligonucleotides having the sequences 5′-GTTGCCAGGAGGAAATCAGT-3′ and 5′-AACAAGCAGGTCGGTGATCT-3′. This cDNA fragment is not part of the MAP2c isoform, which is not restricted to neurons (79). The TH and glutamic acid decarboxylase (GAD67) riboprobes were synthesized from the complete rat TH and GAD67 cDNAs. The choline acetyltransferase (ChAT) riboprobe contains the 3′ end of the ChAT coding sequence and part of the 3′ untranslated region.
Adult and postnatal day 1 (P1) brains were obtained from 225-g and newborn Wistar rats, respectively, perfused with 4% paraformaldehyde (PFA) freshly prepared in phosphate-buffered saline (PBS). Following perfusion, brains were postfixed by overnight incubation in the same solution. Embryos were collected from pregnant Wistar females as previously described (65) and fixed by immersion in 4% PFA for 24 h. Adult brains used for cryosections were cryoprotected by incubation for 48 h in 15% sucrose in PBS and frozen in isopentane. Other tissues were embedded in BSA-gelatin, and 250-μm sections were cut with a Vibratome (Leica VT1000S).
Hybridization experiments were performed essentially as described previously (82). Double in situ staining was carried out with 14-μm cryosections. Probes were detected with alkaline phosphatase-conjugated anti-Dig and anti-Fluo antibodies (Roche). Colorimetric detection was achieved with 5-bromo-4-chloro-3-indolylphosphate (BCIP) and either nitroblue tetrazolium (NBT; Promega) or 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyltetrazolium chloride (INT; Roche). Dig- and Fluo-labeled riboprobes were applied together to the slides. After detection of the Dig-labeled probe with NBT-BCIP, the anti-Dig antibody was removed by washes with 0.1 M glycine (pH 2.2)-0.1% Tween before incubation with the anti-Fluo antibody. No background signal was observed on slides hybridized with the sense control probes and incubated with NBT-BCIP for at least as long as the corresponding antisense probes.
BrdU incorporation assays.
Pregnant Wistar females were injected intraperitoneally with 1 mg of bromodeoxyuridine (BrdU) (Sigma) 2 h before they were killed. Embryos were fixed in 4% PFA and processed for in situ hybridization as described above. BrdU incorporation was visualized by immunohistochemical analysis. Sections were treated for 45 min at room temperature with 2 N HCl in PBS supplemented with 0.25% gelatin (PGT) and 0.1% Triton X-100 and then were incubated for 48 h at 4°C with an anti-BrdU antibody (1:500; Becton Dickinson) diluted in PGT supplemented with 1% Triton X-100. The secondary antibody (biotinylated anti-mouse immunoglobulin G; 1:200 in PGT supplemented with 1% Triton X-100; Vector) was applied to the slides and detected by using diaminobenzidine.
Nucleotide sequence accession number.
The ZENON sequence has been deposited under GenBank accession number AY623002.
RESULTS
Identification of proteins that interact with the rat TH promoter.
We adapted the yeast in vivo one-hybrid genetic screen (80) to isolate cDNAs encoding putative DNA binding domains able to interact with the rat TH promoter. Two yeast strains were generated, one carrying the E-box/dyad motif and the other carrying the oct/hept element. Approximately 1 × 106 and 4 × 106 colonies of each strain were transformed with an adult rat brain library and an E14 rat embryo library, respectively. Ten cDNAs that led to the desired HIS3-positive phenotype when expressed in the strain harboring the E box were isolated (Table 1, dyad cDNAs). The strain carrying the oct/hept element transformed with these cDNAs did not grow in the absence of histidine, suggesting that the encoded proteins interact specifically with the E box/dyad motif. Only one cDNA was able to activate HIS3 gene expression specifically in the strain carrying the oct/hept element (Table 1, oct/hept 4 cDNA).
TABLE 1.
cDNAs isolated by one-hybrid screening with rat TH promoter sequences
| cDNA | Library | Homology | Reference(s) |
|---|---|---|---|
| Dyad 1 | Adult | rITF2 (bHLH) | 27, 85 |
| Dyad 2 | Adult | None | |
| Dyad 7 | Adult | None | |
| Dyad 17a | Adult | Unknown POK protein | |
| Dyad 16 | Embryo (E14) | rITF2 (bHLH) | 27, 85 |
| Dyad 22 | Embryo (E14) | None | |
| Dyad 32 | Embryo (E14) | SREBP2 (bHLH) | 30 |
| Dyad 56 | Embryo (E14) | Sox14 (high mobility group) | 26 |
| Dyad 66 | Embryo (E14) | ATBF1 (zinc finger and homeodomain) | 31 |
| Dyad 100 | Embryo (E14) | None | |
| Oct/hept 4 | Embryo (E14) | Lhx9 (homeodomain) | 10, 67 |
Corresponds to ZENON.
The 11 isolated cDNAs were sequenced. Six encoded the previously characterized DNA binding proteins rITF2, SREBP2, Sox14, ATBF1, and Lhx9. The relationship between these factors and TH gene regulation remains to be analyzed. Four other cDNAs were dissimilar to sequences for all known classes of transcription factors. The corresponding mRNAs could not be found in the brain by in situ hybridization (data not shown). Finally, the sequence of the remaining cDNA (dyad 17) indicated that it encoded a truncated protein containing five putative zinc fingers. SLIC and 3′ anchored PCR strategies (20) were used to elongate the dyad 17 cDNA until a complete open reading frame (ORF), which we named ZENON, was reconstituted. Among the 11 cDNAs isolated, the ZENON cDNA was selected for further studies because it encoded a new protein with putative DNA binding domains.
ZENON is a novel POZ Kruppel-like zinc finger protein.
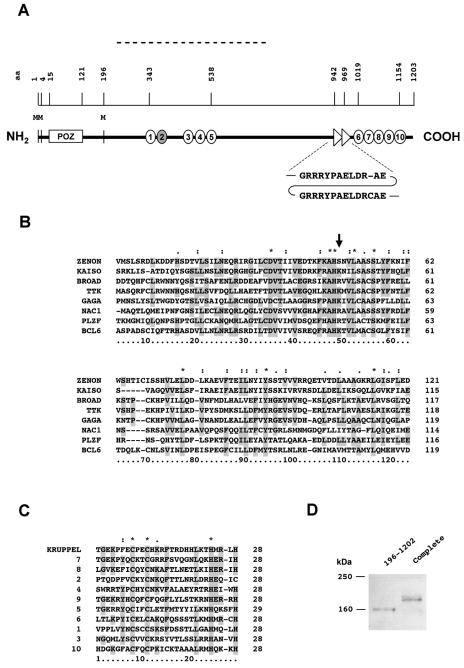
The deduced amino acid sequence of ZENON (1,203 amino acids) (Fig. 1A) is similar to those of members of a growing family of proteins containing a protein-protein interaction domain named the POZ (pox zinc finger) or BTB (broad complex-tramtrack-bric-à-brac) domain (2, 9) (Fig. 1B). ZENON, like many other POZ proteins, contains zinc fingers in addition to the N-terminal POZ domain. Nine are typical C2H2 zinc fingers, and one has a C2HC structure. A single C2HC substitution among several C2H2 zinc fingers was previously reported for Lola, a drosophila POZ protein (23). Four of the C2H2 zinc fingers are preceded by an H-C link that closely matches the characteristic motif TGEKPFE for Kruppel zinc fingers (Fig. 1C). ZENON may therefore be a POK (POZ Kruppel-like zinc finger) protein. The ZENON primary structure is unique among those of POK proteins, since the zinc fingers are clustered into two “hands” of five fingers each, one at the N terminus (“left hand”) and the other at the extreme C terminus (“right hand”). ZENON shares this particularity with the “two-handed” group of zinc finger proteins, which do not contain a POZ domain.
FIG. 1.
