Abstract
Globally, plant-derivatives especially cereals and legumes are the major staple food sources for animals. The seeds of these crops comprise of phytic acid, the major repository form of the phosphorus, which is not digestible by simple-stomached animals. However, it is the most important factor responsible for impeding the absorption of minerals by plants that eventually results in less use of fertilizers that ultimately cause eutrophication in water bodies. Although abundant phosphorus (P) exists in the soils, plants cannot absorb most of the P due to its conversion to unavailable forms. Hence, additional P supplementation is indispensable to the soil to promote crop yields which not only leads to soil infertility but also rapid depletion of non-renewable P reservoirs. Phytase/phosphatase enzyme is essential to liberate P from soils by plants and from seeds by monogastric animals. Phytases are kind of phosphatases which can hydrolyse the indigestible phytate into inorganic Phosphate (Pi) and lower myo-inositol. There are several approaches to mitigate the problems associated with phytate indigestibility. One of the best possible solutions is engineering crops to produce heterologous phytase to improve P utilization by monogastric animals, plant nutrition and sustainable ecological developments. Previously published reviews were focused on either soil phytate or seed-phytate, related issues, but this review will address both the problems as well as phytate related ecological problems. This review summarizes the overall view of engineered phytase crops and their role in sustainable agriculture, animal nutrition and ecological development.
Keywords: Phytase, Animal feed, Phytate, Phosphorus, Transgenic plants
Introduction
Globally, plant derived food products are the basic staple food for animals, which comprising of vital sources of carbohydrates, vitamins, protein, dietary fiber, and non-nutrients (Kumar et al. 2010). Among these food sources, cereals, legumes and oilseed crops are produced over 90% of the world’s harvested area (Reddy et al. 1989). In developing nations, cereals and legumes are the major food sources which contain abundant amount of phytic acid (Gupta et al. 2015). Phytic acid is the major repository form of phosphorus (P) in plant seeds and it has a substantial adhering affinity to cations and forms insoluble phytate salts in solution (Erpel et al. 2016). Phytic acid being negatively charged has a strong chelation property and chelates positively charged minerals such as calcium, magnesium iron and zinc present in food (Kumar et al. 2010; Reddy et al. 2015). This results in micronutrient malnutrition among populations whose staple food constitute grain and legume based diets (Reddy et al. 2013). Ruminant animals have limited ability to digest the phytic acid, therefore, extrinsic phytase is indispensable for swine, poultry, fish, including human (Lei et al. 2013). The livestock industry, such as poultry and swine where the grain and legumes are used as major diet ingredients, often releases excessive amounts of phytates to the environment and causes phosphorus eutrophication (Bohn et al. 2008).
Although abundant amount of P exists in soils, plants cannot acquire it due to its conversion to insoluble phytate forms (Sharma et al. 2013). Phytate (myo-inositol hexakis phosphate) is a major phosphorus repository molecule in the soil, derived from plant remnants and animal compost (Gerke 2015). P is an essential plant macronutrient which limits agricultural production on a global scale (Ramaekers et al. 2010). Hence, to promote crop yield, application of Pi is indispensable even though expensive (Azeem et al. 2015). Seldom plants could use 30% of the applied Pi; High Pi concentrations in water bodies lead to eutrophication when Pi rich topsoil’s are washed away to aquatic bodies (Holford 1997). Repeated application of Pi causes soil infertility as well as rapid depletion of non-renewable limited P reserves (Sharma et al. 2013). Therefore, efficient P utilization is essential for sustainable development and to avert the adverse environmental effects (Scholz et al. 2015). Hence, conversion of existing phytate-P complex of the soil into plant accessible orthophosphate would mitigate P related obstacles.
There are several approaches to mitigate the above problems, but the simple sustainable solution is exudation of phosphatases or phytases, which catalyse the hydrolysis of Phytate-P, is a potentially important way for plants to raise P availability (Richardson et al. 2004). For P bioavailability and to dilute the eutrophication, microbe derived phytase is commonly added to the feed in areas where intense pig and poultry production is carried out (Brinch-Pedersen et al. 2002). However, commercially available phytase is expensive and its manufacturing is time consuming. Thus, to alleviate the mentioned problems, plants can be engineered with an ideal phytase for targeted spatial expression. Transgenic plants expressing adequate phytase could replace additional supplementation of P to monogastric animals as well as it would make P accessible to plant from soil containing phytate-P complex. Engineering crop plants and to employ them as bio reactors, with over-expression of phytase enzyme would address several issues such as sustainable agriculture, monogastric animal nutrition, and ecological development. The review summarizes engineering crop plants with phytase enzyme in various plant tissues, and the potential applications of this knowledge in managing sustainable agriculture animal nutrition and ecological development.
Source and classification of phytases
Phytase enzyme belongs to phosphatase family that dephosphorylates inositol hexakis phosphate (IP6) into lower myo-inositiol and inorganic phosphorus sequentially (Yao et al. 2012). Phytases ubiquitously exist in the nature. It is found in plants, micro-organism and animals and it was first identified in rice bran (Suzuki et al. 1907) and subsequently it was reported in calf blood by McCollum and Hart (1908). Phytases are broadly classified on two basis; depending on the site where dephosphorylation of the phytate molecule initiates and on the basis of pH (Rao et al. 2009). On the basis of hydrolysation initiation site of the phytate, it has been classified into three groups namely, 3-phytases, 6-phytases, and 5-phytases (Yao et al. 2012; Reddy et al. 2013). The 3-phytases (EC 3.1.3.8) are the major categories of phytases and are notably found in fungi and bacteria in the form of histidine acid phosphatases. Fungal phytses have broad substrate specificity and acidic pH profile. Phytases belonging to Aspergillus niger, Neurospora crassa, Pseudomonas, Klebsiella sps, Bacillus sps, M. thermophila, E. nidulan, T. thermophilus start the hydrolysis at third phosphate group (Sajidan et al. 2004; Rao et al. 2009). On the other hand, phytases belonging to most of the plants, E. coli, Paramecium and human lysosome, start hydrolysation of phytate at the sixth phosphate group (EC 3.1.3.26), whereas 5-phytases (EC 3.1.3.72) from Medicago sativa, Phaseolus vulgaris, lilly pollen and Pisum sativum start the hydrolysis at the fifth phosphate group (Rao et al. 2009; Gupta et al. 2015).
On the basis of catalytic mechanism, phytases can be classified into three groups (Tye et al. 2002), i.e., Histidine Acid Phosphatase (HAP) (E.C.3.1.3.2) β-propeller phytases (BPP) (E.C.3.1.3.8) and Purple Acid Phosphatases (PAP) (E.C.3.1.3.2). Histidine Acid Phosphatases are the first commercialized phytases from fungi and it comprises commonly shared active catalytic dipeptide (RHGXRXP) (Singh and satyanarayana 2015). Almost every plant constitutes Purple acid Phosphatases, the name derived due to its colour, when binuclear center of metal ions (Fe–Zn or Fe–Mn) transfer charge from tyrosine residue to chromophoric ferric ion conferring purple colour to the solution (Zamani et al. 2014). PAPs with phyatse activity appear to be restricted in plants. The first PAP gene described (GmPhy) and was isolated from soybean (Gibson and Ullah 1988). Second major characterized phytases are alkaline phytases from Bacillus which are also known as β-propeller phytase, containing 6 blades architecture, and its enzymatic activity is dependent on calcium. These enzymes are also known as alkaline phosphatase due to their alkaline pH (Reddy et al. 2015).
Ideal phytase for transgenic plant expression
GM phytase crops ultimate aim is either to improve the soil P acquisition to the plants or animal food/feed amelioration. Ideal phytase for transgenic plant expression depends on target of application. For example for root expression, it depends on soil pH, isoelectric point, whereas in seed, it is essential to employ the tri or tetra phosphate myo-inositol produced phytase from IP6. If it is in leaf and for animal feed targeting, it should have ideal industrial phytase qualities such as low pH and high thermos ability.
Root expression for soil P targeted application
Phytases have been employed for root expression to mitigate the P related problems like additional P supplementation, eutrophication, and efficient utilization of soil available P. Lower isoelectric point (pI) is recommended for root expression, for example, Peniophora lycii pI (3.6) of phytase was more effective to hydrolyze the soil phytate than higher pI possessing A. niger phytase having pI of 5.0 (George et al. 2007). At lower soil pH, phytate can easily bind to Al3+, Fe3+ whereas, at higher soil pH, phytase will bind to Ca2+ (Gyaneshwar et al. 2002; Azeem et al. 2015). Therefore, Apergillus niger is ideal for root expression in acidic soils while, Bacillus subtilis phytase is preferable to engineer plants that grow on basic soils.
