Abstract
CENP-H is a constitutive centromere component that localizes to the centromere throughout the cell cycle. Because CENP-H is colocalized with CENP-A and CENP-C, it is thought to be an inner centromere protein. We previously generated a conditional loss-of-function mutant of CENP-H and showed that CENP-H is required for targeting of CENP-C to the centromere in chicken DT40 cells. In the present study, we used this mutant to identify the functional region of chicken CENP-H necessary for centromere targeting and cell viability. This region was found by yeast two-hybrid analysis to interact with Hec1, which is a member of the Nuf2 complex that transiently localizes to the centromere during mitosis. Coimmunoprecipitation experiments revealed that CENP-H interacts with the Nuf2 complex in chicken DT40 cells. Photobleaching experiments showed that both Hec1 and CENP-H form stable associations with the centromeres during mitosis, suggesting that Hec1 acts as a structural component of centromeres during mitosis. On the basis of these results and previously published data, we propose that the Nuf2 complex functions as a connector between the inner and outer kinetochores.
The centromere plays a fundamental role in accurate chromosome segregation during mitosis and meiosis in eukaryotes. Its functions include sister chromatid adhesion and separation, microtubule attachment, chromosome movement, formation of the heterochromatin structure, and mitotic checkpoint control (4). Although chromosome segregation errors cause genetic diseases, including some cancers (22), the mechanism by which centromeres interact with microtubules of the spindle apparatus during cell division is not fully understood.
Traditional electron microscopy of chromosomes revealed that the kinetochore of vertebrate cells is a trilaminar button-like structure on the surface of the centromeric heterochromatin. The inner kinetochore plate has an essential role in kinetochore assembly, and the outer kinetochore plate is both a microtubule binding structure and a mitotic checkpoint structure that includes the Bub and Mad complexes (5, 13). The inner kinetochore contains the centromeric DNA as well as centromere proteins (CENPs) A and C (5, 13, 20). CENP-A is a 140-amino-acid (aa) centromere-specific protein in which the C-terminal 90 aa are 60% identical to those of histone H3 (26, 30). CENP-A is found only at active centromeres (38), and it copurifies with nucleosomes, suggesting that it is a component of nucleosomes that form at active centromeres. These fundamental and conserved features of CENP-A for centromere organization suggest that CENP-A is a key determinant for kinetochore assembly (26, 29, 33). CENP-C, another component of the inner kinetochore plate, is known to bind DNA directly (31). Disruption of the CENP-C gene in chicken cells (10) and in mouse (19) revealed that CENP-C is essential for cell growth. A conditional knockout of CENP-C in chicken DT40 cells showed that the absence of CENP-C resulted in mitotic delay, chromosome missegregation, and apoptosis (10, 11). Mitotic arrest has been observed after microinjection of anti-CENP-C antibodies into HeLa cells (37). Antibody microinjection experiments revealed that CENP-C or an associated protein is involved in determination of kinetochore size. Like CENP-A, CENP-C is found only at active centromeres (36) and is needed to form a functional centromere (11). These reports suggest that CENP-C is an important component in kinetochore assembly.
To further investigate the mechanism of kinetochore assembly, we previously identified and characterized CENP-H and CENP-I (12, 28). CENP-H localizes to the centromere throughout the cell cycle, presents at the inner kinetochore plate, and is found only in active centromeres, including neocentromeres (34, 35). Analysis of a conditional knockout of CENP-H in DT40 cells revealed that CENP-H is essential for cell growth and mitotic progression (12). CENP-I is also a constitutive centromere protein that localizes to the centromere throughout the cell cycle (23, 28). A conditional knockout of CENP-I in DT40 cells has been created, and the phenotype of CENP-I knockout cells is similar to that of CENP-H knockout cells. Analyses of both CENP-H and CENP-I knockout cells suggested that CENP-H and CENP-I are mutually interdependent for targeting to the prekinetochore structure and that both are necessary for CENP-C localization to centromeres (28).
In addition to the proteins that are associated with centromeres throughout the cell cycle, there are other centromere proteins that localize transiently to the centromere only during mitosis (5, 13). Of the proteins that localize transiently to the centromere, the Nuf2 complex can participate as a structural component in kinetochore assembly. Candidate structural components of vertebrate kinetochores, including Nuf2 and associated proteins, were identified as homologs of the Saccharomyces cerevisiae Ndc80p complex proteins (1, 6, 14, 18, 24, 25, 27, 40). We hypothesized that the Nuf2 complex is a structural component because it associates with the centromere during G2 phase, whereas proteins that associate only transiently with centromeres usually localize to the centromere after the nuclear envelope breaks down (13, 14).
In the present study, we attempted to identify the functional region of CENP-H by using a CENP-H conditional knockout cell line. We found that a CENP-H derivative that lacks the amino-terminal 72 aa and the carboxy-terminal 10 aa complemented CENP-H function. We also performed yeast two-hybrid analysis using CENP-H as bait and found that CENP-H interacts with Hec1, which is a member of the Nuf2 complex. Photobleaching experiments revealed that both Hec1 and CENP-H proteins are stably associated with the centromere during mitosis, and this finding supports our idea that the Nuf2 complex may be a structural component of the kinetochore. On the basis of these results and previously published data, we propose that the Nuf2 complex functions as a connector between the inner and outer kinetochores.
MATERIALS AND METHODS
Molecular biology, cell culture, and transfection.
All plasmids were constructed by standard methods. All CENP-H mutants were made by PCR and cloned into pEGFP (Clontech). DT40 cells were cultured and transfected as described previously (10, 11). All DT40 cells were cultured at 38°C in Dulbecco's modified medium supplemented with 10% fetal calf serum, 1% chicken serum, penicillin, and streptomycin. Mutant CENP-H plasmids were linearized with ApaLI and transfected with a Gene Pulser II (Bio-Rad). G418 (Sigma) was used at a final concentration of 2 mg/ml to select for stable transfectants. To suppress expression of the tetracycline (TET)-responsive CENP-H transgene, tetracycline (Sigma) was added to the culture medium to a final concentration of 2 μg/ml. A vector expressing histone H2B tagged with monomeric red fluorescent protein (mRFP) (2) was constructed by T. Saiwaki (Osaka University) by replacing enhanced green fluorescent protein (GFP) with mRFP in pEGFP-N1-H2B (20).
Immunocytochemistry.
Immunofluorescent staining of whole cells was performed as described previously (11). Cells were collected onto slides with a cytocentrifuge and fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature, permeabilized in 0.5% NP-40 in PBS for 15 min at room temperature, rinsed three times in 0.5% bovine serum albumin, and incubated for 1 h at 37°C in rabbit anti-CENP-C antibody (1:1,000) (11). Binding of primary antibody was then detected with Cy3-conjugated goat anti-rabbit immunoglobulin G diluted 1:1,000 (Amersham) in PBS-0.5% bovine serum albumin. Chromosomes and nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) at 0.2 μg/ml in Vectashield Antifade (Vector Laboratories). All immunofluorescence images were collected with a cooled charge-coupled-device camera (Cool Snap HQ; Photometrics Image Point) mounted on an Olympus IX71 inverted microscope with a ×60 objective lens (PlanApo 60X; numerical aperture, 1.40) together with a filter wheel. Images were analyzed with IPLab software (Signal Analytics).