ZENON is a POZ Kruppel-like zinc finger protein. (A) Schematic diagram of the primary structure of the ZENON protein. The position of each element is indicated by the amino acid (aa) scale above the structure. The fragment isolated by one-hybrid screening is indicated by a broken line. Amino acids are numbered from the most amino-terminal methionine in frame with the POZ domain. Zinc finger motifs are represented as circles numbered from 1 to 10. Zinc finger 2 is a C2HC zinc finger (shaded circle); the others are C2H2 zinc fingers (white circles). A repeat, GRRRYPAELDR-AE/GRRRYPAELDRCAE, is indicated by two triangles. M, methionine. (B) Amino acid alignment of ZENON and other POZ domains. The alignment was generated with ClustalX software. The most highly conserved amino acids are shaded. Asterisks indicate positions at which 100% identity among all of the sequences is observed. Dots indicate positions at which the amino acids are functionally similar. The arrow indicates a position at which K or R is usually found but is not conserved in the ZENON primary sequence. (C) Amino acid sequence alignment of ZENON zinc fingers 1 to 10 with one Kruppel zinc finger. Note the HX4H spacing in zinc finger 5, with X representing any amino acid. This spacing is thought to provide greater flexibility for DNA binding than the more common HX3H spacing. (D) Western blot analysis of the ZENON translation start site in mammalian cells. 293T cells were transfected with a complete (amino acids 1 to 1202) or a partial (amino acids 196 to 1202) ZENON ORF in frame with a 3′-terminal FLAG sequence. The partial construct includes a methionine surrounded by a consensus Kozak sequence (position 196 in panel A). After separation on a polyacrylamide gel, the ZENON-FLAG fusion proteins were detected with an anti-FLAG antibody. The fusion protein obtained with the complete ZENON coding sequence has a higher molecular mass than the protein encoded by the partial sequence, suggesting that translation can start from methionine residues located upstream from the POZ domain (positions 1 and 4 in panel A), despite the absence of a Kozak sequence.
The ZENON coding sequence has three in-frame ATG codons, the first being preceded by a stop codon (Fig. 1A). The third ATG (methionine 196; GCCATGA) is surrounded by a consensus Kozak sequence (A/G CCATG A/G) (40), unlike the two first ATGs, which are located upstream from the POZ domain (methionine 1, CAGATGA; and methionine 4, GTCATGT). To determine whether translation could be initiated from one of these two 5′-terminal ATG codons, we transformed mammalian cells with a plasmid containing all or part of the ZENON ORF with a 3′-terminal, in-frame tag to allow detection of the polypeptide. Since the expression plasmid did not include any initiation signal, the translation start point was determined by the inserted ZENON sequences. The complete sequence gave rise to a longer peptide than the partial sequence containing only the most 3′-terminal ATG (Fig. 1D). This result indicates that translation is not initiated from methionine 196 when methionines 1 and 4 are present and suggests that the N-terminal part of the ZENON protein contains the complete POZ domain despite the lack of a consensus Kozak sequence at positions 1 and 4.
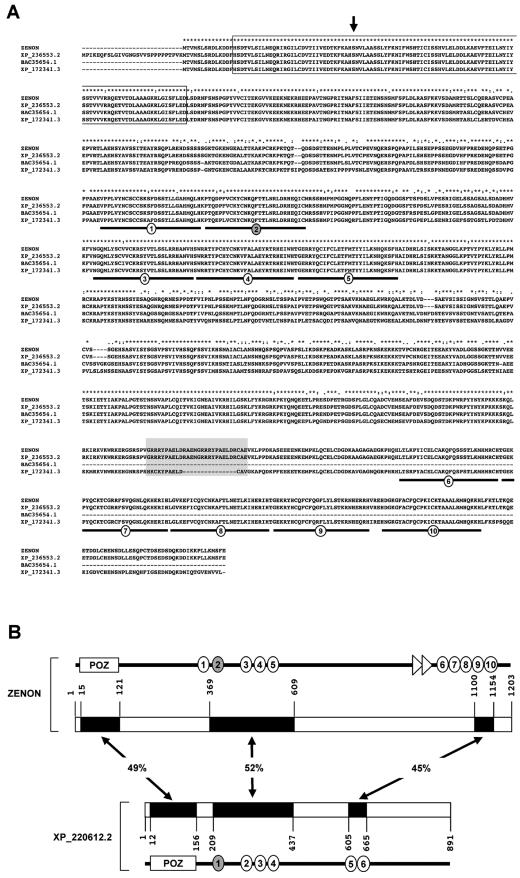
Three predicted polypeptides that are quite similar to ZENON recently appeared in databases (Fig. 2A). Rat sequence XP_236553.2 (RefSeq accession number) is virtually identical to the ZENON sequence. This putative polypeptide was annotated from a genomic database and extends an additional 28 amino acids upstream from the first methionine. In contrast, the mouse and human nucleotide sequences (deduced polypeptide accession numbers BAC35654.1 [EMBL] and XP_172341.2 [RefSeq], respectively) predict the same translation start point as that which we observed experimentally, supporting our assignment of the ORF. Therefore, rat sequence XP_236553.2 can be considered the putative ZENON protein, and BAC35654.1 and XP_172341.2 can be considered mouse and human orthologs. At the actual stage of genome sequence advancement, short ZENON ortholog sequences have been found in cow and dog genomes but not in those of other mammals. No orthologs have been found in chicken, xenopus, zebra fish, drosophila, and nematode databases.
FIG. 2.
Amino acid alignment of ZENON with putative polypeptides present in databases. (A) Alignment of ZENON with putative orthologs. XP_236553.2 and XP_172341.2 were annotated from rat and human genomic sequences, respectively. The BAC35654.1 sequence was predicted from a murine cDNA. The alignment was generated with ClustalX software. Asterisks indicate positions at which 100% identity among all of the sequences is observed. Dots indicate positions at which amino acids are functionally similar. The arrow indicates a position at which K or R is usually found among POZ proteins but is not conserved in the primary sequences of ZENON and its orthologs. The POZ domain (boxed sequence), zinc fingers (underlined sequence), and repeat (shaded sequence) are indicated. Note that the repeat is not evolutionarily conserved because it is not present in humans. (B) Schematic alignment of ZENON with a potential homologous rat polypeptide, XP_220612.2. The related regions are indicated by black boxes, and the percentages of amino acids identical between each region of ZENON and its homolog are indicated.
The rat genomic sequence encoding predicted polypeptide XP_236553.2 maps to chromosome 8q31. A BLAST search with the complete coding sequence for ZENON revealed uninterrupted alignment with this genomic sequence, showing that the ORF consists of a single exon. Although alternative splicing may occur outside the coding region, no polypeptide variants can be generated by this mechanism.
A predicted rat polypeptide distantly related to ZENON was also found in databases (RefSeq accession no. XP_220612.2). This putative protein displays an overall primary structure similar to that of ZENON (Fig. 2B), with an N-terminal POZ domain (49% identity with the ZENON POZ domain, which shares only 30% identity with other POZ domains), a C2HC and C2H2 zinc finger cluster (52% identity with ZENON zinc fingers 2 to 5 and surrounding sequences), and two C-terminal zinc fingers (45% identity with ZENON zinc fingers 9 and 10). Although this predicted polypeptide and ZENON are not closely related, they may belong to the same family. Except for this putative homolog, ZENON did not show similarity outside the conserved domains with any peptide present in the databases, suggesting that it could be a member of a new family of POK proteins.
ZENON is a DNA binding protein.
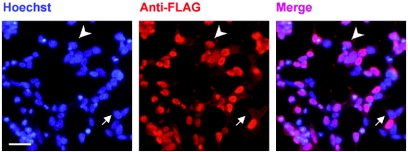
Several POZ proteins (e.g., Kaiso and Bcl-6) are found in nuclei following transient transfection with their full-length cDNAs (17, 19). 293T kidney epithelial cells were transfected with a ZENON-FLAG fusion construct. Immunofluorescence analysis with an anti-FLAG antibody revealed the nuclear distribution of the fusion protein (Fig. 3). This localization was compatible with a function in DNA binding.
FIG. 3.
The ZENON protein is targeted to nuclei. 293T cells were transfected with the complete ZENON coding region (amino acids 1 to 1202) in frame with the FLAG sequence. The ZENON-FLAG fusion protein was detected with an anti-FLAG antibody 48 h after transfection. Nuclei were counterstained with Hoechst reagent. The arrowhead indicates a nucleus containing the ZENON-FLAG fusion protein. The arrow indicates a nucleus in which the ZENON-FLAG fusion protein is not present, probably because the cell was not transfected. Bar, 20 μm.
POK proteins usually interact with DNA through their zinc finger domains. The DNA binding properties of the two ZENON zinc finger clusters were examined separately by EMSA. The complete ZENON polypeptide could not be produced in Escherichia coli because it was subject to proteolytic degradation, as also observed for other large zinc finger proteins (77). Consequently, two partial ZENON polypeptides were generated (Fig. 4A). L-ZENON contains zinc fingers 1 to 5 (left hand) and surrounding regions but not the POZ domain. The cDNA encoding L-ZENON approximates the partial ORF isolated by one-hybrid screening. Bacterial expression of this cDNA gave rise to two products of equal intensities, one full length and one truncated. Both contained the complete left hand of zinc fingers, as indicated by their sizes. R-ZENON contains the C-terminal part of ZENON with zinc fingers 6 to 10 (right hand).
FIG. 4.
