Abstract
Dichomitus squalens is a white-rot fungus that colonizes and grows mainly on softwood and is commonly found in the northern parts of Europe, North America, and Asia. We analyzed the genetic and physiological diversity of eight D. squalens monokaryons derived from a single dikaryon. In addition, an unrelated dikaryon and a newly established dikaryon from two of the studied monokaryons were included. Both growth and lignocellulose acting enzyme profiles were highly variable between the studied monokaryotic and dikaryotic strains, demonstrating a high level of diversity within the species.
Keywords: AFLP, carbon utilization, monokaryon, sexual reproduction, white-rot
INTRODUCTION
Basidiomycete fungi are essential in forest ecology, due to their ability to degrade wood (Eriksson et al. 1990). Wood degrading basidiomycetes have traditionally been divided in two subgroups, white-rot and brown-rot fungi, according to their method of wood decay, although the existence of intermediate species has been suggested (Riley et al. 2014). White-rot fungi are unique in possessing an array of extracellular lignin-modifying enzymes and are therefore able to completely mineralize recalcitrant aromatic lignin polymers (Mäkelä et al. 2014). In addition, they typically harbor a full repertoire of genes encoding enzymes that are targeted for depolymerization of different plant cell wall polysaccharides (Rytioja et al. 2014).
Most basidiomycetes have a sexual reproductive cycle. When suitable substrate and growth conditions are present, haploid (n) spores germinate and a primary mycelium is formed. The primary mycelium can fuse with another primary mycelium of a compatible mating type and form a dikaryotic (n + n, unfused nuclei from different parents) secondary mycelium. The secondary mycelium produces basidiomata where fusion of two haploid nuclei occurs in basidia to give diploid nuclei (2n). After that, meiosis follows and each basidium bears four haploid basidiospores. Therefore, the offspring of two different dikaryons can produce a dikaryon with a different genetic make-up than either parent, which can result in different physiological abilities of the offspring.
There are different mating compatibility systems in fungi. In agaricomycetes, mating is regulated by either a bipolar or a tetrapolar system. Tetrapolar behavior consists of two unlinked genetic complexes, A and B, with four potential mating interactions between spores from one basidiome. In the bipolar case, only compatible or incompatible interactions are found (Raudaskoski & Kothe 2010). In both cases, when hyphae from two compatible mating types find each other and make contact, fusion of the mycelia usually occurs through clamp connections and a dikaryon is formed (Krings et al. 2011, Kim et al. 2014).
In this study, we aimed to dissect the variations in genetic profile, growth, and enzyme production in different monokaryotic strains derived from a dikaryon of the white-rot basidiomycete Dichomitus squalens. This species is commonly found in North America and northern regions of Europe and Asia (Andrews & Gill 1943), and it usually grows on softwood, but is also able to grow and degrade hardwood (Blanchette et al. 1987, Fackler et al. 2007). Dichomitus squalens has a tetrapolar mating system, and in a previous study 20 monokaryotic progeny derived from a single wild-type dikaryon of D. squalens (FBCC312, CBS 432.34) were isolated and their growth rate, cellulose and lignin degrading ability, and isoenzyme patterns were compared (Pham et al. 1990). We performed a more detailed analysis of these strains by studying their genetic variation using amplified fragment length polymorphisms (AFLP), and their ability to grow on different carbon sources and to produce extracellular lignocellulolytic enzymes. In addition, we generated a new dikaryon from two of the monokaryons and compared that to the original parent. The results of this study provide insight into the functional diversity that exists within a single basidiomycete species in nature and how this can evolve in fungi that contain a sexual cycle.
MATERIALS AND METHODS
Fungal strains and cultivations
Dichomitus squalens strains obtained from the CBS collection (Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands) and the Fungal Biotechnology Culture Collection (FBCC, Department of Food and Environmental Sciences, University of Helsinki) were used in this study (Table 1) and maintained on 2 % (w/v) malt extract 1.5 % (w/v) agar (MEA) plates.
Table 1.
Mating type assigned to Dichomitus squalens strains described in Pham et al. (1990) related to their corresponding incompatibility group.