Yeast two-hybrid analysis.
A yeast two-hybrid interaction was performed with the MATCHMAKER GAL4 Two-Hybrid System 3 (Clontech). The entire coding region of human CENP-H was amplified by PCR, and the PCR product was cloned into the pGBKT7 vector downstream of the GAL4 DNA binding domain (BD). This bait fusion construct was transformed into the AH109 host yeast strain. We also prepared a human HeLa cell cDNA library cloned downstream of the GAL4 transcription activation domain in pGADGH and transformed this library into Y187 yeast cells. AH109 cells transformed with the bait construct and Y187 cells transformed with the cDNA library were mated and plated onto synthetic dropout (SD) minimal medium (SD medium without histidine, leucine, and tryptophan [SD−His/−Leu/−Trp]). Colonies were picked, and their growth under stringent conditions (SD−Ade/−His/−Leu/−Trp) was examined. Plasmids containing prey constructs for BD/CENP-H bait-dependent positives were isolated and transformed into Escherichia coli DH5α. Positive clones were then sequenced, and two clones contained the coding sequence of Hec1. Additional results of this two-hybrid screen will be presented elsewhere (Y. Mikami and T. Fukagawa, unpublished data).
To exchange bait and prey and to determine the binding region of CENP-H, PCR products for CENP-H derivatives and Hec1 were cloned into pGBKT7 (for bait construct) or pGADT7 (for prey construct). A bait construct and a prey construct were cotransfected into AH109 yeast cells that were then plated onto SD minimal medium (Clontech).
Immunoprecipitation and Western blot analysis.
DT40 cells (108) were washed with PBS and suspended in 1 ml of lysis buffer (10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 150 mM NaCl, 1% NP-40, and a cocktail of protease inhibitors). Samples were then sonicated on ice for 10 s three times, and the debris was pelleted by centrifugation at 12,000 rpm (Tomy). Lysate (200 μl) and 100 μl of a suspension of M2 Flag beads (Sigma) were mixed and incubated at 4°C overnight. Alternatively, 200 μl of lysate and 5 μl of anti-GFP (MBL) were mixed and incubated at 4°C for 1 h. A 100-μl aliquot of a suspension of protein A-Sepharose beads was added and incubated at 4°C overnight. Beads were then washed three times in lysis buffer. Pelleted beads were suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, boiled for 10 min, and loaded onto SDS-polyacrylamide gels. Proteins were transferred to Hybond P membrane (Amersham). Blots were blocked with 5% skim milk and then incubated with anti-rabbit polyclonal Hec1 (1:2,000) (14), anti-rabbit polyclonal Nuf2 (1:2,000) (14), anti-mouse monoclonal Flag (1:2,000; Sigma), and anti-rabbit polyclonal GFP (1:2,000; MBL) antibodies. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibody (1:15,000; Jackson Laboratories) was used to detect bound primary antibody. Blots were developed with an ECL-Plus kit (Amersham), and protein bands were visualized by STORM (Molecular Dynamics).
Photobleaching.
Inverse fluorescence recovery after photobleaching (iFRAP) experiments were performed with a confocal microscope (LSM510Meta; Carl Zeiss) with a ×63 objective lens (PlanApo 63X; numerical aperture, 1.40). Cells were maintained at 37°C with a heated stage and an objective lens heater. The whole area of each single cell expressed the GFP fusion protein except for the small centromeric region, which was photobleached with the 488-nm laser line (100% transmission, five scans). Cells were monitored in z series (six stacks with 1.0-μm intervals) at 5-s intervals for 25 s before bleaching and for the first 30 s after bleaching and then monitored at 15-s intervals for 7 min (1.0% laser transmission; zoom 5.0; scan speed, 12; 256 by 256 pixels). For quantification, z-series images were subjected to maximum projection with LSM Image Browser (Carl Zeiss), and the fluorescence intensities of the unbleached area, whole-cell area, and background were measured with MetaMorph (Universal Imaging Corporation) software. The net intensity was obtained by subtracting the background intensity. The average signal intensity was obtained from the analysis of at least 10 live cells.
RESULTS
Expression of GFP-tagged CENP-H derivatives in CENP-H-deficient cells.
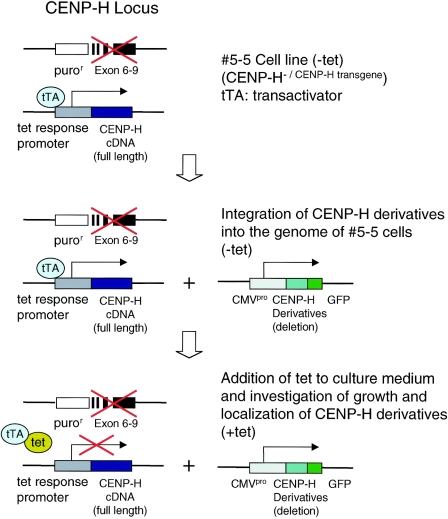
We previously developed the #5-5 CENP-H conditional knockout DT40 cell line in which CENP-H expression is inactivated when TET is added to the culture medium (12), resulting in depletion of CENP-H and rapid cell death. On the basis of TET-induced lethality, we designed a complementation assay to test whether a given fragment of CENP-H is functional (Fig. 1). In brief, #5-5 CENP-H conditional knockout cells were transfected with an expression vector encoding the protein fragment of interest, and growth in medium containing TET was then examined. If the fragment complements the loss of CENP-H, cell growth should be normal. At the same time, we examined subcellular localization of the protein fragment. Localization analysis was performed in medium without TET to avoid the effect of CENP-H inactivation. To express various mutant forms of CENP-H, we generated GFP-tagged CENP-H derivatives in pEGFP, a cytomegalovirus (CMV) promoter-based expression vector. We previously developed a DT40 cell line in which the CENP-H gene was replaced with a gene encoding CENP-H-GFP, and we showed that the CENP-H-GFP fusion protein is functionally identical to CENP-H (12). To confirm this result, in the present, study we introduced vector encoding full-length CENP-H-GFP with the CMV promoter into #5-5 CENP-H conditional knockout cells. CMV-derived CENP-H-GFP localized to centromeres, and #5-5 CENP-H conditional knockout cells that expressed CMV-derived CENP-H-GFP proliferated normally after the addition of TET (Fig. 2), indicating that the GFP tag does not interfere with proper expression and function of CENP-H as previously shown in CENP-H replacement experiments (12).
FIG. 1.
