Abstract
Abstract
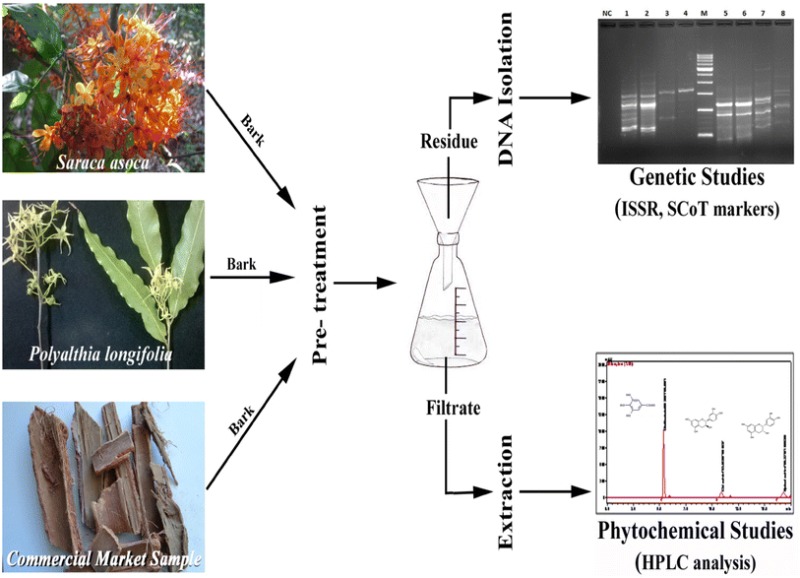
DNA fingerprinting singly or in combination with phytochemical analysis is ideal for quality control of crude plant-based drugs. However, when the source material is tannin rich stem bark, extraction of DNA by conventional methods becomes challenging. In such cases, phytochemical profiling serves as very useful tool for its identification. The work herein described a method for simultaneous DNA isolation and phytochemical extraction for downstream analysis and applications from dried bark powder of Saraca asoca and commercial samples of this crude drug as well as from those of Polyalthia longifolia, its most common adulterant. It is a modified CTAB-based method which involves a pre-extraction step by soaking samples overnight in de-ionized water followed by filtration. The residues in the filter paper were used for DNA isolation and dried filtrate was used for Reverse Phase-High-Performance Liquid Chromatography analysis. Results revealed that genomic DNA isolated was PCR amplifiable with Inter Simple Sequence Repeat and Start Codon Targeted markers. Phenolic compounds of catechin, epicatechin, and gallic acid were detected from the above dried filtrate. The method is simple, reliable and it requires small amount of sample with an option of integrating both phytochemical and DNA-based profiling, from the same starting material. Therefore, the present method could be useful for further potential applications such as quality control assessment of S. asoca products.
Graphical abstract
Keywords: Adulteration, DNA and chemical fingerprinting, HPLC, PCR, Saraca asoca
Introduction
DNA-based identification tools are gaining importance in plant genetic studies. The methods such as DNA barcoding and High-Resolution Melting (HRM) analysis were successfully applied in plant research (Osathanunkul et al. 2015; Simko 2016). Several characterization methods such as Amplified Fragment Length Polymorphism (AFLP), Inter Simple Sequence Repeat (ISSR), Short Tandem Sequence (STS), Simple Sequence Repeat (SSR) etc. have been frequently used techniques (Li et al. 2013; Ganie et al. 2015). For these techniques, quality of DNA assumes prime importance. In recent years, different methods to isolate DNA from various plant sources were reported. However, none of these have been established as a single, universally applicable method. Isolation of quality DNA from wood or bark is challenging due to the presence of polyphenols, proteins, and polysaccharides, which are the major, inhibitors interfering DNA isolation procedures (Rezadoost et al. 2016). Therefore, depending on plant species and part used, the DNA isolation methods are generally modified and optimized.
Saraca asoca (Roxb.) De Wilde (Caesalpiniaceae) is a red listed vulnerable medicinal plant. It is traditionally known for its use in the treatment of a variety of gynaecological disorders (Nadkarni 1976; Hegde et al. 2007; Tandon and Yadav 2017). ‘Ashokrishtam’ and ‘Ashokaghritham’ with few others are major ayurvedic medicinal preparations made from the bark of this plant and extensively traded in India (Singh et al. 2015). As per the reports of the National Medicinal Plant Board (NMPB) and Foundation for Revitalization of Local Health Traditions (FRLHT), India, domestic demand of ‘Ashoka’ bark is ≥100 metric tonnes per year (Ved and Goraya 2007). Ever-increasing demand and dwindling supply due to rapidly vanishing S. asoca populations have resulted in rampant substitution/adulteration of the crude drug to match the demand. Saraca asoca has been substituted/adulterated with various other plant materials (Singh et al. 2015), most commonly with P. longifolia (Sonnerat) Thwait. (Annonaceae), which has earned it the name of “(False) Ashoka” (Sarin 1996; Khatoon et al. 2009). It is perhaps because of this extensive substitution that scarcity of the crude drug is not observed in herbal market.
