Abstract
Recent studies using sequence data from eight sequence loci and coalescent-based species delimitation methods have revealed several species-level lineages of Tulasnella associated with the orchid genera Arthrochilus, Caleana, Chiloglottis, and Drakaea in Australia. Here we formally describe three of those species, Tulasnella prima, T. secunda, and T. warcupii spp. nov., as well as an additional Tulasnella species associated with Chiloglottis growing in Sphagnum, T. sphagneti sp. nov. Species were identified by phylogenetic analyses of the ITS with up to 1.3 % sequence divergence within taxa and a minimum of 7.6 % intraspecific divergence. These new Tulasnella (Tulasnellaceae, Cantharellales) species are currently only known from orchid hosts, with each fungal species showing a strong relationship with an orchid genus. In this study, T. prima and T. sphagneti associate with Chiloglottis, while T. secunda associates with Drakaea and Caleana, and T. warcupii associates with Arthrochilus oreophilus.
Keywords: host specificity, ITS sequencing, orchid mycorrhizas, species delimitation
INTRODUCTION
Tulasnella is a cosmopolitan saprotrophic fungal genus that often forms a mycorrhizal relationship with orchids. There are approximately 90 species epithets in Tulasnella (www.indexfungorum.org) with Kirk et al. (2008) indicating that there are approximately 50 accepted species in the genus. Asexual morphs of Tulasnella were formerly referred to in Epulorhiza. In earlier studies on the genus, Warcup and Talbot (Warcup & Talbot 1967, Warcup 1971, 1981) were able to induce formation of basidia and basidiospores from some Australian orchid-derived cultures by placing a casing of soil over cultures on agar. However, the spore-producing tissues were often slow to form and diffuse. Indeed, sporophores could only be detected by examination under a dissecting microscope. In some cases, such as in T. calospora (Warcup & Talbot 1967), only spores were visible above the casing soil surface. Unfortunately, subsequent studies on Tulasnella have not been able to generate basidiospore formation (Suarez et al. 2006, Cruz et al. 2011). For example, Ma et al. (2003) noted that “despite repeated attempts, none of the epulorhiza-like Rhizoctonia isolates produced hymenia or basidiospores on [various media] after two months”. Although Warcup and Talbot (Warcup & Talbot 1967, Warcup 1971, Warcup & Talbot 1980) utilized morphological characters of the sporophores (such as the size and shape of basidiospores) for taxonomic treatments of Tulasnella from orchids, recent studies on the group have mostly designated operational taxonomic units (OTUs) based solely on phylogenetic analysis of DNA sequence data. Indeed, numerous molecular OTUs have been designated amongst Tulasnella associated with orchids (e.g. Smith et al. 2010, Jacquemyn et al. 2011, 2012, Pandey M et al. 2013, Cruz et al. 2014, Oja et al. 2015) or liverworts (Kottke et al. 2003, Bidartondo & Duckett 2010), without formally naming the species. Formal naming of the species is preferred and essential to prevent confusion of taxonomic units discovered in separate studies (Hibbett & Taylor 2013).
Molecular OTUs within Tulasnella have been designated by two methods. First, application of a sequence divergence threshold for a barcode DNA region such as the ITS; with thresholds ranging from 3–5 % (Suarez et al. 2006, Cruz et al. 2014, Jacquemyn et al. 2014, 2015). Second, application of a multi-gene concordance analysis utilizing coalescent theory that explicitly incorporates gene tree conflicts into a model of phylogenetic history for the populations or species concerned (Yang & Rannala 2010, Fujita et al. 2012) and utilizing a number of independent DNA loci (Linde et al. 2014). The second approach is more rigorous for delimiting species (Taylor et al. 2000) and the similarity within and between species delimited with coalescence can be used to calibrate the cut-off threshold used in the first method.
A study of Tulasnella isolates from Australian terrestrial orchids (Orchidaceae, tribe Diurideae, subtribe Drakaeinae) in the genera Arthrochilus, Chiloglottis, Drakaea, and Paracaleana (Linde et al. 2014), using eight loci analysed by a variety of methods (including phylogenies of individual loci, Bayesian coalescent based species delimitation, and population structure analysis) revealed five phylogenetic species: one associated with Chiloglottis, one with Drakaea and Paracaleana, and three with Arthrochilus (among which one was known from one isolate and another from two isolates). Analysis of the ITS alone recovered the same five phylogenetic species as well-separated and well-supported clades, revealing congruence between the widely used ITS region and the more extensive multi-locus analysis (Linde et al. 2014). The phylogenetic species were not formally named in Linde et al. (2014).
Many of the orchid species associated with Tulasnella are rare or endangered (Hopper & Brown 2006), and the association between orchid and fungus has been and continues to be the subject of much research in Australia (Smith et al. 2010) and elsewhere (McCormick & Jacquemyn 2014). For Tulasnella associated with orchids identification by use of sequences is now the norm, rather than using cultural characters or features of the sporophore. It is therefore appropriate to supply formal names to three of the phylogenetic species (each known from more than two strains) already characterised on sequence data by Linde et al. (2014), along with a further phylogenetic species isolated from Chiloglottis associated with Sphagnum. After assessing information on Tulasnella from Australia, to determine if prior names exist for phylogenetic lineages, we describe four new species of Tulasnella here: T. prima and T. sphagneti spp. nov. from Chiloglottis, T. secunda sp. nov. from Drakaea and Caleana (inclusive of Paracaleana), and T. warcupii sp. nov. from Arthrochilus oreophilus.
MATERIALS AND METHODS
Fungal collections
Taxonomy of the orchid genera, which are all members of the subtribe Drakaeinae, follows Miller & Clements (2014), who accepted the genera Arthrochilus, Caleana (inclusive of Paracaleana), Chiloglottis (inclusive of Simpliglottis), and Drakaea. Tulasnella mycorrhizal associations as identified from previous studies on associations with Australian terrestrial orchids in Arthrochilus, Caleana (as Paracaleana), Chiloglottis and Drakaea (Roche et al. 2010, Phillips et al. 2011, Linde et al. 2014, Phillips et al. 2014), were investigated. Additionally, we treat a Tulasnella isolated from Chiloglottis aff. valida and C. turfosa growing in Sphagnum hummocks within the Kosciuszko National Park, New South Wales (Table 1). Some Chiloglottis orchids growing in Sphagnum were not in flower at the time of collection, and are thus referred to as “Chiloglottis sp.” However, based on previous studies the Chiloglottis species involved are either C. aff. valida, C. valida, or C. turfosa (Peakall et al. 2010, Peakall & Whitehead 2014). Literature on Tulasnella from Australia was reviewed, and this literature along with GenBank and culture collection databases: CBS (CBS-KNAW Fungal Biodiversity Centre culture collection) and MAFF (culture collection, Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Ibaraki, Japan, searched via NIAS [National Institute of Agro-Environmental Sciences] Genebank - http://www.gene.affrc.go.jp/databases-micro_search_en.ph) were searched for isolates of Tulasnella from Australia (Tables 2 and 3).
Table 1.
Tulasnella isolates examined in this study.
| Identity | Isolate no. | Type | Host | GPS, if known | Origin | Habitat | Collectors* | GenBank accession no. | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Tulasnella sphagneti | 12033 (CLM541) | Holotype | Chiloglottis aff. valida | 35.5238S, 148.3656E | Kosciuszko NP, NSW | Alpine Sphagnum hammocks | YT | KY095117 | This study |
| 12030 (CLM583) | Chiloglottis turfosa | 35.5346S, 148.3225E | Kosciuszko NP, NSW | Alpine Sphagnum hammocks | YT | KY445922 | This study | ||
| 13058 | Chiloglottis sp. | 35.5544S, 148.3528E | Kosciuszko NP, NSW | Alpine Sphagnum hammocks | YT | KY445927 | This study | ||
| 13065_1 | Chiloglottis sp. | 35.5544S, 148.3528E | Kosciuszko NP, NSW | Alpine Sphagnum hammocks | YT | KY445926 | This study | ||
| 13065_2 | Chiloglottis sp. | 35.5544S, 148.3528E | Kosciuszko NP, NSW | Alpine Sphagnum hammocks | YT | KY445925 | This study | ||
| 13102_1 | Chiloglottis turfosa | 35.5346S, 148.3225E | Kosciuszko NP, NSW | Alpine Sphagnum hammocks | YT | KY445924 | This study | ||
| 13102_2 | Chiloglottis turfosa | 35.5346S, 148.3225E | Kosciuszko NP, NSW | Alpine Sphagnum hammocks | YT | KY445923 | This study | ||
| 13139 | Chiloglottis sp. | 35.5336S, 148.2647E | Kosciuszko NP, NSW | Alpine Sphagnum hammocks | YT | KY445928 | This study | ||
| 13143 | Chiloglottis sp. | 35.5336S, 148.2647E | Kosciuszko NP, NSW | Alpine Sphagnum hammocks | YT | KY445929 | This study | ||
| Tulasnella prima | CLM159 | Holotype | Chiloglottis trilabra | 34.1385S, 149.9722E | Blue Mountains, NSW | Eucalyptus woodland | RP | KF476556 | (Roche et al. 2010) |