Experimental scheme of complementation assay. We previously created CENP-H conditional knockout cell line #5-5 in which CENP-H expression is inactivated when tetracycline (tet) is added to culture medium. Various deletion derivatives of the CENP-H cDNA are cloned downstream of the CMV promoter (CMVpro) in the pEGFP vector. These constructs integrate at random sites in the genome of #5-5 CENP-H conditional knockout cells. Cells express both full-length CENP-H cDNA and mutant CENP-H. When tetracycline is added to culture medium, expression of full-length CENP-H is turned off, but mutant CENP-H is still expressed. If cells survive after addition of TET, the mutant CENP-H complements CENP-H function. We also investigated subcellular localization of mutant CENP-Hs. puror, puromycin resistance gene.
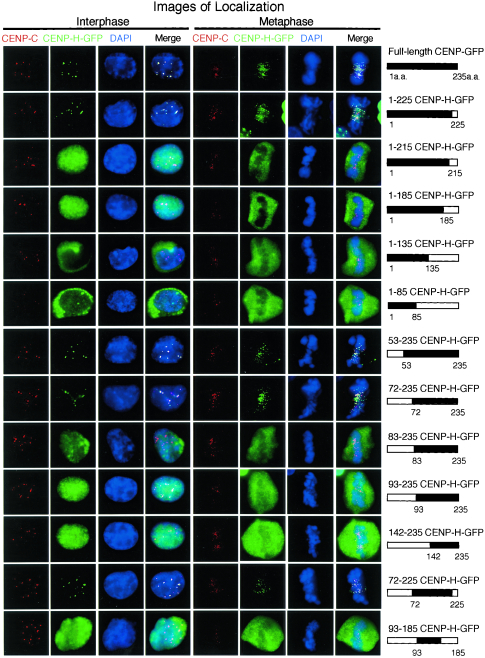
FIG. 2.
Identification of the minimal region required for CENP-H function. A schematic representation of CENP-H-GFP constructs and summary of the complementation and localization analysis are shown. To examine localization of CENP-H derivatives, an anti-CENP-C antibody was used to stain centromeres. Typical images for immunofluorescence are shown. Antibody signals were detected with Cy3-conjugated secondary antibodies (red), and GFP signals appear green. DNA was counterstained with DAPI (blue).
We created a series of GFP-tagged deletion derivatives of CENP-H in pEGFP and used the complementation assay to determine the minimal region required for cell viability. A growth curve was generated for each assay, and cell viability was judged from the growth curve (data not shown). The localization of these GFP-tagged CENP-H fusion proteins during interphase and metaphase was also investigated in the absence of TET by combination with immunostaining with anti-CENP-C antibody as a centromere marker because CENP-C localizes to centromeres throughout the cell cycle (11, 31). We first analyzed CENP-H carboxy-terminal truncation series. The carboxy-terminal 10-aa deletion construct (1-225CENP-H-GFP) complemented CENP-H deficiency, and this CENP-H derivative localized to centromeres (Fig. 2 and Table 1). Other carboxy-terminal truncations (1-215CENP-H-GFP, 1-185CENP-H-GFP, 1-135CENP-H-GFP, and 1-85CENP-H-GFP) did not complement CENP-H deficiency (Table 1). Although 1-215CENP-H-GFP and 1-185CENP-H-GFP localized to the nucleus in interphase, the other two derivatives localized to the cytoplasm, suggesting that the carboxy-terminal region, including aa 135 to 225, is required for nuclear targeting during interphase. However, the 1-215CENP-H-GFP and 1-185CENP-H-GFP derivatives did not associate with chromosomes during mitosis. We then analyzed a CENP-H amino-terminal truncation series. Both the 53-aa and 72-aa deletion constructs (53-235CENP-H-GFP and 72-235CENP-H-GFP) complemented CENP-H deficiency, and these CENP-H derivatives localized to centromeres (Fig. 2). Although other amino-terminal truncation mutants (83-235CENP-H-GFP, 93-235CENP-H-GFP, and 142-235CENP-H-GFP) did not complement CENP-H deficiency (Table 1), all amino-terminal deletion derivatives localized to the nucleus in interphase. These results are consistent with the idea that the carboxy-terminal region is important for nuclear targeting during interphase. Eleven amino acids (residues 72 to 83) are very important for centromere targeting because centromere localization was abolished in CENP-H deletion mutants lacking residues 1 to 82 but observed in a mutant containing aa 72 to 235. The significance of the 11-aa sequence will be discussed below. On the basis of the complementation assay and cellular localization data from CENP-H deletion derivatives, we concluded that the region containing aa 72 to 225 of chicken CENP-H is the minimal region necessary for centromere targeting and that this region is also essential for cell viability.
TABLE 1.
Complementation and localization analysis of CENP-H derivatives
| CENP-H derivative | Compen- sationa | Localization
|
|
|---|---|---|---|
| Interphase | Metaphase | ||
| Full-length CENP-H-GFP | + | Centromere | Centromere |
| 1-225CENP-H-GFP | + | Centromere | Centromere |
| 1-215CENP-H-GFP | − | Nucleus | Cytoplasm, centrosome |
| 1-185CENP-H-GFP | − | Cytoplasm, centrosome | Cytoplasm, centrosome |
| 1-135CENP-H-GFP | − | Nucleus | Cytoplasm, centrosome |
| 1-85CENP-H-GFP | − | Cytoplasm | Cytoplasm, centrosome |
| 53-235CENP-H-GFP | + | Centromere | Centromere |
| 72-235CENP-H-GFP | + | Centromere | Centromere |
| 83-235CENP-H-GFP | − | Nucleus | Cytoplasm |
| 93-235CENP-H-GFP | − | Nucleus | Cytoplasm |
| 142-235CENP-H-GFP | − | Nucleus | Cytoplasm |
| 72-225CENP-H-GFP | + | Centromere | Centromere |
| 93-185CENP-H-GFP | − | Nucleus | Cytoplasm, centrosome |
+ or −, normal growth was or was not observed, respectively, after transfection with the CENP-H derivatives into CENP-H-deficient cells.
CENP-H interacts with Hec1 in yeast two-hybrid analysis.
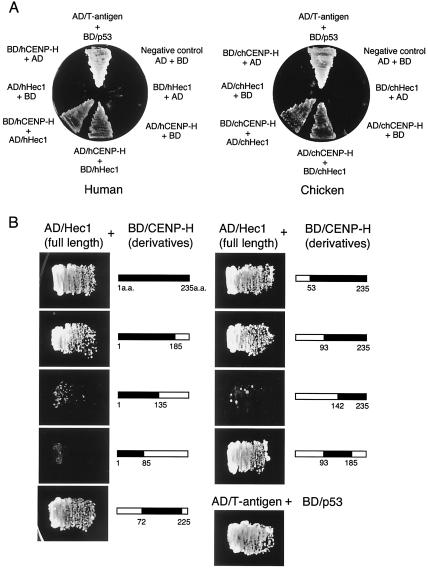
During yeast two-hybrid screening with human CENP-H as bait, human Hec1, a member of the Nuf2 complex (14), was picked up as a positive clone (Fig. 3A). To verify the interaction between CENP-H and Hec1, the constructs were then interchanged. The full-length coding region of Hec1 was cloned into the bait vector, and CENP-H was cloned into the prey vector; this combination also activated the reporter genes (Fig. 3A). We also investigated whether chicken CENP-H interacts with chicken Hec1, as is the case for human CENP-H and Hec1. As shown in Fig. 3B, the combination of chicken CENP-H and chicken Hec1 fused to the GAL4 transcription activation domain (AD/Hec1) induced expression of both reporter genes (His3 and Ade2) in yeast cells, suggesting that the interaction between CENP-H and Hec1 is conserved in vertebrate cells.