Chemoprofiling has generally been a standard practice for species identification and quality control of crude drugs (Avula et al. 2007; Urumarudappa et al. 2016). However, similar morphology and chemical fingerprints of botanicals have often resulted in misidentified (Osathanunkul et al. 2016). DNA fingerprinting analysis in such cases is necessary. However, DNA analysis is often not practicable because of possibility of DNA degradation in commercial samples. Therefore, ideally, combination of both genetic and chemical profiling would be better suited for identification and authentication of plant species (Smillie and Khan 2010; Urumarudappa et al. 2016). Unfortunately, when the crude drug is a bark as in case of S. asoca, DNA isolation becomes very difficult due to the presence of dead cells and several inhibitors of DNA. Herein, we present a simple and easy method that can be used for simultaneous DNA isolation and chemoprofiling from dried bark powder of S. asoca, crude drug samples of S. asoca as well as of its adulterant P. longifolia. The isolated DNA samples were good enough for further downstream applications like ISSR and SCoT fingerprinting analysis as well as RP-HPLC analysis, assays that can be used for quality assessment and/or authentication. In addition, as a tangential utilization of the method, Total Phenolic Content (TPC) and Total Protein Content (TProC) were carried out to analyse the possibility of lowering interference during the isolation of quality DNA.
Materials and methods
Plant sampling
Bark samples of S. asoca (Codes: SA1 and SA2) and P. longifolia (Codes: PL1 and PL2) were collected from Siddapur region (N:14°21.442′E:074°45.684′ Elevation: 520 m) of Western Ghats (Karnataka State) India. Flowering twigs of S. asoca and P. longifolia were morphologically identified and herbarium were deposited at ICMR–NITM, Belagavi, Karnataka, India (Voucher Number: S. asoca: RMRC 997; P. longifolia: RMRC 1256).
Commercial market samples and processing
Commercial bark samples sold as S. asoca were obtained from local markets of Chennai (Tamil Nadu: MA1) and Bengaluru (Karnataka: MA2) India. All the obtained plant materials were thoroughly dried at 45 ± 2 °C in a hot air oven, powdered in a domestic grinder, and used for further processing.
Pre-treatment
Powdered bark samples (250 mg) were soaked in deionised water (10 mL) overnight (14 h) with continuous shaking (110 ± 5 rpm) at 25 ± 2 °C on an orbital shaker (Rivotek, India). The extracts were then filtered through Whatman no. 1 filter paper and the resulted residue was dried in oven at 45 ± 2 °C. The filtrate was kept separately at 40 °C for evaporation.
DNA isolation
The dried residue (powders) were suspended in 1.5 mL of preheated extraction buffer [2% w/v CTAB, 100 mM Tris HCl (pH 8.0), 1.4 M NaCl, 20 mM EDTA, 100 mM polyvinylpyrrolidone (PVP) (w/v), and 1% β-mercaptoethanol (v/v) added immediately before use] in a 15 mL Oakridge tube.
The mixture was incubated at 65 °C for 30 min with intermittent shaking and cooled to room temperature, and 500 µL of 5 M potassium acetate was added, mixed gently, and incubated at −20 °C for 1 h.
Subsequently, 2 mL of chloroform–isoamyl alcohol (24:1) was added and mixed thoroughly followed by centrifugation at 10,000 rpm for 20 min.
The aqueous layer was collected in fresh centrifuge tubes and equal volume of 30% polyethylene glycol (PEG) 600 was added, gently mixed, and incubated at −20 °C for 1 h.
After centrifugation at 10,000 rpm for 25 min, the supernatant was discarded.
500 µL of absolute alcohol was added and incubated at −20 °C for 1 h and centrifuged.
The pellets were washed with 70% ethanol by centrifugation at 10,000 rpm for 10 min, supernatant discarded and pellets were air dried.
The DNA pellet was dissolved in 20 μL TE buffer and stored in −20 °C.
Determination of quality and quantity of isolated DNA
The quality and quantity of isolated DNA were determined using Nanodrop Spectrophotometer (JH BIO, USA) at 260/280, 260/230 nm ratio as well as visually by horizontal electrophoresis on 0.8% agarose gels stained with GelRed (Biotium Inc., USA). Each sample was diluted to 40 ng/μL with TE buffer (10 mM Tris HCI, pH 8.0 and 0.1 mM EDTA, pH 8.0) for PCR amplification and stored at −20 °C.
PCR amplifications
Two primers each of ISSR (UBC, Canada) and SCoT markers (Collard and Mackill 2009) (Sigma Aldrich, India) (Table 1) were used for the study. ISSR amplification was performed in a 25 µL reaction volumes containing 40 ng genomic DNA, 10 μM primer, 200 µM of each dNTP (Merck, India), 3 U/μL of Taq DNA polymerase (Merck, India), along with 10X PCR buffer supplied (TrisHCl, pH 9.0; 15 mM MgCl2). The ISSR–PCR condition consisted of an initial step of denaturation at 94 °C for 5 min, followed by 45 cycles of denaturation at 94 °C for 45 s, annealing at 49 °C for 45 s and extension at 72 °C for 1 min and an additional final extension of 72 °C for 10 min was followed. For SCoT amplification, annealing temperature was raised to 50 °C and the remaining conditions were same as that of ISSR amplification.