| Strain no. (in CBS and/or FBCC collection) | Mating type | Strain no. (Pham et al. 1990) | Incompatibility groupa (Pham et al. 1990) | |
|---|---|---|---|---|
| CBS 453.89 | AB | DS 1 | I | |
| CBS 471.89 | DS 19 | |||
|
| ||||
| CBS 455.89 | ab | DS 3 | II | |
| CBS 456.89 | DS 4 | |||
| CBS 458.89 | DS 6 | |||
| CBS 459.89 | DS 7 | |||
| CBS 462.89 | DS 10 | |||
| CBS 467.89 | DS 15 | |||
| CBS 468.89 | DS 16 | |||
|
| ||||
| CBS 454.89 | Ab | DS 2 | III | |
| CBS 460.89 | DS 8 | |||
| CBS 461.89 | DS 9 | |||
| CBS 463.89 | DS 11 | |||
| CBS 466.89 | DS 14 | |||
| CBS 469.89 | DS 17 | |||
| CBS 470.89 | DS 18 | |||
| CBS 472.89 | DS 20 | |||
|
| ||||
| CBS 457.89 | aB | DS 5 | IV | |
| CBS 464.89 | DS 12 | |||
| CBS 465.89 | DS 13 | |||
|
| ||||
| FBCC312 (CBS 432.34) | AaBb | |||
|
| ||||
| CBS 141873 (cross of CBS 463.89 and CBS 464.89) | AaBb | |||
|
| ||||
| FBCC184 | AaBb | |||
Liquid pre-cultures were performed in 250 mL Erlenmeyer flasks with 50 mL low-nitrogen asparagine-succinate medium (LN-AS) minimal medium (Hatakka & Uusi-Rauva 1983), pH 4.5, amended with 0.05 % glycerol as a carbon source. The medium was inoculated with 5 agar plugs (0.5 cm diam) from MEA plates that were covered with fresh fungal mycelium and incubated for 7 d at 28 °C stationary. From the pre-cultures, 4 mL of homogenized mycelium (Mäkelä et al. 2002) was transferred to 250 mL Erlenmeyer flasks containing 100 mL LN-AS with 2 % powdered wheat bran or Norway spruce (Picea abies) sawdust as carbon source. These cultures were incubated for 16 d at 28 °C under agitation (120 rpm). All cultures were performed in duplicate. Culture medium samples were harvested after 9 and 16 d of growth, centrifuged at 4 °C to remove residual biomass and stored at -20 °C as 2 mL aliquots.
The ability of D. squalens strains to grow on 37 different carbon sources was tested on plates containing LN-AS agar (1.5 % agar-agar (w/v)) amended with: (1) monomeric and oligomeric sugars: D-glucose, D-fructose, D-galactose, D-mannose, D-ribose, D-xylose, L-arabinose, L-rhamnose, D-galacturonic acid, D-glucuronic acid, cellobiose, maltose, lactose, raffinose, or sucrose; (2) polymeric substrates: arabinogalactan, beechwood xylan, birchwood xylan, arabic gum, guar gum, soluble starch, apple pectin, citrus pectin, inulin, calcium lignin, casein, cellulose, or sodium lignin; and (3) powdered complex substrates: wheat bran, sugar beet pulp, citrus pulp, soybean hulls, rice bran, cotton seed pulp, alfalfa meal, or spruce sawdust. All monomeric and oligomeric carbon sources were added to a final concentration of 25 mM, while pure polymeric and complex substrates were added to a final concentration of 1 % and 3 %, respectively. The pH of the medium was adjusted to 4.5. Monosaccharides and cellobiose were filter-sterilized and added to the medium after autoclaving, while the other substrates were added before autoclaving. LN-AS agar plates without any carbon source were used as control.
The strains were inoculated by placing mycelium-covered agar plug (0.5 mm diam) obtained from the edge of 1-wk-old MEA plates in the center of the plates. Incubation time was established for 3 d at 28 °C according to the growth of the first strain reaching the edge of the plate. After this period, pictures were taken and the diameter of the fungal colony was measured for every plate in duplicate.
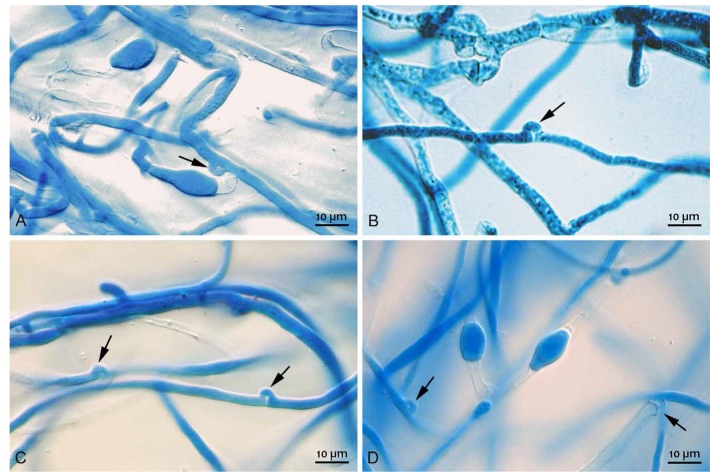
To obtain dikaryons, two agar plugs (0.5 mm diam) covered by mycelia of monokaryons of compatible mating type, CBS 463.89 (Ab) and CBS 464.89 (aB), were placed on a MEA plate at 3 cm distance. After 3 d of growth at 28°C, the mycelia of the different strains made contact, and successful crosses were confirmed by microscopy (Zeiss Axioplan Microscope) by staining a sample of the common mycelium with lactophenol Cotton blue (Leck 1999).