FIG. 3.
CENP-H interacts with Hec1. (A) Two-hybrid analysis of interactions between human CENP-H (hCENP-H) and Hec1 (hHec1) (left) and chicken CENP-H (chCENP-H) and Hec1 (chHec1) (right). As a positive control, the interaction between T antigen (AD/T antigen fusion) and p53 (BD/p53 fusion) is shown. AD indicates the GAL4 transcription activation domain, and BD indicates the DNA binding domain of GAL4. Growth was tested on SD minimal plates (−Ade/−His/−Leu/−Trp). (B) Identification of the region of chicken CENP-H required for interaction with chicken full-length Hec1. As bait, constructs of BD fused with CENP-H deletion derivatives were used. As a prey construct, AD fused with chicken Hec1 was used. Growth was tested on SD minimal plates (−Ade/−His/−Leu/−Trp).
We also used the two-hybrid system to map the region in chicken CENP-H that is responsible for the interaction with Hec1. We made constructs of GAL4 fused to carboxy- and amino-terminal deletion derivatives of CENP-H and tested the interaction with AD/Hec1 (Fig. 3B). Carboxy-terminal truncations of CENP-H did not interact with Hec1 {BD/CENP-H (aa 1 to 135) [BD/CENP-H(1-135)] and BD/CENP-H(1-85)}, whereas the amino-terminal 93-aa deletion of CENP-H [BD/CENP-H(93-235)] interacted with Hec1 (Fig. 3B). However, a longer amino-terminal deletion [BD/CENP-H(142-235)] did not interact with Hec1. From these results, we concluded that the carboxy region (aa 93 to 235) is responsible for the interaction with Hec1. Because the minimal region for CENP-H function is contained within the carboxy region (aa 72 to 225) (Fig. 2), the interaction with Hec1 may play an essential role in CENP-H function. The minimal region identified by complementation assay and localization test (Fig. 2) was slightly different from the region found to be essential for CENP-H-Hec1 interaction in yeast cells. BD/CENP-H(93-185) interacted strongly with AD/Hec1 (Fig. 3), whereas 93-185CENP-H-GFP did not localize to the centromere and did not complement CENP-H function (Fig. 2 and Table 1). The reason for this discrepancy was determined with coimmunoprecipitation experiments in DT40 cells (Fig. 4 and 5).
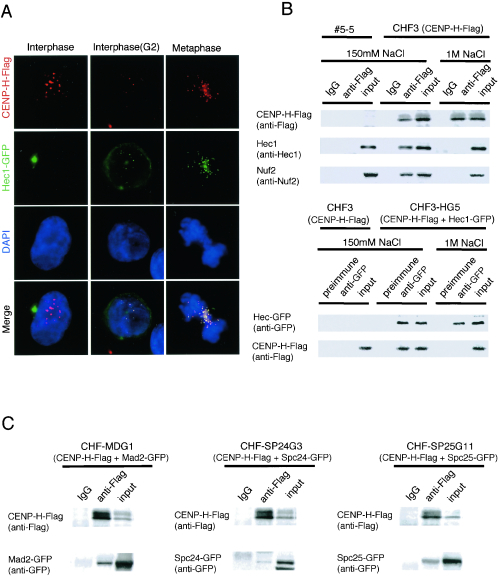
FIG. 4.
CENP-H interacts with the Nuf2 complex in DT40 cells. (A) Localization of Hec1-GFP (green) and CENP-H-Flag (red) at various stages of the cell cycle in DT40 cells that express Hec1-GFP and CENP-H-Flag simultaneously. Nuclei and chromosomes are counterstained with DAPI (blue). As shown in the merged images, colocalization of Hec1-GFP with CENP-H-Flag begins during G2 and is maintained during mitosis. (B) Coimmunoprecipitation analysis to investigate the interaction of CENP-H with Nuf2 and Hec1. CHF3 cells in which expression of CENP-H is replaced with CENP-H-Flag were prepared, and mitotic extracts of CHF3 cells were immunoprecipitated with anti-Flag antibody. Immunoprecipitates were separated by SDS-PAGE and analyzed by Western blotting with anti-Hec1 or anti-Nuf2 antibody. As a control, #5-5 CENP-H conditional knockout cells, which do not express Flag-tagged CENP-H, were used. CHF3-HG5 cells, which express both CENP-H-Flag and Hec1-GFP, were also prepared, and mitotic extracts of CHF3-HG5 cells were immunoprecipitated with anti-GFP antibody. Immunoprecipitateswere separated by SDS-PAGE and analyzed by Western blotting with anti-Flag antibody. One-eighth of anti-Flag immunoprecipitate with anti-Flag was loaded onto gels for CENP-H detection. (C) Coimmunoprecipitation analysis to investigate the interaction of CENP-H with Mad2, Spc24, and Spc25. CHF-MDG1 cells that express both CENP-H-Flag and Mad2-GFP, CHF-SP24G cells that express both CENP-H-Flag and Spc24-GFP, and CHFSP25G11 cells that express both CENP-H-Flag and Spc25-GFP were created. Mitotic extracts of these three cell lines were immunoprecipitated with anti-Flag antibody. Immunoprecipitates were separated by SDS-PAGE and analyzed by Western blotting with anti-Flag and anti-GFP antibodies. IgG, immunoglobulin G.
FIG. 5.
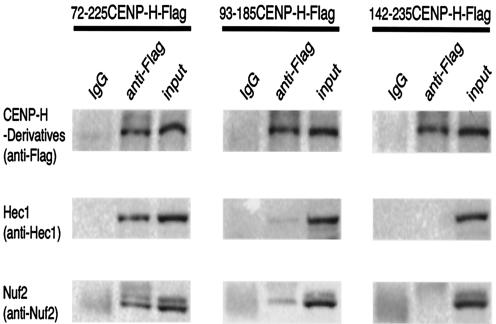
Coimmunoprecipitation analysis to investigate the interaction of CENP-H derivatives with Nuf2 complex. Cell lines expressing several CENP-H deletion derivatives were created. Mitotic extracts of these cell lines were immunoprecipitated with anti-Flag antibody. Immunoprecipitates were separated by SDS-PAGE and analyzed by Western blotting with anti-Nuf2 and Hec1 antibodies. One-eighth of each anti-Flag immunoprecipitate was separated by SDS-PAGE and detected with CENP-H as described in the legend of Fig. 4B. IgG, immunoglobulin G.