Table 1.
Primers used for ISSR and SCoT amplifications during the study
| Primer name | Sequence (5′–3′) |
|---|---|
| ISSR primers | |
| UBC 834 | AGAGAGAGAGAGAGAGYTa |
| UBC 880 | GGAGAGGAGAGGAGA |
| SCoT primers | |
| S-2 | CAACAATGGCTACCACCCT |
| S-3 | CAACAATGGCTACCACCGC |
aY = C + T
ISSR and SCoT marker amplifications were performed in Mastercycler® Nexus (Eppendorf AG, Germany) thermal cycler. The electrophoresis of the PCR products was carried out using 1.5% agarose gel in 1X TAE buffer and GelRed (Biotium Inc, USA) as a staining dye. Agarose gels were photographed using gel documentation system (Syngene, UK). In both amplifications, negative controls were maintained as reactions without template DNA. To ensure reproducibility, each PCR amplification was repeated at least twice.
TPC and TProC
The extract obtained as filtrate from bark samples soaked in deionised water was reduced to dryness on a water bath (85 ± 5 °C), re-dissolved in water (0.1% w/v), and used for TPC estimation. Total phenolic content was estimated using previously reported modified Folin–Ciocalteu method (Pawar et al. 2011). The absorbance of the blue colour was read at 760 nm on Thermo, 60G scan, microplate reader (spectrophotometer). The extracts prepared (0.1% w/v in water) were quantified and the results were compared with standard curves of tannic acid. The results were expressed as gram tannic acid equivalent per 100 g dry powdered samples (g/100 g or %). The above extracts were used to estimate total proteins using Lowry’s method (Lowry et al. 1951). The absorbance at 660 nm was read for five different concentrations (40, 60, 80, 100, and 120 µg/mL) of Bovine Serum Albumin (BSA) as standard and were compared with values of bark extracts to estimate total protein. The results were expressed in mg/g dried bark powder.
Chemoprofiling using RP-HPLC analysis
Sample and standard preparation for RP-HPLC analysis
A known amount of dried extract obtained as filtrate from bark samples soaked in de-ionized water was dissolved in the solvent system to obtain mg/mL concentration samples. Similarly, different concentrations of gallic acid (0.012, 0.1, 1, and 5 µg/mL), catechin (0.02, 0.1, 1, 5, 10, and 20 µg/mL), and epicatechin (0.02, 0.1, 1, 5, 10, and 20 µg/mL) were prepared in methanol and were used to obtain calibration and linearity. All test samples were passed through 0.20 µm NY-membrane (Sartorius, Germany) before injecting into HPLC.
Instrumentation and RP-HPLC chromatographic conditions
RP-HPLC analysis was performed on a Shimadzu chromatographic system (Model no. LC-20AD) consisting of a quaternary pump, auto injector (SIL20-ACHT), degasser (DGU-20A5), and dual λ ultraviolet (UV) absorbance diode array detector (Model no. SPD-M20A). The built in LC (liquid chromatography)-solution software system was used for data processing. Chromatographic separation was achieved on a CAPCELL PAK C18 MG II S5 250–4.6 mm (5 μm) column. A mobile phase consisting of “A” (acetonitrile), “B” (water), and “C” (glacial acetic acid) was used for separation with 12:85:3 in an isocratic mode with injection volume of 10 μL. The flow rate was 0.7 mL/min and the detection wavelength of photodiode array (PDA) was set to 280 nm with 20 min run time for both standard and sample. Using the above, calibration curve for the standard was made and unknown concentration of catechin, gallic acid, and epicatechin in the bark samples of S. asoca was determined.
Statistical analysis
The experiment was performed in triplicate and the data were reported as means and ±standard deviation. GraphPad Prism (Evaluation version) was used for statistical analysis.
Results and discussion
DNA isolation and analysis
The isolated DNA was confirmed by 0.8% agarose gel (Fig. 1) as PCR amplifiable genomic bands. The average yield of DNA isolated using the present protocol ranged from 0.550 ± 0.028 to 1.557 ± 0.078 µg/g dry powders and the ratio of OD 260/230 and 260/280 is presented in Table 2. The ratio of absorbance at 260/280 between 1.7 and 1.9 is used as a standard for determining a pure DNA sample. Anything below 1.7 indicates the presence of protein and to a lesser extent latent phenol or carbohydrate, contamination and above 1.9 indicates contamination with RNA (Llongueras et al. 2013).
Fig. 1.