| 07033-45.II.2 | Chiloglottis seminuda | 34.6295S, 150.1539E | Exeter, NSW | Eucalyptus woodland | RP | HM196800 | (Roche et al. 2010) | ||
| CLM306 | Chiloglottis formicifera | 34.6537S, 150.6016E | Upper Kangaroo Valley, NSW | Eucalyptus woodland | RP | KF476550 | (Roche et al. 2010) | ||
| CLM308 | Chiloglottis formicifera | 34.6537S, 150.6016E | Upper Kangaroo Valley, NSW | Eucalyptus woodland | RP | KF476551 | (Roche et al. 2010) | ||
| CLM309 | Chiloglottis formicifera | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | KF476543 | (Roche et al. 2010) | ||
| CLM310.1 | Chiloglottis aff. jeanesii | 35.5056S, 149.5351E | Tallaganda State Forest, NSW | Eucalyptus woodland | RP | HM196792 | (Roche et al. 2010) | ||
| CLM310.2 | Chiloglottis aff. jeanesii | 35.5056S, 149.5351E | Tallaganda State Forest, NSW | Eucalyptus woodland | RP | HM196791 | (Roche et al. 2010) | ||
| CLM316.1 | Chiloglottis seminuda | 34.6295S, 150.1539E | Exeter, NSW | Eucalyptus woodland | RP | HM196797 | (Roche et al. 2010) | ||
| CLM316.2 | Chiloglottis seminuda | 34.6295S, 150.1539E | Exeter, NSW | Eucalyptus woodland | RP | HM196798 | (Roche et al. 2010) | ||
| CLM346 | Chiloglottis reflexa | 33.5211S, 150.3707E | Mt Wilson, NSW | Eucalyptus woodland | RP | HM196805 | (Roche et al. 2010) | ||
| CLM361 | Chiloglottis diphylla | 33.5154S, 150.4886E | Bilpin, NSW | Eucalyptus woodland | RP | HM196803 | (Roche et al. 2010) | ||
| CLM366 | Chiloglottis trapeziformis | 35.2749S, 149.0976E | Black Mountain, ACT | Eucalyptus woodland | CCL | HM196794 | (Roche et al. 2010) | ||
| CLM371 | Chiloglottis trapeziformis | 35.2749S, 149.0976E | Black Mountain, ACT | Eucalyptus woodland | CCL | HM196799 | (Roche et al. 2010) | ||
| CLM372 | Chiloglottis trapeziformis | 35.2749S, 149.0976E | Black Mountain, ACT | Eucalyptus woodland | CCL | HM196789 | (Roche et al. 2010) | ||
| CLM377 | Chiloglottis aff. jeanesii | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196782 | (Roche et al. 2010) | ||
| CLM380.1 | Chiloglottis aff. jeanesii | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196779 | (Roche et al. 2010) | ||
| CLM380.2 | Chiloglottis aff. jeanesii | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196788 | (Roche et al. 2010) | ||
| CLM381.1 | Chiloglottis aff. jeanesii | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196783 | (Roche et al. 2010) | ||
| CLM381.2 | Chiloglottis aff. jeanesii | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196784 | (Roche et al. 2010) | ||
| CLM388 | Chiloglottis aff. jeanesii | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196787 | (Roche et al. 2010) | ||
| CLM389 | Chiloglottis aff. jeanesii | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196795 | (Roche et al. 2010) | ||
| CLM390 | Chiloglottis aff. jeanesii | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196785 | (Roche et al. 2010) | ||
| CLM391 | Chiloglottis aff. jeanesii | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196786 | (Roche et al. 2010) | ||
| CLM393 | Chiloglottis valida | 35.5056S, 149.5351E | Tallaganda State Forest, NSW | Eucalyptus woodland | RP | HM196804 | (Roche et al. 2010) | ||
| CLM395 | Chiloglottis valida | 33.9409S, 150.0552E | Kanangra Boyd NP, NSW | Eucalyptus woodland | RP | HM196801 | (Roche et al. 2010) | ||
| CLM405 | Chiloglottis trapeziformis | 35.2751S, 149.1097E | ANBG, ACT | Eucalyptus woodland | SR | HM196796 | (Roche et al. 2010) | ||
| SRBG01.II.3 | Chiloglottis trapeziformis | 35.2751S, 149.1097E | ANBG, ACT | Eucalyptus woodland | SR | HM196790 | (Roche et al. 2010) | ||
| CLM407 | Chiloglottis trapeziformis | 35.2751S, 149.1097E | ANBG, ACT | Eucalyptus woodland | SR | HM196793 | (Roche et al. 2010) | ||
| CLM416 | Chiloglottis trapeziformis | 35.2749S, 149.0976E | Black Mountain, ACT | Eucalyptus woodland | SR | HM196809 | (Roche et al. 2010) | ||
| SRBG03.I.8 | Chiloglottis trapeziformis | 35.2751S, 149.1097E | ANBG, ACT | Eucalyptus woodland | SR | HM196807 | (Roche et al. 2010) | ||
| SRBG03.III.12 | Chiloglottis trapeziformis | 35.2751S, 149.1097E | ANBG, ACT | Eucalyptus woodland | SR | HM196810 | (Roche et al. 2010) | ||
| SRBG03.IV.5 | Chiloglottis trapeziformis | 35.2751S, 149.1097E | ANBG, ACT | Eucalyptus woodland | SR | HM196806 | (Roche et al. 2010) | ||
| CLM068 | Chiloglottis diphylla | 33.5154S, 150.4886E | Blue Mountains, NSW | Eucalyptus woodland | RP | KF476552 | (Roche et al. 2010) | ||
| Tulasnella secunda | CLM009 | Holotype | Drakaea elastica | NA | Karnup, WA | Kunzea woodland | RDP | KF476575 | (Linde et al. 2014) |
| CLM222 | Paracaleana minor | 35.2690S 149.0920E | Black Mountain, ACT | Eucalyptus woodland | CCL | KF476568 | (Linde et al. 2014) | ||
| CLM251 | Drakaea concolor | NA | Mt Gregory, WA | Sandplain heath | RDP | KF476588 | (Linde et al. 2014) | ||
| CLM253 | Drakaea confluens | NA | Lake Gnartaminny, WA | Jarrah forest | RDP | KF476592 | (Linde et al. 2014) | ||
| CLM255 | Drakaea livida | NA | Walpole WA | Jarrah forest | RDP | KF476590 | (Linde et al. 2014) | ||
| CLM257 | Drakaea glyptodon | NA | Moore River, WA | Banksia woodland | RDP | KF476583 | (Linde et al. 2014) | ||
| CLM258 | Drakaea glyptodon | NA | Margaret River, WA | Eucalyptus woodland | RDP | KF476586 | (Linde et al. 2014) | ||
| CLM259 | Drakaea glyptodon | NA | Ruabon, WA | Kunzea woodland | RDP | KF476577 | (Linde et al. 2014) | ||
| CLM260 | Drakaea elastica | NA | Nambeelup, WA | Kunzea woodland | RDP | KF476593 | (Linde et al. 2014) | ||
| CLM261 | Drakaea gracilis | NA | Westdale, WA | Eucalyptus woodland | RDP | KF476591 | (Linde et al. 2014) | ||
| CLM266 | Drakaea confluens | NA | Lake Gnartaminny, WA | Jarrah forest | RDP | KF476579 | (Linde et al. 2014) | ||
| CLM267 | Paracaleana hortiorum | NA | Talbot West, WA | Eucalyptus woodland | RDP | KF476584 | (Linde et al. 2014) | ||
| CLM268 | Paracaleana terminalis | NA | Mt Gregory, WA | Sandplain heath | RDP | KF476574 | (Linde et al. 2014) | ||
| CLM272 | Paracaleana lyonsii | NA | Eurardy, WA | Sandplain heath | RDP | KF476573 | (Linde et al. 2014) | ||
| CLM273 | Drakaea livida | NA | Canning Mills, WA | Jarrah forest | RDP | KF476576 | (Linde et al. 2014) | ||
| CLM274 | Paracaleana triens | NA | Talbot West, WA | Kunzea woodland | RDP | KF476580 | (Linde et al. 2014) | ||
| CLM276 | Drakaea isolata | NA | Lake Chinocup, WA | Mallee woodland | RDP | KF476585 | (Linde et al. 2014) | ||
| CLM277 | Drakaea gracilis | NA | Westdale, WA | Eucalyptus woodland | RDP | KF476578 | (Linde et al. 2014) | ||
| I3DLIsl6 | Drakaea livida | NA | Walpole, WA | Jarrah forest | RDP | HQ386778 | (Phillips et al. 2011) | ||
| B9DGDen1 | Drakaea glyptodon | NA | Denbarker WA | Jarrah forest | RDP | HQ386743 | (Phillips et al. 2011) | ||
| JS4 | Drakaea glyptodon | NA | Southern WA | JX138567 | (Sommer et al. 2012) | ||||
| Tulasnella warcupii | CLM027 | Holotype | Arthrochilus oreophilus | 17.2697S, 145.4535E | Atherton Tablelands, Qld | Eucalyptus woodland | DG | KF476596 | (Linde et al. 2014) |
| CLM007 | Arthrochilus oreophilus | 17.3402S, 145.4210E | Atherton Tablelands, Qld | Eucalyptus woodland | DG | KF476600 | (Linde et al. 2014) | ||
| CLM022 | Arthrochilus oreophilus | 17.3402S, 145.4210E | Atherton Tablelands, Qld | Eucalyptus woodland | DG | KF476601 | (Linde et al. 2014) | ||
| CLM028 | Arthrochilus oreophilus | 17.3402S, 145.4210E | Atherton Tablelands, Qld | Eucalyptus woodland | DG | KF476599 | (Linde et al. 2014) | ||
| CLM091 | Arthrochilus oreophilus | 17.2697S, 145.4535E | Atherton Tablelands, Qld | Eucalyptus woodland | DG | KF476598 | (Linde et al. 2014) | ||
| CLM092 | Arthrochilus oreophilus | 17.3402S, 145.4210E | Atherton Tablelands, Qld | Eucalyptus woodland | DG | KF476597 | (Linde et al. 2014) | ||
| (Linde et al. 2014) | |||||||||
| Unassigned | CLM084 | Arthrochilus oreophilus | 17.3311E 145.4131S | Atherton Tablelands, Qld | Eucalyptus woodland | DG | KF476594 | (Linde et al. 2014) | |
| Unassigned | CLM085 | Arthrochilus oreophilus | 17.3311E 145.4131S | Atherton Tablelands, Qld | Eucalyptus woodland | DG | KF476595 | (Linde et al. 2014) | |
| Unassigned | CLM031 | Arthrochilus oreophilus | 17.3311E 145.4131S | Atherton Tablelands, Qld | Eucalyptus woodland | DG | KF476602 | (Linde et al. 2014) | |
| Unassigned | BB0002_2_A | Terrestrial orchid | Australia | Howard,C.G. and Clements,M.A. | JN015192 | Unpublished |
ANBG = Australian National Botanic Gardens; NP = National Park.
*CCL = Celeste Linde; YT = Yann Triponez; RP = Rod Peakall; RDP = Ryan Phillips; DG = Don Gomez.
Table 2.
Isolates of Tulasnella (some as Tulasnellaceae or Epulorhiza) from Australian orchids, additional to those analysed from orchid hosts in Drakaeinae by Roche et al. (2010) and Linde et al. (2014). All isolates collected by Warcup currently present in culture collections are shown, along with all isolates from which sequences have been obtained. Orchid hosts in Drakaeinae are in bold. Note that AY643803 derived from isolate PN1 from Pterosylis nutans is given in GenBank as “asexual morph: Epulorhiza repens”, but Bougoure et al. (2005) considered the isolate was most likely a Thanatephorus (and it is therefore omitted below).