CENP-H interacts with the Nuf2 complex in DT40 cells.
Because CENP-H interacts with Hec1 in the yeast two-hybrid system, we investigated cellular localization of both proteins during the cell cycle. We made a DT40 cell line in which CENP-H-Flag and Hec1-GFP were expressed simultaneously, and we performed immunostaining with anti-Flag antibody during various cell cycle stages (Fig. 4A). As previously reported (14), Hec1 localized to the centrosome but not to the centromere during G1 and S phases, whereas CENP-H was localized to the centromere throughout the cell cycle (Fig. 4A). Hec1 translocates to the nucleus and localizes to centromeres in G2, and it remains associated with the centromere during mitosis. During G2 and mitosis, CENP-H-Flag and Hec1-GFP signals were perfectly colocalized at centromeres. Therefore, we used mitotic extracts to investigate interactions between CENP-H and Hec1 in DT40 cells by coimmunoprecipitation assay. Because Hec1 associates strongly with Nuf2 throughout the cell cycle (14), we also investigated the interaction of CENP-H with Nuf2. We prepared a cell line in which expression of CENP-H was replaced by expression of CENP-H-Flag (CHF3 cells). We isolated the soluble fraction from mitotic extracts of CHF3 cells in lysis buffer containing 150 mM NaCl and performed immunoprecipitation with anti-Flag antibody. Immunoprecipitates were then separated by SDS-PAGE and analyzed by Western blotting with anti-Hec1 or anti-Nuf2 antibody (Fig. 4B). Hec1 and Nuf2 were detected in immunoprecipitates with anti-Flag antibody. This association was abolished in extract buffer containing 1 M NaCl (Fig. 4B). As a control, we used #5-5 CENP-H conditional knockout cells, which do not express Flag-tagged CENP-H. Nuf2 and Hec1 were not detected in anti-Flag immunoprecipitates of #5-5 CENP-H conditional knockout cells (Fig. 4B). We also performed coimmunoprecipitation experiments with anti-GFP antibody using cells in which CENP-H-Flag and Hec1-GFP are expressed simultaneously (CHF3-HG5 cells). We analyzed the immunoprecipitates by Western blotting with anti-Flag antibody and detected CENP-H-Flag (Fig. 4B). As a control, we used CHF3 cells, which do not express Hec1-GFP but express CENP-H-Flag. CENP-H-Flag was not detected in anti-GFP immunoprecipitates of CHF3 cells (Fig. 4B). Recently, other members of the Nuf2 complex, Spc24 and Spc25, were identified in human cells (1). We cloned chicken homologs of Spc24 and Spc25 and created cell lines in which CENP-H-Flag and Spc24-GFP or CENP-H-Flag and Spc25-GFP are expressed simultaneously (CHF-SP24G3 and CHF-SP25G11, respectively). We performed coimmunoprecipitation experiments with anti-Flag antibody using these cells and analyzed the immunoprecipitates by Western blotting with anti-GFP antibody. We detected Spc24-GFP and Spc25-GFP signals (Fig. 4C). These results indicate that CENP-H interacts with the Nuf2 complex at centromeres in DT40 cells. It was previously reported that the Nuf2 complex interacts with the Mad2 complex, which is essential for the mitotic checkpoint pathway (14, 24). Therefore, we performed coimmunoprecipitation experiments with CENP-H and Mad2. We generated cells that expressed CENP-H-Flag and Mad2-GFP simultaneously (CHF-MDG1 cells) and performed immunoprecipitation experiments with anti-Flag antibody. We analyzed the immunoprecipitates by Western blotting with anti-GFP antibody and detected a Mad2-GFP signal (Fig. 4C).
As mentioned above, there is a discrepancy between the CENP-H minimal region necessary for centromere targeting and cell viability and that required for interaction with Hec1 in yeast two-hybrid analyses (Fig. 2 and 3). The CENP-H truncated mutant (aa 93 to 185) strongly interacted with Hec1 in yeast cells but did not target to the centromere during both interphase and metaphase. Therefore, we performed coimmunoprecipitation experiments with CENP-H truncated mutants (Fig. 5). We created cell lines that express 72-225CENP-H-Flag, 93-185CENP-H-Flag, and 142-235CENP-H-Flag, respectively. We then performed immunoprecipitation experiments with anti-Flag antibody. Immunoprecipitates were separated by SDS-PAGE and analyzed by Western blotting with anti-Hec1 or anti-Nuf2 antibody (Fig. 5). Immunoprecipitate with 72-225CENP-H gave Hec1 and Nuf2 signals, which are similar to that from immunoprecipitate with full-length CENP-H (Fig. 4B and 5). In contrast, immunoprecipitates with 93-185CENP-H or 142-235CENP-H gave weak or no signals for Hec1 and Nuf2 (Fig. 5). Theses results suggest that the 93-185CENP-H region can interact with Hec1 in yeast cells but does not bind strongly to Hec1 in DT40 cells because this region cannot target to the centromere. We observed that 93-185CENP-H-GFP was weakly localized to the centrosome during mitosis (Fig. 2). Hec1 was also weakly associated with the centrosome during mitosis (14). Therefore, we think that the weak interaction of 83-185CENP-H-Flag and Hec1 occurred at the centrosome. In conclusion, the interaction of CENP-H with Hec1 occurs mainly at the centromere during mitosis, and aa 72 to 225 of CENP-H are essential for CENP-H function.
Both CENP-H and Hec1 are stably associated with centromeres during mitosis.
We observed an interaction between CENP-H and Hec1 in yeast cells and chicken DT40 cells and were interested in examining the stability of each protein at the centromeres. We previously reported that the Hec1 association with the centrosome is dynamic but is stable with the centromere (14). Stability of CENP-H has not been previously described, although CENP-H is predicted to be a structural component of centromeres. We used iFRAP to study the kinetics of GFP-tagged CENP-H and Hec1 in living cells. With iFRAP, the entire chromosome or nucleus, with the exception of a small region of interest containing the CENP-H or Hec1 signals, is photobleached with a pulsed laser. The loss of the fluorescent signal is then monitored by time lapse microscopy. iFRAP, as opposed to FRAP, is the method of choice for such experiments because it provides relatively direct evidence of a protein's residence time in the chromosome or nucleus, and the measurement is independent of the size of these structures (7, 8). We used cell lines in which expression of CENP-H-GFP or Hec1-GFP was replaced with that of wild-type CENP-H or Hec1, respectively. To determine the stage of the cell cycle, we visualized the chromatin by histone H2B-mRFP expression. We measured the fluorescence intensity of at least 10 live cells for each sample.