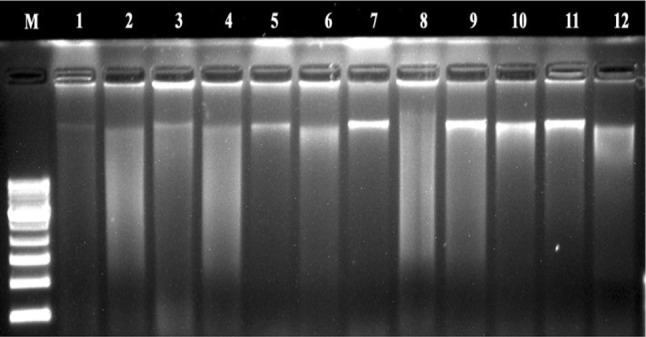
DNA samples isolated in duplicates (M mol. wt. markers, lanes 1–4: two individuals of S. asoca bark samples (SA1&SA2); lanes 5–8: MA2 and MA1 market samples; and lanes 9–12: two individuals of P. longifolia samples (PL1&PL2)
Table 2.
Average yields and 260/230, 260/280 absorbance ratios of isolated DNA
| Code | 260/230a | 260/280a | Concentrationa (ng/µL) | Yielda (µg/0.1 g) |
|---|---|---|---|---|
| SA1 | 0.86 ± 0.04 | 1.58 ± 0.08 | 110.80 ± 5.52 | 0.886 ± 0.044 |
| SA2 | 1.23 ± 0.06 | 1.73 ± 0.09 | 194.61 ± 9.73 | 1.557 ± 0.078 |
| MA1 | 0.55 ± 0.03 | 1.48 ± 0.07 | 134.00 ± 6.76 | 1.072 ± 0.054 |
| MA2 | 0.72 ± 0.04 | 0.95 ± 0.05 | 68.82 ± 3.44 | 0.550 ± 0.028 |
| PL1 | 0.74 ± 0.03 | 1.71 ± 0.9 | 127.18 ± 6.34 | 1.017 ± 0.051 |
| PL2 | 1.37 ± 0.07 | 1.88 ± 0.08 | 118.67 ± 5.94 | 0.949 ± 0.047 |
aValues in table represent mean of three extractions ±SD
The isolated DNA samples were amplified using ISSR (UBC 834, UBC 880) (Fig. 2a) and SCoT (S2, S3) markers (Fig. 2b). Amplification occurred with the use of both the markers with prediction of consistent bands revealed good quality of DNA. It was also interesting to note that isolated DNA obtained from commercial market sample MA2 had lower purity (0.95 ± 0.05 in 260/280) and concentration (068.82 ± 3.44 ng/µL) compared to others. Sample MA2 resulted in poor PCR amplification and showed sheared bands when resolved on 1.5% agarose gel (Fig. 2c). The observations of the present study are in accordance with previous reports on adulteration, wherein it was reported that longer storage and adulteration/substitution affect the drug including its DNA quality (Li et al. 2013; Smillie and Khan 2010; Kazi et al. 2013).
Fig. 2.
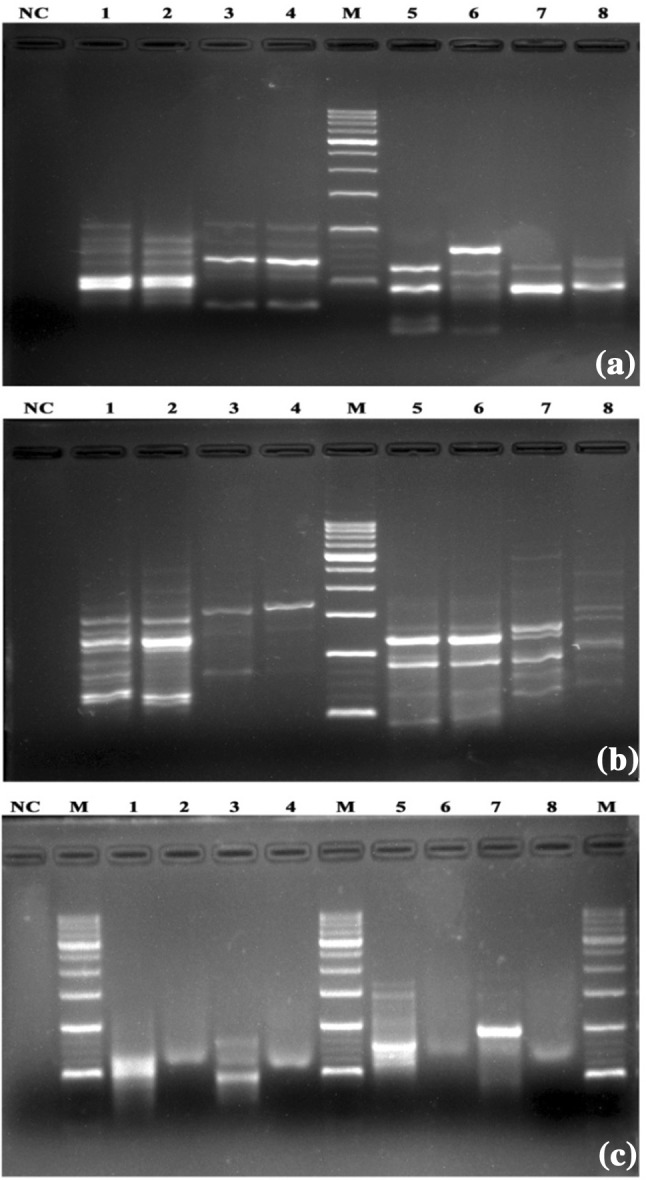
a ISSR profile of S. asoca (primer UBC 834: lanes 1–2; primer UBC 880: lanes 5–6) and P. longifolia (primer UBC 834: lanes 3–4; primer UBC 880: lanes 7–8). b SCoT profile of S. asoca (primer S-2: lanes 1–2; primer S-3: lanes 5–6) and P. longifolia (primer S-2: lanes 3–4; primer S-2: lanes 7–8). c ISSR and SCoT profile of MA1 (primer UBC 834: lane 1 and UBC 880: lane 3; primer S-2: lane 5 and S-3: lane 7) MA2 samples (primer UBC 834: lane 2 and UBC 880: lane 4; primer S-2: lane 6 and primer S-3 lane 8). a–c M standard molecular weight marker (100 + 500 bp), NC negative control
Isolation of quality DNA from dried, powdered plant material and from highly differentiated tissue is challenging. To overcome this issue, the present method was designed and optimized from the methods previously described for recalcitrant Piper nigrum berries and the market samples of dry tea (Singh et al. 1999; Dhanya et al. 2007). The major problem in using bark as source for DNA is that they usually contain higher amount of tannins and polyphenols that inhibit DNA isolation and PCR. Thus, to reduce the interference of such bio-compounds during extraction, all the dried bark powders were soaked overnight in de-ionized water. This pre-extraction step was found essential, without which DNA yield was lower, and there was shear in the DNA during agarose gel electrophoresis, which, in turn affected PCR amplification. The elimination of polyphenols was also ensured by adding potassium acetate, PVP, and PEG treatments during isolation of DNA. This step is necessary to remove secondary metabolites and coloured pigments to some extent (Dhanya et al. 2007). It was observed in the present study that use of potassium acetate instead of sodium acetate significantly improved the quality of DNA isolated with better amplification.