Seed expression for animal nutrition targeted application
For animal nutrition, phytase is majorly expressed in seed and leaf and it should retain industrial phytase qualities and ensure degradation of phytate till tri- or di-inositol phosphate. Complete degradation of seed phytate would lead to seed in-viability, therefore, Bacillus phytase is suitable as it dephosphorylates till tri-my inositol. Lower myo-inositol (tri-myo-inositol) plays a crucial role in many metabolic signaling pathways. Thus, complete degradation of phytate may affect plant growth and ultimately crop yield (Rao et al. 2009). Phytase from A. niger is suitable for gastric pH but it will hydrolyze the phytate completely. Poultry feed comprises abundant of Ca; hence Bacillus subtilis phytase is opted due to its strict substrate specificity to Ca++ (Lei et al. 2013) and for aquaculture (agastric fish) target also it is ideal due to its functioning, in alkaline pH (Kumar et al. 2012). However, single phytase may not satisfy the all desired qualities, hence it is essential to look for new phytases or tailoring existed phytases with desired qualities. Therefore, for expressing phytase in crop plants it is essential to employ them according to the target of application.
Prominence of transgenic phytase plants in sustainable animal nutrition, agriculture and ecological development
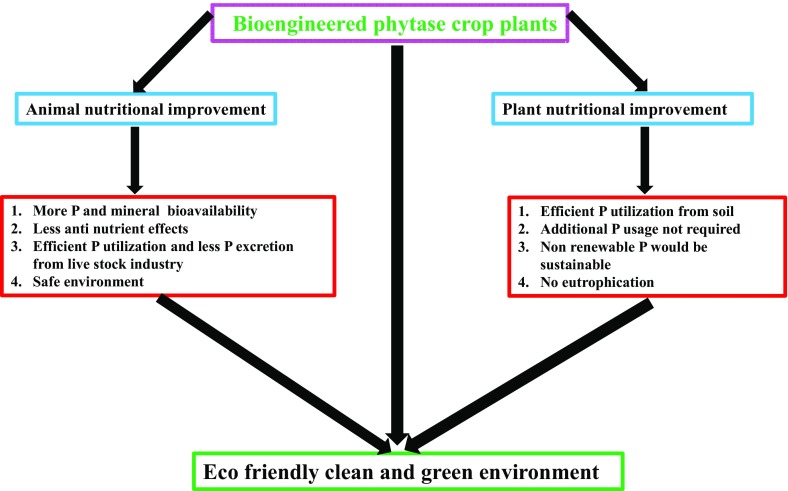
There are several approaches to improve P bioavailability for plant, animals and to mitigate the P pollution. The strategies include, phytase enzyme addition to the animal feed (Lei et al. 2013), feed soaking in water (Lott et al. 2000), breeding of lower phytic acid crop varieties (Raboy et al. 2000), production of transgenic animals that excrete phytate (Golovan et al. 2001), engineering plants with phytase enzyme (Brinch-Pedersen et al. 2002; Wang et al. 2013). Solubilization of soil phytate by microorganism is another strategy for P solubilization in soil (Sharma et al. 2013). Except creation of transgenic plants over-expressing phytase, all strategies are either laborious, tedious or economically not feasible. To alleviate the above issues, the best possible solution is creating transgenic plants. Transgenic plants can act as bioreactors which are cost effective and will ensure uninterrupted production of phytase. Application of phytase over-expressing transgenic plants is depicted in (Fig. 1).
Fig. 1.
Genetically modified phytase crops applications in plants in animal and plant nutrition
Role of phytase expressing transgenic plants in sustainable plant nutrition and P recycling
P is the one of the key element in the plant nutrition and it plays crucial role in almost all metabolic processes such as respiration, photosynthesis, signal transduction, macromolecular biosynthesis, N fixation in legumes, energy metabolism, and enzyme regulation (Singh and Satyanarayana 2011; Sharma et al. 2013; Azeem et al. 2015). P is the non-renewable resource and its absorption by plants is poor due to its conversion to unavailable predominant organic (P o) P form. The prevalent form of P o in the agriculture soil is inositol hexa and penta phosphates (phytates) and it represents approximately 60% of P in the soil (Tarafdar and Jungk 1987; Mudge et al. 2003; Stutter et al. 2015). Due to poor solubilizing property of phytate, it cannot be directly absorbed by plants from the soil, thus repeated application of inorganic P is required to promote crop yield. Less than 30% of the applied P can be used by plants (Norrish and Rosser 1983) and inefficient utilization of P eventually leads to soil infertility (Gyaneshwar et al. 2002) and environmental imbalance. Inorganic P is an exhaustive phosphate reservoir and is expected to be depleted by 2050. Hence, P availability would become a serious issue in future (Vance et al. 2003; Abelson 1999; Singh and Satyanarayana 2011). Thus, it is essential to utilize the existing P sources judiciously for sustainable development. It has been proposed that if we use the globally accumulated P in agricultural soils in sustainable fashion, it would be sufficient to sustain crop production for about 100 years (Khan et al. 2009). Globally, commercial crop seeds and fruits collectively produce approximately fifty-one million metric tons of phytate annually (Lei et al. 2013) and it is equivalent to about 67% of the annual P utilization in manufacturing P fertilizers (Mullaney et al. 2007). The statistical facts related to phytate revealed the prominence of phytate-P for P recycling in the soil.
There are various strategies to improve the P availability from soil to the plants, such as addition of P solubilizing microbes to the soil, addition inorganic P, etc. But these processes are tedious, laborious and expensive. Hence, development of transgenic plants over-expressing phytase in roots would be the best solution to address reduced availability of P in most soils. Therefore, several attempts have been made by researchers to reclaim the sustainable P utility, plant nutrition and ecological balance (Fig. 2a). Model plant tobacco has been engineered to express BsPhyC and AnPhy genes encoding phytase. BsPhyC over-expressing transgenic plants exhibited improved P assimilation and better yield in terms of increased flower, fruit number. There was improvent in IP6/IP5 ratio but the transgenic plants produced small sized seeds (Yip et al. 2003). AnPhy gene harboring tobacco plants acquired significant P (more than 52% than control) from soil in the presence of phyate ameliorate condition (George et al. 2005). However, it was concluded that exudation of phytase by plant roots is alone may not be sufficient to improve plant nutrition and it would require supplementation of P fertilizers. Another model species, Arabidopsis thalina, expressing extracellular phytase (PhyA) was as effective as Pi supplied control plants in yielding comparatively which indicated that there was no requirement of additional P supplementation (Richardson et al. 2001). In another study, Mudge et al. (2003) showed tissue specific promoter would be ideal for driving gene expression rather than constitutive promoter. They proved Arabisopsis plants expressing PhyA gene driven by phosphate transporter1 promoter is effective than CaMV35 promoter in performing better in P deficiency conditions and show improved plant nutrition. Li et al. (2009) also expressed AfPhy gene under the regulation of root specific pyk10 promoter in soybean. The transgenic plants exhibited phytase activity which can mitigate P related problems. Expression of Arabisopsis purple acid phosphatase15 in soybean also improved P acquisition efficiency which improved plant dry weight and P content (Wang et al. 2009). Potato and sweet potato expressing secretory synthesized phytase and E. coli APPA, respectively, improved P utilization efficiency and resulted in 40% P accumulation in potato leaves and improved vegetative growth and yield in sweet potato (Zimmermann et al. 2003; Hong et al. 2008). Extracellularly expressed phyA in cotton significantly enhanced P utilization from soil phytate (Liu et al. 2011). Over-expression of AnPhyA and EcAppA in Brassica napus greatly boosted phosphorus uptake, plant biomass and seed yields in the presence of sole P source (Wang et al. 2013), whereas in natural soil, organic P utilization efficacy was enhanced in Medicago truncatula when the same genes were over-expressed in it (Ma et al. 2012).
Fig. 2.