Enzymatic activity assays
Liquid pre-cultures and transfer conditions were performed according to Rytioja et al. (2017), but in this study 2 g of powdered wheat bran and spruce sawdust were used for 100 mL LN-AS cultures, respectively. The cultures were performed as two biological replicates and sampled after 9 and 16 d. All p-nitrophenol (pNP) assays (Table 2) were performed in triplicate in 96-well plates using a FLUORstar OPTIMA microplate reader (BMG). For pNP assays, 0.1 % of the corresponding substrate was used in 50 mM sodium acetate buffer (pH 5.0). Reaction mixtures for α-L-arabinofuranosidase (ABF), α-1,4-D-galactosidase (AGL), β-1,4-D-glucosidase (BGL) and β-1,4-D-galactosidase (LAC) activity were incubated for 1 h, while reaction mixtures for β-xylosidase (BXL), cellobiohydrolase (CBH) and β-mannosidase (MND) activity were incubated for 4 h. All reactions were incubated at 28 °C and were stopped by adding 100 μL 0.25 M sodium carbonate. A pNP standard curve was generated to calculate the concentration of the released pNP. The activities are expressed in nmol/min/mL.
Table 2.
Substrates assayed for polysaccharide degrading enzymatic activities, corresponding enzymes and their abbreviation, and suppliers of the substrates.
| Substrate | Enzyme | Abbreviation | Supplier |
|---|---|---|---|
| 4-Nitrophenyl α-L-arabinofuranoside | α-L-Arabinofuranosidase | ABF | Sigma |
| 4-Nitrophenyl α-D-galactopyranoside | α-1,4-D-Galactosidase | AGL | Sigma |
| 4-Nitrophenyl β-D-glucopyranoside | β-1,4-D-Glucosidase | BGL | Sigma |
| 4-Nitrophenyl β-D-xylopyranoside | β-Xylosidase | BXL | Sigma |
| 4-Nitrophenyl β-D-cellobioside | Cellobiohydrolase | CBH | Sigma |
| 4-Nitrophenyl α-D-glucopyranoside | α-1,4-D-Glucosidase | AGD | Koch-light |
| 4-Nitrophenyl β-D-galactopyranoside | β-1,4-D-Galactosidase | LAC | Sigma |
| 4-Nitrophenyl β-D-mannopyranoside | β-Mannosidase | MND | Sigma |
| 4-Nitrophenyl maltoside | Glucoamylase | GLA | Acros |
Laccase and manganese peroxidase (MnP) activities were measured by following the oxidation of 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonate) (ABTS); ϵ420=36×104 M−1 cm−1; Sigma) at 440 nm using a Biochrom Libra S22 UV/Vis Spectrophotometer. Laccase activity was determined in 50 mM sodium malonate buffer (pH 4.5) for 30 s at 25 °C (Eggert et al. 1996). After this first measurement 0.5 mM manganese sulfate and 0.1 mM hydrogen peroxide were added to the same reaction to start the MnP reaction, and change of absorbance was followed again for 30 s (Hofrichter & Fritsche 1997, Hofrichter et al. 1998). Three technical replicates were performed on biological duplicates and these were averaged in the graphs.
SDS-PAGE
Protein profiles were obtained from the same culture liquids that were used to measure enzyme activities. Sixteen μl of each culture liquid sample was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10 % (wt/vol) acrylamide/30 % bisacrylamide gels and a molecular weight marker (Bio-Rad unstained marker) was used to identify the molecular mass of the protein bands. The gels were stained using the Silver staining method (Chevallet et al. 2006) and documented using the HP scanner 4400c.
AFLP
To describe how related the D. squalens strains are at the genetic level, they were clustered in a phylogenetic tree based on amplified fragment length polymorphisms (AFLPs). DNA isolation from mycelium grown on MEA plates was performed with a Microbial DNA isolation kit (MO BIO Laboratories, Carlsbad, USA) according to the manufacturer’s recommendations. The experimental procedure was adapted from Illnait-Zaragozi et al. (2012). To each AFLP-reaction, 0.2 μL of Orange DNA size standard (MCLAB, DSMO-101) was added and samples were processed on a 3730xl DNA Analyzer (Applied Biosystems). Raw data was normalized and further analyzed with Bionumerics (v. 4.61, Applied Maths, Sint-Martens-Latem, Belgium) using the Pearson correlation coefficient and UPGMA clustering method.
RESULTS
Growth profiling and mating type determination
Most Dichtomitus squalens strains used in this study have previously been described (Pham et al. 1990), but their genotypes and phenotypes had not been analyzed in detail. The mating type of the strains was verified by crossing them with each other, and resulted with strains of mating type AB, ab, Ab and aB fitting in the previously suggested groups I, II, III and IV, respectively (Table 1).
All strains from the study of Pham et al. (1990) were initially tested by growth on glucose, fructose, xylose, galacturonic acid, cellobiose, beechwood xylan, guar gum, apple pectin, and citrus pectin (data not shown). Based on phenotypic differences that suggested different carbon source utilization abilities and compatibility of mating types, a selection of eight monokaryotic strains was made that included representatives of all mating types (AB, Ab, aB, and ab). In addition, the parental dikaryotic strain (FBCC312) of the monokaryons, and a dikaryon FBCC184 which was a separate natural isolate, were included. Finally, another dikaryon, D. squalens CBS 141873, was formed by successfully crossing the compatible monokaryons CBS 463.89 and CBS 464.89 (Fig. 1), and was included in the study.
Fig. 1.
