Abstract
Lactase has excellent applications in dairy industry and commercially this enzyme is produced from bacterial sources but not in high yields. In this work, the production of lactase was improved by designing of nutrient components in fermentation medium by one factor at a time. Lactose and yeast extract were selected as preferable carbon and nitrogen sources for lactase production with tryptophan and MgSO4 showing enhanced production. Statistical analysis proved to be a useful and powerful tool in developing optimum fermentation conditions. The individual and interactive role of lactose, yeast extract, magnesium sulfate, and tryptophan concentration on lactase production was examined by central composite design. Submerged fermentation with Bacillus subtilis strain VUVD001 produced lactase activity of 63.54 U/ml in optimized medium. The activity was threefold higher in comparison to an unoptimized medium. This result confirmed that the designed medium was useful for producing higher yields of lactase.
Keywords: Lactase, B. subtilis strain VUVD001, Yeast extract, Shake flask culture, Response surface methodology
Introduction
Lactase has catalytic activity for hydrolysis of lactose, cellobiose, and cellotriose. Lactase has a potential application in dairy industry as a solubilizing agent for milk products and uses members of the bacterial genera Lactobacillus (reuteri and plantarum), Bifidobacterium (adolescentis, infantis), B.circulans, etc., for this purpose as they are generally regarded as safe (GRAS) (Ichikawa et al. 1992; Batra et al. 2002). Therefore, the lactase produced by members of these genera can be consumed without excessive purification by lactose intolerance patients (Somkuti et al. 1998; He et al. 2008). Reduction in lactase activity may cause lactose intolerance (Karasova et al. 2002). The probiotic nature of bacterial genera Lactobacillus and Bifidobacterium enhances digestion capacity in lactose intolerance patients (Vinderola and Reinheimer 2003). It has been demonstrated that bacterial sources are highly preferable for production of lactase due to simple fermentation, high enzyme activity, and stability (Goodman and Pederson 1975; Picard et al. 2005). The yeast species, Kluyveromyces lactis and Kluyveromyces marxianus and Saccharomyces fragilis and molds such as Aspergillus and Rhizomucor species have also been used in the production of lactase (Shaikh et al. 1997; Santos et al. 1998). Designing of the suitable fermentation medium is a crucial step in bioprocess industries to improve the yield. The extracellular product concentration and microbial growth levels are strongly influenced by nutritional components of the medium (Garg and Jain 1995; Swift et al. 2000). The production and activity of enzymes are affected by different parameters such as type of strain, culture conditions, namely temperature, pH, agitation, incubation period, and the ratio of carbon and nitrogen sources in medium (Schneider et al. 2001; Jurado et al. 2004). The molar concentration of ions namely Ca2+, Mg2+, Na+, NH4+, and K+ also influences the activity and stability of lactase (Garman et al. 1996). Response surface methodology (RSM) is a more convenient tool for designing experiments, plotting models, evaluating the effects of factors and exploring optimum conditions of factors for significant responses. RSM is also used for optimization of prominent varieties of fermentation media and studying interactions among various bioprocess parameters with the minimum number of experiments (Amid et al. 2011; Gajdhane et al. 2016). In the past, production of lactase by RSM has been reported for various fungal strains (Liu et al. 2007; Anisha and Prema 2006). But till date, no previous work has used RSM for medium optimization for production of lactase from bacterial species. In the present experimentation, production of lactase in shake flask culture with B. subtilis strainVUVD001 was found to be higher when compared to other bacterial species. Therefore, this strain is considered as a potential bacterium for lactase production and hence, we aimed to develop a suitable medium for enhanced production of lactase through optimization of nutrient components concentrations by response surface methodology (RSM).
Materials and methods
Microorganism
The organism VUVD001 used in this study was isolated from dairy effluent, Sangam dairy, Vadlamudi, Guntur dist. India. Based on 16S rRNA gene sequence analysis strain VUVD001 was found to be most similar (99%) to a cluster represented by B. subtilis and relatives. The sequencing data of this strain has deposited National Centre for Biotechnology Information (NCBI) under accession number KT894158. The culture was maintained in our laboratory at room temperature and preserved at 4 °C on nutrient agar medium.