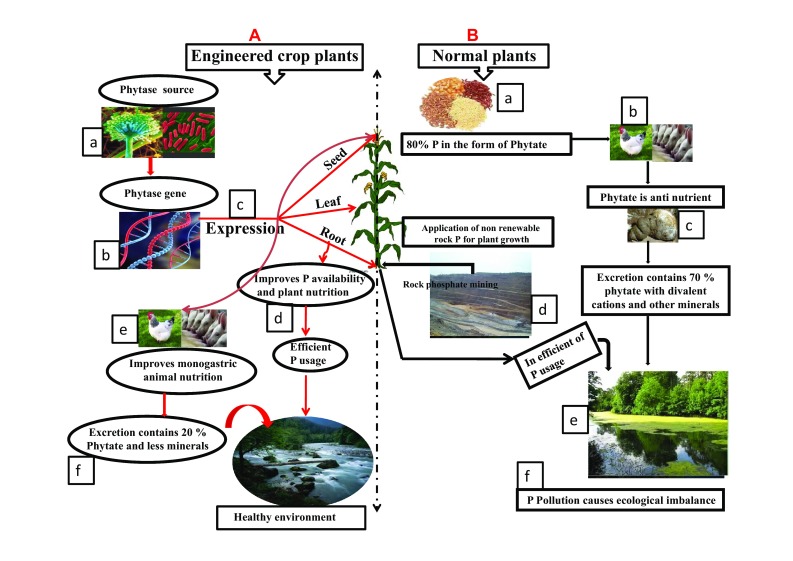
A Effects of phytase expressing transgenic plants: a Common source of phytase: microbe and plants. b Ideal phytase gene identification for spatial targeted application. c The phytase expression in root, leaf and seed according to required application. d Root specific expression improves, P acquisition from soil; and mitigate P related issues. e Seed phytase expression improves monogastric animal nutrition and reduces P pollution. f Through this approach we could achieve sustainable plant, animal nutrition and ecological development. B Normal crop plant effects: a normal seed contains PA content 1.5% of dry weight of the seed and it accounts 80% of P in this form. b Enriched phytate seed, consumed monogastric animals lose P and other essential minerals. c Inefficient assimilation of P excrete through feces d normal plant cannot absorb directly organic P, thus it required additional P, which is nonrenewable source. f Inefficient utilized P runoff nearby water bodies and cause water pollution
So far 12 various crop plants have been engineered with phytase gene and heterologous expression of phytase enzyme in plants demonstrated that it is an ingenious way for sustainable P utilization (Fig. 2a). Bio farming of crop plants with phytase is economically feasible and expression of recombinant phytase has lofty potential in elevating plant P acquisition from soil phytate. Numerous experiments proved enhanced phytate-P availability in soil by expressing phytase gene in transgenic plants (List—Table 1).
Table 1.
Transgenic phytase crops and their results
| Source | Vector- promoter | Plant system | Result | References |
|---|---|---|---|---|
| Fungi | ||||
| Aspergillus niger | pM0G413- CaMV35s | Tobacco seed | Improved broiler growth | Pen et al. (1993) |
| Aspergillus ficuum | pBI121-CaMV35s | Tobacco | Showed thermo-stability | Wang et al. (2007) |
| Aspergillus niger | pPLEX502 -CaMV 35S & AtPt | Tobacco | Improved P acquisition from soil | George et al. (2009) |
| Aspergillus niger | pAER02 CaMV35s | Tobacco | Proved phytate also is required P acquisition from soil | George et al. (2005) |
| Aspergillus niger | pMOG413 CaMV35s P | Tobacco leaf | Proved phytase stability and can be used as alternative to commercial phytase | Verwoerd et al. (1995) |
| Aspergillus ficuum | pTZ117-CaMV35S | Tobacco leaf | Proved it is equal to commercial phytase | Ullah et al. (1999) |
| Aspergillus ficuum | pPC-KSA-Pky10 | Soybean root | P absorbance efficiency improved | Li et al. (2009) |
| Aspergillus awamori | pBI121-CaMV35s | Soybean | Thermo-stability | Gao et al. (2007) |
| Aspergillus niger | pPhy35s, CaMV35s, seed specific | Soybean seed | Alternative to commercial phytase | Li et al. (1997) |
| Aspergillus niger | pPLEX502 AtPt | Arabidopsis root | Improved P acquisition from soil | Mudge et al. (2003) |
| Aspergillus niger | pUBARN& pUPhyN, Maize ubiquitin-1 | Wheat grain | Phytate degraded into lower forms | Brinch-Pedersen et al. (2000) |
| Aspergillus fumigatus | p1DX5SPConPhyN Wheat HMW 1DX5 GS | Wheat endosperm | Broiler growth was improved like with commercial phytase | Brinch-Pedersen et al. (2006) |
| Aspergillus niger | pSPHP3303T-Phy Maize globulin-1 P | Maize embryo | Phytate content degraded without disturbing seed germination | Chen et al. (2008) |
| Aspergillus niger | pLPL-Rice glutelin-1 | Maize endosperm | 95% P availability increased | Drakakaki et al. (2005) |
| Aspergillus niger | pYU159, Maize ubiquitin | Rice seed | Enhanced inorganic P content | Liu et al. (2006) |
| Aspergillus | pYP46 D35s omega | Canola seed | Proved it can be used as an alternative to the commercial phytase | Peng et al. (2006) |
| Aspergillus ficuum | pTZ117-CaMV35S | Alfalfa leaf | Proved best alternative to the commercial phytase | Ullah et al. (2002) |
| Aspergillus niger | pMOG413-CaMV 35Ps | Sesamame root | Improved P acquisition | Jin et al. (2004) |
| Aspergillus ficuum | pTZ117 CaMV35s promoter | Potato leaves | Proved best alternative to the commercial phytase | Ullah et al. (2003) |
| Aspergillus niger | pBI121-CaMV35s | Brassica napus | Proved recombinant phytase can be best alternative to commercial phytase and improved P acquisition from soil | Wang et al. (2013) |
| Aspergillus niger | p3HB-Kan | Chlamydomonas | It can be best alternative to commercial phytase | Erpel et al. (2016) |
| Bacteria | ||||
| Bacillus subtilus | pCAMBIA1300- CaMV35s | Tobacco | Improved shoot root biomass and IP4/IP5 | Yip et al. (2003) |
| Escherichia coli | pZY101-Soybean lectin | Soybean seed | To mitigate P related problems | Bilyeu et al. (2008) |
| Peniophora lycii | pPLEX502 -CaMV 35S & AtPt | Tobacco | Improved P acquisition from soil | George et al. (2009) |
| Escherichia coli | pCAMBIA2301 Sporamin promoter | Sweet Potato tubers | Improved Plant and animal nutrition | Hong et al. (2008) |
| Escherichia coli | pATPA | Chlamydomonas | Phytate content significantly reduced in broilers excretion | Yoon et al. (2011) |
| Escherichia coli | pBI121-CaMV35s | Brassica napus | Proved recombinant phytase can be best alternative to commercial phytase and Improved P acquisition from soil | Wang et al. (2013) |
| Escherichia coli | Rice | Improved winstar pig nutrition | Cheng-Chih et al. (2008) | |
| Bacillus subtilus | PCx-CAS, 35S Promoter | Tobacco, Arabidopsis | Increased shoot biomass and P acquisition | Lung et al. (2005) |
| Plant and others | ||||
| Arabidopsis thaliana | pBa002a, AtPAP15 | Tobacco | Improved P acquisition from soil | Kuang et al. (2009) |
| Glycine max | pPHY35P-DCaMV 35s | Soybean | To mitigate P related problems | Cheira et al. (2004) |
| Arabidopsis thaliana | pTF101.1-CaMV35S, MtPT1 | Soybean | Improved internal P, plant yield | Wang et al. (2009) |
| Medicago trunculata | pCAMBIA3301-MtPT1 | Arabidopsis root | Fourfold increased fresh and dry weight of the plant | Xiao et al. (2006) |
| Arabidopsis thaliana | pBa002a-AtPAP15 | Arabidopsis | Improved P acquisition from soil | Kuang et al. (2009) |
| Arabidopsis thaliana | pTF101.1-CaMV35S, MtPT1 | Clover | Improved P acquisition | Ma et al. (2009) |
| Oryza sativa | Ole-18 | Rice | Low phytate produced | Ali et al. (2013) |
| Saccharomyces cerevisiae | pOriF, pModF, pOriT-Rice chlorophyll a/b binding (cab) | Rice leaf | Phytase was stable even after 12 weeks of ensilage treatment | Hamada et al. (2005) |
Role of phytase expressing transgenic plants in sustainable animal nutrition
Phytic acid synthesis starts at cytoplasm and it is transported to vacuole at seed maturation stage (Raboy 2007). The seed contains almost 80% P in the form of phytic acid and it accounts 1.5% of dry weight (Bohn et al. 2008). Phytic acid thus get incorporated in diets of people who consume cereals and legumes as their staple food especially in the developing and underdeveloped nations (Kumar et al. 2010). Monogastric animals including humans have limited ability to digest phytic acid and it can easily bind to positively charged cations and form insoluble complexes (Raboy 2001; Joshi and Satyanarayana 2015). Additionally, it shows negative impact on protein, lipid and carbohydrate digestion as well as their utilization (Cheryan and Rackis 1980; Vohra and satyanarayana 2003; Lee et al. 2006; Kumar et al. 2010). The daily consumption of phytate is approximately 2000–2600 mg (Reddy 2002). Therefore, monotonous consumption of phytate containing cereals and legumes cause micronutrient malnutrition. Addition of inorganic P to the animal feed is common practice in livestock industry especially in swine and poultry to accelerate animal growth, which is not only expensive but also results in the poor utility of P by the animals. The excreta of these animals containing phytate become the leading cause of eutrophication when the same enters aquatic ecosystem via surface runoff (Joshi and Satyanarayana 2015). To mitigate the above problems and for sustainable development, exogenous addition of phytase to the food and feed is recommended. Unfortunately, effective phytases derived from microorganism is not acceptable to be added into food and feed without purification and since the purification process is laborious and time consuming, it becomes quite infeasible. Thus, heterologous expression of phytase in crops plants would alleviate above all problems (Fig. 3).