Interaction of ZENON zinc finger domains with the E box. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of purified L-ZENON and R-ZENON proteins. L-ZENON was generated from a sequence containing zinc fingers 1 to 5 (left hand), whereas R-ZENON was produced from a sequence containing zinc fingers 6 to 10 (right hand). Approximately 20 μg of L-ZENON and 5 μg of R-ZENON were run on a 9% polyacrylamide gel and stained with Coomassie blue. The L-ZENON sequence gave rise to two products of equal intensities, one full length and one truncated. Both contained the complete left hand of zinc fingers, as indicated by their size. (B) EMSA with a 32P-labeled pTH probe spanning the TH proximal promoter. The pTH probe is shown alone (first lane) or in the presence of 1 μg each of L-ZENON and R-ZENON recombinant proteins (following lanes). Arrows indicate free probe and complexes I and II observed with L-ZENON. (C) EMSA with a 32P-labeled E probe covering the E box and surrounding dyad element. The E probe is shown alone (first lane) or with 1 μg of L-ZENON alone (second lane) or in the presence of orthophenanthroline (o-phenanth; following lanes). (D) EMSA with the 32P-labeled E probe incubated with 1 μg of L-ZENON alone or in the presence of various competitors (molar excess, 50- or 200-fold). Complex intensity represents the sum of complex I and complex II intensities quantified with Gelanalyst software. The results shown are representative of four independent gel retardation assays.
The interactions between DNA and some POZ zinc finger proteins as well as two-handed zinc finger proteins are highly flexible, with large variations in orientation and spacing of the target sequences (35, 62, 66). As L-ZENON was initially found to interact with the E box in yeast, the right hand of zinc fingers may recognize a sequence located outside the immediate vicinity of the E box. R-ZENON, however, failed to bind a large radioactive probe (pTH probe) covering 200 bp of the TH proximal promoter, including the E box (Fig. 4B). In contrast, two complexes, denoted I and II, were resolved in the presence of L-ZENON, suggesting that only the left hand of zinc fingers interacts with the TH promoter. Complexes I and II may contain the full-length and truncated versions of L-ZENON, respectively. Two L-ZENON-containing complexes were also obtained with a shorter probe spanning only the E-box/dyad element (E probe). These complexes were disrupted by the zinc chelator orthophenanthroline, confirming the involvement of the zinc fingers in DNA binding (Fig. 4C). The specificity of L-ZENON binding to DNA was assessed by competition experiments (Fig. 4D). No complex was observed when a cold probe was used as the competitor (complex intensity was reduced by 79% with a 50-fold molar excess of E probe competitor). Oligonucleotides with a mutated E box (ΔE) or an intact E box surrounded by unrelated flanking sequences (PAN) were less effective competitors (23 and 58% complex intensity reductions, respectively).
In summary, the N-terminal zinc finger cluster in ZENON is a putative DNA binding domain that interacts preferentially with the E-box/dyad element. Taken together with the nuclear localization, these results suggest that ZENON is a DNA binding protein.
ZENON functions as a transcriptional activator independently of the POZ domain.
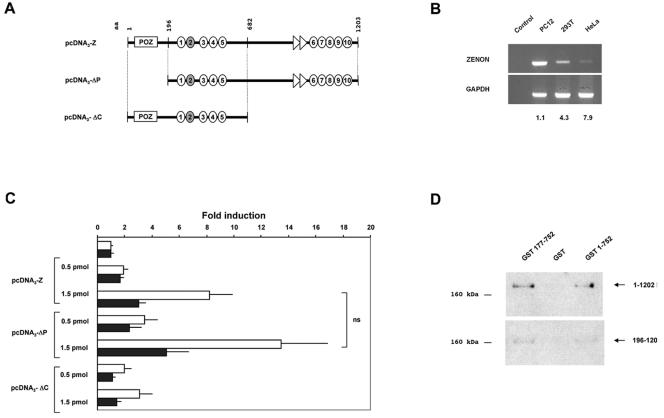
To investigate whether ZENON could act as a transcriptional regulator, we next conducted overexpression experiments with three cell lines: the TH-positive neuroendocrine cell line PC12 and TH-negative 293T and HeLa cells. Cells were cotransfected with reporter plasmids and various amounts of expression vectors carrying all or part of the ZENON coding sequence. The potential DNA binding domain, as defined by binding experiments, was included in all ZENON constructs: pcDNA3-Z contained the whole coding sequence, pcDNA3-ΔP lacked the POZ domain, and pcDNA3-ΔC lacked the C-terminal zinc finger cluster (Fig. 5A). Reporter plasmids contained the luciferase gene driven by the 5-kb rat TH promoter with either a wild-type or a mutant E box. In 293T and HeLa cells, TH promoter activity was induced in the presence of pcDNA3-Z (4.3- and 7.9-fold increases relative to activity obtained with empty pcDNA3, respectively) (Fig. 5B). In contrast, the overexpression of ZENON had no effect in PC12 cells. Induction factors were correlated with the level of expression of endogenous ZENON in the various cell lines, as revealed by a comparative RT-PCR analysis. Endogenous ZENON is highly expressed in PC12 cells, explaining why ZENON overexpression had no effect on TH promoter activity in these cells. ZENON is weakly expressed in 293T cells and hardly detectable in HeLa cells. Therefore, the latter cell line constitutes an appropriate cellular model for ZENON overexpression experiments (Fig. 5B). In HeLa cells, ZENON induced TH promoter activity in a dose-dependent manner (Fig. 5C). Consistent with the results of the binding experiments, the integrity of the E box was required for the full effect. All three ZENON constructs activated transcription, but maximal induction was observed when the C-terminal part was included (13.5-, 8.2-, and 3.1-fold increases obtained with pcDNA3-ΔP, pcDNA3-Z, and pcDNA3-ΔC, respectively). In contrast, the absence of the POZ domain did not significantly affect the induction of TH promoter activity, although a modest increase in transcriptional induction was observed with pcDNA3-ΔP. Similar results were obtained for 293T cells (data not shown).
FIG. 5.
ZENON modulates TH transcriptional activity independently of the POZ domain. (A) ZENON effectors used in cotransfection experiments. The indicated fragments of the ZENON ORF were introduced into pcDNA3 downstream from a cytomegalovirus promoter to generate pcDNA3-Z, pcDNA3-ΔP, and pcDNA3-ΔC. (B) Comparative RT-PCR analyses of ZENON expression in PC12, 293T, and HeLa cells. The first lane shows a negative control (PC12 cells in the absence of reverse transcriptase). Amplification of GAPDH was used to normalize for RT efficiency in each cell line. Values (induction factors) represent the induction of wild-type TH promoter activity (5kbTH-Luc) by 1 pmol of pcDNA3-Z in each cell line relative to the control (1 pmol of empty pcDNA3). (C) Cotransfection experiments with ZENON effectors in HeLa cells. Reporter plasmids contained the luciferase gene driven by the rat TH promoter, either intact (5kbTH-Luc; white bars) or bearing a mutated E box [5kbTH(ΔE)-Luc; black bars]. Cells were transfected with various amounts of ZENON effector constructs (0 to 1.5 pmol), the total amount of DNA being brought to 1.5 pmol by the addition of insert-free pcDNA3. In the absence of ZENON effectors, the transcriptional activity of 5kbTH-Luc was approximately twice that of 5kbTH(ΔE)-Luc; results are shown as induction of TH promoter activity to facilitate interpretation. Values represent the means ± 95% confidence intervals for five independent experiments performed in triplicate with various plasmid preparations. 5kbTH-Luc induction by ZENON effectors was significant compared to that obtained with empty pcDNA3 (P < 10−4). For each ZENON effector, 5kbTH-Luc induction and 5kbTH(ΔE)-Luc induction were significantly different (P = 0.001). 5kbTH-Luc induction by pcDNA3-ΔC was significantly lower than 5kbTH-Luc induction by pcDNA3-Z and pcDNA3-ΔP (P < 10−5). ns, no significant difference in the induction of TH promoter activity was obtained with pcDNA3-Z and pcDNA3-ΔP (P > 0.01). (D) GST pull-down assay with GST-ZENON fusion proteins GST 177-752 and GST 1-752 and GST alone. GST 177-752 does not include the POZ domain. The fusion proteins and GST were incubated with cell extracts containing the complete or partial ZENON-FLAG polypeptide and then were subjected to Western blot analysis with an anti-FLAG antibody. (Upper panel) Complete ZENON-FLAG polypeptide (1 to 1202). (Lower panel) ZENON-FLAG polypeptide lacking the POZ domain (196 to 1202).