Clamp connections of the Dichomitus squalens compatible monokaryons (CBS 463.89 and CBS 464.89) crossing to form the dikaryon (CBS 141873) as visualized by lactophenol cotton blue staining. Clamp connections are indicated by arrows (A–D).
Growth profiling of all the studied monokaryotic and dikaryotic strains was performed on 37 carbon sources to reveal the phenotypic differences between them (Supplementary Material Fig. 1). Monokaryon CBS 471.89 (AB) produced a larger colony diameter than the other monokaryons and dikaryons on all monosaccharides, but not on complex substrates, where both the parental strain FBCC312 and monokaryon CBS 471.89 (AB) showed a more similar pattern of good growth. On some substrates, such as alfalfa meal, the best growth was observed for the dikaryon FBCC312 (Supplementary Material Fig. 1). Monokaryons CBS 462.89 (ab), CBS 463.89 (Ab) and CBS 464.89 (aB) also showed relatively good growth in most of the substrates compared to the other monokaryotic strains. Conversely, CBS 453.89 (AB), CBS 465.89 (aB) and CBS 466.89 (Ab) showed poor growth in all substrates. The new dikaryon formed in this study (CBS 141873) had a similar growth profile to the monokaryons from which it was generated (CBS 463.89 and CBS 464.89). The parental dikaryon (FBCC312) grew better on most of the carbon sources than the newly formed dikaryon (CBS 141873) (Supplementary Fig. 1). These two dikaryons (FBCC312 and CBS 141873) both grew overall better than the non-related dikaryon FBCC184.
AFLP analysis reveals high genetic variation between the monokaryotic strains
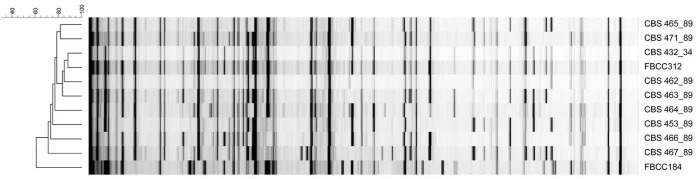
AFLP analysis was conducted to analyze the relationship and genetic variability between the Dichomitus squalens monokaryotic strains and also between the three dikaryons (Fig. 2). Two batches of a single dikaryon from two culture collections, FBCC312 and CBS 432.34, clustered together confirming that they are in fact the same strain, which is the parent of the progeny. Monokaryons CBS 463.89, CBS 463.89, CBS 464.89, and CBS 471.89, which showed good growth on most carbon sources (Supplementary Fig. 1) were clustered close to each other and to the parental dikaryon FBCC312. Monokaryons that showed poor growth were clustered more distantly to the other monokaryons and the dikaryon FBCC312. The dikaryon FBCC184 positioned further away from the monokaryons and their parental dikaryon (FBCC312), which reflects its origin as it is a separate isolate of this fungal species.
Fig. 2.
Clustering of AFLP patterns of Dichomitus squalens isolates. Similarity of the patterns is given in percentage.
Monokaryotic progeny demonstrated high diversity in their enzyme activity profile
Since Dichomitus squalens uses wood as its main carbon source in nature, several enzyme activities related to plant biomass degradation were measured in the cultures of the strains grown on spruce sawdust or powdered wheat bran containing liquid medium. The conditions were chosen to compare the behavior of the fungus growing in wheat bran, which is rich in easily degradable polysaccharides, to spruce, which in addition to polysaccharides contains a high amount of lignin and is the natural substrate of D. squalens. These two substrates were also compared in detail in a recent study of FBCC312 (Rytioja et al. 2017) and conditions were chosen to mimic those of the previous study.