Fig. 3.
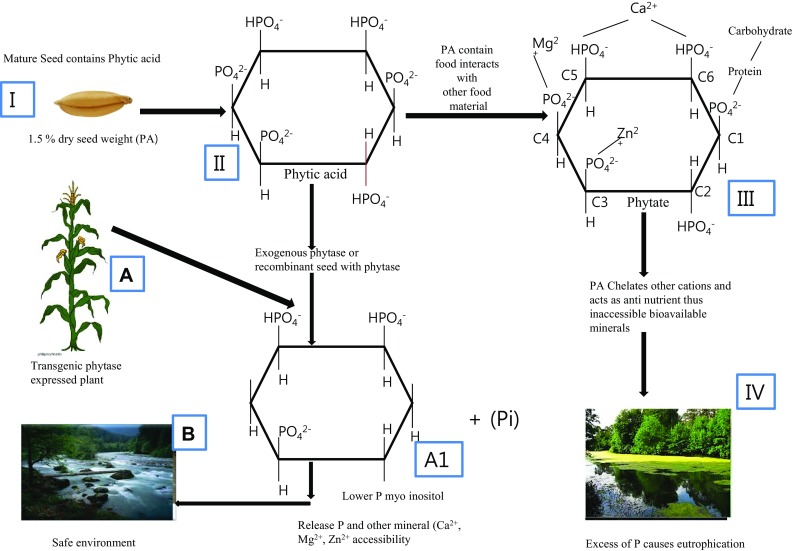
I The mature seed contains 1.5% dry weight of phytate content and it holds 80% of unavailable form of P. II Phytic acid (PA) chemical structure. III PA easily binds to divalent cat ions (Phytate) and as well as protein and carbohydrate. IV Undigested phytase excretion causes eutrophication or ecological imbalance A Inclusion of transgenic phytase in seed A1 Phytic acid liberates P and it would not binds to divalent cations additionally very limited fecal P excretion B As a result safe environment would be maintained
Several attempts have been made to improve monogastric animal’s nutrition and to mitigate eutrophication. All the experiments were carried out mostly with microbial phytase from A. niger, Bacillus subtilis E. coli. The first attempt made by Pen et al. (1993) demonstrated that the transgenic tobacco seed expressing A. niger phytase has beneficial effects on P liberation and the chicken fed trails showed enhanced broiler growth rate than commercial phytase or Pi inclusion. Same gene when expressed with KDEL signal peptide in transgenic canola seed and fed to chicks, there was comparable improvement in broilers weight similar to the control birds that were fed with feed containing commercial phytase (Peng et al. 2006). Similar results were noticed by Denbow et al. (1998), Brinch-Pedersen et al. (2006) in their studies where they employed fungal phytase expressing transgenic soybean and wheat, respectively, to feed broilers. Recombinant corn expressing E. coli APPA when fed to broiler chicks exhibited increased mineral retention including (P) (Ca) (N). The results concluded that transgenic corn is as effective as the commercial microbial phytase and thus it minimizes the additional supplementation of dietary P (Nyannor et al. 2009). Six microbial phytases were expressed in Chlamydomonas reinhardtii and the results concluded that efficient expression depends on N-terminal signal peptide and the codon optimization. The aim of the study was to investigate whether microalgae produced recombinant phytase can be fed to chicks to reduce the P faecal excretion and to reduce the chances of eutrophication. They fed Chlamydomonas expressing E. coli phytase to young chicks and the results indicated significant reduction in phytate excretion and inorganic P in the bodies of the birds increased by 43 and 41% (Yoon et al. 2011). Chunqi et al. (2014) fed the plant transgenic corn expressing AnphyA to hens and it resulted in increased P digestibility than the conventional corn without posing any deleterious health effects. Genetically modified corn expressing AnphyA was fed to growing pigs; the results showed elevated P absorbance and decreased phytate content in excreta (Li et al. 2013). Maize over -expressing AnPhyA in seeds, when fed to human intestine cacao-2 cells, exhibited improved iron uptake and decreased phytate content (Drakakaki et al. 2005). In another study, the introduction of E. coli phytase in rice also improved P assimilation and other nutrients availability (Cheng-Chih et al. 2008). All these above findings indicate that expression of phytase enzyme in plants can be the best alternative to the commercial phytase which would improve ruminant nutrition, ensure safe environment and sustainable P management.
Role of transgenic plants expressing phytase in ecological development and sustainable P management
P is crucial for life on earth and it is an indispensable element in nucleic acids, phospholipids, phosphoproteins and takes part in cell signaling. P is essential macronutrient for plants that ensures universal food security (Cordell and white. 2015). Despite the fact that mined P led to green revolution, repeated use of P fertilizers will deprive the P reservoirs and the year with peak P utilization has been estimated to be 2033 by Global Phosphorus Research Initiative (GPRI) (Cordell 2010). After Peak P, the production of P will gradually decrease and P repositories will diminish within a century (Cordell and white 2014). Thus, appropriate action is essential to maintain nonrenewable P resources for future generations and to better crop yield, otherwise, global population would be affected by acute hunger (FAO 2015). In 2007–2008, cost of di calcium P was $200–$250 per ton whereas currently its cost is around $1200, which means that it has increased by six times (Lei et al. 2013). Applied P to the plant hardly utilized (approximately, 30%) while the rest is leading cause of eutrophication. P is the most frequent limiting mineral in the livestock feed, and fodder as approximately 3 g kg−1 P exists in the form of phytate (Selle et al. 2007). Due to poor digestibility, phytate is not accessible for ruminants thus additional supplementation of Pi is common practice to promote swine and poultry growth. One pig can excrete 2.3 kg of P in their life cycle, for instance, US producing 600 million pigs annually can cause huge loss of P. Another serious concern of excessive P leaching is eutrophication which causes serious problems to aquatic animals through the formation of algal blooms and dissolved oxygen concentrations; known as dead zones and such dead zones as of now are around 400 across the globe (Diaz et al. 2008; Scholz et al. 2015). Supplementing commercial phytase to animal feed reduces and decreases 50% fecal excretion of P. It took around 90 years to launch commercial phytase into the market and currently it accounts for huge market value of approximately $350 million annually (Lei et al. 2013). Therefore, the inclusion of phytase to animal feed would reduce supplementing of Pi, excretion of P and as well as it would maintain sustainable P utilization. Repeated addition of commercial phytase is not economically feasible and it requires labour for production and for addition to the animal feed. Thus, we propose that if we generate transgenic plants over-expressing phytase it would address various issues associated with phytate. Several studies elucidated the transgenic phytase crops role in sustainable P management and ecological development, for example, transgenic maize and Chlamydomonas over-expressed E. coli appa phytase feed trails showed improved broilers growth, less P excretion and minimized P requirement (Nyannor and adeola 2008; Yoon et al. 2011). In another study, Anphy over-expressed corn was exhibited greatly utilized phytate content in human gastric CaCo2 cells which means less phytate excretion was observed (Drakakaki et al. 2005). Transgenic phytase expressed canola feed trails showed it could be alternative to commercial Natophus phytase (Zhang et al. 2000). P deficient conditions plant may adapt specific mechanism to obtain P from soil by secreting acid phosphatases and root hair elongation, etc. (Reddy et al. 2017). Numerous studies also proved acid phosphatase or phytase over-expressing transgenic crops greatly utilized phytate content from soil (Mudge et al. 2003; Jin et al. 2004; Lung et al. 2005; Xiao et al. 2006; Hong et al. 2008; George et al. 2009; Kuang et al. 2009) (Table 1) i.e., transgenic phytase crops may require less P as well as farmer need not to apply much commercial P. Thus, nonrenewable P mining will be limited therefore P can be utilized sustainable manner. Hence there might be less or no P leaching that would help to maintain healthy environment and also attenuates eutrophication (Fig. 2b–e).