The observation that the ZENON POZ domain is not involved in transcriptional induction was surprising, as dimerization through this domain is thought to be necessary for transcriptional regulation (49). To evaluate the capacity of the ZENON POZ domain to trigger homodimerization, we performed GST pull-down assays with two GST-ZENON fusion proteins, GST 1-752 and GST 177-752 (Fig. 5D). GST 1-752 spans the POZ domain and N-terminal zinc finger cluster, and GST 177-752 lacks the POZ domain. Intriguingly, the full-length ZENON-FLAG polypeptide was found associated with both GST fusion proteins, demonstrating that the POZ domain is not essential for a ZENON self-interaction in vitro (Fig. 5D, upper panel). Consistent with this observation, a partial ZENON-FLAG polypeptide lacking the POZ domain was also bound by both GST 177-752 and GST 1-752 (Fig. 5D, lower panel). No ZENON protein could be detected with GST alone.
Altogether, these results suggest that ZENON stimulates TH gene transcription via a mechanism independent of oligomerization involving the POZ domain. The ZENON POZ domain rather may be involved in heteromeric interactions, which could mediate other functions in addition to TH gene regulation. To begin to examine these functions, we next determined the organs and cells which express ZENON.
ZENON is expressed in neurons in the adult rat brain.
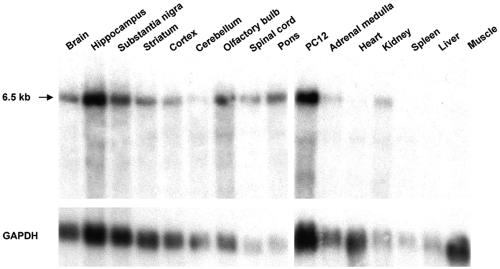
The distribution of ZENON mRNA was first investigated by high-stringency Northern blot analysis of various adult rat organs (Fig. 6). A single 6.5-kb mRNA was detected in all brain regions examined, in cell line PC12, and in the adrenal medulla. No signal was detected in nonneural tissues, including the heart, spleen, liver, and muscle, although mRNA was found in the kidneys. ZENON expression thus appears to be restricted mainly to neural and neuroendocrine tissues. The mRNA detected was much longer than the 3,609-bp ORF, probably due to a long 3′ untranslated region, as is common for POZ mRNAs (17).
FIG. 6.
Northern blot analysis showing the tissue distributions of ZENON mRNA. (Upper panel) Poly(A)+ RNAs from various rat tissues (0.5 to 2 μg/lane) hybridized with a ZENON riboprobe. The arrow indicates the estimated size of ZENON mRNA. (Lower panel) Hybridization with a GAPDH probe to normalize for loading differences.
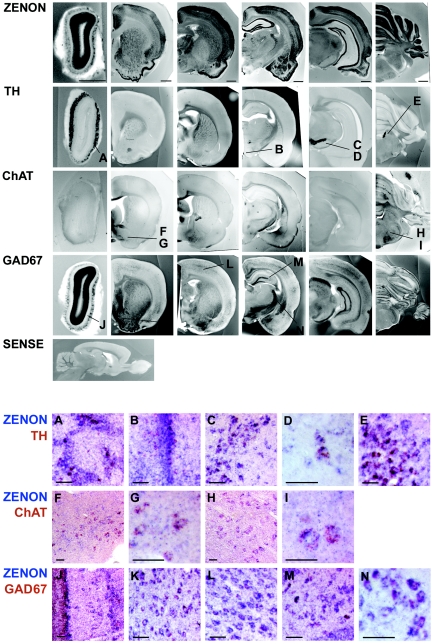
We next investigated whether ZENON could be linked to a particular class of neurotransmitters. In situ hybridization with serial adult rat brain sections showed that ZENON expression domain contains catecholaminergic, cholinergic, and γ-aminobutyric acid (GABA)-ergic nuclei, as revealed by TH, ChAT, and GAD67 stainings, respectively (Fig. 7, upper panels). To further characterize the cell types expressing ZENON, we performed double in situ hybridization with a Dig-labeled ZENON riboprobe and several Fluo-labeled riboprobes. Double-labeling experiments with a Fluo-TH probe revealed ZENON expression in all catecholaminergic neurons of the olfactory bulb, substantia nigra, and locus coeruleus. In the hypothalamus, colocalization was less obvious because of the high density of ZENON-positive cells in the region (Fig. 7A to E). When a Fluo-ChAT probe was used, doubly labeled cells were detected in the septum, diagonal band, and pons. These results suggest that most cholinergic neurons express ZENON, although the existence of ZENON-negative cholinergic cells cannot be excluded (Fig. 7F to I). The Dig-ZENON probe also colocalized with a Fluo-GAD67 probe in most GABA-ergic neurons of the olfactory bulb, septum, cerebral cortex, hippocampus, and thalamus (Fig. 7J to N). ZENON mRNA was thus found in several neuronal subtypes.
FIG. 7.
ZENON mRNA is present in various neuronal subtypes in adult brain. (Upper panels) Adult rat sections (250 μm) were hybridized with ZENON, TH, ChAT, GAD67, and sense riboprobes, which were detected by NBT-BCIP. Bars, 1 mm. The letters indicate the regions from which doubly stained sections A to N were taken. (Lower panels) Adult rat brain sections (14 μm) were hybridized with Dig- and Fluo-labeled riboprobes, which were consecutively detected by incubation with NBT-BCIP (blue) and INT-BCIP (red) substrates. Bars, 50 μm. (A to E) Dig-ZENON and Fluo-TH riboprobes. A, olfactory bulb; B, hypothalamus; C and D, substantia nigra; E, locus coeruleus. (F to I) Dig-ZENON and Fluo-ChAT riboprobes. F and G, diagonal band; H and I, pons. (J to N) Dig-ZENON and Fluo-GAD67 riboprobes. J, olfactory bulb; K, septum; L, cerebral cortex; M, hippocampus; N, reticulate nucleus of thalamus.
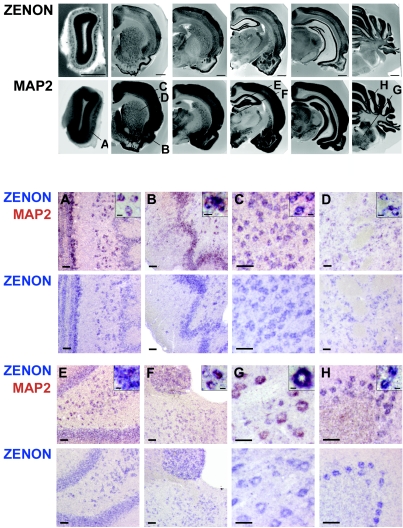
To determine whether ZENON was expressed in all neurons, a panneuronal MAP2 probe was used. ZENON and MAP2 expression domains completely overlapped on serial sections (Fig. 8, upper panels). The more intense signal observed with the MAP2 probe was due to the presence of MAP2 mRNA in the dendrites. Double-labeling experiments with Dig-ZENON and Fluo-MAP2 probes revealed colocalization in virtually all neurons of the olfactory bulb, pyriform cortex, cerebral cortex, striatum, hippocampus, thalamus, pons, and cerebellum (Fig. 8A to H). All ZENON-positive cells expressed MAP2, demonstrating that in the adult brain, ZENON expression is restricted mainly to neurons.
FIG. 8.
ZENON mRNA colocalizes with MAP2 mRNA in adult brain. (Upper panels) Serial adult rat sections (250 μm) were hybridized with Dig-ZENON and Dig-MAP2 riboprobes, which were detected by NBT-BCIP. Bars, 1 mm. The letters indicate the regions from which doubly stained sections A to H were taken. (Lower panels) Adult rat brain sections (14 μm) were hybridized with Dig-ZENON and Fluo-MAP2 riboprobes, which were consecutively detected by incubation with NBT-BCIP (blue) and INT-BCIP (red) substrates. For each brain region examined, a double-labeling experiment and in situ hybridization with the Dig-ZENON riboprobe only performed on adjacent sections are shown in parallel. Bars, 50 μm. Insets show higher magnifications (bars, 10 μm) of the doubly labeled cells. A, olfactory bulb; B, pyriform cortex; C, cerebral cortex; D, striatum; E, hippocampus; F, thalamus; G, pons; H, cerebellum.
ZENON is present in mature neurons.
We next investigated ZENON expression in rat embryos (Fig. 9). The developing CNS presents the advantage that several differentiation stages can be visualized in the same transverse section of the neural tube. The dividing progenitors are located within the ventricular zone. When a progenitor exits the cell cycle, it enters the adjacent subventricular zone to initiate differentiation and migrates laterally into the marginal zone as differentiation progresses. Hence, at least during early developmental stages, the position of a cell is determined by the time at which the last division was completed, with mature cells being located in the lateral part of the marginal zone.
FIG. 9.