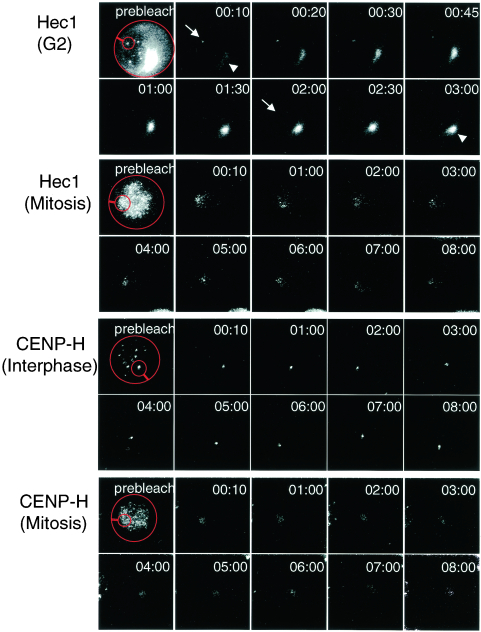
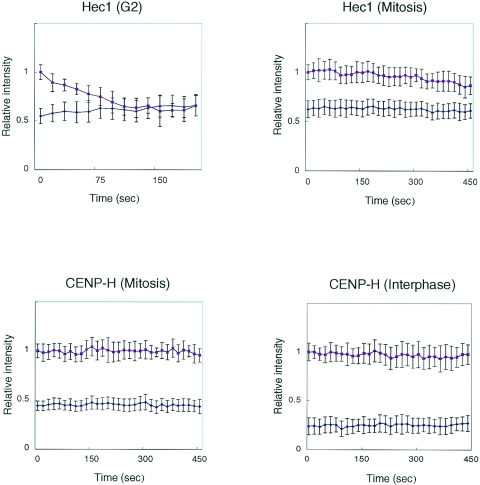
Because Hec1 localizes to the centromere during G2 and remains there during mitosis, we first performed iFRAP analysis to examine the stability of the Hec1-GFP association with centromeres. We measured the fluorescence intensities of the unbleached area and of the whole cells across time. During G2, the fluorescence intensity of the unbleached region decreased gradually, and the fluorescence intensity of the bleached region recovered (Fig. 6 ; see also http://spinner.lab.nig.ac.jp/∼tfukagaw/supplement_mcb/index.html). After approximately 150 s, the signal intensities of the whole cells and of the unbleached region became equivalent (Fig. 6 and 7; see also http://spinner.lab.nig.ac.jp/∼tfukagaw/supplement_mcb/index.html). In contrast, we observed little loss of fluorescence intensity of Hec1-GFP in the unbleached region during mitosis, and the fluorescence intensity of the bleached region remained constant (Fig. 6 and 7; see also http://spinner.lab.nig.ac.jp/∼tfukagaw/supplement_mcb/index.html). These findings indicate that Hec1-GFP associates stably with the centromere during mitosis. We investigated the stability of CENP-H-GFP by using iFRAP analysis during both mitosis and interphase (Fig. 6). The fluorescence intensity remained unchanged in both bleached and unbleached regions for at least 30 min (Fig. 6 and 7; see also http://spinner.lab.nig.ac.jp/∼tfukagaw/supplement_mcb/index.html), suggesting that CENP-H is stable at centromeres throughout the cell cycle. On the basis of the results of iFRAP analysis, we concluded that CENP-H and Hec1 form a stable association with the centromere during mitosis.
FIG. 6.
Images of photobleaching experiments. Turnover of Hec1-GFP and CENP-H-GFP was analyzed. With the exception of a small region of the centromere (small red circle), the entire cell (big red circle) expressing a GFP fusion protein was photobleached with a laser, and imageswere obtained by fluorescence time lapse microscopy. Prebleaching and recovery images are shown. In iFRAP for Hec1-GFP, unbleached centromere signals (arrow) decrease and fluorescence intensities of cytoplasm (arrowhead) and bleached centromeres increase during G2. The fluorescence intensity of unbleached regions for both Hec1-GFP during mitosis and CENP-H-GFP during both interphase and mitosis remains almost unchanged. The average intensity is plotted and shown in Fig. 7.
FIG. 7.
Graphic representation of iFRAP data. Quantification of iFRAP kinetics is shown. Red lines indicate loss of fluorescence in unbleached areas. Blue lines indicate relative fluorescence intensities of whole-cell regions. In iFRAP analysis of Hec1-GFP during G2, fluorescence intensities of whole cells and the unbleached region had equalized by ∼150 s. In contrast, the fluorescence intensity in unbleached regions for both Hec1-GFP during mitosis and CENP-H-GFP during both interphase and mitosis changed little.
DISCUSSION
Identification of the functional region of CENP-H.
We identified the functional domain of CENP-H by using a genetic system previously described (12). We previously showed that CENP-H is necessary for kinetochore assembly and is required for centromere targeting of CENP-C, which is an essential component of the kinetochore (12). Therefore, we believe that CENP-H is a structural component of the kinetochore assembly and that the functional domain of CENP-H should contain a centromere-targeting signal. Thus, we also investigated localization of GFP fusion forms of mutant CENP-H proteins. Deletions of the carboxy-terminal region (aa 1 to 135 and 1 to 85) lead to cytoplasmic localization of CENP-H, indicating that the carboxy-terminal region of CENP-H is responsible for nuclear localization. Putative nuclear localization sequence (NLS) (RKKR) was predicted from sequences of aa 158 to 161 and aa 152 to 155 of human and mouse CENP-H, respectively (35). The NLS is conserved in chicken CENP-H, which contains the RRKR sequence at aa 144 to 147. Because deletion of this sequence leads to cytoplasmic localization of CENP-H, these four amino acids appear to function as the NLS. Further analyses showed that a 153-amino-acid region of chicken CENP-H (aa 72 to 225) carries information sufficient to specify centromere localization of the protein. This centromere-targeting region can complement CENP-H deficiency. It is notable that this region (aa 72 to 225) shows relatively high similarity (59.4%) between human and chicken CENP-H, whereas the amino-terminal region (aa 1 to 71), which is not essential for centromere targeting and cell viability, shows lower sequence similarity (29.1%). When we deleted an additional 11 aa in the CENP-H (aa 83 to 235) mutant lacking residues 1 to 82, centromere localization was abolished, suggesting that these 11 amino acids (aa 72 to 83) contain important information for centromere targeting of CENP-H. This sequence is well conserved between human and chicken and may be a binding site for other kinetochore components and/or a site for modification such as phosphorylation. We indeed found a serine residue in the 11-aa sequence. When we examined the sequence with phosphoBase, a software tool used to predict phosphorylation (21), we found that this serine residue may be phosphorylated. It will be important to analyze these sequences further to understand the role of CENP-H in detail. It was previously reported that CENP-H forms multimers (35), and CENP-H deletion may result in the loss of multimerization of CENP-H, leading to a loss of centromere binding. Therefore, we examined multimerization of CENP-H by yeast two-hybrid analysis and coimmunoprecipitation experiments. However, we did not observe multimerization (see http://spinner.lab.nig.ac.jp/∼tfukagaw/supplement_mcb/index.html).