DNA profiles obtained for S. asoca, commercial market samples, and those of P. longifolia exhibited clear differences. Even SCoT markers proved a clear polymorphism within S. asoca distinct from P. longifolia. Although commercial sample (MA2) showed shearing of bands after PCR amplification, the method was found to hold good for the rest of the samples. The different procedures of processing of raw drugs or adulterations might be possible reasons for the poor amplification of MA2 (Smillie and Khan 2010; Kazi et al. 2013).
TPC and TProC analysis
Barks are rich in polyphenols especially tannic acid, which retards DNA isolation. Soaking of the bark powder in de-ionized water for 12–14 h (overnight) removed hydrolysable polyphenols and tannins. The extractive yield from the filtrate of overnight soaked de-ionized water was calculated (Table 3). The total phenolic content of all these samples was determined in equivalence of tannic acid (Table 3). The ranges of extractive yields were from 06.24 ± 0.312 to 18.92 ± 0.946 g/100 g. TPC ranged from highest in MA1 to the lowest in PL2 (Table 3). Saraca asoca bark samples (SA1 and SA2) and commercial market samples (MA1 and MA2) showed on an average similar TPC when compared with that of P. longifolia samples (PL1 and PL2).
Table 3.
Phytochemical attributes for filtrates of soaked samples of S. asoca, market and P. longifolia
| Sample | Code | Yield g/100 g ± SD | TPCa % ± SD | GA mg/g ± SD | CA mg/g ± SD | EC mg/g ± SD | Protein (mg/g) ± SD |
|---|---|---|---|---|---|---|---|
| S. asoca | SA1 | 12.48 ± 0.624 | 29.41 ± 1.47 | 0.190 ± 0.009 | 0.270 ± 0.013 | 0.057 ± 0.003 | 0.343 ± 0.017 |
| SA2 | 11.20 ± 0.560 | 23.73 ± 1.19 | 0.011 ± 0.003 | 0.039 ± 0.002 | 0.010 ± 0.005 | 0.232 ± 0.012 | |
| Market | MA1 | 10.92 ± 0.546 | 30.23 ± 1.51 | 0.189 ± 0.009 | 0.062 ± 0.003 | 0.007 ± 0.000 | 0.302 ± 0.015 |
| MA2 | 06.92 ± 0.346 | 28.90 ± 1.45 | 0.992 ± 0.050 | ND | 0.018 ± 0.003 | 0.130 ± 0.006 | |
| P. longifolia | PL1 | 06.24 ± 0.312 | 17.67 ± 0.88 | 1.374 ± 0.069 | ND | 0.087 ± 0.004 | 0.060 ± 0.003 |
| PL2 | 18.92 ± 0.946 | 04.14 ± 0.21 | 1.385 ± 0.076 | ND | 0.016 ± 0.007 | 0.062 ± 0.003 |
ND not detected, GA gallic acid, CA catechin, EC epicatechin
aTPC was represented as % tannic acid equivalent (TAE) and total proteins as mg BSA equivalent/g sample
The total protein estimated using Lowry’s method is presented in Table 3. The protein values ranged from 0.060 ± 0.003 to 0.343 ± 0.017 mg equivalent BSA/g dried bark powder. Interestingly, it was observed that protein values of both the P. longifolia samples were more or less similar and these values were lower than that observed in other samples. Higher amount of protein was observed in SA1 followed by MA1, SA2, and MA2 (Table 3).