ZENON gene expression during development. Rat sections (250 μm) were hybridized with Dig-ZENON and Dig-MAP2 riboprobes, which were detected by NBT-BCIP. Bars, 1 mm. E12 (A to C), E13 (D to F), and E14 (G to I) panels show whole-embryo sections at the E12, E13, and E14 developmental stages, respectively. The plan and level of each section are indicated on the embryo schemes. The arrow indicates ZENON gene expression in the ventral spinal cord. The arrowhead indicates ZENON gene expression in the lateral telencephalon and diencephalon. Asterisks indicate expression in the trigeminal and dorsal root ganglia. E17 (J to K) and P1 (L to N) panels show coronal hemisections of the forebrain and hindbrain at the E17 and P1 developmental stages, respectively.
ZENON mRNA was never observed within the ventricular zone of the embryonic CNS. ZENON mRNA was first detected in the ventral spinal cord at E12, most likely in motoneurons (Fig. 9C). At later stages, the signal in the spinal cord was more intense and extended along the ventral-dorsal differentiation gradient (Fig. 9E to I). No clear signal was detected in the heads of E12 and E13 embryos; therefore, ZENON expression appears later in the brain than in the spinal cord. ZENON mRNA was first observed in the brain at E14 as lateral staining in the telencephalon and diencephalon (Fig. 9G). In E17 brains, a strong signal was observed in the cortex, thalamus, and hindbrain; this signal extended to virtually all brain regions by P1 (Fig. 9J to N). Although the signal was concentrated mainly in the CNS, ZENON was also expressed in the PNS. ZENON mRNA was never observed in the migratory neural crest. In the PNS ZENON mRNA was first detected at E13 as a weak signal in postmigratory neural crest cells of the trigeminal and dorsal root ganglia (Fig. 9D to F). ZENON expression was not observed in placodally derived neurons at early stages. Outside the nervous system, ZENON was present in the limbs (Fig. 9F and I), transiently in somites (Fig. 9C) and, in contrast to our findings for adult rats, in the embryonic liver (Fig. 9F and I).
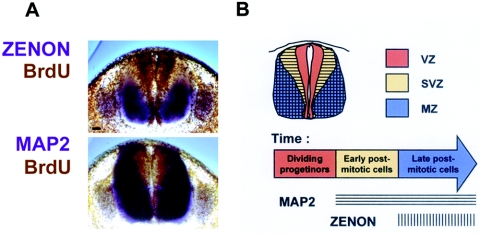
The onset of ZENON expression in the CNS of E12 embryos was delayed compared to neuronal differentiation, as assessed on the basis of MAP2 expression. In E13 and E14 neural tubes, the ZENON signal was restricted to the ventral-lateral part of the MAP2 territory. In contrast, ZENON expression was essentially the same as that of MAP2 in P1 brain regions. We performed BrdU incorporation assays to investigate why ZENON and MAP2 expression patterns were different during embryogenesis but overlapped in P1 brain regions (Fig. 10A). In the spinal cord of E14 embryos, the MAP2 expression domain was complementary to BrdU staining, indicating that progenitors exhibit neuronal features shortly after they exit the cell cycle. In contrast, an unlabeled region was observed between the areas that were BrdU and ZENON positive. This physical gap corresponds to a temporal delay in the expression of ZENON: early postmitotic neurons first express only MAP2 and then begin to express ZENON as they progress into differentiation (Fig. 10B). Ventricular and subventricular zones become residual, and the marginal zone, containing mature neurons, extends as development progresses. This explains why the ZENON expression pattern is similar to that of MAP2 only during late developmental stages.
FIG. 10.
ZENON is a marker of late postmitotic neurons in the developing spinal cord. (A) E14 rat sections (250 μm) were hybridized with Dig-ZENON and Dig-MAP2 riboprobes, which were detected by NBT-BCIP. Samples were subsequently subjected to immunohistochemical analysis to determine BrdU incorporation. Bar, 50 μm. (B) Interpretation correlating ZENON and MAP2 expression patterns with the timing of differentiation. VZ, ventricular zone; SVZ, subventricular zone; MZ, marginal zone.
These observations demonstrate that ZENON is a marker of late postmitotic neurons in the embryonic CNS. ZENON is not expressed in the migratory neural crest or in the postmigratory neural crest at early stages, suggesting that it is also a marker of late postmitotic neurons in the PNS. ZENON expression persists during adulthood; thus, we conclude that ZENON is a marker of mature neurons.
DISCUSSION
The rat TH proximal promoter contains several sequences potentially bound by transcription factors believed to be important for the development of the nervous system. An E-box/dyad motif is required for the appropriate expression of a reporter gene in a catecholaminergic cell line and is potentially recognized by bHLH and zinc finger transcription factors (34, 55). Several classes of homeodomain proteins may bind juxtaposed oct/hept elements (18, 87). In a one-hybrid genetic screen, we used the E-box/dyad and oct/hept elements as targets to identify novel transcription factors present in the brain. We isolated ZENON, a novel POZ Kruppel-like zinc finger protein that has features of a transcription factor. The most striking property of ZENON is that its expression is confined mainly to mature neurons, suggesting a function in the differentiation and/or survival of late postmitotic neurons.
ZENON may be the first identified member of a novel family of POK transcription factors.
The POZ domain is a conserved protein-protein interaction motif found in many transcription factors, oncogenic proteins, and other biologically important proteins (2, 9, 15, 14, 48). The ZENON POZ domain is similar to that of Kaiso, a transcription factor that was first identified as a p120 catenin-interacting protein and that exhibits methylation-dependent DNA binding (17, 63). However, the two proteins are not closely related, since they are dissimilar outside the POZ domain. Databases contain a putative rat ORF identical to the ZENON ORF (and the putative human and mouse orthologs) and a similar predicted polypeptide. Otherwise, except for the conserved domains, the ZENON sequence is not similar to any other sequences, suggesting that ZENON could be the first identified member of a small family of POK proteins. Strikingly, ZENON appears late during evolution, since no related ORF was found in the zebra fish, drosophila, and nematode genomes.
A particularity of ZENON family members is the presence of two hands of five zinc fingers each. The left hand was encoded by the partial cDNA isolated by the one-hybrid system. That this domain binds the E-box/dyad element was confirmed in vitro, and we demonstrated that the zinc fingers were responsible for the binding. E boxes of the CAGGTG type, such as the rat TH E box, are targets for zinc finger proteins of several families, including Snail-related proteins (34) and two-handed proteins (21). The comparison with two-handed proteins is particularly relevant, since they exhibit a primary structure close to that of ZENON, with two separate zinc finger clusters. A model was proposed for DNA binding by two-handed proteins; in this model, the two hands of zinc fingers independently interact with two distant AGGTG sequences (66). In addition to the E box, an AGGTG sequence is present within the rat TH proximal promoter (68 bp upstream from the transcription start site). However, R-ZENON failed to bind the probe containing this motif; therefore, ZENON does not appear to show the same binding properties as two-handed zinc finger proteins. In agreement with this observation, the sequence of ZENON zinc fingers is not closely related to that of two-handed proteins.
ZENON is an atypical POZ protein: the activation of transcription is independent of the POZ domain.
Consistent with the enhancer function of the E-box element (86), ZENON strongly induces TH promoter activity in a cell line. This finding is in contrast to the transcriptional repression by the POZ domain reported for most POZ proteins (32, 38, 43, 45, 58). The ZENON POZ domain includes serine at a key position at which lysine or arginine is usually found. This substitution may well be responsible for the absence of repression, as suggested by mutagenesis studies of three POZ proteins having different effects on transcription (49, 50): the repressor PLZF, the repressor Bcl-6, and FAZF, whose POZ domain is transcriptionally inert. Based on the observation that the FAZF POZ domain contains a serine instead of lysine or arginine, the authors of those studies showed that lysine or arginine was necessary for repression, allowing the formation of a charged pocket that mediates interactions with corepressors. Replacement of lysine or arginine with serine was sufficient to abolish repression. This finding may explain the absence of repression by the FAZF and ZENON POZ domains.
A striking feature of ZENON is that the POZ domain appears to be nonessential for transcriptional regulation. Other POZ factors, including GAGA and ZF5, behave as transcriptional activators, but their POZ domains are necessary for DNA binding or transcriptional activation. The POZ domain of ZF5 mediates both activation and repression, depending on the promoter context (33, 57). A DNA binding model has been proposed for GAGA, a strong transcriptional activator of several drosophila genes (11, 59, 81). This model involves oligomerization through the POZ domain and suggests a key role for the POZ domain in selecting the target genes regulated by GAGA (35). POZ factors often contain other transcriptional regulation domains outside the POZ domain (45, 52, 58). The activation of transcription by ZENON is probably mediated by another domain, possibly located in the C-terminal part of the protein, because the removal of this part substantially reduced the induction of TH promoter activity.