| Tulasnella species | Host | Original isolate | Reference for original isolate | Culture collection strain | Sequence ITS* | Sequence LSU | Reference for sequence |
|---|---|---|---|---|---|---|---|
| T. asymmetrica | Thelymitra nuda | JH Warcup 0267 | (Warcup 1973) | MAFF 305807 AFTOL ID 1678 | DQ520101 | Garnica & Weiss (unpub.) | |
| T. asymmetrica | Thelymitra luteocilium | JH Warcup 085 | (Warcup & Talbot 1967) | MAFF 305806 (ex-type) | DQ388046 | (Suarez et al. 2006) | |
| KC152339–44 [clones c001–c006] | (Cruz et al. 2014) | ||||||
| T. asymmetrica | Thelymitra epipactoides | JH Warcup 0302 | (Warcup 1973) | MAFF305808 | DQ388047 | (Suarez et al. 2006) | |
| KC152347–49,51,52,56 [clones c001– c005, c009] | (Cruz et al. 2014) | ||||||
| T. asymmetrica | Thelymitra epipactoides | JH Warcup 0591 | (Warcup 1973) | NIAES 5809 | (Bidartondo et al. 2003) | ||
| MAFF P305809 = NIAES 5809 | DQ388048 | (Suarez et al. 2006) | |||||
| NIAES 5809 | KC152345–46, KC152350, KC152353–55, [clones c001, c003, c005, c008–c010] | (Cruz et al. 2014) | |||||
| T. calospora | Acianthus exsertus | JH Warcup 07 | (Warcup & Talbot 1967) | MAFF305801 | no sequences | ||
| T. calospora | Diuris maculata | JH Warcup 0388 | (Warcup 1973) | MAFF305802 | no sequences | ||
| T. calospora | Thelymitra aristata | JH Warcup 0584 | (Warcup 1973) | MAFF305803 | no sequences | ||
| T. calospora | Thelymitra sp. | JH Warcup 0638 | (Warcup 1973) | MAFF305804 | no sequences | ||
| T. calospora | host not specified | JH Warcup 0689 | (Warcup 1973) | MAFF305805 | no sequences | ||
| T. calospora | Caladenia reticulata | JH Warcup 062 | CBS 573.83 | AY243521 | (Taylor et al. 2003) | ||
| T. irregularis | Dendrobium dicuphum | JH Warcup 0632 | CBS 574.83 [ex type] = JCM 9996 | AF345560 | (Kristiansen et al. 2001) | ||
| AY243519 | (Taylor et al. 2003) | ||||||
| T. calospora | Microtis parviflora | TM1 | (Perkins et al. 1995) | AY643804 | (Bougoure et al. 2005) | ||
| Tulasnella sp. | [presume from orchid] | JT Otero 306 | DQ061110 | Otero (unpub.) | |||
| Tulasnella sp. | [presume from orchid] | JT Otero 307 | DQ061111 | Otero (unpub.) | |||
| Epulorhiza ‘possibly’ [in GenBank as ‘Fungi’] | Acianthus pusillus | AP2 | AY643806 | (Bougoure et al. 2005) | |||
| Epulorhiza sp. | Diuris corymbosa | Kings_Park_Dm01 | EF160068 | (Bonnardeaux et al. 2007) | |||
| Epulorhiza sp. | Prasophyllum giganteum | Kings_Park_Pg01 | EF160067 | (Bonnardeaux et al. 2007) | |||
| Epulorhiza sp. | Pyrorchis nigricans | 7 isolates | EF176464–66, 69–72 | (Bonnardeaux et al. 2007) | |||
| Epulorhiza sp. | Disa bracteata | 11 isolates | EF176473–77, 79–83, 85 | (Bonnardeaux et al. 2007) | |||
| Tulasnella sp. | ‘terrestrial orchid’ | BB0002_2_A | JN015192 | Howard & Clements (unpub.) | |||
| T. calospora | Diuris magnifica | DR88 | KT601561 | Davis et al. (unpub) | |||
| T. calospora | Disa bracteata | DR28 | KT601562 | Davis et al. (unpub) | |||
| T. calospora | Microtis media | DR126 | KT601563 | Davis et al. (unpub) | |||
| Tulasnellaceae sp. RP-2011 | Drakaea elastica, D. glyptodon, D. livida, D. micrantha, D. thynniphila | 50 isolates | HQ386734–83 | (Phillips et al. 2011) | |||
| Tulasnellaceae sp. 1–3 | Thelymitra macrophylla | JP15, JP44, JP49 | JX138557–59 | (Sommer et al. 2012) | |||
| Tulasnellaceae sp. 4–5 | Disa bracteata | JP24, JP26 | JX138560–61 | (Sommer et al. 2012) | |||
| Tulasnellaceae sp. 6–7 | Pyrorchis nigricans | JP33, JP37 | JX138562–63 | (Sommer et al. 2012) | |||
| Tulasnellaceae sp. 8–9 | Diuris magnifica | JP40, JP60 | JX138564–65 | (Sommer et al. 2012) | |||
| Tulasnellaceae sp. 10 | Microtis sp. | JP63 | JX138566 | (Sommer et al. 2012) | |||
| Tulasnellaceae sp. 11 | Drakaea glyptodon | JS4 | JX138567 | (Sommer et al. 2012) | |||
| Tulasnellaceae sp. 12 | Spiculaea ciliata | JS43 | JX138568 | (Sommer et al. 2012) | |||
| Tulasnellaceae sp. 13 | Lyperanthus serratus | JS64 | JX138569 | (Sommer et al. 2012) | |||
| Tulasnellaceae sp. 14 | Microtis capularis | JS66, JS68 | JX138570–71 | (Sommer et al. 2012) | |||
| Tulasnellaceae sp. 16 | Microtis media | JS163 | JX138572 | (Sommer et al. 2012) | |||
| Tulasnella sp. [as ‘Uncultured mycorrhizal fungus’] | Diuris fragrantissima | 30 isolates | DQ790719–38,86-95 | DQ790751–60,84-85 | (Smith et al. 2010) | ||
| Tulasnella sp. [as ‘Uncultured mycorrhizal fungus’] | Diuris punctata | 7 isolates | DQ790798 | DQ790763,65, 72, 77,79,82,98 | (Smith et al. 2010) | ||
| Tulasnella sp. [as ‘Uncultured mycorrhizal fungus’] | Diuris punctata var. daltonii | 2 isolates | DQ790804,08 | DQ790769,73 | (Smith et al. 2010) | ||
| Tulasnella sp. [as ‘Uncultured mycorrhizal fungus’] | Diuris dendrobioides | 1 isolate | DQ790802 | DQ790767 | (Smith et al. 2010)) | ||
| Tulasnella sp. [as ‘Uncultured mycorrhizal fungus’] | Diuris chryeopsis | 3 isolates | DQ790796,99 | DQ790761,64, 80 | (Smith et al. 2010) | ||
| DQ790815 |
*some sequences are ITS plus partial LSU.
Table 3.
Tulasnella species isolated from Australian orchids by JH Warcup and PHB Talbot and other studies. Sporophore morphology was from sporophores (i.e. the “perfect state”) initiated from cultures. Tulasnella species in bold were newly described by Warcup and Talbot. Orchid genera in Drakaeinae are given in bold. References do not include publications where original isolates of Warcup were later sequenced (as cited in Table 2). Unpublished observations derive from sequences in GenBank as detailed in Table 2.
| Tulasnella species | Orchid genera | Identification method | Comments | References |
|---|---|---|---|---|
| T. allantospora | Chiloglottis, Corybas | sporophore morphology | (Warcup & Talbot 1971, Warcup 1981) | |
| T. asymmetrica | Chiloglottis, Cryptostylis, Dendrobium, Thelymitra | sporophore morphology | also as Tulasnella sp., isolate 086 (Warcup & Talbot 1967); see Warcup & Talbot (1971) | (Warcup & Talbot 1967, 1971, Warcup 1973, 1981) |
| T. calospora | Acianthus, Caladenia, Corybas, Cymbidium, Dendrobium, Diuris, Eriochilus, Lyperanthus, Microtis, Orthoceras, Thelymitra | sporophore morphology | (Warcup & Talbot 1967, Warcup 1971, 1973, 1981, 1990) | |
| Disa, Diuris, Microtis | sequence | (Bougoure et al. 2005) | ||
| T. cruciata | Acianthus, Chiloglottis, Thelymitra | sporophore morphology | (Warcup & Talbot 1971, Warcup 1973, 1981, 1990) | |
| T. deliquescens [as Epulorhiza repens] | Acianthus, Microtis | culture morphology | (Perkins et al. 1995) | |
| T. irregularis | Dendrobium | sporophore morphology | also as Tulasnella sp. 1, isolate 0632 (Warcup 1973); see Warcup & Talbot (1980) | (Warcup & Talbot 1980, Warcup 1981) |
| T. violea | Drakaea | culture morphology | (Warcup 1981) | |
| Thelymitra | sporophore morphology | (Warcup & Talbot 1971, Warcup 1973, 1990) | ||
| Tulasnella sp. (some as Epulorhiza sp. or Tulasnellaceae sp.) | Arthrochilus, Caladenia, Caleana, Calochilus, Chiloglottis, Corybas, Cryptostylis, Cymbidium, Dendrobium, Dipodium, Drakaea, Microtis, Thelymitra | culture morphology | “culturally seem Tulasnella, perfect states not seen” | Warcup 1973, 1981, 1990, Perkins & McGee 1995, Perkins et al. 1995) |
| Acianthus, Caleana (as Paracaleana), Disa, Diuris, Drakaea, Lyperanthus, Microtis, Prasophyllum, Pyrorchis, Spiculaea, Thelymitra | sequence | (Bougoure et al. 2005, Bonnardeaux et al. 2007, Smith et al. 2010, Phillips et al. 2011, Sommer et al. 2012) |
Fungal isolation
Isolations were made within 7 d of the field collection of the plant tissue using a modified version of the protocol of Roche et al. (2010). We used two types of isolation media to grow mycorrhizal isolates: Fungal Isolation Media (FIM; Clements & Ellyard 1979) and 3MN+A-Z, which is a Melin-Norkrans medium (low CN MMN) (Wright et al. 2010) modified with 15g/L agar and human vitamin and mineral supplements (Centrum “Balanced Formula”, Wyeth Consumer Healthcare, Baulkham Hills, NSW, Australia) instead of thiamine. One Centrum tablet was dissolved in 100 mL water, filter sterilised, and 10 mL added per litre of 3 MN medium (post autoclaving). Peloton-rich tissues (collars) of orchids were washed several times with sterile distilled water after which the tissue was macerated in sterile distilled water to release pelotons, which were plated onto agar plates containing antibiotics (FIM + tetracycline 25 mg/mL, and 3MN+A-Z + streptomycin 50 mg/mL). Germinating pelotons were transferred to either FIM or 3MN+A-Z media after 3–10 d. The medium chosen depended on which the pelotons germinated. After 3–4 wk all colonies were hyphal-tipped and subcultured to ensure colonies consisted of a single genotype. Cultures were stored at 5 °C on FIM or 3MN+A-Z agar slants covered with mineral oil. Voucher specimens of the fungi, as dried-down liquid cultures, are lodged at the National Herbarium of Victoria (MEL) and ex-type cultures are stored in the culture collection of the Department of Agriculture, Victoria (VPRI).
DNA extraction, sequencing and phylogenetic analysis
Small agar blocks cut from colony edges of isolates were briefly homogenised in 2 mL screw-cap tubes containing sterilise distilled water and glass beads. The blocks were homogenised in a FP120 (Thermo Scientific, Milford, MA) homogenizer for 5 s at 5.5 m/s. Petri dishes containing either half strength FIM or 3MN+A-Z broths were inoculated with the homogenised agar blocks and incubated at room temperature (approximately 23 °C) in the dark. Mycelium was harvested, stored at -4 °C, and lyophilized prior to DNA extraction. DNA extractions of the lyophilized-mycelium were performed using Qiagen DNeasy Plant Mini Kit or DNeasy 96 Plant Kit according to the manufacturer’s instructions (Amersham Biosciences, Hilden, Germany).
As previously noted, in a comprehensive evaluation of eight nuclear and mitochondrial loci, Linde et al. (2014) sequenced Tulasnella isolates from orchids in the genera Arthrochilus, Caleana (as Paracaleana), Chiloglottis, and Drakaea. That study showed that within Tulasnella a single locus, ITS (nucR ITS), revealed congruent species delimitation and phylogenetic outcomes. Therefore, for the phylogenetic analysis of additional Tulasnella isolates from Chiloglottis, we only employed ITS. ITS sequences were amplified with the primers ITS1 and ITS4 (White et al. 1990) following methods described in Roche et al. (2010) for the PCR reaction, thermal cycling, purification of PCR and extension products. Products were sequenced bi-directionally with ABI PRISM BigDye Terminator v. 3.1 sequencing kit (Applied Biosystems, Foster City, CA) on an ABI-3130 automated sequencer. Sequences were edited using the program Sequencher v. 4.7 (GeneCodes, Ann Arbor, MI to correct for base read ambiguities. Our sequences were aligned with the most similar sequences available from GenBank (http://www.ncbi.nlm.nih.gov). Alignments were performed in Geneious v. 8 (http://www.geneious.com; Kearse et al. 2012) using ClustalW followed by manual adjustments to optimise indel locations.
Sequences for phylogenetic analysis included representatives of species-level clades in one of the two main subclades of phylogenetic group IV of the phylogeny of Tulasnella constructed by Cruz et al. (2011). This subclade contains Tulasnella sp. ECU 6 and T. eichleriana. To these sequences were added a selection of previously sequenced isolates from Australian orchids (Table 1) representing the phylogenetic breadth of the OTUs identified by Linde et al. (2014) along with new sequences from Chiloglottis associated with Sphagnum (Table 1). BLAST matches were carried out for representative sequences of putative OTUs from Australian orchids from our analysis to recover related sequences in GenBank. Phylogenies were estimated using Bayesian inference with MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003) and maximum likelihood (ML) analysis through the RAxML Blackbox (Stamatakis et al. 2008). Support for the nodes was assessed with Bayesian Posterior Probabilities (BPP) in MrBayes and for ML trees using 1000 pseudoreplicates of nonparametric bootstrapping. A GTR+G substitution model was used for all analyses as other models are nested within these. Trees were visualised using FigTree v. 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) and rooted to Tulasnella eichleriana sequences. Trees include identical sequences from different isolates; however the identical sequences were removed when nodes support was assessed. Pairwise sequence divergence of the ITS sequences within and among lineages were estimated with the Kimura-2-parameter distances with gap deletion in MEGA5 (Tamura et al. 2011).