Constraints of phytase engineered crop plants
Although phytase expressing transgenic plants are beneficial for plant and animal nutrition and ecological development; there are issues like loss of seed viability, yield, susceptibility for environmental stress, non-acceptance of Genetically Modified Organisms (GMOs). Among all phytases, fungal phytases are best suitable for generation of transgenic crops for animal nutrition improvement. Unfortunately, it liberates all the Ps (6P) from my inositol ring, which might lead to seed inviability. Low phytate or lower inositol molecules are the key messengers in cell signaling and transduction. Thus, complete degradation of phytate would lead to stress susceptibility and lower yield (Raboy 2009). Bacillus subtilis and plant originated phytases dephosphorylate only 3 and 2 phosphates from inositol ring, respectively, but Bacillus phytase works at basic pH which is not suitable for ruminant gastric pH. Further, plant phytases activity is very low (Rao et al. 2009; Reddy et al. 2013) with poses constrain in their utilization. Expression of phytase in plants can alter the plant or seed morphology. Yip et al. (2003) reported small seed syndrome which resulted in lose seed viability in tobacco. Field trials involving low phytic acid (lpa1-1) maize mutant showed 6% yield lose and delayed flowering and huge effect on germination and stress tolerance (Raboy 2009). In barley, low phytate (lpa-M955) condition (developed by blocking of phytic acid biosynthesis) inhibited cytokinin, ethylene pathway related genes thus altering signal transduction (Bowen et al. 2007). Phytate is associated to biotic abiotic stress tolerance, for example heterologous expression of mammalian Type 1 Ins polyphosphate 5- phosphatase in Arabidopsis, revealed elevated drought tolerance (Perera et al. 2008) and Dhole and Reddy (2016) showed PA is also essential in addition to stress responsive genes for Yellow mosaic virus as well as powdery mildew disease tolerance. Thus, tailoring phytic acid in crop plants would lead to susceptibility to biotic and abiotic stress. Biotechnology based GM crops cultivation provides numerous benefits to farmers by increasing yield, profit and declined pesticide application to the tune of (28%), (68%), (39%), respectively, and it meets the food requirement for emerging global population (Lucht. 2015). Although GM crops are beneficial, the protest or skeptical attitude from environmental nongovernmental organizations (NGO), societal frame workers and policy makers, affect the consumer attitude towards GM crops (Zilberman et al. 2013). For example, most of the nations from Europe are strictly opposing GM food crops, thus, implementing this technology is not that much easy. Globally more than 70% of the GM crops are used as animal feed to feed 100 billion animals, but there is no detrimental effect against health and performance (Fernandez-Cornejo et al. 2014). Therefore, production of transgenic crops expressing phytase may not be an issue. Phytate being an anti-nutrient, efficiently bind to free radicals such as iron ions and act as an antioxidant (Raboy 2003; Kumar et al. 2010) and rescue cells from oxidative stress. Numerous studies revealed it works as an anti-cancer agent by suppressing cell proliferation (Midorikawa et al. 2001; Shamsuddin 2002; Somasundar et al. 2005). Since phytate helps in by suppressing viral replication, it might work as an anti-HIV agent (Otake et al. 1998). Phytate intake is negatively correlated to blood glucose level and works as an anti-diabetic agent (Larsson et al. 1997) and helps in reducing serum cholesterol, triglyceride level phytate thus protecting human and animals against coronary diseases (Jariwalla et al. 1990; Persson et al. 1998). If we degrade phytate in seed, human or ruminants may lose these beneficial effects. However, extensive clinical studies are essential to support these above mentioned positive effects of phytate. Numerous studies on the other hand have proven negative effects of phytate in plant, animal, and environment. Hence, it is mandatory to pursue further research to improve the phytate related problems through genetic engineering.
Conclusions
Phytatic acid is the major repository form of the P, which is not digestible by simple stomached animals. The estimated global commercial crop seeds and fruits collectively produce fifty-one million metric tons of phytate annually equivalent to 67% of the annual P utilization in manufacturing P fertilizers. This phytate is considered as anti-nutrient due to its strong negative charge, which can be easily attracted nearby positive ions. Thus, animal and plant nutrition are greatly affected, and also it is not only an anti-nutrient but also causes eutrophication due to improper P utilization. The enzyme phytase was introduced for sustainable P utilization. After launching commercial phytase, the phytase market value enormously increased due to its potential applications in nutritional improvement and reduced P related problems. Several nations (22) mandated that phytase could be added in animals feed to resolve the eutrophication. However, phytase purification is expensive as well as laborious. On the contrary, engineering of crop plants with phytase gene is inexpensive, economically feasible and is also amenable to widespread acceptance and utilization. Bioengineered plants over-expressing phytase, not only improve animal and plant nutrition but will also be useful for protecting the environment in addition to paving way for sustainable P management. Hence, several attempts have been made using this approach and better results have been obtained in terms of improved animal nutrition and P utilization. However, further studies are necessary to assess the feasibility of transgenic based technology for addressing the problems associated with phytate consumption.
Acknowledgements
This study was supported by 2017 the RDA fellowship program of National Institute of Horticultural and Herbal Science, Rural development of Administration, Republic of Korea.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
References
- Abelson PH. A potential phosphate crisis. Science. 1999;283:2015. doi: 10.1126/science.283.5410.2015. [DOI] [PubMed] [Google Scholar]
- Ali N, Paul S, Gayen D, Sarkar SN, Datta K, Datta SK. Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1,3,4,5,6 pentakis phosphate 2-kinase gene (IPK1) PLoS One. 2013;8:e68161. doi: 10.1371/journal.pone.0068161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azeem M, Riaz A, Chaudhary AN, Hayat R, Hussain Q, Tahir MI, Imran M. Microbial phytase activity and their role in organic P mineralization. Arch Agronom Soil Sci. 2015;61:751–766. doi: 10.1080/03650340.2014.963796. [DOI] [Google Scholar]
- Bilyeu KD, Zeng P, Coello P, Zhang ZJ, Krishnan HB, Bailey A, Beuselinck PR, Polacco JC. Quantitative conversion of phytate to inorganic phosphorus in soybean seeds expressing a bacterial phytase. Plant Physiol. 2008;146:468–477. doi: 10.1104/pp.107.113480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn L, Meyer AS, Rasmussen SK. Phytate: impact on environment and human nutrition. A challenge for molecular breeding. J Zhejiang Univ Sci B. 2008;9:165–191. doi: 10.1631/jzus.B0710640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen DE, Souza EJ, Guttieri MJ, Raboy V, Fu J. A low phytic acid barley mutation alters seed gene expression. Crop Sci. 2007;47:149. doi: 10.2135/cropsci2006.07.0456tpg. [DOI] [Google Scholar]
- Brinch-Pedersen H, Hatzack F, Stoger E, Arcalis E, Pontopidan K, Holm PB. Generation of transgenic wheat (Triticum aestivum L.) for constitutive accumulation of an Aspergillus phytase. Mol Breed. 2000;6:195–206. doi: 10.1023/A:1009690730620. [DOI] [Google Scholar]