The function of the ZENON POZ domain remains to be established. POZ domains are interfaces for protein-protein interactions, mediating homomeric and heteromeric associations with other POZ proteins. The ZENON POZ domain may be involved in interactions with other POZ factors, as it was not required for self-association. A combinatorial code for gene expression has been suggested to be mediated by POZ domains. This scenario is illustrated by the POZ protein Bach2, a repressor of the immunoglobulin heavy-chain 3′ enhancer. After association with MAZR through the POZ domain, both factors strongly upregulate fgf4 gene transcription in a POZ domain-dependent manner (39). Another example is TTK, which interacts with GAGA and represses GAGA-mediated activation. POZ domains of both proteins appear to be necessary and sufficient for TTK-GAGA interactions (59, 60). Thus, depending on the partner, POZ factors regulate different target genes and exhibit opposite effects on transcription. Heteromeric association provides the potential for an increased repertoire of DNA sequence interactions. ZENON may be recruited on different promoters by interacting with other POZ proteins through the POZ domain.
Association with differentiation and/or survival of late postmitotic neurons.
ZENON is found in many neuronal subtypes in the adult brain, including catecholaminergic, cholinergic, and GABA-ergic neurons. These data indicate that the functions of ZENON are not restricted to TH gene regulation. Two attempts to isolate E-box/dyad element-interacting proteins did not lead to the identification of a catecholaminergic cell-specific transcription factor, although different methods and cDNA libraries were used (85; this study). This observation suggest that the E-box/dyad element, crucial for cell-specific TH gene expression in transient transfection experiments, does not have the same role in vivo. The E-box/dyad element may participate in the maintenance of TH gene expression once the first steps of catecholaminergic differentiation have been initiated from other, as-yet-unidentified regulatory sites. Such a maintenance function does not necessarily require interactions with catecholaminergic cell-specific transcription factors. Alternatively, rather than a unique catecholaminergic cell-specific protein, a combination of widely expressed transcription factors recruited by different regulatory elements could account for cell-specific TH gene expression.
Unlike other POZ proteins (7, 16), XENON expression shows no obvious regionalization in the developing CNS and is largely similar to MAP2 expression at late stages, suggesting a panneuronal function, at least in the CNS. Most importantly, the onset of ZENON expression in the CNS is delayed compared to that of MAP2, a marker classically used to characterize differentiated neurons. This observation contrasts with the expression patterns reported for other POZ proteins. For example, XKaiso, a POZ protein widely present in the xenopus CNS, is supplied maternally and expressed throughout early development (38). The expression of Lola, a drosophila POZ protein implicated in axon pathfinding, decreases sharply during late embryogenesis (23, 24). In the neural tube, the restriction of ZENON to the lateral marginal zone could indicate a role in late differentiation events. In addition, persistent expression in neurons of the adult brain suggests that ZENON may be required for the maintenance of neuronal features and/or for the survival of neurons. Unlike most other cell types that are constantly being renewed, neurons, as postmitotic cells generated during embryogenesis, have to survive throughout the life of the organism. This unique longevity may involve specific antiapoptotic mechanisms, particularly in species with a long life expectancy. Several POZ proteins, including Bcl-6 (reviewed in reference 3), are implicated in cell cycle regulation and can inhibit apoptosis, and the absence of POZ protein Miz1 leads to increased apoptosis in the ectoderm of knockout mice (1). ZENON has not been found in drosophila and nematode, which are species with a short life expectancy. Based on this observation, it is attractive to speculate that ZENON regulates the expression of antiapopototic genes, preventing mature neurons from undergoing apoptosis.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, Université Pierre et Marie Curie, Fondation pour la Recherche Médicale, Institut de Recherche sur la Moelle Epinière, Association de Recherche sur le Cancer, and Aventis Pharma.
The yeast strains and vectors used in the one-hybrid system were kindly provided by Dominique Thomas. We thank Francine Côté and Jeannette Nardelli for helpful advice and critical reading of the manuscript and Rolando Meloni for the dissection of brain subregions.
REFERENCES
- 1.Adhikary, S., K. Peukert, H. Karsunky, V. Beuger, W. Lutz, H. P. Elsasser, T. Moroy, and M. Eilers. 2003. Miz1 is required for early embryonic development during gastrulation. Mol. Cell. Biol. 23:7648-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albagli, O., P. Dhordain, C. Deweindt, G. Lecocq, and D. Leprince. 1995. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 6:1193-1198. [PubMed] [Google Scholar]
- 3.Albagli-Curiel, O. 2003. Ambivalent role of BCL6 in cell survival and transformation. Oncogene 22:507-516. [DOI] [PubMed] [Google Scholar]
- 4.Albanèse, V., N. F. Biguet, H. Kiefer, E. Bayard, J. Mallet, and R. Meloni. 2001. Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum. Mol. Genet. 10:1785-1792. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 6.Anderson, D. J., J. F. Carnahan, A. Michelsohn, and P. H. Patterson. 1991. Antibody markers identify a common progenitor to sympathetic neurons and chromaffin cells in vivo and reveal the timing of commitment to neuronal differentiation in the sympathoadrenal lineage. J. Neurosci. 11:3507-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avantaggiato, V., P. P. Pandolfi, M. Ruthardt, N. Hawe, D. Acampora, P. G. Pelicci, and A. Simeone. 1995. Developmental analysis of murine promyelocyte leukemia zinc finger (PLZF) gene expression: implications for the neuromeric model of the forebrain organization. J. Neurosci. 15:4927-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee, S. A., S. Roffler-Tarlov, M. Szabo, L. Frohman, and D. M. Chikaraishi. 1994. DNA regulatory sequences of the rat tyrosine hydroxylase gene direct correct catecholaminergic cell-type specificity of a human growth hormone reporter in the CNS of transgenic mice causing a dwarf phenotype. Brain Res. Mol. Brain Res. 24:89-106. [DOI] [PubMed] [Google Scholar]
- 9.Bardwell, V. J., and R. Treisman. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8:1664-1677. [DOI] [PubMed] [Google Scholar]
- 10.Bertuzzi, S., F. D. Porter, A. Pitts, M. Kumar, A. Agulnick, C. Wassif, and H. Westphal. 1999. Characterization of Lhx9, a novel LIM/homeobox gene expressed by the pioneer neurons in the mouse cerebral cortex. Mech. Dev. 81:193-198. [DOI] [PubMed] [Google Scholar]
- 11.Biggin, M. D., and R. Tjian. 1988. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell 53:699-711. [DOI] [PubMed] [Google Scholar]
- 12.Blaiseau, P. L., A. D. Isnard, Y. Surdin-Kerjan, and D. Thomas. 1997. Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol. Cell. Biol. 17:3640-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castillo, S. O., J. S. Baffi, M. Palkovits, D. S. Goldstein, I. J. Kopin, J. Witta, M. A. Magnuson, and V. M. Nikodem. 1998. Dopamine biosynthesis is selectively abolished in substantia nigra/ventral tegmental area but not in hypothalamic neurons in mice with targeted disruption of the Nurr1 gene. Mol. Cell. Neurosci. 11:36-46. [DOI] [PubMed] [Google Scholar]
- 14.Cha, X. Y., R. C. Pierce, P. W. Kalivas, and S. A. Mackler. 1997. NAC-1, a rat brain mRNA, is increased in the nucleus accumbens three weeks after chronic cocaine self-administration. J. Neurosci. 17:6864-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, T., J. R. Stone, and A. J. Williams. 2001. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 21:3609-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook, M., A. Gould, N. Brand, J. Davies, P. Strutt, R. Shaknovich, J. Licht, S. Waxman, Z. Chen, S. Gluecksohn-Waelsch, R. Krumlauf, and A. Zelent. 1995. Expression of the zinc-finger gene PLZF at rhombomere boundaries in the vertebrate hindbrain. Proc. Natl. Acad. Sci. USA 92:2249-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel, J. M., and A. B. Reynolds. 1999. The catenin p120ctn interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol. 19:3614-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson, S. J., S. O. Yoon, D. M. Chikaraishi, K. A. Lillycrop, and D. S. Latchman. 1994. The Oct-2 transcription factor represses tyrosine hydroxylase expression via a heptamer TAATGARAT-like motif in the gene promoter. Nucleic Acids Res. 22:1023-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhordain, P., O. Albagli, S. Ansieau, M. H. Koken, C. Deweindt, S. Quief, D. Lantoine, A. Leutz, J. P. Kerckaert, and D. Leprince. 1995. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene 11:2689-2697. [PubMed] [Google Scholar]
- 20.Dumas Milne Edwards, J. B., P. Ravassard, C. Icard-Liepkalns, and J. Mallet. 1995. cDNA cloning by RT-PCR, p. 89-118. In M. J. McPherson, B. D. Hames, and G. R. Taylor (ed.), PCR2: a practical approach. IRL Press, Oxford, United Kingdom.