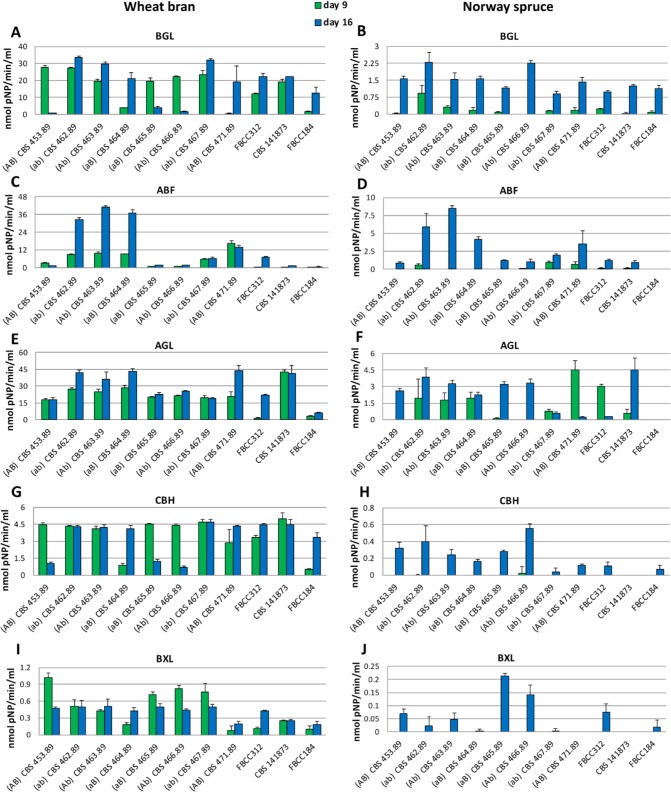
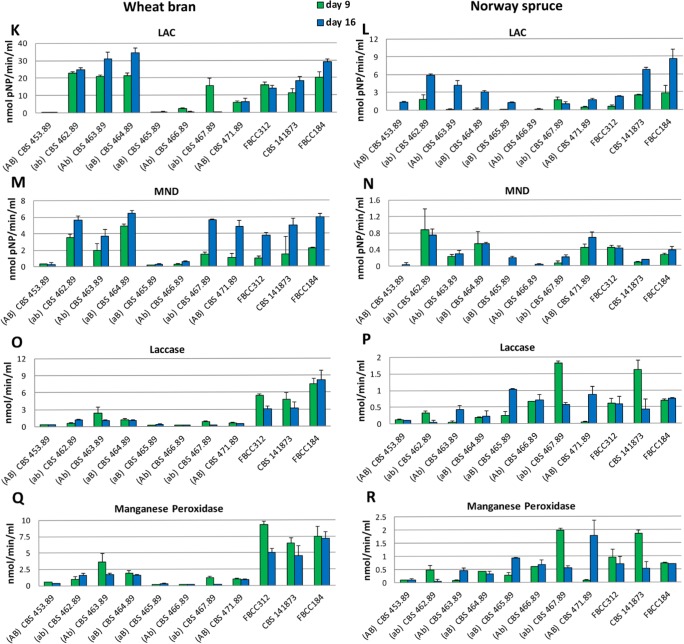
In spruce medium, most of the enzyme activities related to polysaccharide degradation increased from 9 to 16 d of cultivation (Fig. 3B, D, H, J). The only exceptions were AGL (Fig. 3F), for which the monokaryons CBS 467.89 and CBS 471.89 and dikaryon FBCC312 presented higher activity at the earlier time point, and MND (Fig. 3N), for which monokaryon CBS 462.89 and dikaryon FBCC312 showed higher activity at the earlier time point. In contrast, more variation in the production of the hydrolytic polysaccharide active enzyme activities between the strains was observed during growth on wheat bran. Interestingly, the variation in activity was larger for some enzymes (e.g. ABF, LAC; Fig. 3C–D and 3K–L) than for others (e.g. BGL, CBH, Fig 3A–B and 3G–H), suggesting a different fine tuning of the regulation in the different strains. Comparing both media, in wheat bran all the activities measured were much higher (from 4 to 10 times higher depending on the enzyme) than in spruce sawdust, including the ligninolytic enzymes (laccase and MnP).
Fig. 3.
Extracellular enzyme activities detected in the culture liquids of the Dichomitus squalens strains after 9 and 16 days of growth. BGL in wheat bran (A) and Norway spruce (B); ABF in wheat bran (C) and Norway spruce (D); AGL in wheat bran (E) and Norway spruce (F); CBH in wheat bran (G) and Norway spruce (H); BXL in wheat bran (I) and Norway spruce (J); LAC in wheat bran (K) and Norway spruce (L); MND in wheat bran (M) and Norway spruce (N); laccase in wheat bran (O) and Norway spruce (P); MnP in wheat bran (Q) and Norway spruce (R). Vertical bars represent standard deviation of at least four replicate measurements.
The activity patterns of the ligninolytic activities differed from those of the polysaccharide active enzymes (Fig. 3). On both wheat bran and spruce sawdust amended cultures, the laccase and MnP activities were higher at the earlier time point for most of the strains (Fig. 3O–R). However, a more notable difference was detected on wheat bran cultures, where much higher activity levels of these two enzymes were produced by the dikaryons than the monokaryons, reaching three-fold higher in some cases, for example dikaryon CBS 141873 at the early time point compared to monokaryons CBS 453.89, 462.89 and 463.89 (Fig. 3P, R).
The activities detected for the newly made dikaryon (CBS 141873) differed from those of FBCC312, which was especially evident for AGL where CBS 141873 had significantly higher activity in wheat bran medium at both time points (Fig. 3E–F). Also in spruce sawdust it demonstrated a much higher increase of activity over time compared to FBCC312 for which AGL activity decreased. For laccase and MnP, the difference was more evident in Norway spruce sawdust where CBS 141873 showed higher activity at the earlier time point (Fig. 3P–R). This indicates that the two monokaryons derived from FBCC312 did not re-combine to a strain with the same capabilities as FBCC312. Differences were observed in the activity level as well as the relative activity between the two time points studied. Finally, there was generally higher ligninolytic activity for five of the monokaryons (except CBS 464.89, CBS 465.89 and CBS 466.89) and the three dikaryons at the early time point compared to polysaccharide active enzymes.