Media and fermentation conditions
The original fermentation medium comprised (in g/l): lactose, 4; yeast extract, 4; MgSO4·7H2O, 1 and tryptophan, 0.1. The shake flask fermentation was carried out by inoculating 5 ml seed culture in a conical flask containing 100 ml of production medium. The inoculated flask is kept for incubation for 36 h at pH 7, 36 °C on a rotary shaker (150 rpm)
Effect of various nutrient sources
The suitable carbon and nitrogen sources were identified for the production of enzyme by allowing bacterial strain proliferation in production medium. The medium was supplemented with different carbon sources such as lactose, glucose, sucrose, fructose, and starch in the concentration range of 0.4% (w/v) and 0.4% of nitrogen sources like yeast extract, urea, sodium nitrate, ammonium nitrate to investigate their effect on enzyme production. Similarly, the metal ions (0.1%) such as MgSO4, MnSO4 and ZnSO4 and different amino acids (0.01%) namely glycine, tryptophan, and cysteine were added to the medium to investigate its significant effect on production (Sharma and Singh 2014).
Lactase assay
The lactase activity was determined using ortho-nitrophenyl-β-galactoside (ONPG) as substrate. The ONPG solution was prepared with phosphate buffer and used for the assay. 0.5 ml of enzyme source was added with 2.0 ml of substrate and incubated for 30 min. The reaction was stopped by addition of 0.5 ml of 1 M Na2CO3 and absorbance was recorded at 420 nm. Activity of lactase was determined from ONP standard graph. One unit (U) of activity is defined as the amount of enzyme that liberates 1 micromole of ONP from the substrate per minute under assay conditions (Ghosh et al. 2012).
RSM for optimization of medium and model validation
The Design Expert software (Version 7.0.0, Stat-Ease Inc., Minneapolis, USA), was used to optimize the production medium variables, namely lactose (A), yeast extract (B), tryptophan (C), and MgSO4 (D) as shown in Table 1. The variables from low (−1) to high (+1) were used to study the effect of independent variables on production. Regression analysis of experimental data was performed and response surface graphs were drawn. The model was validated by conducting experiment in triplicates at best-predicted optimal variables of the medium.
Table 1.
Variable ranges used in experimental design
| Symbol | Name of the variable | Range | Code level | ||
|---|---|---|---|---|---|
| −1 | 0 | +1 | |||
| A | Lactose, g/L | 5–20 | 5 | 12.5 | 20 |
| B | Yeast extract, g/L | 5–15 | 5 | 10 | 15 |
| C | Tryptophan, g/L | 0.2–0.6 | 0.2 | 0.4 | 0.6 |
| D | MgSO4, g/L | 2–6 | 2 | 4 | 6 |
Results and discussion
Effect of carbon sources on enzyme production
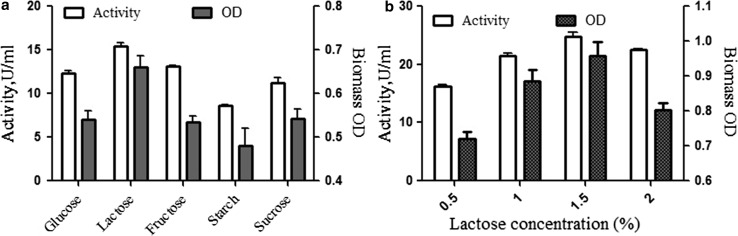
The amount of carbon source in fermentation media is the primary energy source and essential for bacterial growth and production of lactase in submerged fermentation. Carbon source may regulate the biosynthesis of lactase in different microorganisms (Alazzeh et al. 2009). The fermentation medium was supplemented with 0.4% of different carbon sources to study their individual influence on production by traditional optimization. It was found that glucose and fructose enhanced the levels of biomass and yield but they were less efficient as compared to lactose (15.14 U/ml) (Fig. 1a). Further, the lactose quantity was optimized by varying the concentrations from 0.5 to 2%. The results revealed a significant improvement in lactase activity (24.84 U/ml) and biomass at 1.5% of lactose (Fig. 1b). Sriphannam et al. (2012) and Pulicherla et al. (2013) have reported that lactose may enhance the production of lactase by probiotic strains Lactobacillus fermentum CM33 and Thalassospira frigidphilosprofundus, respectively, in submerged process. The present findings are in corroboration with these reports.