- 21.Genetta, T., D. Ruezinsky, and T. Kadesch. 1994. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol. Cell. Biol. 14:6153-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giniger, E., K. Tietje, L. Y. Jan, and Y. N. Jan. 1994. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development 120:1385-1398. [DOI] [PubMed] [Google Scholar]
- 24.Goeke, S., E. A. Greene, P. K. Grant, M. A. Gates, D. Crowner, T. Aigaki, and E. Giniger. 2003. Alternative splicing of lola generates 19 transcription factors controlling axon guidance in Drosophila. Nat. Neurosci. 6:917-924. [DOI] [PubMed] [Google Scholar]
- 25.Guillemot, F., L. C. Lo, J. E. Johnson, A. Auerbach, D. J. Anderson, and A. L. Joyner. 1993. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75:463-476. [DOI] [PubMed] [Google Scholar]
- 26.Hargrave, M., A. Karunaratne, L. Cox, S. Wood, P. Koopman, and T. Yamada. 2000. The HMG box transcription factor gene Sox14 marks a novel subset of ventral interneurons and is regulated by sonic hedgehog. Dev. Biol. 219:142-153. [DOI] [PubMed] [Google Scholar]
- 27.Henthorn, P., M. Kiledjian, and T. Kadesch. 1990. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science 247:467-470. [DOI] [PubMed] [Google Scholar]
- 28.Hirsch, M. R., M. C. Tiveron, F. Guillemot, J. F. Brunet, and C. Goridis. 1998. Control of noradrenergic differentiation and Phox2a expression by MASH1 in the central and peripheral nervous system. Development 125:599-608. [DOI] [PubMed] [Google Scholar]
- 29.Howard, M., D. N. Foster, and P. Cserjesi. 1999. Expression of HAND gene products may be sufficient for the differentiation of avian neural crest-derived cells into catecholaminergic neurons in culture. Dev. Biol. 215:62-77. [DOI] [PubMed] [Google Scholar]
- 30.Hua, X., C. Yokoyama, J. Wu, M. R. Briggs, M. S. Brown, J. L. Goldstein, and X. Wang. 1993. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. USA 90:11603-11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ido, A., Y. Miura, M. Watanabe, M. Sakai, Y. Inoue, T. Miki, T. Hashimoto, T. Morinaga, S. Nishi, and T. Tamaoki. 1996. Cloning of the cDNA encoding the mouse ATBF1 transcription factor. Gene 168:227-231. [DOI] [PubMed] [Google Scholar]
- 32.Kang, S. I., W. J. Chang, S. G. Cho, and I. Y. Kim. 2003. Modification of promyelocytic leukemia zinc finger protein (PLZF) by SUMO-1 conjugation regulates its transcriptional repressor activity. J. Biol. Chem. 278:51479-51483. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan, J., and K. Calame. 1997. The ZiN/POZ domain of ZF5 is required for both transcriptional activation and repression. Nucleic Acids Res. 25:1108-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kataoka, H., T. Murayama, M. Yokode, S. Mori, H. Sano, H. Ozaki, Y. Yokota, S. Nishikawa, and T. Kita. 2000. A novel snail-related transcription factor Smuc regulates basic helix-loop-helix transcription factor activities via specific E-box motifs. Nucleic Acids Res. 28:626-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katsani, K. R., M. A. Hajibagheri, and C. P. Verrijzer. 1999. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 18:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, K. S., C. H. Kim, D. Y. Hwang, H. Seo, S. Chung, S. J. Hong, J. K. Lim, T. Anderson, and O. Isacson. 2003. Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. J. Neurochem. 85:622-634. [DOI] [PubMed] [Google Scholar]
- 37.Kim, K. S., M. K. Lee, J. Carroll, and T. H. Joh. 1993. Both the basal and inducible transcription of the tyrosine hydroxylase gene are dependent upon a cAMP response element. J. Biol. Chem. 268:15689-15695. [PubMed] [Google Scholar]
- 38.Kim, S. W., X. Fang, H. Ji, A. F. Paulson, J. M. Daniel, M. Ciesiolka, F. van Roy, and P. D. McCrea. 2002. Isolation and characterization of XKaiso, a transcriptional repressor that associates with the catenin Xp120(ctn) in Xenopus laevis. J. Biol. Chem. 277:8202-8208. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi, A., H. Yamagiwa, H. Hoshino, A. Muto, K. Sato, M. Morita, N. Hayashi, M. Yamamoto, and K. Igarashi. 2000. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol. 20:1733-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazaroff, M., S. Patankar, S. O. Yoon, and D. M. Chikaraishi. 1995. The cyclic AMP response element directs tyrosine hydroxylase expression in catecholaminergic central and peripheral nervous system cell lines from transgenic mice. J. Biol. Chem. 270:21579-21589. [DOI] [PubMed] [Google Scholar]
- 42.Lazaroff, M., Y. Qi, and D. M. Chikaraishi. 1998. Differentiation of a catecholaminergic CNS cell line modifies tyrosine hydroxylase transcriptional regulation. J. Neurochem. 71:51-59. [DOI] [PubMed] [Google Scholar]
- 43.Lee, D. K., D. Suh, H. J. Edenberg, and M. W. Hur. 2002. POZ domain transcription factor, FBI-1, represses transcription of ADH5/FDH by interacting with the zinc finger and interfering with DNA binding activity of Sp1. J. Biol. Chem. 277:26761-26768. [DOI] [PubMed] [Google Scholar]
- 44.Levitt, M., S. Spector, A. Sjoerdsma, and S. Udenfriend. 1965. Elucidation of the rate-limiting step in norepinephrine biosynthesis in the perfused guinea-pig heart. J. Pharmacol. Exp. Ther. 148:1-8. [PubMed] [Google Scholar]
- 45.Li, J. Y., M. A. English, H. J. Ball, P. L. Yeyati, S. Waxman, and J. D. Licht. 1997. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J. Biol. Chem. 272:22447-22455. [DOI] [PubMed] [Google Scholar]
- 46.Lim, K. C., G. Lakshmanan, S. E. Crawford, Y. Gu, F. Grosveld, and J. D. Engel. 2000. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat. Genet. 25:209-212. [DOI] [PubMed] [Google Scholar]
- 47.Liu, J., J. P. Merlie, R. D. Todd, and K. L. O'Malley. 1997. Identification of cell type-specific promoter elements associated with the rat tyrosine hydroxylase gene using transgenic founder analysis. Brain Res. Mol. Brain Res. 50:33-42. [DOI] [PubMed] [Google Scholar]
- 48.Mackler, S. A., L. Korutla, X. Y. Cha, M. J. Koebbe, K. M. Fournier, M. S. Bowers, and P. W. Kalivas. 2000. NAC-1 is a brain POZ/BTB protein that can prevent cocaine-induced sensitization in the rat. J. Neurosci. 20:6210-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melnick, A., K. F. Ahmad, S. Arai, A. Polinger, H. Ball, K. L. Borden, G. W. Carlile, G. G. Prive, and J. D. Licht. 2000. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 20:6550-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melnick, A., G. Carlile, K. F. Ahmad, C. L. Kiang, C. Corcoran, V. Bardwell, G. G. Prive, and J. D. Licht. 2002. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell. Biol. 22:1804-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meloni, R., V. Albanèse, P. Ravassard, F. Treilhou, and J. Mallet. 1998. A tetranucleotide polymorphic microsatellite, located in the first intron of the tyrosine hydroxylase gene, acts as a transcription regulatory element in vitro. Hum. Mol. Genet. 7:423-428. [DOI] [PubMed] [Google Scholar]
- 52.Miaw, S. C., A. Choi, E. Yu, H. Kishikawa, and I. C. Ho. 2000. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity 12:323-333. [DOI] [PubMed] [Google Scholar]
- 53.Min, N., T. H. Joh, K. S. Kim, C. Peng, and J. H. Son. 1994. 5′ upstream DNA sequence of the rat tyrosine hydroxylase gene directs high-level and tissue-specific expression to catecholaminergic neurons in the central nervous system of transgenic mice. Brain Res. Mol. Brain Res. 27:281-289. [DOI] [PubMed] [Google Scholar]
- 54.Morin, X., H. Cremer, M. R. Hirsch, R. P. Kapur, C. Goridis, and J. F. Brunet. 1997. Defects in sensory and autonomic ganglia and absence of locus coeruleus in mice deficient for the homeobox gene Phox2a. Neuron 18:411-423. [DOI] [PubMed] [Google Scholar]