DISCUSSION
Fungi with a sexual reproductive cycle have the possibility to alter their genetic information through recombination during meiosis (Kothe 1996). Therefore, dikaryotic species, such as Dichomitus squalens, can produce monokaryotic offspring with diverse genetic combinations. Strain improvement by cycles of basidiome production and crosses, without the need for mutagenesis, is a feasible approach to improve the ability of white-rot fungi to degrade lignin (Wyatt & Broda 1995). In this study, we explored this variation by analyzing the genetic variation, carbon source usage profile and enzyme activity profile of eight monokaryotic strains derived from a single dikaryon of D. squalens and compared these to the parental dikaryon, a new dikaryon generated from two of these monokaryons, and an unrelated dikaryon that was isolated in nature.
Regarding the growth profile, D. squalens monokaryon CBS 471.89 (AB) seems to have inherited an improved system to use mono- and oligosaccharides compared to all the other tested strains, including the dikaryons, which is especially evident for the disaccharide cellobiose. This observation is in agreement with a previous study on another white-rot species, Trametes versicolor, which demonstrated that a monokaryon presented a better growth rate than its parental dikaryon on glucose-soy agar and hardwood kraft pulp (Addleman & Archibald 1993). However, the monokaryotic strains of the white-rot fungus Pleurotus ostreatus have been reported to grow markedly slower than their dikaryotic parental strains (Eichlerová & Homolka 1999). No correlation between the mating type, appearance of the colony, and the production of ligninolytic enzyme activity, was detected between the P. ostreatus monokaryons (Eichlerová & Homolka 1999). This is in line with our observations, since D. squalens strains did not have an apparent relation between the improved ability for carbon source utilization and mating type genes since the strains showing better growth on different carbon sources belong to all four mating types: CBS 471.89 (AB), CBS 462.89 (ab), CBS 463.89 (Ab), and CBS 464.89 (aB). These strains clustered close to each other in the AFLP analysis and to the parental strain (FBCC312), suggesting that they inherited common genetic abilities that cause the improved growth. Moreover, monokaryon CBS 453.89 (AB) showed poor growth in all carbon sources and, to a lesser extent this was also observed for monokaryons CBS 465.89 (aB) and CBS 466.89 (Ab), indicating that poor growth is also not linked to the mating type. These strains also clustered more distantly from the parental strain in the AFLP analysis. Despite the improved ability to utilize mono- and oligosaccharides by the monokaryon CBS 471.89 (AB), its abilities to grow on complex substrates were similar to the parental strain. The newly made D. squalens dikaryon (CBS 141873) showed a different enzyme activity profile than its parental dikaryon FBCC312. Previously, significantly improved properties, including enzyme activity, growth rate, and chemical conversion abilities, compared to the parental dikaryon have been reported from white-rot fungal dikaryons derived from compatible monokaryons (Eichlerová & Homolka 1999).
In addition to lignin-active enzymes, white-rot fungi also possess a full arsenal of enzymes that decompose the other wood polymers (Nagy et al. 2017). This was also evident from our data, since during growth in liquid media with wheat bran (low lignin content) the D. squalens strains produced much higher levels of polysaccharide-related enzymes compared to liquid medium with spruce sawdust (high lignin content). The observation confirms a previous report that lignin can be a barrier for the utilization of the polysaccharides, and that initially the energy of the fungus is directed towards enzymes necessary to degrade the lignin to enable access to polysaccharides (Rytioja et al. 2017). Partial degradation of easily accessible hemicelluloses likely provides the energy for lignin degradation (Blanchette 1984), while production of polysaccharide degrading enzymes increases gradually when these polysaccharides become more accessible by removal of lignin (Pandey & Singh 2014). This has also been observed for other basidiomycete white-rot fungi (Patyshakuliyeva et al. 2015, Kuuskeri et al. 2016). The results of our study also show that white-rot fungi in both monokaryotic and dikaryotic forms use a similar strategy for plant biomass degradation.
Our results also suggest a highly diverse ability of the mono- and dikaryotic strains of D. squalens to degrade plant biomass related substrates. This is in line with results from monokaryotic strains of the white-rot fungi Pycnoporus cinnabarinus (Herpoël et al. 2000) and P. sanguineus (Lomascolo et al. 2002) producing higher laccase activity, and P. ostreatus (Eichlerová et al. 2002) and Trametes hirsuta (Li et al. 2012) producing higher laccase and MnP activity than the parental dikaryon. This not only has implications for the efficiency of different strains of a fungal species in biomass decomposition in natural biotopes, but also indicates the importance of strain selection for biotechnological applications (Eichlerová et al. 2003). This has been shown in previous studies by demonstrating that, for example, isolates of the white-rot fungus Ceriporiopsis subvermispora were superior to those of Phanerochaete chrysosporium for biopulping (Blanchette et al. 1992). In fact, isolation of basidiospore-derived monokaryotic strains has been an efficient method to obtain new white-rot fungal strains with high variation in the production of enzymes involved in lignin modification and degradation (Eichlerová-Voláková & Homolka 1997, Santoyo et al. 2008). In future studies, we plan to further dissect the mechanisms underlying these differences by analyzing genomes, transcriptomes, and proteomes, of selected D. squalens strains. This may reveal whether the observed differences are mainly at the level of genome content, for example differences in plant cell wall active enzyme encoding gene repertoire, or at the post-genomic or regulatory level. The number of genome-sequenced white-rot fungi has increased significantly in the last few years (Rytioja et al. 2014, Floudas et al. 2015, Nagy et al. 2016, 2017) and in several, but not all, cases monokaryotic strains have been sequenced. However, the variation between two genome-sequenced monokaryotic strains of Pleurotus ostreatus that were produced by dedikaryotization of the parental strain was exemplified in their transcriptome analysis. The functionally annotated genes were categorized into groups of unknown functions, glycosyl hydrolases, and redox enzymes, and the relative importance of these groups was shown to differ between the monokaryotic strains (Alfaro et al. 2016). However, none of these or the subsequent transcriptome or proteome studies addressed the differences in abilities between monokaryotic and dikaryotic strains in-depth.