RESULTS
Tulasnella species from Australian orchids
In placing formal names on phylogenetic species of Tulasnella, it is necessary to consider any names from previous work on the genus. Essentially, type specimens (that anchor names) need to be placed into the phylogenetic framework. However, given the lack of diagnostic morphological characters for recently isolated strains, a significant issue is whether types or suitable reference material exists and if sequence data are available for that material. Three species of Tulasnella have been described from Australian orchids: T. cruciata, originally from Acianthus and Dendrobium; T. irregularis from Dendrobium; and T. asymmetrica originally from Thelymitra. May et al. (2003) collated records of Tulasnella from all sources from Australia, including reports of a further four species: Tulasnella allantospora, T. calospora, T. deliquescens, and T. violea. According to Roberts (1994), T. asymmetrica was morphologically indistinguishable from T. pinicola, and was listed by Roberts (1999) as a synonym of the latter species. Furthermore, Roberts (1999) noted that the Australian report of T. allantospora by Warcup & Talbot (1971) was possibly misidentified, and might represent T. rubropallens; and T. calospora in the sense of Warcup & Talbot (1967) was deemed to be T. deliquescens. In making redispositions of Australian Tulasnella names, Roberts (1999) noted that he had not examined type material or voucher collections for reports by Warcup & Talbot (1967, 1971) of T. allantospora and T. calospora. Indeed, type material of T. cruciata or T. irregularis could not be located in ADW (Roberts 1999), and although the type of T. asymmetrica is listed by Roberts (1999) as housed at ADW, it was not examined. Warcup’s collections were originally in ADW and subsequently transferred to AD (macrofungi) and DAR (microfungi). There are ex-type cultures of T. asymmetrica (Warcup 085, MAFF 305806) and T. irregularis (Warcup 0632, CBS 574.83 = JCM 9996), but apparently none of T. cruciata.
Tulasnella isolates have been obtained from 21 terrestrial orchid genera and one lithophytic/epiphytic orchid genus (Dendrobium) in Australia (Table 3) (Warcup & Talbot 1967, 1971, 1980, Warcup 1971, 1973, 1981, 1990). For the genera Arthrochilus, Caleana (or from Paracaleana, under which name Caleana was formerly placed), Chiloglottis, and Drakaea that were the source of the apparently novel phylogenetic species delimited by Linde et al. (2014), the only previous reports are of unidentified Tulasnella isolates. An exception is a report of Tulasnella violea from Drakaea, identified only from cultural characteristics (Warcup 1981). However, for Chiloglottis there are reports of T. allantospora, T. asymmetrica, T. cruciata and also an unidentified species (Warcup 1973, 1981). For the two Tulasnella species described from Australia (T. asymmetrica and T. cruciata), the types are from other orchid genera, and the isolates of these two species from Chiloglottis were collected after the species were described.
Phylogenetic analysis of isolates from Arthrochilus, Caleana, Chiloglottis, and Drakaea
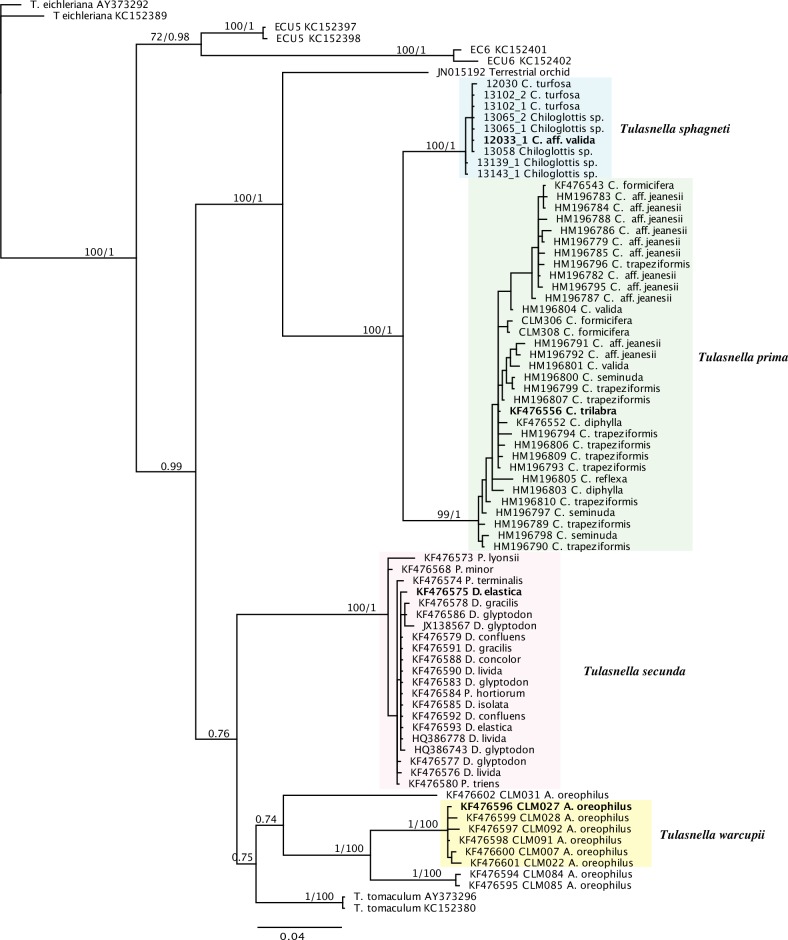
GenBank BLAST searches using query ITS sequences of Tulasnella isolates in this study revealed these sequences were related to T. eichleriana, T. tomaculum, and two Tulasnella lineages (ECU 5 and ECU 6) isolated from decaying branches in Ecuador. Representative sequences of these four lineages were added to the 72 fungal sequences from the Australian orchid genera Arthrochilus, Chiloglottis, Drakaea, and Caleana. The resulting phylogram show high bootstrap (100 %) and posterior probability (1) support for the three phylogenetic species of Tulasnella from (a) Arthrochilus oreophilus, (b) Chiloglottis, and (c) Drakaea and Caleana, that had previously been delimited on multi-gene data by Linde et al. (2014). Further sequences from Chiloglottis (exclusively associated with Chiloglottis growing in Sphagnum) formed a well-separated clade, sister to the other sequences from Chiloglottis (Fig. 1). ITS sequences from the ex-type culture of T. asymmetrica fall outside of the clades depicted in Fig. 1, as do all other sequences from Australian orchids (data not shown) with the exception of HQ386778, HQ386743 and JX138567 from Drakaea, that fell within the clade of isolates from Drakaea and Caleana and JN015192 from an Australian terrestrial orchid that is sister to the two clades consisting of isolates from Chiloglottis. A number of additional sequences from Drakaea (Phillips et al. 2011) all cluster within the clade from Drakaea with 100% bootstrap support. Those matching sequences were subsequently excluded from the final analysis. The LSU sequence from the ex-type culture of T. irregularis is only distantly related to LSU sequences from Australian isolates of Tulasnella from Arthrochilus, Chiloglottis, Drakaea, and Caleana (data not shown).
Fig. 1.
Rooted MrBayes tree for Tulasnella obtained for ITS. The tree with the highest log likelihood is shown. The numbers above the branches are maximum likelihood bootstrap values/Bayesian posterior probabilities. Bootstrap values of ≥ 70 % and Bayesian posterior probabilities of ≥ 0.70 are shown. The branch length is proportional to the inferred divergence level. Host from which the Tulasnella isolate was collected from is indicated after the isolate number or GenBank number. Sequences from the holotype of each species is indicated in bold.
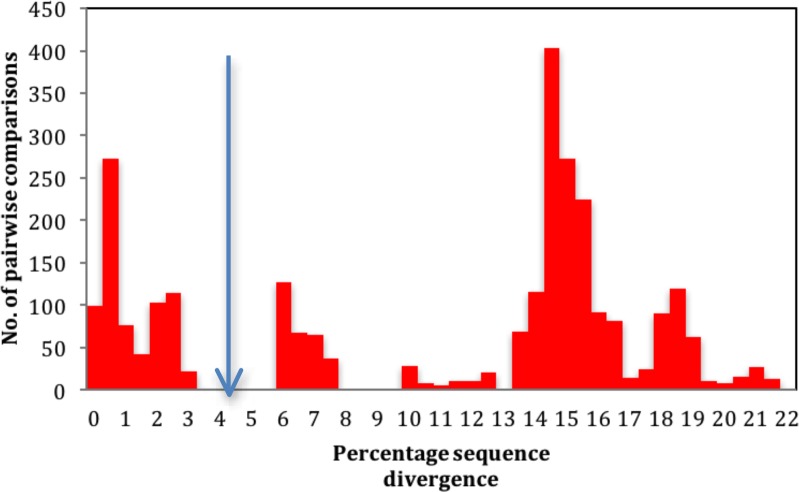
The percentage sequence divergence between the two lineages from Chiloglottis was 6.3 %. Sequence divergence between all other Australian Tulasnella lineages and close relatives ranged from 9.8–20.6 % (Table 4). The natural ITS barcode gap between all Tulasnella lineages studied here is between 4–6 % sequence divergence (Fig. 2).
Table 4.
Within host group and between host group Kimura -2P distances for Tulasnella as calculated from ITS. All positions containing gaps and missing data were eliminated. There were a total of 601 positions in the final dataset.
| Within taxa | T. prima | T. sphagneti | T. warcupii | T. secunda | T. tomaculum | T. eichleriana | T. ECU5 | |
|---|---|---|---|---|---|---|---|---|
| Tulasnella prima | 1.2 ± 0.3 | |||||||
| Tulasnella sphagneti | 0.1 ± 0.1 | 6.3 ± 1.0 | ||||||
| Tulasnella warcupii | 3.8 ± 0.4 | 18.1 ± 1.7 | 16.7 ± 1.6 | |||||
| Tulasnella secunda | 0.2 ± 0.1 | 14.8 ± 1.6 | 15.1 ± 1.5 | 13.9 ± 1.5 | ||||
| Tulasnella tomaculum | 0 | 15.6 ± 1.6 | 13.3 ± 1.5 | 11.6 ± 1.3 | 9.8 ± 1.3 | |||
| Tulasnella eichleriana | 2.2 ± 0.6 | 15.4 ± 1.6 | 15.1 ± 1.6 | 15.0 ± 1.5 | 12.7 ± 1.5 | 11.4 ± 1.4 | ||
| Tulasnella ECU5 | 0.2 ± 0.2 | 15.2 ± 1.6 | 13.8 ± 1.6 | 14.9 ± 1.6 | 14.3 ± 1.7 | 10.5 ± 1.4 | 11.2 ± 1.4 | |
| Tulasnella ECU6 | 1.5 ±0.5 | 20.6 ± 2.0 | 18.3 ± 1.9 | 17.6 ± 1.7 | 18.6 ± 1.9 | 14.6 ± 1.6 | 17.0 ± 1.7 | 14.5 ± 1.6 |
Fig. 2.
Barcode gap; percentage sequence divergence among Tulasnella isolates. The vertical arrow indicates the ~3.3 to 5.7 % ITS sequence divergence threshold for this dataset.
Recognition of novel taxa
Support for three of the novel taxa was high across the eight loci analysed by Linde et al. (2014) (Table 5) and all clades had long subtending basal stems in the phylogenies generated. Base-pair differences and their positions for each lineage are given in Table 6. Therefore we conclude that each can be regarded as a well-supported phylogenetic species. The additional clade consisting of isolates from Chiloglottis associated with Sphagnum was also well-supported in the ITS tree (Fig. 1) and well-separated from the sister clade, above the divergence established between the three phylogenetic species delimited on multi-locus concordance, and is therefore recognised as a fourth phylogenetic species.
Table 5.