- Brinch-Pedersen H, Sørensen LD, Holm PB. Engineering crop plants: getting a handle on phosphate. Trends Plant Sci. 2002;7:118–125. doi: 10.1016/S1360-1385(01)02222-1. [DOI] [PubMed] [Google Scholar]
- Brinch-Pedersen H, Hatzack F, Stoger E, Arcalis E, Pontopidan K, Holm PB. Heat-stable phytases in transgenic wheat (Triticum aestivum L.) deposition pattern, thermo stability and phytate hydrolysis. J Agric Food Chem. 2006;54:4624–4632. doi: 10.1021/jf0600152. [DOI] [PubMed] [Google Scholar]
- Cheira JM, Finer JJ, Grabau EA. Ectopic expression of a soybean phytase in developing seeds of Glycinemax to improve phosphorus availability. Plant Mol Biol. 2004;56:895–904. doi: 10.1007/s11103-004-5293-6. [DOI] [PubMed] [Google Scholar]
- Chen R, Xue G, Chen P, Yao B, Yang W, Ma Q, Fan Y, Zhao Z, Tarczynski MC, Shi J. Transgenic maize plants expressing a fungal phytase gene. Transgen Res. 2008;17:633–643. doi: 10.1007/s11248-007-9138-3. [DOI] [PubMed] [Google Scholar]
- Cheng-Chih T, Lai CH, Yang CS, Lin CK, Tsen HY. Toxicological evaluation of transgenic rice flour with an Escherichia coli phytase gene appA by subchronic feeding study in Wistar rats. J Sci Food Agric. 2008;3:382–388. [Google Scholar]
- Cheryan M, Rackis JJ. Phytic acid interactions in food systems. Crit Rev Food Sci Nutr. 1980;13:297–335. doi: 10.1080/10408398009527293. [DOI] [PubMed] [Google Scholar]
- Chunqi G, Qiugang M, Lihong Z, Jianyun Z, Cheng J. Effect of dietary phytase transgenic corn on physiological characteristics and the fate of recombinant plant DNA in laying hens. Asian Aust J Animal Sci. 2014;27:77–82. doi: 10.5713/ajas.2013.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell D (2010) The Story of Phosphorus: Sustainability implications of global phosphorus scarcity for food security
- Cordell D, White S. Life’s bottleneck: sustaining the world’s phosphorus for a food secure future. Annu Rev Environ Resour. 2014;39:161–188. doi: 10.1146/annurev-environ-010213-113300. [DOI] [Google Scholar]
- Cordell D, White S. Tracking phosphorus security: indicators of phosphorus vulnerability in the global food system. Food Secur. 2015;7:337–350. doi: 10.1007/s12571-015-0442-0. [DOI] [Google Scholar]
- Denbow DM, Grabau EA, Lacy GH, Kornegay ET, Russell DR, Umbeck PF. Soybeans transformed with a fungal phytase gene improve phosphorus availability for broilers. Poult Sci. 1998;77:878–881. doi: 10.1093/ps/77.6.878. [DOI] [PubMed] [Google Scholar]
- Dhole VJ, Reddy KS (2016) Association of phytic acid content with biotic stress tolerance in mungbean (Vigna radiata L. Wilczek). Phytoparasitica 1-7
- Diaz J, Ingall E, Benitez-Nelson C, Paterson D, de Jonge MD, McNulty I, Brandes JA. Marine polyphosphate: a key player in geologic phosphorus sequestration. Science. 2008;320:652–655. doi: 10.1126/science.1151751. [DOI] [PubMed] [Google Scholar]
- Drakakaki G, Marcel S, Glahn RP, Lund EK, Pariagh S, Fischer R, Christou P, Stoger E. Endospermspecific co-expression of recombinant soybean ferritin and Aspergillus phytase in maize results in significant increases in the levels of bio available iron. Plant Mol Biol. 2005;59:869–880. doi: 10.1007/s11103-005-1537-3. [DOI] [PubMed] [Google Scholar]
- Erpel F, Restovic F, Arce-Johnson P. Development of phytase-expressing chlamydomonas reinhardtii for monogastric animal nutrition. BMC Biotechnol. 2016;16:1. doi: 10.1186/s12896-016-0258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2015) The state of food insecurity in the World. Food and Agricultural Organisation of the United Nations
- Fernandez-Cornejo J, Wechsler S, Livingston M, Mitchell L (2014) Genetically engineered crops in the United States. USDA-ERS Economic Research Report (162)
- Gao XR, Wang GK, Su Q, Wang Y, An LJ. Phytase expression in transgenic soybeans: stable transformation with a vector-less construct. Biotechnol Lett. 2007;29:1781–1787. doi: 10.1007/s10529-007-9439-x. [DOI] [PubMed] [Google Scholar]
- George TS, Richardson AE, Li SS, Gregory PJ, Daniell TJ. Extracellular release of a heterologous phytase from roots of transgenic plants: does manipulation of rhizosphere biochemistry impact microbial community structure? FEMS Microbiol Ecol. 2009;70:433–445. doi: 10.1111/j.1574-6941.2009.00762.x. [DOI] [PubMed] [Google Scholar]
- George TS, Simpson RJ, Hadobas PA, Richardson AE. Expression of a fungal phytase gene in Nicotiana tabacum improves phosphorus nutrition of plants grown in amended soils. Plant Biotechnol J. 2005;3:129–140. doi: 10.1111/j.1467-7652.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- George TS, Simpson RJ, Gregory PJ, Richardson AE. Differential interaction of Aspergillus niger and Peniophora lycii phytases with soil particles affects the hydrolysis of inositol phosphates. Soil Biol Biochem. 2007;39:793–803. doi: 10.1016/j.soilbio.2006.09.029. [DOI] [Google Scholar]
- Gerke J. The acquisition of phosphate by higher plants: effect of carboxylate release by the roots. A critical review. J Plant Nutr Soil Sci. 2015;178:351–364. doi: 10.1002/jpln.201400590. [DOI] [Google Scholar]
- Gibson DM, Ullah AH. Purification and characterization of phytase from cotyledons of germinating soybean seeds. Arch Biochem Biophys. 1988;260:503–513. doi: 10.1016/0003-9861(88)90475-4. [DOI] [PubMed] [Google Scholar]
- Golovan SP, Meidinger RG, Ajakaiye A, Cottrill M, Wiederkehr MZ, Barney DJ, Laursen J. Pigs expressing salivary phytase produce low-phosphorus manure. Nat Biotechnol. 2001;19:741–745. doi: 10.1038/90788. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Gangoliya SS, Singh NK. Reduction of phytic acid and enhancement of bioavailable micronutrients in food grains. J Food Sci Technol. 2015;52:676–684. doi: 10.1007/s13197-013-0978-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyaneshwar P, Kumar GN, Parekh LJ, Poole PS (2002) Role of soil microorganisms in improving P nutrition of plants. In: Food security in nutrient-stressed environments: exploiting plants’ genetic capabilities, Springer, Amsterdam, pp 133–143
- Hamada A, Yamaguchi K, Ohnishi N, Harada M, Nikumaru S, Honda H. High-level production of yeast (Schwanniomyces occidentalis) phytase in transgenic rice plants by a combination of signal sequence and codon modification of the phytase gene. Plant Biotechnol J. 2005;3:43–55. doi: 10.1111/j.1467-7652.2004.00098.x. [DOI] [PubMed] [Google Scholar]
- Holford ICR. Soil phosphorus its measurement and its uptake by plants. Aust J Soil Res. 1997;35:227–239. doi: 10.1071/S96047. [DOI] [Google Scholar]
- Hong YF, Liu CY, Cheng KJ, Hour AL, Chan MT, Tseng TH, Chen KY, Shaw JF, Yu SM. The sweet potato sporamin promoter confers high-level phytase expression and improves organic phosphorus acquisition and tuber yield of transgenic potato. Plant Mol Biol. 2008;67:347–361. doi: 10.1007/s11103-008-9324-6. [DOI] [PubMed] [Google Scholar]
- Jariwalla RJ, Sabin R, Lawson S, Herman ZS. Lowering of serum cholesterol and triglycerides and modulation of divalent cations by dietary phytate. J Appl Nutr. 1990;42:18–28. [Google Scholar]
- Jin UH, Chun JA, Lee JW, YiYB LS, Chung CH. Expression and characterization of extracellular fungal phytase in transformed sesame hairy root cultures. Protein Exp Purif. 2004;37:486–492. doi: 10.1016/j.pep.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Joshi S, Satyanarayana T. Heterologous expression of yeast and fungal phytases: developments and future perspectives. Indian J Biotechnol. 2015;14:293–311. [Google Scholar]
- Khan AA, Jilani G, Akhtar MS, Naqvi SMS, Rasheed M. Phosphorus solubilizing bacteria: occurrence, mechanisms and their role in crop production. J Agric Biol Sci. 2009;1:48–58. [Google Scholar]
- Kuang R, Chan KH, Yeung E, Lim BL. Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol. 2009;151:199–209. doi: 10.1104/pp.109.143180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Sinha AK, Makkar HPS, Becker K. Dietary roles of phytate and phytase in human nutrition review. Food Chem. 2010;120:945–959. doi: 10.1016/j.foodchem.2009.11.052. [DOI] [Google Scholar]