Supplementary Material
REFERENCES
- Addleman K, Archibald F. (1993) Kraft pulp bleaching and delignification by dikaryons and monokaryons of Trametes versicolor. Applied and Environmental Microbiology 59: 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro M, Castanera R, Lavín JL, Grigoriev IV, Oguiza JA, et al (2016) Comparative and transcriptional analysis of the predicted scretome in the lignocellulose-degrading basidiomycete Pleurotus ostreatus. Environmental Microbiology 18: 4710–4726. [DOI] [PubMed] [Google Scholar]
- Andrews SR, Gill LS. (1943) Western red rot in immature Ponderosa pine in the Southwest. Journal of Forestry 41: 565–573. [Google Scholar]
- Blanchette RA. (1984) Screening wood decayed by white rot fungi for preferential lignin degradation. Applied and Environmental Microbiology 48: 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette RA, Otjen L, Carlson MC. (1987) Lignin distribution in cell walls of birch wood decayed by white rot basidiomycetes. Phytopathology 77: 684–690. [Google Scholar]
- Blanchette RA, Burnes TA, Eerdmans MM, Akhtar M. (1992) Evaluating isolates of Phanerochaete chrysosporium and Ceriporiopsis subvermispora for use in biological pulping processes. Holzforschung 46: 109–115. [Google Scholar]
- Chevallet M, Luche S, Rabilloud T. (2006) Silver staining of proteins in polyacrylamide gels. Nature Protocols 1: 1852–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert C, Temp U, Dean JF, Eriksson KE. (1996) A fungal metabolite mediates degradation of non-phenolic lignin structures and synthetic lignin by laccase. FEBS Letters 391: 144–148. [DOI] [PubMed] [Google Scholar]
- Eichlerová I, Homolka L. (1999) Preparation and crossing of basidiospore-derived monokaryons – a useful tool for obtaining laccase and other ligninolytic enzyme higher-producing dikaryotic strains of Pleurotus ostreatus. Antonie van Leeuwenhoek 75: 321–327. [DOI] [PubMed] [Google Scholar]
- Eichlerová I, Homolka L, Nerud F. (2002) Decolorization of synthetic dyes by Pleurotus ostreatus isolates differing in ligninolytic properties. Folia Microbiologica 47: 691–695. [DOI] [PubMed] [Google Scholar]
- Eichlerová I, Homolka L, Nerud F. (2003) Decolorization of orange G by Pleurotus ostreatus monokaryotic isolates with different laccase activity. Folia Microbiologica 48: 775–779. [DOI] [PubMed] [Google Scholar]
- Eichlerová-Voláková I, Homolka L. (1997) Variability of ligninolytic enzyme activities in basidiospore isolates of the fungus Pleurotus ostreatus in comparison with that of protoplast-derived isolates. Folia Microbiologica 42: 583–588. [Google Scholar]
- Eriksson K-E, Blanchette RA, Ander P. (1990) Microbial and Enzymatic Degradation of Wood and Wood Components. Berlin: Springer-Verlag. [Google Scholar]
- Fackler K, Gradinger C, Schmutzer M, Tavzes C, Burgert I, et al (2007) Biotechnological wood modification with selective white-rot fungi and its molecular mechanisms. Food Technology and Biotechnology 45: 269–276. [Google Scholar]
- Floudas D, Held BW, Riley R, Nagy LG, Koehler G, et al (2015) Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genetics and Biology 76: 78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakka AI, Uusi-Rauva AK. (1983) Degradation of 14C-labelled poplar wood lignin by selected white-rot fungi. European Journal of Applied Microbiology and Biotechnology 17: 235–242. [Google Scholar]
- Herpoël I, Moukha S, Lesage-Meessen L, Sigoillot J-C, Asther M. (2000) Selection of Pycnoporus cinnabarinus strains for laccase production. FEMS Microbiology Letters 183: 301–306. [DOI] [PubMed] [Google Scholar]
- Hofrichter M, Fritsche W. (1997) Depolymerization of low-rank coal by extracellular fungal enzyme systems. III. In vitro depolymerization of coal humic acids by a crude preparation of manganese peroxidase of the white-rot fungus Nematoloma frowardii b19. Applied Microbiology and Biotechnology 47: 566–571. [Google Scholar]
- Hofrichter M, Ziegenhagen D, Vares T, Friedrich M, Jäger MG, et al (1998) Oxidative decomposition of malonic acid as basis for the action of manganese peroxidase in the absence of hydrogen peroxide. FEBS Letters 434: 362–366. [DOI] [PubMed] [Google Scholar]
- Illnait-Zaragozí MT, Martínez-Machín GF, Fernández-Andreu CM, Perurena-Lancha MR, Theelen B, et al (2012) Environmental isolation and characterisation of Cryptococcus species from living trees in Havana city, Cuba. Mycoses 55: e138–e144. [DOI] [PubMed] [Google Scholar]