Presence of and support for clades of six phylogenetic species of Tulasnella from Australian orchids in the genera Arthrochilus, Chiloglottis, Drakaea and Caleana in phylogenetic trees constructed separately for each of eight loci, as indicated on trees presented as Fig. 2 and Supporting information figures S2-S8 of Linde et al. (2014). Values are bootstrap/Bayesian posterior probability. +: clade present (support less than BS 70% and BPP 0.80); *: one isolate (CLM417) fell outside of the clade, basal to all other sequences; **: support values are from Fig. 1 of the present work (all other clades in this tree representing the phylogenetic species are also 100/1.00); n=1: one isolate only, falls outside of other clades, and separate to any other singletons; na: not present in analysis.
| no. isolates | ITS | mtLSU | C4102 | C12424 | C14436 | C3304 | C4722 | C10499 | |
|---|---|---|---|---|---|---|---|---|---|
| T. prima | 33 | 100/1.00 | 97/1.00 | 100/1.00 | 100/1.00 | 85/0.99* | 100/1.00 | 100/1.00 | 99/1.00 |
| T. sphagneti | 9 | 100/1.00** | na | na | na | na | na | na | na |
| T. secunda | 21 | 100/1.00 | + | 95/1.00 | 86/0/96 | 96/1.00 | 100/1.00 | 99/1.00 | 94/1.00 |
| T. warcupii | 6 | 100/1.00 | 94/0.88 | 100/1.00 | 100/1.00 | -/0.94 | 99/1.00 | n=1 | 100/1.00 |
| T. sp. Arthrochilus II | 2 | 100/1.00 | 98/0.99 | 100/1.00 | 100/1.00 | 98/1.00 | + | n=1 | na |
| T. sp. Arthrochilus III | 1 | n=1 | n=1 | n=1 | n=1 | n=1 | n=1 | na | n=1 |
Table 6.
Basepair differences and their positions, among Tulasnella tomaculum, T. sphagneti, T. prima, T. secunda and T. warcupii. Polymorphic basepair differences in two or more isolates of a species are given by the most common basepair/alternative basepair.
| 1 | 5 | 6 | 7 | 8 | 9 | 15 | 16 | 17 | 18 | 19 | 24 | 27 | 28 | 30 | 37 | 41 | 44 | 51 | 62 | 63 | 64 | 74 | 90 | 92 | |
|
| |||||||||||||||||||||||||
| T. tomaculum | G | G | T | G | C | T | C | G | T | T | T | A | G | C | C | G | T | - | - | T | C | G | - | T | T |
| T. sphagneti | T | . | C | T | G | A | T | C | . | C | C | C | T | . | . | . | C | T | C | C | . | T | - | A | . |
| C. prima | T | . | C | T | G | A | T | T | . | C | . | T | T | . | . | . | C | T | T | . | . | T | - | A | . |
| T. secunda | . | . | C | A/. | . | . | . | . | A | . | C | C | . | . | . | A | . | - | - | . | T | . | - | . | . |
| T. warcupii | . | A | C | . | . | C | . | C | . | . | . | . | . | ./T | T | . | . | - | - | . | . | . | C | . | C |
|
| |||||||||||||||||||||||||
| 97 | 99 | 105 | 107 | 111 | 113 | 117 | 120 | 121 | 122 | 123 | 124 | 125 | 126 | 129 | 131 | 135 | 136 | 144 | 145 | 146 | 147 | 149 | 150 | 152 | |
|
| |||||||||||||||||||||||||
| T. tomaculum | C | A | C | T | C | G | C | G | G | C | - | T | G | G | C | A | A | G | C | C | G | - | T | C | T |
| T. sphagneti | T | C | . | . | . | A | T | . | - | T | C | C | . | A | . | T | T | . | T | . | . | G | . | . | C |
| T. prima | T | . | C | C | . | A | T | T | . | T | - | C | . | ./A | . | T | T | T | T | . | . | G | C | A | . |
| T. secunda | . | . | . | . | . | . | . | A | . | A | - | . | T | . | T/. | . | . | . | . | T | T | G | C | T | C |
| T. warcupii | . | . | . | . | T | . | . | . | T | T | - | . | . | . | . | . | . | . | . | . | T | - | . | . | C |
|
| |||||||||||||||||||||||||
| 153 | 155 | 160 | 162 | 163 | 172 | 174 | 180 | 182 | 183 | 184 | 195 | 196 | 197 | 209 | 212 | 214 | 215 | 218 | 231 | 232 | 233 | 234 | 259 | 376 | |
|
| |||||||||||||||||||||||||
| T. tomaculum | G | A | T | C | T | G | C | T | A | A | C | C | T | T | T | - | A | - | T | C | A | T | A | T | T |
| T. sphagneti | A | G | C | . | C | A | . | . | T | . | T | T | C | C | . | A | T | T | C | . | . | . | . | . | C |
| T. prima | . | G | C | . | ./C | A/T | . | . | C | . | T | T | C | . | ./C | A | T | T | C | A | . | . | . | C | C |
| T. secunda | . | . | C | T | C | A | T | . | C | . | . | A | . | . | . | A | - | - | C | . | . | C | . | . | . |
| T. warcupii | . | . | C | . | C | T | . | C | . | G | . | T | . | A | C | A | . | T | . | . | G | . | T | . | . |
|
| |||||||||||||||||||||||||
| 378 | 391 | 410 | 411 | 414 | 421 | 422 | 423 | 424 | 425 | 426 | 431 | 432 | 434 | 435 | 436 | 438 | 445 | 447 | 448 | 449 | 452 | 454 | 455 | 456 | |
|
| |||||||||||||||||||||||||
| T. tomaculum | T | C | T | C | C | C | C | T | G | A | T | C | T | G | G | C | G | C | G | T | G | C | C | T | C |
| T. sphagneti | C | T | . | . | A | . | . | C | T | C | . | T | C | A/. | . | . | . | . | . | . | . | . | T | C | T |
| T. prima | C | T | . | . | A | . | . | C | A/T | T/C | . | T | C | A/. | . | T | . | . | . | C | A/. | T | T | C | T |
| T. secunda | . | . | . | . | . | T | . | . | . | T | . | T | . | A | A | T | . | . | A | . | . | . | T | C | . |
| T. warcupii | . | . | C | G | . | . | T | . | A | . | A | . | . | . | . | . | A | T | . | C | . | . | T | C | . |
|
| |||||||||||||||||||||||||
| 457 | 458 | 459 | 460 | 463 | 469 | 470 | 471 | 472 | 475 | 481 | 482 | 484 | 486 | 490 | 492 | 493 | 494 | 497 | 501 | 503 | 504 | 505 | 506 | 508 | |
|
| |||||||||||||||||||||||||
| T. tomaculum | G | C | G | T | T | A | C | C | A | T | - | - | - | G | T | G | C | A | G | C | C | G | T | T | G |
| T. sphagneti | . | . | A | . | C | G | . | . | G | . | C | A | - | C | C | . | . | G | . | T | T | . | G | . | . |
| T. prima | . | T | . | . | . | G | . | T | G | C | A | T | - | . | C | . | T | G | T | T | . | A | A | A | ./A |
| T. secunda | A | . | A | C | . | G | . | . | G/. | C | C | T | C/T | . | . | . | ./T | G | . | T | T | . | . | . | A |
| T. warcupii | . | T | . | C | C | G | T | . | G | - | C | T | - | . | C | A | . | G | . | . | . | . | . | . | A |
|
| |||||||||||||||||||||||||
| 509 | 510 | 511 | 513 | 514 | 515 | 516 | 517 | 518 | 519 | 521 | 522 | 523 | 525 | 526 | 527 | 532 | 540 | 542 | 545 | 548 | 549 | 550 | 551 | 553 | |
|
| |||||||||||||||||||||||||
| T. tomaculum | C | G | G | A | G | A | T | G | T | G | T | A | - | G | T | T | T | C | G | C | C | A | T | C | C |
| T. sphagneti | . | . | T | G | C | T | C | C | A | T | A | G | T | . | C | . | C | . | T | T | . | G | G | T | . |
| T. prima | . | A | ./T | G | T | T | C | T | G/A | C | . | G | T | . | C | . | C | . | T | T | . | G | A | T/. | . |
| T. secunda | . | . | . | . | A | . | C | C | . | T | . | G | - | A | C | . | . | T | . | . | A | G | A | - | . |
| T. warcupii | T | . | . | G | . | G | C | T | . | T | . | G | - | . | C | G | C | . | . | T | . | G | A | . | A |
|
| |||||||||||||||||||||||||
| 554 | 555 | 557 | 558 | 562 | 565 | 566 | 571 | 577 | 587 | 588 | 590 | 592 | 594 | 595 | 598 | 599 | 600 | 606 | 607 | 623 | 644 | 645 | 646 | 647 | |
|
| |||||||||||||||||||||||||
| T. tomaculum | C | C | - | - | G | T | G | C | C | C | - | T | C | T | G | A | G | A | T | T | T | C | T | C | T |
| T. sphagneti | T | A | C | C | A | A | . | . | . | . | G | C | T | G | T | G | C | G | . | . | G | . | C | . | . |
| T. prima | T | A | - | C | A | G | . | T | ./T | . | G | C | T | G | T | . | T | . | . | . | G | . | C | T | G |
| T. secunda | T | T | C | C | - | C | A | . | . | T | G | . | . | . | . | . | . | . | . | ./C | G | . | C | T | G |
| T. warcupii | T | . | - | C | A | C | . | . | . | . | G | . | - | C | . | . | . | . | C | C | . | T | C | . | . |
|
| |||||||||||||||||||||||||
| 657 | 658 | 659 | 660 | 662 | 663 | 664 | 665 | 667 | 668 | 669 | 670 | 671 | 675 | 680 | 681 | 686 | 687 | 688 | 689 | 690 | 695 | 696 | 698 | 700 | |
|
| |||||||||||||||||||||||||
| T. tomaculum | T | C | G | G | T | C | - | G | G | T | G | C | C | G | C | T | C | T | C | G | - | G | A | C | T |
| T. sphagneti | . | G | . | A | . | . | - | T | . | A | C | . | T | A | . | . | . | . | . | . | - | A | . | A | - |
| T. prima | . | G | . | A | . | . | - | T | . | A | . | . | T | . | . | . | . | . | . | . | - | A | C | A | - |
| T. secunda | C | T | . | A | . | . | T | T | . | C | . | . | . | . | A | C | T | . | . | . | - | A | . | T | G |
| T. warcupii | G | A | T | . | C | G | - | A | A | C | . | T | . | . | . | C | T | C | T | A | C | A | . | . | - |
|
| |||||||||||||||||||||||||
| 703 | 707 | 708 | 709 | 710 | 711 | 712 | 713 | 714 | 717 | 719 | 720 | 721 | 723 | 724 | 725 | 726 | 727 | 728 | 746 | 749 | 750 | 756 | |||
|
| |||||||||||||||||||||||||
| T. tomaculum | - | G | C | T | G | T | G | C | - | - | A | - | A | - | C | - | T | C | A | T | G | T | G | ||
| T. sphagneti | T | T | T | C | C | . | C | G | - | T | G | G | G | - | - | T | . | . | . | C | A | . | T | ||
| T. prima | T | T | T | C | A | . | T | G | - | C | G | - | - | - | T | T | . | . | . | C | A | C | T | ||
| T. secunda | A | T | T | - | T | G | T | - | - | T | G | - | . | G | . | T | C | . | . | . | . | . | . | ||
| T. warcupii | - | T | T | C | - | G | T | G | C | G/- | G | - | . | - | . | - | C | . | T | . | . | . | . | ||
None of the clades for these four phylogenetic species contain sequences from material of Tulasnella previously described from Australia, or indeed any other sequences of described species in GenBank. In addition, the ITS sequences from ex-type cultures of T. asymmetrica and T. irregularis do not cluster with or are close to any of the four phylogenetic species described here. Therefore, we conclude that these four phylogenetic species are previously unrecognized, and consequently they are formally described below. Two further putative new phylogenetic species (Tulasnella sp. Arthrochilus II and Tulasnella sp. Arthrochilus III) (Linde et al. 2014) that were represented by only two and one isolates respectively, are not formally described here pending discovery of further isolates. Previous morphology-based identifications of various Tulasnella species from hosts in Drakaeinae (Table 3) will all need to be re-visited and confirmed with sequence data, if voucher specimens or cultures still exist.