Centromere-targeting region of CENP-H interacts with Hec1.
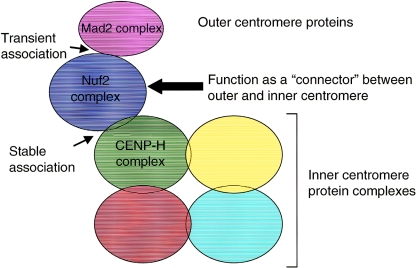
We identified the minimal region of CENP-H responsible for centromere targeting and cell viability. During screening for molecules that interact with CENP-H by a yeast two-hybrid analysis, we isolated Hec1 and focused on this protein in the present study. Hec1 was originally identified as a retinoblastoma protein-associated protein (9), and microinjection of Hec1 antibodies into cultured cells disrupts mitotic progression (3). Ndc80p, the yeast homologue of Hec1, makes a complex with Nuf2p, Spc24p, and Spc25p in S. cerevisiae (18, 40); we and other groups showed that this complex is evolutionarily conserved (1, 6, 14, 24, 25, 27). Martin-Lluesma et al. (24) reported that human Hec1 interacts with Mad1 and is required for recruitment of spindle checkpoint components, including Mps1 and Mad1/Mad2 complexes, to centromeres. Although detailed localization of the Mad2 complex has not been mapped to the kinetochore plate, another checkpoint protein, BubR1, has been mapped to the outer kinetochore (17). Because checkpoint proteins must associate transiently with the kinetochore, it is reasonable to expect that they would localize to the outer kinetochore plate. FRAP analysis of Mad2-GFP at the centromere revealed that turnover of this protein at the kinetochore is rapid (15, 16, 32). In contrast, the iFRAP analysis reported here revealed that localization of Hec1 at the centromere is stable during mitosis (Fig. 6 and 7). Considering the results of these FRAP analyses, we propose that the Nuf2 complex, including Hec1, is stably associated with the centromere and provides a site for checkpoint proteins (outer kinetochore proteins) to work in regulation of cell cycle progression. At the same time, the Nuf2 complex associates with CENP-H, which is an inner kinetochore protein. CENP-H is a constitutive centromere protein and is predicted to be a structural component for kinetochore assembly (12). Our iFRAP analysis revealed that localization of CENP-H-GFP at the centromere is quite stable throughout the cell cycle, and this result supported our idea that CENP-H is a structural component of the inner kinetochore. There are two classes of centromere proteins. One class comprises structural components of the inner kinetochore, including CENP-C, -H, and -I. The other class includes proteins, such as mitotic checkpoint proteins, that transiently associate with the kinetochore. In the present study, we showed that the Nuf2 complex interacts with CENP-H; there have been reports that the Nuf2 complex interacts with the Mad2 complex, which associates transiently with the kinetochore (15, 16, 32). Here, we propose a model in which the Nuf2 complex functions as a connector between the inner and outer kinetochores (Fig. 8), although we could not complete this model until electron microscope data were available. Our iFRAP data suggest that the Nuf2 complex is tightly associated with CENP-H during mitosis, and in our model, the Nuf2 complex is anchored to the inner kinetochore and provides the site for outer kinetochore proteins, including Mad2 complex (Fig. 8). We previously reported that during interphase, a prekinetochore structure is created with constitutive centromere proteins, including CENP-A, -C, -H, and -I and other centromere proteins assembled into prekinetochore after nuclear envelope breakdown (mitosis). The Nuf2 complex associates with the centromere during interphase (G2 phase), and this timing is earlier than that of other centromere proteins that associate transiently with the centromere (Fig. 4) (14). This temporal localization is consistent with our model in which the Nuf2 complex is integrated into the inner kinetochore in late G2 and functions as a connector between the inner and outer kinetochores during mitosis.
FIG. 8.
Model of the relation between inner and outer kinetochore structures. The inner kinetochore structure is divided into several subcomplexes that may interact with each other. The CENP-H complex is one such subcomplex. The Nuf2 complex is targeted to the inner kinetochore during G2 and forms a stable association with the CENP-H complex during mitosis. The Nuf2 complex also interacts with the Mad2 complex, which is a component of the outer kinetochore and is associated transiently with centromeres. In this model, we propose that the Nuf2 complex functions as a connector between inner and outer kinetochores.
Although we found a physical relation between CENP-H and the Nuf2 complex, there is a discrepancy in the CENP-H and Nuf2/Hec1 knockout phenotype, which was described previously (12, 14). If CENP-H interacts with the Nuf2 complex at the kinetochore, it is not clear why CENP-H-deficient cells delay mitosis and proceed to the next cell cycle before cell death, in contrast to Hec1- and Nuf2-deficient cells, which show mitotic arrest. We think that CENP-H may be a structural component in the kinetochore, whereas Hec1/Nuf2 may be involved in a signaling pathway that regulates cell cycle progression. In CENP-H-deficient cells, the kinetochore structure was disrupted. However, weak Nuf2/Hec1 signals were still visible, although a majority of proteins were reduced (14). A small amount of Nuf2/Hec1 protein may be involved a signaling pathway to some extent. However, this signaling pathway is not active in Nuf2/Hec1-deficient cells, which leads to mitotic arrest. We believe that the discrepancy in knockout phenotypes is due to a difference in the functional role of each protein in the kinetochore.
Here, we report the interaction of CENP-H with the Nuf2 complex. However, we estimated that only 12.5% of Hec1 was recovered by immunoprecipitation of CENP-H because only one-eighth of the anti-Flag immunoprecipitate was used for CENP-H detection (Fig. 4B). This finding suggests that CENP-H interacts with other molecules. Sugata et al. (35) previously reported that human CENP-H interacts with MCAK in vitro. MCAK is a mitotic centromere-associated kinesin and is thought to localize to centromeric heterochromatin and the inner kinetochore (41). In addition, CENP-H should associate with constitutive centromere components. We are presently studying the mechanism of assembly of the kinetochore using the DT40 knockout system (13). Recent proteomics approaches have revealed that the kinetochore structure is divided into several subcomplexes in S. cerevisiae (39). Therefore, there should be a CENP-H core complex, and this core complex should interact with components of other subcomplexes, including the Nuf2 complex in the vertebrate kinetochore (Fig. 8).
Acknowledgments
We are very grateful to K. Kita for technical assistance and M. Okada for discussion. We thank T. Kanda, G. M. Wahl, R. Y. Tsien, and T. Saiwaki for plasmids.