- 55.Murre, C., P. S. McCaw, H. Vaessin, M. Caudy, L. Y. Jan, Y. N. Jan, C. V. Cabrera, J. N. Buskin, S. D. Hauschka, A. B. Lassar, H. Weintraub, and D. Baltimore. 1989. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58:537-544. [DOI] [PubMed] [Google Scholar]
- 56.Nagatsu, T., M. Levitt, and S. Udenfriend. 1964. Tyrosine hydroxylase. The initial step in norepinephrine biosynthesis. J. Biol. Chem. 239:2910-2917. [PubMed] [Google Scholar]
- 57.Numoto, M., K. Yokoro, and J. Koshi. 1999. ZF5, which is a Kruppel-type transcriptional repressor, requires the zinc finger domain for self-association. Biochem. Biophys. Res. Commun. 256:573-578. [DOI] [PubMed] [Google Scholar]
- 58.Okabe, S., T. Fukuda, K. Ishibashi, S. Kojima, S. Okada, M. Hatano, M. Ebara, H. Saisho, and T. Tokuhisa. 1998. BAZF, a novel Bcl6 homolog, functions as a transcriptional repressor. Mol. Cell. Biol. 18:4235-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pagans, S., M. Ortiz-Lombardia, M. L. Espinas, J. Bernues, and F. Azorin. 2002. The Drosophila transcription factor tramtrack (TTK) interacts with trithorax-like (GAGA) and represses GAGA-mediated activation. Nucleic Acids Res. 30:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pagans, S., D. Pineyro, A. Kosoy, J. Bernues, and F. Azorin. 2004. Repression by TTK69 of GAGA-mediated activation occurs in the absence of TTK69 binding to DNA and solely requires the contribution of the POZ/BTB domain of TTK69. J. Biol. Chem. 279:9725-9732. [DOI] [PubMed] [Google Scholar]
- 61.Pattyn, A., X. Morin, H. Cremer, C. Goridis, and J. F. Brunet. 1999. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature 399:366-370. [DOI] [PubMed] [Google Scholar]
- 62.Pessler, F., and N. Hernandez. 2003. Flexible DNA binding of the BTB/POZ-domain protein FBI-1. J. Biol. Chem. 278:29327-29335. [DOI] [PubMed] [Google Scholar]
- 63.Prokhortchouk, A., B. Hendrich, H. Jorgensen, A. Ruzov, M. Wilm, G. Georgiev, A. Bird, and E. Prokhortchouk. 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Puelles, L., and C. Verney. 1998. Early neuromeric distribution of tyrosine-hydroxylase-immunoreactive neurons in human embryos. J. Comp. Neurol. 394:283-308. [DOI] [PubMed] [Google Scholar]
- 65.Ravassard, P., F. Chatail, J. Mallet, and C. Icard-Liepkalns. 1997. Relax, a novel rat bHLH transcriptional regulator transiently expressed in the ventricular proliferating zone of the developing central nervous system. J. Neurosci. Res. 48:146-158. [DOI] [PubMed] [Google Scholar]
- 66.Remacle, J. E., H. Kraft, W. Lerchner, G. Wuytens, C. Collart, K. Verschueren, J. C. Smith, and D. Huylebroeck. 1999. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 18:5073-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Retaux, S., M. Rogard, I. Bach, V. Failli, and M. J. Besson. 1999. Lhx9: a novel LIM-homeodomain gene expressed in the developing forebrain. J. Neurosci. 19:783-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saucedo-Cardenas, O., J. D. Quintana-Hau, W. D. Le, M. P. Smidt, J. J. Cox, F. De Mayo, J. P. Burbach, and O. M. Conneely. 1998. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc. Natl. Acad. Sci. USA 95:4013-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schimmel, J. J., L. Crews, S. Roffler-Tarlov, and D. M. Chikaraishi. 1999. 4.5 kb of the rat tyrosine hydroxylase 5′ flanking sequence directs tissue specific expression during development and contains consensus sites for multiple transcription factors. Brain Res. Mol. Brain Res. 74:1-14. [DOI] [PubMed] [Google Scholar]
- 70.Smidt, M. P., C. H. Asbreuk, J. J. Cox, H. Chen, R. L. Johnson, and J. P. Burbach. 2000. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat. Neurosci. 3:337-341. [DOI] [PubMed] [Google Scholar]
- 71.Smits, S. M., T. Ponnio, O. M. Conneely, J. P. Burbach, and M. P. Smidt. 2003. Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur. J. Neurosci. 18:1731-1738. [DOI] [PubMed] [Google Scholar]
- 72.Son, J. H., N. Min, and T. H. Joh. 1996. Early ontogeny of catecholaminergic cell lineage in brain and peripheral neurons monitored by tyrosine hydroxylase-lacZ transgene. Brain Res. Mol. Brain Res. 36:300-308. [DOI] [PubMed] [Google Scholar]
- 73.Specht, L. A., V. M. Pickel, T. H. Joh, and D. J. Reis. 1981. Light-microscopic immunocytochemical localization of tyrosine hydroxylase in prenatal rat brain. I. Early ontogeny. J. Comp. Neurol. 199:233-253. [DOI] [PubMed] [Google Scholar]
- 74.Specht, L. A., V. M. Pickel, T. H. Joh, and D. J. Reis. 1981. Light-microscopic immunocytochemical localization of tyrosine hydroxylase in prenatal rat brain. II. Late ontogeny. J. Comp. Neurol. 199:255-276. [DOI] [PubMed] [Google Scholar]
- 75.Tinti, C., C. Yang, H. Seo, B. Conti, C. Kim, T. H. Joh, and K. S. Kim. 1997. Structure/function relationship of the cAMP response element in tyrosine hydroxylase gene transcription. J. Biol. Chem. 272:19158-19164. [DOI] [PubMed] [Google Scholar]
- 76.Trocmé, C., C. Sarkis, J. M. Hermel, R. Duchateau, S. Harrison, M. Simonneau, R. Al-Shawi, and J. Mallet. 1998. CRE and TRE sequences of the rat tyrosine hydroxylase promoter are required for TH basal expression in adult mice but not in the embryo. Eur. J. Neurosci. 10:508-521. [DOI] [PubMed] [Google Scholar]
- 77.Verschueren, K., J. E. Remacle, C. Collart, H. Kraft, B. S. Baker, P. Tylzanowski, L. Nelles, G. Wuytens, M. T. Su, R. Bodmer, J. C. Smith, and D. Huylebroeck. 1999. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J. Biol. Chem. 274:20489-20498. [DOI] [PubMed] [Google Scholar]
- 78.Vitalis, T., O. Cases, D. Engelkamp, C. Verney, and D. J. Price. 2000. Defect of tyrosine hydroxylase-immunoreactive neurons in the brains of mice lacking the transcription factor Pax6. J. Neurosci. 20:6501-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vouyiouklis, D. A., and P. J. Brophy. 1995. Microtubule-associated proteins in developing oligodendrocytes: transient expression of a MAP2c isoform in oligodendrocyte precursors. J. Neurosci. Res. 42:803-817. [DOI] [PubMed] [Google Scholar]
- 80.Wang, M. M., and R. R. Reed. 1993. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature 364:121-126. [DOI] [PubMed] [Google Scholar]
- 81.Wilkins, R. C., and J. T. Lis. 1997. Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res. 25:3963-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilkinson, D. G. 1994. Whole mount in situ hybridization of vertebrate embryos, p. 75-83. In D. G. Wilkinson (ed.), In situ hybridization: a practical approach. IRL Press, Oxford, United Kingdom.
- 83.Wilson, T. E., T. J. Fahrner, M. Johnston, and J. Milbrandt. 1991. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science 252:1296-1300. [DOI] [PubMed] [Google Scholar]
- 84.Yang, C., H. S. Kim, H. Seo, and K. S. Kim. 1998. Identification and characterization of potential cis-regulatory elements governing transcriptional activation of the rat tyrosine hydroxylase gene. J. Neurochem. 71:1358-1368. [DOI] [PubMed] [Google Scholar]
- 85.Yoon, S. O., and D. M. Chikaraishi. 1994. Isolation of two E-box binding factors that interact with the rat tyrosine hydroxylase enhancer. J. Biol. Chem. 269:18453-18462. [PubMed] [Google Scholar]
- 86.Yoon, S. O., and D. M. Chikaraishi. 1992. Tissue-specific transcription of the rat tyrosine hydroxylase gene requires synergy between an AP-1 motif and an overlapping E box-containing dyad. Neuron 9:55-67. [DOI] [PubMed] [Google Scholar]
- 87.Zellmer, E., Z. Zhang, D. Greco, J. Rhodes, S. Cassel, and E. J. Lewis. 1995. A homeodomain protein selectively expressed in noradrenergic tissue regulates transcription of neurotransmitter biosynthetic genes. J. Neurosci. 15:8109-8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zetterstrom, R. H., L. Solomin, L. Jansson, B. J. Hoffer, L. Olson, and T. Perlmann. 1997. Dopamine neuron agenesis in Nurr1-deficient mice. Science 276:248-250. [DOI] [PubMed] [Google Scholar]