- Kumar V, Sinha AK, Makkar HPS, De Boeck G, Becker K. Phytate and phytase in fish nutrition. J Anim Physiol Anim Nutr. 2012;96:335–364. doi: 10.1111/j.1439-0396.2011.01169.x. [DOI] [PubMed] [Google Scholar]
- Larsson O, Barker CJ, Sjöholm Å, Carlqvist H, Michell RH, Bertorello A, Berggren PO. Inhibition of phosphatases and increased Ca2+ channel activity by inositol hexakisphosphate. Science. 1997;278:471–474. doi: 10.1126/science.278.5337.471. [DOI] [PubMed] [Google Scholar]
- Lee SH, Park HJ, Chun HK, Cho SY, Cho SM, Lillehoj HS. Dietary phytic acid lowers the blood glucose level in diabetic KK mice. Nutr Res. 2006;26:474–479. doi: 10.1016/j.nutres.2006.06.017. [DOI] [Google Scholar]
- Lei XG, Weaver JD, MullaneyE UA, Azain MJ. Phytase a new life for an old enzyme. Annual Rev Anim Biosci. 2013;1:283–309. doi: 10.1146/annurev-animal-031412-103717. [DOI] [PubMed] [Google Scholar]
- Li J, Hegeman CE, Hanlon RW, Lacy GH, Denbow MD, Grabau EA. Secretion of active recombinant phytase from soybean cell-suspension cultures. Plant Physiol. 1997;114:1103–1111. doi: 10.1104/pp.114.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Yang S, Li M, Qiao Y, Wang J. Functional analysis of an Aspergillus ficuum phytase gene in Saccharomyces cerevisiae and its root-specific secretory expression in transgenic soybean plants. Biotechnol Lett. 2009;31:1297–1303. doi: 10.1007/s10529-009-9992-6. [DOI] [PubMed] [Google Scholar]
- Li SF, Niu YB, Liu JS, Lu L, Zhang LY, Ran CY, Feng MS, Du B, Deng JL, Luo XG. Energy, amino acid and phosphorus digestibility of phytase transgenic corn for growing pigs. J Anim Sci. 2013;91:298–308. doi: 10.2527/jas.2012-5211. [DOI] [PubMed] [Google Scholar]
- Liu Q, Quan LQ, Feng J, Li Z, Jiang W, Hmei GM, Hong HY. Transgenic expression of the recombinant phytase in rice (Oryza sativa) Rice Sci. 2006;13:79–84. [Google Scholar]
- Liu JF, Wang XF, Li QL, Li X, Zhang GY, Li MG, Ma ZY. Biolistic transformation of cotton (Gossypium hirsutum L.) with the phyA gene from Aspergillus ficuum. Plant Cell Tissue Organ Cult. 2011;106:207–214. doi: 10.1007/s11240-010-9908-0. [DOI] [Google Scholar]
- Lott JN, Ockenden I, Raboy V, Batten GD. Phytic acid and phosphorus in crop seeds and fruits: a global estimate. Seed Sci Res. 2000;10:11–33. [Google Scholar]
- Lucht JM. Public acceptance of plant biotechnology and GM crops. Viruses. 2015;7:4254–4281. doi: 10.3390/v7082819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung SC, Chan WL, Yip W, Wang L, Yeung EC, Lim BL. Secretion of beta-propeller phytase from tobacco and Arabidopsis roots enhances phosphorus utilization. Plant Sci. 2005;169:341–349. doi: 10.1016/j.plantsci.2005.03.006. [DOI] [Google Scholar]
- Ma XF, Wright E, Ge Y, Bell J, Xi J, Bouton JH, Wang ZY. Improving phosphorus acquisition of white clover (Trifolium repens L.) by transgenic expression of plant-derived phytase and acid phosphatase genes. Plant Sci. 2009;176:479–488. doi: 10.1016/j.plantsci.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Ma XF, Tudor S, Butler T, Ge Y, Xi Y, Bouton J, Harrison M, Wang ZY. Transgenic expression of phytase and acid phosphatase genes in alfalfa (Medicagosativa) leads to improved phosphate uptake in natural soils. Mol Breed. 2012;1:377–391. doi: 10.1007/s11032-011-9628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum EV, Hart EB. On the occurrence of a phytin-splitting enzyme in animal tissues. J Biol Chem. 1908;4:497–500. [Google Scholar]
- Midorikawa K, Murata M, Oikawa S, Hiraku Y, Kawanishi S. Protective effect of phytic acid on oxidative DNA damage with reference to cancer chemoprevention. Biochem Biophys Res Commun. 2001;288:552–557. doi: 10.1006/bbrc.2001.5808. [DOI] [PubMed] [Google Scholar]
- Mudge SR, Smith FW, Richardson AE. Root-specific and phosphate-regulated expression of phytase under the control of a phosphate transporter promoter enables Arabidopsis to grow on phytate as a sole P source. Plant Sci. 2003;165:871–878. doi: 10.1016/S0168-9452(03)00286-3. [DOI] [Google Scholar]
- Mullaney EJ, Ullah AH, Turner B, Richardson A, Mullaney E (2007) Phytases: attributes, catalytic mechanisms and applications. Inositol phosphates: linking agriculture and the environment, pp 97–110
- Norrish K, Rosser H (1983) Mineral phosphate. Soils: an Australian viewpoint, pp 335–361
- Nyannor EK, Adeola O. Corn expressing an Escherichia coli-derived phytase gene: comparative evaluation study in broiler chicks. Poult Sci. 2008;87:2015–2022. doi: 10.3382/ps.2007-00501. [DOI] [PubMed] [Google Scholar]
- Nyannor EKD, Bedford MR, Adeola O. Corn expressing an Escherichia coli-derived phytase gene: residual phytase activity and microstructure of digesta in broiler chicks. Poult Sci. 2009;88(7):1413–1420. doi: 10.3382/ps.2009-00003. [DOI] [PubMed] [Google Scholar]
- Otake T, Mori H, Morimoto M, Miyano K, Ueba N, Oishi I, Kurimura T. Anti-HIV-1 activity of myo-inositol hexaphosphoric acid (IP6) and myo-inositol hexasulfate (IS6) Anticancer Res. 1998;19:3723–3726. [PubMed] [Google Scholar]
- Pen J, Venvoerd TC, van Paridon PA, Beudeker RF, VandenElzen PJM, Geerse K, van der Klis JD, Versteegh AJ, van Ooyen AJJ, Hoekema A. Phytase containing transgenic seeds as a novel feed additive for improved phosphorus utilization. BioTechnology. 1993;11:811–814. [Google Scholar]
- Peng RH, Yao QH, Xiong AS, Cheng ZM, Li Y. Codon modifications and an endoplasmic reticulumtargeting sequence additively enhance expression of an Aspergillus phytase gene in transgenic canola. Plant Cell Rep. 2006;25:124–132. doi: 10.1007/s00299-005-0036-y. [DOI] [PubMed] [Google Scholar]
- Perera IY, Hung CY, Moore CD, Stevenson-Paulik J, Boss WF. Transgenic Arabidopsis plants expressing the type 1 inositol 5-phosphatase exhibit increased drought tolerance and altered abscisic acid signaling. Plant Cell. 2008;20:2876–2893. doi: 10.1105/tpc.108.061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H, Türk M, Nyman M, Sandberg AS. Binding of Cu2+, Zn2+, and Cd2+ to inositol tri-, tetra-, penta-, and hexaphosphates. J Agric Food Chem. 1998;46:3194–3200. doi: 10.1021/jf971055w. [DOI] [Google Scholar]
- Raboy V. Seeds for a better future: ‘low phytate’ grains help to overcome malnutrition and reduce pollution. Trends Plant Sci. 2001;6:458–462. doi: 10.1016/S1360-1385(01)02104-5. [DOI] [PubMed] [Google Scholar]
- Raboy V. myo-Inositol-1, 2, 3, 4, 5, 6-hexakisphosphate. Phytochemistry. 2003;64:1033–1043. doi: 10.1016/S0031-9422(03)00446-1. [DOI] [PubMed] [Google Scholar]
- Raboy V. The ABCs of low-phytate crops. Nat Biotechnol. 2007;25:874–875. doi: 10.1038/nbt0807-874. [DOI] [PubMed] [Google Scholar]
- Raboy V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009;177:281–296. doi: 10.1016/j.plantsci.2009.06.012. [DOI] [Google Scholar]
- Raboy V, Gerbasi PF, Young KA, Stoneberg SD, Pickett SG, Bauman AT, Ertl DS. Origin and seed phenotype of maize low phytic acid 1–1 and low phytic acid 2–1. Plant Physiol. 2000;124:355–368. doi: 10.1104/pp.124.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaekers L, Remans R, Rao IM, Blair MW, Vanderleyden J. Strategies for improving phosphorus acquisition efficiency of crop plants. Field Crop Res. 2010;117:169–176. doi: 10.1016/j.fcr.2010.03.001. [DOI] [Google Scholar]
- Rao DE, Rao KV, Reddy TP, Reddy VD. Molecular characterization physicochemical properties known and potential applications of phytases An overview. Crit Rev Biotechnol. 2009;29:182–198. doi: 10.1080/07388550902919571. [DOI] [PubMed] [Google Scholar]
- Reddy NR (2002) Occurrence distribution content and dietary intake of phytate. Food Phytates 25–51
- Reddy CS, Vani K, Pandey S, Vijaylakshmi M, Reddy PCO, Kaul T. Manipulating microbial phytases for heterologous expression in crops for sustainable nutrition. Ann Plant Sci. 2013;2:436–454. [Google Scholar]