Here we diagnose the new species on the basis of both sequence-based synapomorphies and clade-based definitions from molecular phylogenies (Hibbett et al. 2011, Renner 2016). This is because it is not possible to be certain as to which morphological characters are actually diagnostic. In time, certain morphological features may turn out to be unique for particular taxa, but this can only be known if morphological data are comprehensive across known species of the genus. In addition, it is sequence data that are routinely used to identify isolates of Tulasnella, and hence we are providing both rigorous species delimitation and the means to identify further isolates with certainty. Therefore, our descriptions of the morphology of cultures and of hyphal characters are provided for completeness, rather than as species characteristics.
TAXONOMY
Tulasnella prima Linde & T.W. May, sp. nov. MycoBank MB817404
(Fig. 3A)
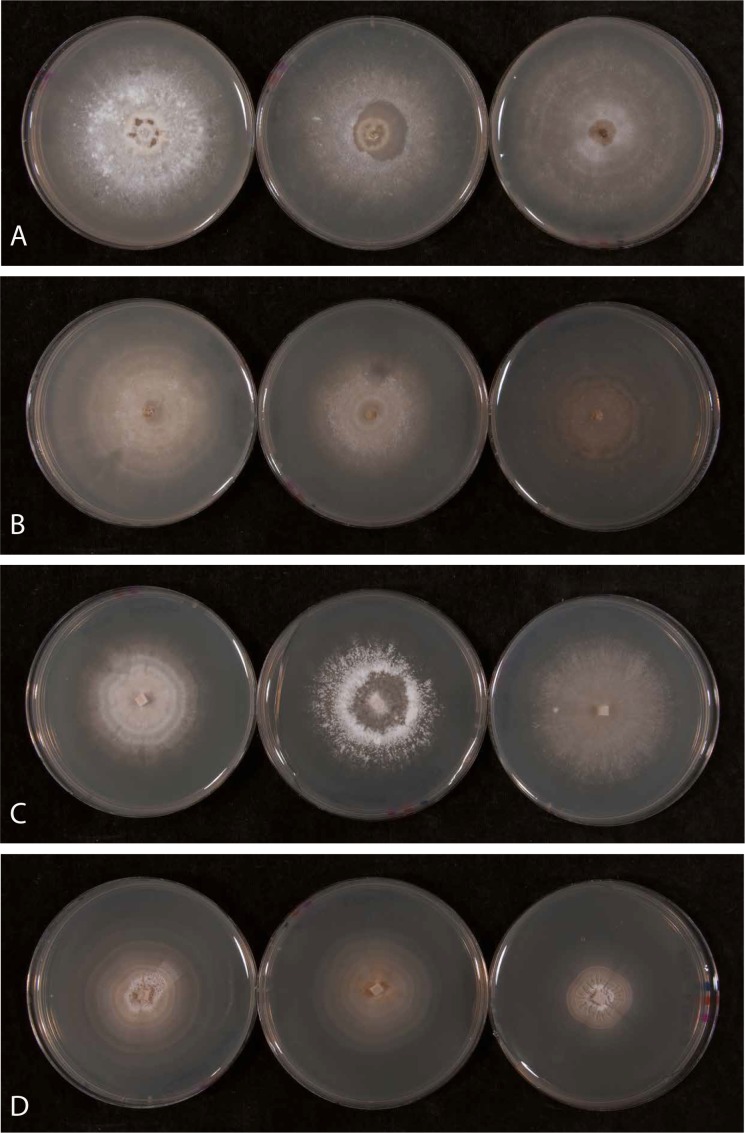
Fig. 3.
Tulasnella cultures on quarter strength PDA (left), half strength FIM (middle) and 3MN +A-Z (right) media. A. Tulasnella prima (CLM159); B. T. sphagneti (CLM541); C. T. secunda (CLM009) and D. T. warcupii (CLM027).
Etymology: Referring to the first Tulasnella found in the host, Chiloglottis.
Type: Australia: New South Wales: Blue Mountains, Mt Werong, Ranger Fire Trail, isolated from Chiloglottis trilabra, 22 Mar. 2007, C.C. Linde & R. Peakall CLM159 (MEL 2402822 – holotype; ex-type culture VPRI 42810).
Diagnosis: Tulasnella prima can be diagnosed by the following nucleotide characters, which are fixed between T. tomaculum and T. prima respectively: Locus ITS: ITS1 upstream from the 18S at position 18 (G:T), 23-25 (TGCT:CTGA), 32-33 (CG:--), 38 (G:T/A), 41 (A:T/C), 44 (G:T), 58 (T:C), 61 (-:T), 68 (-:T), 80 (G:T), 101 (T:A), 117 (T:C), 123 (G:A), 127 (C:T), 130 (G:T), 132-133 (CT:TC), 140 (A:T), 144-145 (AG:TT), 152 (C:T), 156 (T:G), 158 (-:A/G), 163 (A:G), 165 (C:-), 168 (T:C), 180 (G:A/T), 190 (A:T/C), 192 (C:T), 203-204 (CT:TC), 214-215 (AC:GT), 217-218 (TG:CT), 224-225 (TA:--), 237 )C:A/G). 5.8S starting from ITS1 end: Position 4 (-:T/-), 22 (T:C), 139 (T:C), 141 (T:C), 154 (C:T). ITS2 starting from 5.8S end: Position 14 (T:-), 16 (C:A), 25 (T:C), 27 (A:T/C), 32-33 (CT:TC), 37 (C:T), 49 (T:C), 55-56 (CT:TC), 59 (C:T), 69 (A:G), 71-72 (CA:TG), 75 (T:C), 77-81 (TCTGA:CTAT/CG), 84 (T:C), 88 (A:G), 91 (G:C/T), 93 (C:T), 96-98 (GTT:AAA), 105 (A:-), 107 (A:T), 109-110 (--:CT), 116-117 (TA:--), 120 (T:C), 126 (T:C), 136 (G:T), 139 (C:T), 143-144 (AT:GA), 147-148 (CC:TA/G), 150 (-:C/T), 154 (G:A), 157 (T:G), 163 (C:T), 180 (T:G), 187 (-:T/C), 191 (G:T/C), 215 (T:G), 222 (T:-), 237 (T:-), 241 (-:G/A), 251 (C:G), 253 (G:A), 256 (G:T, 259 (T:A), 262 (C:T), 285 (G-:AC), 290-293 (TCCG:CTGC), 295-296 (CG:TT), 298-299 (TG:AT), 302-303 (CG:GT/C), 305 (A:G), 307-308 (AC:TT), 328 (T:C), 331 (G:A), 338 (G:T).
Clade-based diagnosis: The least inclusive clade in the ITS phylogeny in Fig. 1 containing HM196790 and HM196786.
Substrate or host: Roots and underground stem-collars of Chiloglottis orchid species.
Distribution: High rainfall parts of south-eastern Australia and Tasmania in Eucalyptus woodlands and forests. Current known distribution coincides with that of the Chiloglottis hosts.
Notes: Cultures on quarter strength PDA show fine concentric rings. Culture edges lack concentric rings, are broad and diffused. Culture appearance is quite variable, with some cultures showing aerial mycelium. Not all cultures grow on 3MN +A-Z. Hyphae from cultures are cylindrical, 2–5 μm diam, branched, often at right angles, septate, lacking clamp connections; wall slightly thickened (to 0.25 μm); rarely with refractive internal bodies, and then small; sometimes uneven (with undulate outline); sometimes with swollen elements to 9.5 μm diam that are thick-walled (to <0.5 μm thick), clavate, terminal or intercalary, sometimes in short chains.
Additional material examined: See Table 1.
Tulasnella secunda Linde & T.W. May, sp. nov. MycoBank MB817406
(Fig. 3C)
Etymology: Referring to the second Tulasnella described that associates with Drakaeinae orchids.
Type: Australia: Western Australia: Paganoni Swamp Reserve, Karnup, isolated from Drakaea elastica, 2008, R.D. Phillips [C.C. Linde CLM009], (MEL 2402819 – holotype; ex-type culture VPRI 42808).
Diagnosis: Tulasnella secunda can be diagnosed by the following nucleotide characters, which are fixed between T. tomaculum and T. secunda respectively. Locus ITS: ITS1 upstream from the 18S at position 34-35 (TT:-A), 41 (A:C/T), 54 (G:A), 77 (C:T), 128 (G:A), 130 (C:A), 132 (G:T), 152-153 (-C:TT), 155-156 (TC:CT), 158 (T:C), 163 (C:-), 166 (T:-), 170 (-:C), 179 (G:A), 181 (C:T), 189 (A:C), 202 (C:A), 211-212 (AC:--), 219-220 (CA:AC), 223 (T:C), 238 (T:C). 5.8S no differences. ITS2 starting from 5.8S end: Position 23 (C:T), 27 (A:T), 32 (C:T), 35-37 (GGC:AAT), 48 (G:A), 55-56 (CT:T/AC), 58 (G:A), 60-61 (GT:AC), 69 (A:G), 73 (T:C), 76 (T:C), 80 (-:C/T), 89 (A:G), 94 (C:T), 96 (C:T), 101 (G:A), 107 (G:A), 109-110 (TG:CC), 112 (G:-), 115 (A:T/C), 118-119 (-T:AC), 133 (C:T), 141 (C:-), 143-147 (TCCC:--GA), 149 (-:T), 151 (T:C), 156-157 (TG:CA), 178-179 (-C:TG), 214 (T:G), 235-237 (TCT:CTG), 248-249 (TC:CT), 251 (G:A), 254-255 (G-:TT), 258 (T:C), 270-271 (CT:AC), 276 (C:T), 284 (G:A), 287 (CT:TG), 291 (G:A), 294-295 (GC:TT), 300 (C:T), 305 (A:G), 308 (-:C).
Clade-based diagnosis: The least inclusive clade in the ITS phylogeny in Fig. 1 containing KF476573 and JX138567.
Substrate or host: Underground stem-collars of Caleana and Drakaea orchid species.
Distribution: South-western and south-eastern Australia, extending from high rainfall areas to the margin of the arid zone, occurring in open areas within eucalypt forests and woodlands, Banksia woodlands and sandplain heath. Most records are from well-drained grey sandy soils, but also known from yellow sands, laterite, sandy clay soil, etc. Current known distribution coincides with that of Caleana (inclusive of Paracaleana) and Drakaea.
Notes: This taxon was referred to as “Tulasnellaceae sp. RP-2011” by Phillips et al. (2011). Cultures often have a rose-pink colour due to bacterial associates that are not affected by streptomycin in the isolation medium. Application of tetracyclin eliminates bacteria and cultures then assume an off-white colour. On quarter strength PDA, cultures show some aerial mycelium giving it a velvety look. Cultures also have concentric rings with culture edges diffused. Cultures often show scalloped edges. Hyphae from cultures are cylindrical, 2-5 μm diam, frequently branched, often at right angles, septate, lacking clamp connections; wall slightly thickened to thickened (to 0.25 μm); often with refractive internal bodies; sometimes uneven (with undulate outline); often with swollen elements to 10.5 μm diam that are thick-walled (to 0.5 μm thick) and globose to clavate when terminal, and globose to ellipsoid when intercalary; when terminal, subtended by one or two swollen, clavate elements, but not in chains. Refractive bodies within the hyphae are more common and obvious in this and T. warcupii than in the other two species.
Additional material examined: See Table 1.
Tulasnella sphagneti Linde & T.W. May, sp. nov. MycoBank MB817405
(Fig. 3B)
Etymology: Referring to the Sphagnum habitat of the orchid host.