Quantification of gallic acid, catechin, and epicatechin using RP-HPLC analysis
In the present study, the major chemical compounds of S. asoca, such as (+)-catechin, (−)-epicatechin, and gallic acid were quantified (Saha et al. 2013; Shirolkar et al. 2013). Previously used RP-HPLC protocol was employed during the study (Hegde et al. 2017). Calibration curves for gallic acid, catechin, and epicatechin produced equation y = 32538x + 8078.5, y = 8240.5x + 851.1, and y = 9378.8x + 756.1, respectively, with coefficient of determination (R 2) not less than 0.995 (Fig. 3b). Chromatographic peaks were confirmed by comparing retention times and UV spectra with reference standards. The amount of gallic acid, catechin, and epicatechin ranged from 0.011 ± 0.003 to 1.385 ± 0.076, 0.039 ± 0.002 to 0.270 ± 0.013, and 0.007 ± 0.000 to 0.087 ± 0.004 mg/g, respectively (Table 3; Fig. 3). Interestingly, gallic acid content was highest in PL1 and PL2 compared to other samples. Catechin was present in SA1, SA2, and MA1 and was not detected in MA2, PL1, and PL2. Epicatechin content was observed in lower amount compared to gallic acid and catechin. It is pertinent to discuss here that the phytochemical composition of a plant is influenced by genetic as well as environmental factors and further variation might also be due to the extraction method used (Pai et al. 2011; Shukla et al. 2017).
Fig. 3.
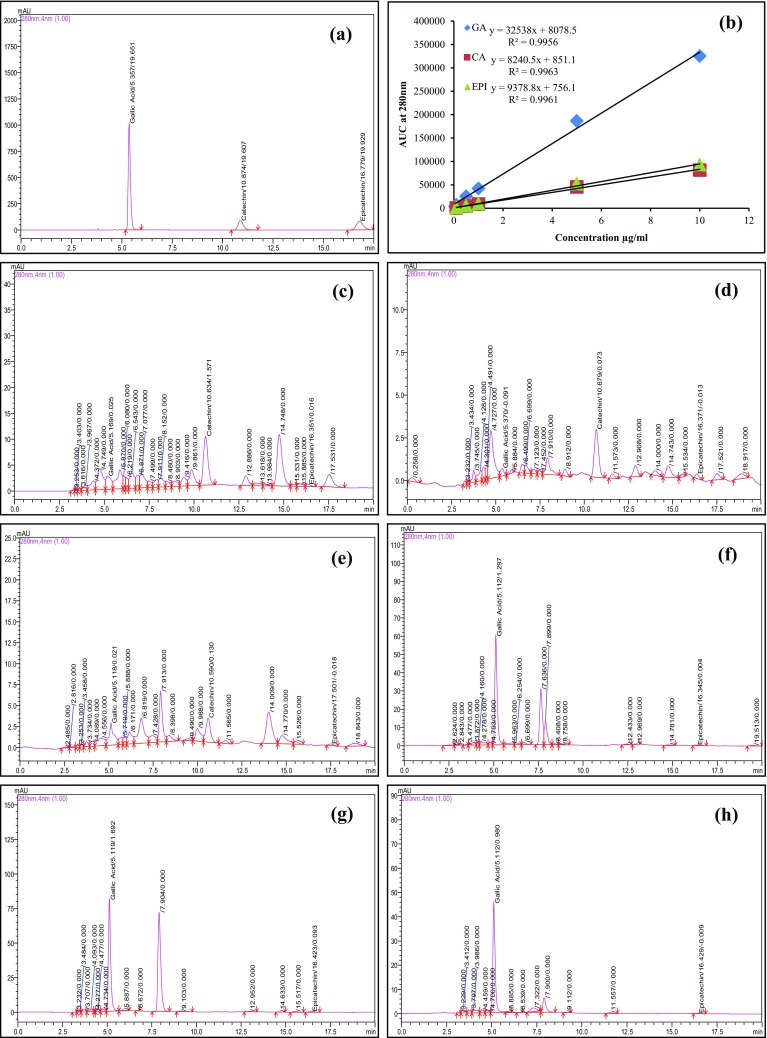
HPLC profiles of a standard gallic acid, catechin, and epicatechin (20 µg/mL); b five-point calibration curve; c SA1; d SA2; e MA1; f MA2; g PL1; and h PL2
Use of authentic medicinal plant is the most crucial aspect in preparation of any herbal medicine. Adulteration is a major concern both with domestic and international markets (Williamson et al. 2015; Urumarudappa et al. 2016; Pendkar et al. 2016). With increasing demand of herbal products, the importance of authentication of botanicals gains utmost importance (Smillie and Khan 2010; Xin et al. 2015). To the best of our knowledge, there has been no report of any easy method of simultaneous DNA and phytochemical extraction from the bark samples used as crude drug of S. asoca and its most common adulterant P. longifolia. In the present study, we have optimized a method to serve this purpose despite the difficulty of isolating DNA from the bark with contains inhibitors. This methodology would provide researchers and industry with an easy way to simultaneously isolate DNA for PCR-based assays and extract photoconstituents for chemoprofiling for various downstream applications including assays for identification and authentication and maintaining quality standards for S. asoca, one of the most commercially important medicinal plants of India.