This work was supported by PRESTO of JST and a grant-in-aid for Scientific Research on Priority Areas “Genome Biology” and “Cell Cycle” from the Ministry of Education, Science, Sports, and Culture of Japan.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bharadwaj, R., W. Qi, and H. Yu. 2004. Identification of two novel components of the human Ndc80 kinetochore complex. J. Biol. Chem. 279:13076-13085. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Y., D. J. Riley, P. L. Chen, and W. H. Lee. 1997. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol. Cell. Biol. 17:6049-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 5.Craig, J. M., W. C. Earnshaw, and P. Vagnarelli. 1999. Mammalian centromeres: DNA sequence, protein composition, and role in cell cycle progression. Exp. Cell Res. 246:249-262. [DOI] [PubMed] [Google Scholar]
- 6.DeLuca, J. G., B. J. Howell, J. C. Canman, J. M. Hickey, G. Fang, and E. D. Salmon. 2003. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 13:2103-2109. [DOI] [PubMed] [Google Scholar]
- 7.Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. I. Rothblum, R. D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298:1623-1626. [DOI] [PubMed] [Google Scholar]
- 8.Dundr, M., M. D. Hebert, T. S. Karpova, D. Stanek, H. Xu, K. B. Shpargel, U. T. Meier, K. M. Neugebauer, A. G. Matera, and T. Misteli. 2004. In vivo kinetics of Cajal body components. J. Cell Biol. 164:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durfee, T., K. Becherer, P. L. Chen, S. H. Yeh, Y. Yang, A. E. Kilburn, W. H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 10.Fukagawa, T., and W. R. A. Brown. 1997. Efficient conditional mutation of the vertebrate CENP-C gene. Hum. Mol. Genet. 6:2301-2308. [DOI] [PubMed] [Google Scholar]
- 11.Fukagawa, T., C. Pendon, J. Morris, and W. Brown. 1999. CENP-C is necessary but not sufficient to induce formation of functional centromere. EMBO J. 18:4196-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukagawa, T., Y. Mikami, A. Nishihashi, V. Regnier, T. Haraguchi, Y. Hiraoka, N. Sugata, K. Todokoro, W. Brown, and T. Ikemura. 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20:4603-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukagawa, T. 2004. Assembly of kinetochore in vertebrate cells. Exp. Cell Res. 296:21-27. [DOI] [PubMed] [Google Scholar]
- 14.Hori, T., T. Haraguchi, Y. Hiraoka, H. Kimura, and T. Fukagawa. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347-3362. [DOI] [PubMed] [Google Scholar]
- 15.Howell, B. J., D. B. Hoffman, G. Fang, A. W. Murray, and E. D. Salmon. 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150:1233-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howell, B. J., B. Moree, E. M. Farrar, S. Stewart, G. Fang, and E. D. Salmon. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953-964. [DOI] [PubMed] [Google Scholar]
- 17.Jablonski, S. A., G. K. T. Chan, C. A. Cooke, W. C. Earnshaw, and T. J. Yen. 1998. The hBUB1 and BUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma 107:386-396. [DOI] [PubMed] [Google Scholar]
- 18.Janke, C., J. Ortiz, J. Lechner, A. Shevchenko, A. Shevchenko, M. M. Magiera, C. Schramm, and E. Schiebel. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalitsis, P., K. J. Fowler, E. Earle, J. Hill, and K. H. A. Choo. 1998. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc. Natl. Acad. Sci. USA 95:576-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanda, T., K. F. Sullivan, and G. M. Wahl. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377-385. [DOI] [PubMed] [Google Scholar]
- 21.Kreegipuu, A., N. Blom, and S. Brunak. 1999. PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res. 27:237-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 23.Liu, S. T., J. C. Hittle, S. A. Jablonski, M. S. Campbell, K. Yoda, and T. J. Yen. 2003. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 5:341-345. [DOI] [PubMed] [Google Scholar]
- 24.Martin-Lluesma, S., V. M. Stucke, and E. A. Nigg. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297:2267-2270. [DOI] [PubMed] [Google Scholar]
- 25.McCleland, M. L., M. J. Kallio, G. A. Barrett-Wilt, C. A. Kestner, J. Shabanowitz, D. F. Hunt, G. J. Gorbsky, and P. T. Stukenberg. 2004. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr. Biol. 14:131-137. [DOI] [PubMed] [Google Scholar]
- 26.Mellone, B. G., and R. C. Allshire. 2003. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Cell Biol. 13:191-198. [DOI] [PubMed] [Google Scholar]
- 27.Nabetani, A., T. Koujin, C. Tsutsumi, T. Haraguchi, and Y. Hiraoka. 2001. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma 110:322-334. [DOI] [PubMed] [Google Scholar]
- 28.Nishihashi, A., T. Haraguchi, Y. Hiraoka, T. Ikemura, V. Regnier, H. Dodson, W. C. Earnshaw, and T. Fukagawa. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell 2:463-476. [DOI] [PubMed] [Google Scholar]
- 29.Palmer, D. K., and R. L. Margolis. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pluta, A. F., A. M. Mackay, A. M. Ainsztein, I. G. Goldberg, and W. C. Earnshaw. 1995. The centromere: hub of chromosomal activities. Science 270:1591-1594. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh, H., J. Tomkiel, C. A. Cooke, H. Ratrie, M. Maure, N. F. Rothfield, and W. C. Earnshaw. 1992. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70:115-125. [DOI] [PubMed] [Google Scholar]
- 32.Shah, J. V., E. Botvinick, Z. Bonday, F. Furnari, M. Berns, and D. W. Cleveland. 2004. Dynamics of centromere and kinetochore proteins: implications for checkpoint signaling and silencing. Curr. Biol. 14:942-952. [DOI] [PubMed] [Google Scholar]
- 33.Shelby, R. D., O. Vafa, and K. F. Sullivan. 1997. Assembly of CENP-A into centromere chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugata, N., E. Munekata, and K. Todokoro. 1999. Characterization of a novel kinetochore protein, CENP-H. J. Biol. Chem. 274:27343-27346. [DOI] [PubMed] [Google Scholar]
- 35.Sugata, N., S. Li, W. C. Earnshaw, T. J. Yen, K. Yoda, H. Masumoto, E. Munekata, P. E. Warburton, and K. Todokoro. 2000. Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere-kinetochore complexes. Hum. Mol. Genet. 9:2919-2926. [DOI] [PubMed] [Google Scholar]
- 36.Sullivan, B. A., and S. Schwartz. 1995. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 4:2189-2197. [DOI] [PubMed] [Google Scholar]
- 37.Tomkiel, J., C. A. Cooke, H. Saitoh, R. L. Bernat, and W. C. Earnshaw. 1994. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol. 125:531-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warburton, P. E., C. A. Cooke, S. Bourassa, O. Vafa, B. A. Sullivan, G. Stetten, G. Gimelli, D. Warburton, C. Tyler-Smith, K. F. Sullivan, G. G. Poirier, and W. C. Earnshaw. 1997. Immunolocalization of CENP-A suggests a novel nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7:901-904. [DOI] [PubMed] [Google Scholar]
- 39.Westermann, S., I. M. Cheeseman, S. Anderson, J. R. Yates III, D. G. Drubin, and G. Barnes. 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163:215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wordeman, L., and T. J. Mitchison. 1995. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]