Type: Australia: New South Wales: Kosciuszko NP, alongside Tantangara Road, isolated from Chiloglottis aff. valida growing in a Sphagnum hummock, 19 Jan. 2012, C.C. Linde CLM541 & E. Triponez (MEL 2402823 – holotype; ex-type culture VPRI 42811).
Diagnosis: Tulasnella sphagneti can be diagnosed by the following nucleotide characters, which are fixed between between T. tomaculum and T. sphagneti respectively: Locus ITS: ITS1 upstream from the 18S at position 18 (G:T), 23 (-:C), 26 (C:-), 27 (T:A), 33 (C:-), 34 (G:), 39 (G:C), 42 (A:C), 45 (G:T), 59 (T:C), 62 (-:T), 69 (:C), 79 (T:C), 81 (G:T), 102 (T:A), 108 (C:T), 110 (A:C), 124 (G:A), 128 (C:T), 132 (G:T), 134 (T:C), 136 (G:A), 141 (A:T), 145 (A:T), 154 (C:T), 156 (-:G), 161 (:-C), 162 (-:A), 166 (A:-), 167 (T:-), 168 (C:-), 171 (T:C), 174 (T:C), 183 (G:A), 193 (A:T), 195 (C:T), 206 (C:T), 207 (T:C), 216 (C:T), 217 (A:G), 218 (C:T), 220 (T:C), 221 (G:T), 227 (T:), 228 (A:). 5.8S starting from ITS1 end: Position 138 (T:C), 140 (T:C), 153 (C:T). ITS2 starting from 5.8S end: Position 14 (T:), 16 (C:A), 25 (T:C), 26 (G:T), 27 (A:C), 32 (C:T), 33 (T:C), 55 (C:T), 56 (T:C), 57 (G:T), 60 (G:A), 64 (T:C), 69 (A:G), 72 (A:G), 77 (-:C), 80 (T:A), 81 (G:C), 85 (T:C), 89 (A:G), 94 (C:T), 96 (C:T), 98 (T:G), 104 (G:T), 106-115 (AGATGTGTTA:GCTCCATAGT), 118 (T:C), 124 (T:C), 134 (G:T), 137 (C:T), 141-142 (-A:GG), 145-147 (-CC:TAT), 149 (T:C), 153 (G:A), 156 (T:A), 179 (T:G), 187-188 (CA:TC), 190-191 (A-:CG), 214 (T:G), 221 (T:), 236 (T:C), 249 (C:G), 251 (G:A), 254 (G:T), 257-258 (TG:AC), 260 (C:T), 264 (G:A), 283 (G:A), 286-287 (CT:AC), 293-295 (G--:TTC), 298-299 (GT:CG), 301 (C:T), 304-305 (AA:GG), 307 (C:T), 327 (T:C), 330 (G:A), 337 (G:T).
Clade-based diagnosis: The least inclusive clade in the ITS phylogeny in Fig. 1 containing 13143 and 12030.
Substrate or host: Roots and collars of Chiloglottis valida, C. aff. valida, and C. turfosa growing in Sphagnum hammocks in alpine areas in eastern Australia.
Distribution: South-eastern Australia, occurring in alpine habitats associated with Sphagnum hummocks. Current known distribution coincides with that of Chiloglottis hosts within this particular habitat.
Notes: Cultures on quarter strength PDA show fine concentric rings. Culture edges lack concentric rings, are broad and diffused. Hyphae from cultures cylindrical, 2–5.5 μm diam, branched, often at right angles, septate, lacking clamp connections; wall slightly thickened (to 0.25 μm); rarely with refractive internal bodies, and then in narrower hyphae; sometimes uneven (with undulate outline); rarely with swollen elements to 7 μm diam that are slightly thick-walled, subglobose and terminal. Swollen elements are less common in this species than in the other three.
Additional material examined: See Table 1.
Tulasnella warcupii Linde & T.W. May, sp. nov. MycoBank MB817407
(Fig. 3D)
Etymology: After J. H. Warcup who was instrumental in studying mycorrhizal fungi associated with orchids in Australia.
Type: Australia: Queensland: Atherton Tablelands, Herberton Range State Forest, Atherton, isolated from Arthrochilus oreophilus, 1 Apr. 2010, C.C. Linde & D. Gomez CLM027 (MEL 2402821 – holotype; MEL 2402820 – isotype; ex-holotype culture VPRI 42809).
Diagnosis: Tulasnella warcupii can be diagnosed by the following nucleotide characters, which are fixed between T. tomaculum and T. warcupii respectively: Locus ITS: ITS1 upstream from the 18S at position 22-23 (GT:AC), 26 (T:C), 33 (G:C), 37 (C:-), 47 (C:T), 101 (T:C), 120 (C:T), 130 (G:T/A), 131 (C:T), 154 (G:T), 158 (T:C), 163 (C:-), 166 (T:C), 169 (T:C), 178 (G:T), 186 (T:C), 189 (A:G), 201 (C:T), 203 (T:A), 212-213 (--:G/AT), 218-221 (GTCA:----), 238 (A:G), 240 (A:T). 5.8S starting from ITS1 end: Position 1 (T:-). ITS2 starting from 5.8S end: Position 13 (C:G), 24 (C:T), 26 (G:A), 28 (T:A), 39 (G:A), 46 (C:T), 49 (T:C), 55-56 (CT:TC), 59 (C:T), 61 (T:C), 64 (T:C), 69-70 (AC:GT), 72 (A:G), 76 (T:C), 84 (T:C), 86 (G:A), 88 (A:G), 92 (-:-/G), 93 (-:-/C), 102-103 (GC:AT), 107 (A:G), 109-111 (ATG:GCT), 113 (G:T), 116 (A:G), 119-120 (TT:CG), 125 (T:C), 138 (C:T), 142-143 (AT:GA), 145-146 (CC:AT), 149 (-:C), 153 (G:A), 156 (T:C), 179 (T:G), 181 (C:T), 184 (T:C), 196-197 (TT:CC), 234-235 (CT:TC), 247-249 (TCG:GAT), 251-253 (TCG:CGA), 255-256 (GT:AC), 258 (C:T), 269 (T:C), 274 (-:T), 277 (C:A), 278 (G:C/T), 283 (G:A/-), 287 (T:-), 293-295 (GCT:TTC), 302 (-:-/G), 303 (A:-/G), 307 (T:C), 309 (A:T).
Clade-based diagnosis: the least inclusive clade in the ITS phylogeny in Fig. 1 containing KF476596 and KF476601.
Substrate or host: Roots and collars of Arthrochilus oreophilus.
Distribution: Atherton Tablelands in Queensland, Australia, in association with Arthrochilus oreophilus in Eucalyptus woodland.
Notes: On PDA cultures show fine concentric rings with a velvety edge. Usually no aerial mycelium is visible. Cultures are off-white to yellowish. Of the four Tulasnella species described here, it is the slowest growing. Hyphae from cultures are cylindrical, 1–2.5(–4) μm diam, branched, often at right angles, septate, lacking clamp connections; wall slightly thickened to thickened (to 0.5 μm); often with refractive internal bodies; sometimes uneven (with undulate outline) to distinctly monilioid, with short, repeated, globose to subglobose elements to 7 μm diam. The minimum diameter of hyphae is noticeably thinner than in the other three species, and this is the only one of the four species to show chains of globose elements.
Additional material examined: See Table 1.
DISCUSSION
Here we describe four new species of Tulasnella that are found in association with Australian terrestrial orchids belonging to the Diurideae, using diagnostic DNA characters as advocated by Renner (Renner 2016). Three of these species (T. prima, T. secunda, and T. warcupii) were initially revealed by an in depth study using eight sequence loci and three different methods of species delimitation (Linde et al. 2014). These three species were shown to successfully germinate seed of members of the orchid genus they associate with (Linde et al. 2014). Our addition of Tulasnella isolates from Chiloglottis growing in Sphagnum hummocks in alpine areas in eastern Australia revealed the fourth species, T. sphagneti. This represents the second Tulasnella species to be found associated with Chiloglottis orchids. Based on the widely accepted 3 % sequence divergence cut-off value for species delimitation (Nilsson et al. 2008), or the 3–5 % divergence proposed for delimiting Tulasnella species (Girlanda et al. 2011, Jacquemyn et al. 2011), the sequence divergence between the four new species described in this study, exceeds these cut-off thresholds (6.3 %).
Tulasnella is representative of the complexity of contemporary taxonomic mycology. Some species are rigorously defined on multi-gene data, or on the single region (ITS) that has been confirmed as having utility as a barcode in this genus, while other species have been and are being described with excellent details of morphological characters. Unfortunately, few species are well known from both morphology and molecular sequence data. Ideally, all type material should be sequenced, which would allow integration of the two approaches. However, Cruz et al. (2016) point out that “sequencing of old fungarium specimens of Tulasnella spp. has been unsuccessful probably due to inappropriate conservation of DNA” and they consider this could well remain the case even with improvements in techniques. Therefore, sequenced epitypes will need to be designated where the strict application of names without sequences is ambiguous, but the challenge will be to match modern cultures or collections to old names.
Apart from their association with orchids, the ecology and distribution of the new Tulasnella species described here remains poorly known. Interestingly, all orchid species investigated, within the orchid genera Chiloglottis, Drakaea, and Caleana, associate with a single Tulasnella species with one exception. The one exception is in Chiloglottis where both T. prima and T. sphagneti associate with Chiloglottis orchids, but T. sphagneti is so far only found in Chiloglottis species growing in Sphagnum. Where orchids in the Drakaeinae are host to multiple Tulasnella species, the fungi are closely related. Tulasnella prima and T. sphagneti from Chiloglottis are sister taxa and the three Tulasnella species from Arthrochilus form a clade. However, the overall phylogeny of the Tulasnella species from Drakaeinae does not appear to match that of the hosts (Miller & Clements 2014), where Chiloglottis is sister to Drakaea, and these form a clade sister to the remaining genera, including Arthrochilus and Caleana. Remarkably, in Caleana, the only orchid genus in this group to be found in both eastern and western Australia, this association extends across the continent. In contrast to the orchid genus-wide association of most Tulasnellas in this study, mycorrhizal associations of the tropical Arthrochilus oreophilus appear far more diverse. Previously, three Tulasnella OTUs were shown to occur in a narrow sample of this subtropical species (Linde et al. 2014). Because two of the OTUs are represented by only one or two sequences, and lack living cultures, only one (T. warcupii) is described here. Our findings raise the question of why only a small diversity of Tulasnella fungi associates with a large number of orchid species across a vast geographic range.
The pattern of one fungal species to many orchid species appears to be in stark contrast to studies of orchid-mycorrhizal interactions outside Australia, which have consistently found a number of mycorrhizal OTUs associating with sympatric as well as allopatric orchid congeners (Jacquemyn et al 2015). For example, 15 OTUs from Tulasnellaceae were associated with four species of Anacamptis orchids. Of those 15 OTUs, 13 associated with seven species of Ophrys and two Orchis species, whereas nine OTUs associated with three Serapias species (Pellegrino et al. 2014). The high diversity of Tulasnella was such that within sites up to 15 OTUs were co-occurring and 85 % of plants associated with more than three different OTUs (Pellegrino et al. 2014). A corresponding result was found along a single 1000 m transect with the same orchid genera where 16 Tulasnellaceae OTUs were recovered for 20 species of orchids (Jacquemyn et al. 2015). The same pattern is found in Andean tropical rainforests where up to six Tulasnella OTUs may associate with Stelis orchid species and Pleurothallis lilijae (Suarez et al. 2006, Kotte et al. 2008). Consistent among these studies and ours, is the finding that multiple species of an orchid genus can share the same fungal OTU. However, the ability to germinate orchid seed was not shown in other studies, making it difficult to ascertain the real mycorrhizal diversity associating with the orchids.