Conclusions
The method of simultaneous DNA isolation and phytochemical extraction from bark of S. asoca is simple, time saving, and useful for various downstream application having enormous advantage of using only small quantity of raw botanical drugs. The method also provides a benefit of using the same sample for phytochemical profiling by chromatographic fingerprints using HPLC and genetic profiling by PCR-based applications. The method was found to be equally good for use with commercially available crude bark samples of S. asoca and with its most common adulterant P. longifolia and, therefore, has potential for utilization in research and industry for identification, authentication, and quality assessment.
Acknowledgements
Authors are indebted to Director-in-Charge, Scientific Advisory Committee Members of RMRC–ICMR, Belagavi, for providing critical scientific inputs and Dr. Harsha V. Hegde, RMRC–ICMR, Belagavi, India for identification of plant specimens. The authors are also thankful to Dr. Sunil Jalalpure, Dy, Director of Dr. Prabhakar Kore Basic Sciences Research Centre of KLE University Belagavi for help with instrumentation and retesting. SH is grateful to ICMR, New Delhi (Govt. of India) and KLE University, Belagavi for funding.
Compliance with ethical standards
Funding
This study was funded by grant from the ICMR, New Delhi, Govt. of India and KLE University (Grant No. 45/53/2013/BMS/TRM; KLEU/Accs/12-13/D-1552).
Conflict of interest
Authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the authors.
References
- Avula B, Ali Z, Khan IA. Chemical fingerprinting of Actaea racemosa (black cohosh) and its comparison study with closely related Actaea species (A. pachypoda, A. podocarpa, A. rubra) by HPLC. Chromatographia. 2007;66:757–762. doi: 10.1365/s10337-007-0384-6. [DOI] [Google Scholar]
- Collard BCY, Mackill DJ. Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol Report. 2009;27:86–93. doi: 10.1007/s11105-008-0060-5. [DOI] [Google Scholar]
- Dhanya K, Kizhakkayil J, Syamkumar S, Sasikumar B. Isolation and amplification of genomic DNA from recalcitrant dried berries of black pepper (Piper nigrum L.)—a medicinal spice. Mol Biotechnol. 2007;37:165–168. doi: 10.1007/s12033-007-0044-y. [DOI] [PubMed] [Google Scholar]
- Ganie SH, Upadhyay P, Das S, Sharma MP. Authentication of medicinal plants by DNA markers. Plant Gene. 2015;4:83–99. doi: 10.1016/j.plgene.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde HV, Hegde GR, Kholkute SD. Herbal care for reproductive health: ethno medicobotany from Uttara Kannada district in Karnataka, India. Complement Ther Clin Pract. 2007;13:38–45. doi: 10.1016/j.ctcp.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Hegde S, Hegde HV, Jalalpure SS, Peram MR, Pai SR, Roy S (2017) Resolving identification issues of Saraca asoca from its adulterant and commercial samples using phytochemical markers. Pharmacogn Mag (in press) [DOI] [PMC free article] [PubMed]
- Kazi T, Hussain N, Bremner P, Slater A, Howard C. The application of a DNA-based identification technique to over-the-counter herbal medicines. Fitoterapia. 2013;87:27–30. doi: 10.1016/j.fitote.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Khatoon S, Singh N, Kumar S, Srivastava N, Rathi A, Mehrotra S. Authentication and quality evaluation of an important ayurvedic drug-Ashoka bark. J Sci Ind Res. 2009;68:393–400. [Google Scholar]
- Li M, Pui-Hay P, Pang-Chui S (2013) Molecular Identification of Traditional Medicinal Materials. In: Lu-qi Huang (ed) Molecular pharmacognosy. Springer, Dordrecht, pp 45–66
- Llongueras JP, Nair S, Salas-Leiva D, Schwarzbach AE. Comparing DNA extraction methods for analysis of botanical materials found in anti-diabetic supplements. Mol Biotechnol. 2013;53:249–256. doi: 10.1007/s12033-012-9520-0. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough JJ, Farr AL, Randal RJ. Protein measurements with the polyphenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Nadkarni KM. Indian materia medica. Bombay: Popular Prakashan; 1976. pp. 469–470. [Google Scholar]
- Osathanunkul M, Suwannapoom C, Ounjai S, Rora JA, Madesis P, de Boer H. Refining DNA barcoding coupled high resolution melting for discrimination of 12 closely related croton species. PLoS One. 2015;10:e0138888. doi: 10.1371/journal.pone.0138888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osathanunkul M, Suwannapoom C, Osathanunkul K, Madesis P, de Boer H. Evaluation of DNA barcoding coupled high resolution melting for discrimination of closely related species in phytopharmaceuticals. Phytomedicine. 2016;23:156–165. doi: 10.1016/j.phymed.2015.11.018. [DOI] [PubMed] [Google Scholar]