- Reddy CS, Achary VMM, Manna M, Singh J, Kaul T, Reddy MK. Isolation and molecular characterization of thermostable phytase from Bacillus subtilis (BSPhyARRMK33) Appl Biochem Biotechnol. 2015;175:3058–3067. doi: 10.1007/s12010-015-1487-4. [DOI] [PubMed] [Google Scholar]
- Reddy CS, Kim KM, James D, Varakumar P, Reddy MK. PgPAP18 a heat-inducible novel purple acid phosphatase 18-like gene (PgPAP18-like) from Pennisetum glaucum, may play a crucial role in environmental stress adaptation. Acta Physiol Plant. 2017;39:54. doi: 10.1007/s11738-017-2348-2. [DOI] [Google Scholar]
- Richardson AE, Hadobas PA, Hayes JE. Extracellular secretion of Aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant J. 2001;25:641–649. doi: 10.1046/j.1365-313x.2001.00998.x. [DOI] [PubMed] [Google Scholar]
- Richardson AE, George TS, Hens M, Simpson RJ (2004) Utilization of soil organic phosphorus by higher plants in organic phosphorus in the environment Walling ford. CABI Publishing pp, 165:184
- Sajidan A, Farouk A, Greiner R, Jungblut P, Müller EC, Borriss R. Molecular and physiological characterisation of a 3-phytase from soil bacterium Klebsiella sp. ASR1. Appl Microbiol Biotechnol. 2004;65:110–118. doi: 10.1007/s00253-003-1530-1. [DOI] [PubMed] [Google Scholar]
- Reddy NR, Pierson MD, Sathe SK, Salunkhe DK. Phytates in cereals and legumes. Florida: CRC Press; 1989. [Google Scholar]
- Scholz RW, Hellums DT, Roy AA. Global sustainable phosphorus management: a transdisciplinary venture. Curr Sci India. 2015;108:1237–1246. [Google Scholar]
- Selle PH, Ravindran V, Ravindran G, Bryden WL. Effects of dietary lysine and microbial phytase on growth performance and nutrient utilisation of broiler chickens. Asian Aust J Anim Sci. 2007;20:1100–1107. doi: 10.5713/ajas.2007.1100. [DOI] [Google Scholar]
- Shamsuddin AM. Anti-cancer function of phytic acid. Int J Food Sci Technol. 2002;37:782. doi: 10.1046/j.1365-2621.2002.00620.x. [DOI] [Google Scholar]
- Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA. Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus. 2013;2:1. doi: 10.1186/2193-1801-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Satyanarayana T. Microbial phytases in phosphorus acquisition and plant growth promotion. Physiol Mol Biol Plants. 2011;17:93–103. doi: 10.1007/s12298-011-0062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Satyanarayana T. Fungal phytases: characteristics and amelioration of nutritional quality and growth of non-ruminants. J Anim Physiol Anim Nutr. 2015;99:646–660. doi: 10.1111/jpn.12236. [DOI] [PubMed] [Google Scholar]
- Somasundar P, Riggs DR, Jackson BJ, Cunningham C, Vona-Davis L, McFadden DW. Inositol hexaphosphate (IP6): a novel treatment for pancreatic cancer 1. J Surg Res. 2005;126:199–203. doi: 10.1016/j.jss.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Stutter MI, Shand CA, George TS, Blackwell MS, Dixon L, Bol R, Haygarth PM. Land use and soil factors affecting accumulation of phosphorus species in temperate soils. Geoderma. 2015;257:29–39. doi: 10.1016/j.geoderma.2015.03.020. [DOI] [Google Scholar]
- Suzuki U, Yoshimura K, Takaishi M. Ueberein enzyme phytase das anhydro-oxy-methilen diphos-phorusa ure spaltet, Bulletin of the College of Agriculture. Tokyo Imperial University. 1907;7:503–512. [Google Scholar]
- Tarafdar JC, Jungk A. Phosphatase activity in the rhizosphere and its relation to the depletion of soil organic phosphorus. Biol Fertil Soils. 1987;3:199–204. doi: 10.1007/BF00640630. [DOI] [Google Scholar]
- Tye A, Siu F, Leung T, Lim B. Molecular cloning and the biochemical characterization of two novel phytases from B. subtilis 168 and B. licheniformis. Appl Microbiol Biotechnol. 2002;59:190–197. doi: 10.1007/s00253-002-1033-5. [DOI] [PubMed] [Google Scholar]
- Ullah AH, Sethumadhavan K, Mullaney EJ, Ziegelhoffer T, Austin-Phillips S. Characterization of recombinant fungal phytase (phyA) expressed in tobacco leaves. Biochem Biophys Res Commun. 1999;264:201–206. doi: 10.1006/bbrc.1999.1501. [DOI] [PubMed] [Google Scholar]
- Ullah AH, Sethumadhavan K, Mullaney EJ, Ziegelhoffer T, Austin-Phillips S. Cloned and expressed fungal phyA gene in alfalfa produces a stable phytase. Biochem Biophys Res Commun. 2002;290:1343–1348. doi: 10.1006/bbrc.2002.6361. [DOI] [PubMed] [Google Scholar]
- Ullah AH, Sethumadhavan K, Mullaney EJ, Ziegelhoffer T, Phillips AS. Fungal phyA gene expressed in potato leaves produces active and stable phytase. Biochem Biophys Res Commun. 2003;306:603–609. doi: 10.1016/S0006-291X(03)01002-7. [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003;157:423–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Van Paridon PA, van Ooyen AJ, van Lent JW, Hoekema A, Pen J. Stable accumulation of Aspergillus niger phytase in transgenic tobacco leaves. Plant Physiol. 1995;109:1199–1205. doi: 10.1104/pp.109.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohra A, Satyanarayana T. Phytases: microbial sources, production, purification, and potential biotechnological applications. Crit Rev Biotechnol. 2003;23:29–60. doi: 10.1080/713609297. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gao X, Su Q, Wu W, An L. Expression of a heat stable phytase from Aspergillus fumigatus in tobacco (Nicotiana tabacum L. cv. NC89) Indian J Biochem Biophys. 2007;44:26–30. [PubMed] [Google Scholar]
- Wang X, Wang Y, Tian J, Lim BL, Yan X, Liao H. Over expressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol. 2009;151:233–240. doi: 10.1104/pp.109.138891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ye X, Ding G, Xu F. Over expression of phyA and appA genes improves soil organic phosphorus utilization and seed phytase activity in Brassica napus. PLoS One. 2013;8:e60801. doi: 10.1371/journal.pone.0060801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Harrison MJ, Wang ZY. Transgenic expression of a novel M. truncatula phytase gene results in improved acquisition of organic phosphorus by Arabidopsis. Planta. 2006;222:27–36. doi: 10.1007/s00425-005-1511-y. [DOI] [PubMed] [Google Scholar]
- Yao MZ, Zhang YH, Lu WL, Hu MQ, Wang W, Liang AH. Phytases: crystal structures protein engineering and potential biotechnological applications. J Appl Microbiol. 2012;112:1–14. doi: 10.1111/j.1365-2672.2011.05181.x. [DOI] [PubMed] [Google Scholar]
- Yip W, Wang L, Cheng C, Wu W, Lung S, Lim BL. The introduction of a phytase gene from Bacillus subtilis improved the growth performance of transgenic tobacco. Biochem Biophys Res Commun. 2003;310:1148–1154. doi: 10.1016/j.bbrc.2003.09.136. [DOI] [PubMed] [Google Scholar]
- Yoon SM, Kim SY, Li KF, Yoon BH, Choe S, Kuo MM. Transgenic microalgae expressing Escherichia coli AppA phytase as feed additive to reduce phytate excretion in the manure of young broiler chicks. Appl Microbiol Biotechnol. 2011;91:553–563. doi: 10.1007/s00253-011-3279-2. [DOI] [PubMed] [Google Scholar]
- Zamani K, Lohrasebi T, Sabet MS, Malboobi MA, Mousavi A. Expression pattern and subcellular localization of Arabidopsis purple acid phosphatase AtPAP9. Gene Exp Pattern. 2014;14:9–18. doi: 10.1016/j.gep.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang ZB, Kornegay ET, Radcliffe JS, Wilson JH, Veit HP. Comparison of phytase from genetically engineered Aspergillus and canola in weanling pig diets. J Anim Sci. 2000;78:2868–2878. doi: 10.2527/2000.78112868x. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Kaplan S, Kim E, Hochman G, Graff G. Continents divided: understanding differences between Europe and North America in acceptance of GM crops. GM Crop Food. 2013;4:202–208. doi: 10.4161/gmcr.26981. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Zardi G, Lehmann M, Zeder C, Amrhein N, Frossard E, Bucher M. Engineering the root-soil interface via targeted expression of a synthetic phytase gene in trichoblasts. Plant Biotechnol J. 2003;1:353–360. doi: 10.1046/j.1467-7652.2003.00033.x. [DOI] [PubMed] [Google Scholar]