Our description of four new species of Tulasnella, all associated with Australian orchids, extends the number of formally described species known as mycorrhizal agents of orchids. However, it is evident that more Tulasnella species await DNA analysis and formal description. For example, previous studies on Tulasnella ITS diversity associated with Diuris orchids have uncovered a large number of OTUs (Smith et al. 2010), likely to represent many undescribed Tulasnella species. It is further evident that earlier morphologically based studies by Warcup and co-workers prior to the advent of DNA sequencing grossly underestimated the Tulasnella species diversity associated with orchids in Australia.
Although Tulasnella is most commonly detected in association with orchids, orchids are not essential for Tulasnella existence. To understand issues such as the ecology, habitat, and geographic range of these fungi, it is essential to develop detection methods that are independent of the orchid, such as a metagenomic approach. This may not only uncover further Tulasnella diversity, but will also shed light on the lives of these fungi independent of orchids.
Acknowledgments
The research was supported by the Australian Research Council (LP098338 and LP110100408) to CCL and RP.
REFERENCES
- Bidartondo MI, Bruns TD, Weiss M, Sergio C, Read DJ. (2003) Specialized cheating of the ectomycorrhizal symbiosis by an epiparasitic liverwort. Proceedings of the Royal Society, B-Biological Sciences 270: 835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidartondo MI, Duckett JG. (2010) Conservative ecological and evolutionary patterns in liverwort-fungal symbioses. Proceedings of the Royal Society, B-Biological Sciences 277: 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnardeaux Y, Brundrett M, Batty A, Dixon K, Koch J, et al. (2007) Diversity of mycorrhizal fungi of terrestrial orchids: compatibility webs, brief encounters, lasting relationships and alien invasions. Mycological Research 111: 51–61. [DOI] [PubMed] [Google Scholar]
- Bougoure JJ, Bougoure DS, Cairney JWG, Dearnaley JDW. (2005) ITS-RFLP and sequence analysis of endophytes from Acianthus, Caladenia and Pterostylis (Orchidaceae) in southeastern Queensland. Mycological Research 109: 452–460. [DOI] [PubMed] [Google Scholar]
- Clements MA, Ellyard RK. (1979) The symbiotic germination of Australian terrestrial orchids. American Orchid Society Bulletin 48: 810–816. [Google Scholar]
- Cruz D, Suarez JP, Kottke I, Piepenbring M. (2014) Cryptic species revealed by molecular phylogenetic analysis of sequences obtained from basidiomata of Tulasnella. Mycologia 106: 708–722. [DOI] [PubMed] [Google Scholar]
- Cruz D, Suarez JP, Kottke I, Piepenbring M, Oberwinkler F. (2011) Defining species in Tulasnella by correlating morphology and nrDNA ITS-5.8S sequence data of basidiomata from a tropical Andean forest. Mycological Progress 10: 229–238. [Google Scholar]
- Fujita MK, Leaché AD, Burbrink FT, McGuire JA, Moritz C. (2012) Coalescent-based species delimitation in an integrative taxonomy. Trends in Ecology and Evolution 27: 480–488. [DOI] [PubMed] [Google Scholar]
- Girlanda M, Segreto R, Cafasso D, Liebel HT, Rodda M, et al. (2011) Photosynthetic mediterranean meadow orchids feature partial mycoheterotrophy and specific mycorrhizal associations. American Journal of Botany 98: 1148–1163. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, et al. (2011) Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fungal Biology Reviews 25: 38–47. [Google Scholar]
- Hibbett DS, Taylor JW. (2013) Fungal systematics: is a new age of enlightenment at hand? Nature Reviews, Microbiology 11: 129–133. [DOI] [PubMed] [Google Scholar]
- Hopper SD, Brown AP. (2006) Australia’s wasp-pollinated flying duck orchids revised (Paracaleana: Orchidaceae). Australian Systematic Botany 19: 211–244. [Google Scholar]
- Jacquemyn H, Brys R, Merckx VSFT, Waud M, Lievens B, et al. (2014) Coexisting orchid species have distinct mycorrhizal communities and display strong spatial segregation. New Phytologist 202: 616–627. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Deja A, De hert K, Cachapa Bailarote B, Lievens B. (2012) Variation in mycorrhizal associations with tulasnelloid fungi among populations of five Dactylorhiza species. PLoS One 7: e42212. doi:42210.41371/journal.pone.0042212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemyn H, Merckx V, Brys R, Tyteca D, Cammue BPA, et al. (2011) Analysis of network architecture reveals phylogenetic constraints on mycorrhizal specificity in the genus Orchis (Orchidaceae). New Phytologist 192: 518–528. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Waud M, Merckx VSFT, Lievens B, Brys R. (2015) Mycorrhizal diversity, seed germination and long-term changes in population size across nine populations of the terrestrial orchid Neottia ovata. Molecular Ecology 24: 3269–3280. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottke I, Beiter A, Weiss M, Haug I, Oberwinkler F, et al. (2003) Heterobasidiomycetes form symbiotic associations with hepatics: Jungermanniales have sebacinoid mycobionts while Aneura pinguis (Metzgeriales) is associated with a Tulasnella species. Mycological Research 107: 957–968. [DOI] [PubMed] [Google Scholar]
- Kristiansen KA, Taylor DL, Køller R, Rasmussen HN, Rosendahl S. (2001) Identification of mycorrhizal fungi from single pelotons of Dactylorhiza majalis (Orchidaceae) using single-strand conformation polymorphism and mitochondrial ribosomal large subunit DNA sequences. Molecular Ecology 10: 2089–2093. [DOI] [PubMed] [Google Scholar]
- Linde CC, Phillips RD, Crisp MD, Peakall R. (2014) Congruent species delineation of Tulasnella using multiple loci and methods. New Phytologist 201: 6–12. [DOI] [PubMed] [Google Scholar]
- Ma M, Tan TK, Wong SM. (2003) Identification and molecular phylogeny of Epulorhiza isolates from tropical orchids. Mycological Research 107: 1041–1049. [DOI] [PubMed] [Google Scholar]
- May TW, Milne J, Shingles S, Jones RH. (2003) Fungi of Australia. Vol. 2B. Catalogue and bibliography of Australian fungi, Basidiomycota & Myxomycota. Melbourne: ABRS/CSIRO Publishing. [Google Scholar]
- McCormick MK, Jacquemyn H. (2014) What constrains the distribution of orchid populations? New Phytologist 202: 392–400. [Google Scholar]
- Miller JT, Clements MA. (2014) Molecular phylogenetic analyses of Drakaeinae: Diurideae (Orchidaceae) based on DNA sequences of the internal transcribed spacer region. Australian Systematic Botany 27: 3–22. [Google Scholar]
- Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson K-H. (2008) Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evolutionary Bioinformatics 4: 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oja J, Kohout P, Tedersoo L, Kull T, Koljalg U. (2015) Temporal patterns of orchid mycorrhizal fungi in meadows and forests as revealed by 454 pyrosequencing. New Phytologist 205: 1608–1618. [DOI] [PubMed] [Google Scholar]
- Pandey M, Sharma J, Taylor DL, Yadon VL. (2013) A narrowly endemic photosynthetic orchid is non-specific in its mycorrhizal associations. Molecular Ecology 22: 2341–2354. [DOI] [PubMed] [Google Scholar]
- Peakall R, Ebert D, Poldy J, Barrow RA, Francke W, et al. (2010) Pollinator specificity, floral odour chemistry and the phylogeny of Australian sexually deceptive Chiloglottis orchid: implications for pollinator-driven speciation. New Phytologist 188: 437–450. [DOI] [PubMed] [Google Scholar]
- Peakall R, Whitehead MR. (2014) Floral odour chemistry defines species boundaries and underpins strong reproductive isolation in sexually deceptive orchids. Annals of Botany 113: 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins AJ, Masuhara G, Mcgee PA. (1995) Specificity of the associations between Microtis parviflora (Orchidaceae) and Its mycorrhizal fungi. Australian Journal of Botany 43: 85–91. [Google Scholar]
- Perkins AJ, McGee PA. (1995) Distribution of the orchid mycorrhizal fungus, Rhizoctonia solani, in relation to its host, Pterostylis acuminata, in the field. Australian Journal of Botany 43: 565–575. [Google Scholar]
- Phillips RD, Barrett MD, Dixon KW, Hopper SD. (2011) Do mycorrhizal symbioses cause rarity in orchids? Journal of Ecology 99: 858–869. [Google Scholar]
- Phillips RD, Peakall R, Hutchinson MF, Linde CC, Xu T, et al. (2014) Specialized ecological interactions and plant species rarity: the role of pollinators and mycorrhizal fungi across multiple spatial scales. Biological Conservation 169: 285–295. [Google Scholar]
- Renner SS. (2016) A return to Linnaeus’s focus on diagnosis, not description: the use of DNA characters in the formal naming of species. Systematic Biology: syw032 [epub in advance of print]. [DOI] [PubMed] [Google Scholar]
- Roberts P. (1994) Globose and ellipsoid-spored Tulasnella species from Devon and Surrey, with a key to the genus in Europe. Mycological Research 98: 1431–1452. [Google Scholar]
- Roberts P. (1999) Rhizoctonia-forming Fungi: a taxonomic guide. Kew: Royal Botanic Gardens. [Google Scholar]
- Roche S, Carter R, Peakall R, Smith L, Whitehead M, et al. (2010) A narrow group of monophyletic Tulasnella (Tulasnellaceae) symbiont lineages are associated with multiple species of Chiloglottis (Orchidaceae): Implications for orchid diversity. American Journal of Botany 97: 1313–1327. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Smith ZF, James EA, McLean CB. (2010) Mycorrhizal specificity of Diuris fragrantissima (Orchidaceae) and persistence in a reintroduced population. Australasian Journal of Botany 58: 97–106. [Google Scholar]
- Sommer J, Pausch J, Brundrett MC, Dixon KW, Bidartondo MI, et al. (2012) Limited carbon and mineral nutrient gain from mycorrhizal fungi by adult Australian orchids. American Journal of Botany 99: 1133–1145. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Suarez JP, Weiss M, Abele A, Garnica S, Oberwinkler F, et al. (2006) Diverse tulasnelloid fungi form mycorrhizas with epiphytic orchids in an Andean cloud forest. Mycological Research 110: 1257–1270. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DL, Bruns TD, Szaro TM, Hodges SA. (2003) Divergence in mycorrhizal specialization within Hexalectris spicata (Orchidaceae), a nonphotosynthetic desert orchid. American Journal of Botany 90: 1168-1179. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, et al. (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Warcup JH. (1971) Specificity of mycorrhizal association in some Australian terrestrial orchids. New Phytologist 70: 41–46. [Google Scholar]
- Warcup JH. (1973) Symbiotic germination of some Australian terrrestrial orchids. New Phytologist 72: 387–392. [Google Scholar]
- Warcup JH. (1981) The mycorrhizal relationships of Australian orchids. New Phytologist 87: 371–381. [Google Scholar]
- Warcup JH. (1990) Mycorrhizas. In: Orchids of South Australia (Bates RJ, Weber JZ, eds): 21–26. Adelaide: Flora and Fauna of South Australia Handbook Committee. [Google Scholar]
- Warcup JH, Talbot PHB. (1967) Perfect states of rhizoctonias associated with orchids. New Phytologist 66: 631–641. [Google Scholar]
- Warcup JH, Talbot PHB. (1971) Perfect states of rhizoctonias associated with orchids. II. New Phytologist 70: 35–40. [Google Scholar]
- Warcup JH, Talbot PHB. (1980) Perfect states of rhizoctonias associated with orchids. III. New Phytologist 86: 267–272. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds): 315–322. San Diego: Academic Press. [Google Scholar]
- Wright MM, Cross R, Cousens RD, May TW, McLean CB. (2010) Taxonomic and functional characterisation of fungi from the Sebacina vermifera complex from common and rare orchids in the genus Caladenia. Mycorrhiza 20: 375–390. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Rannala B. (2010) Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences, USA 107: 9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]