- Kim KH, Kang YM, Im CH, Ali A, Kim SY, et al (2014) Identification and functional analysis of pheromone and receptor genes in the B3 mating locus of Pleurotus eryngii. PLoS One 9: e104693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe E. (1996) Tetrapolar fungal mating types: sexes by the thousands. FEMS Microbiology Reviews 18: 65–87. [DOI] [PubMed] [Google Scholar]
- Krings M, Dotzler N, Galtier J, Taylor TN. (2011) Oldest fossil basidiomycete clamp connections. Mycoscience 52: 18–23. [Google Scholar]
- Kuuskeri J, Häkkinen M, Laine P, Smolander OP, Tamene F, et al. (2016) Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: growth on spruce wood and decay effect on lignocellulose. Biotechnology for Biofuels 9: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leck A. (1999) Preparation of lactophenol cotton blue slide mounts. Community Eye Health 12: 24. [PMC free article] [PubMed] [Google Scholar]
- Li J, Sun F, Li X, Yan Z, Yuan Y, et al. (2012) Enhanced saccharification of corn straw pretreated by alkali combining crude ligninolytic enzymes. Journal of Chemical Technology & Biotechnology 87: 1687–1693. [Google Scholar]
- Lomascolo A, Cayol J-L, Roche M, Guo L, Robert J-L, et al. (2002) Molecular clustering of Pycnoporus strains from various geographic origins and isolation of monokaryotic strains for laccase hyperproduction. Mycological Research 106: 1193–1203. [Google Scholar]
- Mäkelä MR, Galkin S, Hatakka A, Lundell TK. (2002) Production of organic acids and oxalate decarboxylase in lignin-degrading white rot fungi. Enzyme and Microbial Technology 30: 542–549. [Google Scholar]
- Mäkelä MR, Hildén KS, de Vries RP. (2014) Degradation and modification of plant biomass by fungi. In: The Mycota. Vol. 13. Fungal Genomics (Nowrousian M. ed.): 175–208. 2nd edn Berlin: Springer-Verlag. [Google Scholar]
- Nagy LG, Riley R, Bergmann PJ, Krizsán K, Martin FM, et al. (2017) Genetic bases of fungal white rot wood decay predicted by phylogenomic analysis of correlated gene-phenotype evolution. Molecular Biology and Evolution 34: 35–44. [DOI] [PubMed] [Google Scholar]
- Nagy LG, Riley R, Tritt A, Adam C, Daum C, et al. (2016) Comparative genomics of early-diverging mushroom-forming fungi provides insights into the origins of lignocellulose decay capabilities. Molecular Biology and Evolution 33: 959–970. [DOI] [PubMed] [Google Scholar]
- Pandey VK, Singh MP. (2014) Biodegradation of wheat straw by Pleurotus ostreatus. Cellular and Molecular Biology 60: 29–34. [PubMed] [Google Scholar]
- Patyshakuliyeva A, Post H, Zhou M, Jurak E, Heck AJR, et al. (2015) Uncovering the abilities of Agaricus bisporus to degrade plant biomass throughout its life cycle. Environmental Microbiology 17: 3098–3109. [DOI] [PubMed] [Google Scholar]
- Pham TTT, Maaroufi A, Odier E. (1990) Inheritance of cellulose- and lignin- degrading ability as well as endoglucanase isozyme pattern in Dichomitus squalens. Applied Microbiology and Biotechnology 33: 99–104. [Google Scholar]
- Raudaskoski M, Kothe E. (2010) Basidiomycete mating type genes and pheromone signaling. Eukaryotic Cell 9: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, et al. (2014) Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proceedings of the National Academy of Sciences, USA 27: 9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytioja J, Hildén K, Di Falco M, Zhou M, Aguilar-Pontes MV, et al. (2017) The molecular response of the white-rot fungus Dichomitus squalens to wood and non-woody biomass as examined by transcriptome and exoproteome analyses. Environmental Microbiology. DoI: 10.1111/1462-2920.13652 [DOI] [PubMed] [Google Scholar]
- Rytioja J, Hildén KS, Yuzon J, Hatakka A, de Vries RP. et al. (2014) Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiology and Molecular Biology Reviews 78: 614–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo F, González AE, Terrón MC, Ramírez L, Pisabarro AG. (2008) Quantitative linkage mapping of lignin-degrading enzymatic activities in Pleurotus ostreatus. Enzyme and Microbial Technology 43: 137–143. [Google Scholar]
- Wyatt AM, Broda P. (1995) Informed strain improvement for lignin degradation by Phanerochaete chrysosporium. Microbiology 141: 2811–2822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.