- Pai SR, Nimbalkar MS, Pawar NV, Dixit GB. Optimization of extraction techniques and quantification of betulinic acid (BA) by RP-HPLC method from Ancistrocladus heyneanus Wall. Ex Grah. Ind Crops Prod. 2011;34:1458–1464. doi: 10.1016/j.indcrop.2011.05.006. [DOI] [Google Scholar]
- Pawar N, Pai S, Nimbalkar M, Dixit G. RP-HPLC analysis of phenolic antioxidant compound 6-gingerol from different ginger cultivars. Food Chem. 2011;126:1330–1336. doi: 10.1016/j.foodchem.2010.11.090. [DOI] [Google Scholar]
- Pendkar SK, Hegde S, Nayak SU, Hegde H, Kholkute SD, Roy S. Detection of adulteration by Wedelia calendulacea in Eclipta alba through ISSR and RAPD markers. Planta Med Int Open. 2016;3:e43–e46. doi: 10.1055/s-0042-108742. [DOI] [Google Scholar]
- Rezadoost MH, Kordrostami M, Kumleh HH. An efficient protocol for isolation of inhibitor-free nucleic acids even from recalcitrant plants. 3 Biotech. 2016;6:61. doi: 10.1007/s13205-016-0375-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha J, Mukherjee S, Gupta K, Gupta B. High-performance thin-layer chromatographic analysis of antioxidants present in different parts of Saraca asoca (Roxb.) de Wilde. J Pharm Res. 2013;7:798–803. [Google Scholar]
- Sarin YK (1996) Illustrated herbs of ayurveda. New Delhi, India: Council of Scientific and Industrial Research (CSIR) and Indian Council of Medical Research (ICMR), India, p 263
- Shirolkar A, Gahlaut A, Chhillar AK, Dabur R. Quantitative analysis of catechins in Saraca asoca and correlation with antimicrobial activity. J Pharm Anal. 2013;3:421–428. doi: 10.1016/j.jpha.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Hegde S, Kumar A, Chaudhary G, Tewari SK, Upreti DK, Pal M. Fatty acid composition and antibacterial potential of Cassia tora (leaves and stem) collected from different geographic areas of India. J Food Drug Anal. 2017 doi: 10.1016/j.jfda.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simko I. High-resolution DNA melting analysis in plant research. Trends Plant Sci. 2016;21:528–537. doi: 10.1016/j.tplants.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Singh M, Bandana, Ahuja PS. Isolation and PCR amplification of genomic DNA from market samples of dry tea. Plant Mol Biol Report. 1999;17:171–178. doi: 10.1023/A:1007562802361. [DOI] [Google Scholar]
- Singh S, Krishna THA, Kamalraj S, Kuriakose GC, Valayil JM, Jayabaskaran C. Phytomedicinal importance of Saraca asoca (Ashoka): an exciting past, an emerging present and a promising future. Curr Sci. 2015;109:1790–1801. doi: 10.18520/cs/v109/i10/1790-1801. [DOI] [Google Scholar]
- Smillie TJ, Khan IA. A comprehensive approach to identifying and authenticating botanical products. Clin Pharmacol Ther. 2010;87:175–186. doi: 10.1038/clpt.2009.287. [DOI] [PubMed] [Google Scholar]
- Tandon N, Yadav SN. Contributions of Indian Council of Medical Research (ICMR) on Medicinal Plants. J Ethnopharmacol. 2017;197:39–45. doi: 10.1016/j.jep.2016.07.064. [DOI] [PubMed] [Google Scholar]
- Urumarudappa SKJ, Gogna N, Newmaster SG, Venkatarangaiah K, Subramanyam R, Saroja SG, Gudasalamani R, Dorai K, Ramanan US. DNA barcoding and NMR spectroscopy-based assessment of species adulteration in the raw herbal trade of Saraca asoca (Roxb.) Willd, an important medicinal plant. Int J Legal Med. 2016;130:1457–1470. doi: 10.1007/s00414-016-1436-y. [DOI] [PubMed] [Google Scholar]
- Ved DK, Goraya GS (2007) Demand and supply of medicinal plants in India. National Medicinal Plant Board (NMPB), New Delhi and Foundation for Revitalization of Local Health Traditions (FRLHT), Bengaluru, India. http://nmpb.nic.in/FRLHT/Contents.pdf. Accessed 22 May 2017
- Williamson EM, Chan K, Xu Q, Nachtergael A, Bunel V, Zhang L, Ouedraogo M, Nortier J, Qu F, Shaw D, Liu X, Stévigny C, Kahumba J, Pelkonen O, Duez P. Evaluating the safety of herbal medicines: integrated toxicological approaches. Science. 2015;347:S47–S49. [Google Scholar]
- Xin T, Li X, Yao H, Lin Y, Ma X, Cheng R, Song J, Ni L, Fan C, Chen S. Survey of commercial Rhodiola products revealed species diversity and potential safety issues. Sci Rep. 2015;5:8337. doi: 10.1038/srep08337. [DOI] [PMC free article] [PubMed] [Google Scholar]