Fig. 1.
Effect of various carbon sources on Lactase production and biomass. a Different carbon sources of glucose, lactose, fructose, starch, sucrose, and b different concentrations of lactose in percent
Effect of nitrogen sources
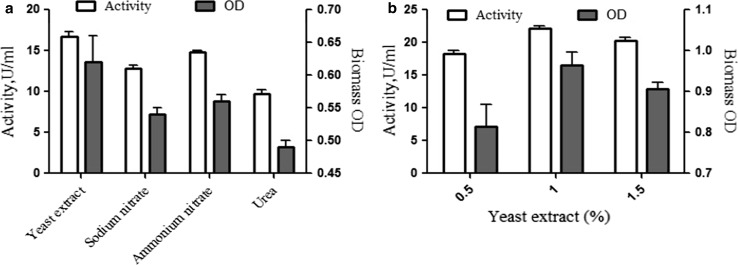
The nitrogen is an important factor which affects microbial biosynthesis of lactase (Shaikh et al. 1997). The effect of different nitrogen sources namely sodium nitrate, ammonium nitrate, yeast extract, and urea on cell growth and lactase production was investigated. The results indicated the enhancement in lactase activity with yeast extract and ammonium nitrate. However, the effectiveness of ammonium nitrate is less when compared to yeast extract (Fig. 2a). Further, the concentration of yeast extract for production was evaluated and maximum lactase activity of 22.19 U/ml was obtained at one percent of yeast extract (Fig. 2b). Hsu et al. (2007) used Bifidobacterium longum CCRC15708 for production of lactase in shake flask culture system and found highest lactase production with yeast extract as nitrogen source. Similarly, Devi et al. (2012) also reported highest activity of lactase with one percent of yeast extract in the submerged fermentation with Bacillus sp. MNJ23.
Fig. 2.
Effect of nitrogen sources on lactase production and biomass. a Different sources, i.e., yeast extract, sodium nitrate, ammonium nitrate, urea, and b yeast extract concentrations in percent
Effect of tryptophan induction on lactase activity
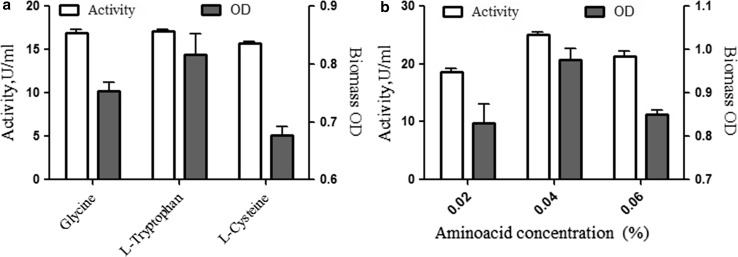
The amino acid in the medium acts as a stimulator for biosynthesis and excretion of enzymes. Therefore, production medium with amino acid may be considered as an experimental tool for significant enhancement of enzyme production (Gupta et al. 2003). Based on this, in the present study, tryptophan, glycine, and cysteine were selected to study their impacts on biomass and lactase production. It was found that among the three amino acids tested, tryptophan resulted in highest activity (Fig. 3a). Its effect on production was further evaluated for improving lactase production. It was observed that the medium fed with 0.04 percent tryptophan significantly enhanced the enzyme production to 25.10 U/ml in the shake flask fermentation (Fig. 3b). Similarly, Akcan (2011) suggested that the addition of l-tryptophan may enhance the biosynthesis of lactase by Bacillus licheniformis ATCC12759 in submerged fermentation.
Fig. 3.
Effect of amino acids on lactase production and biomass. a Different amino acids, i.e., glycine, l-tryptophan, l-cysteine, and b tryptophan concentrations in percent
Effect of ionic sources on lactase activity
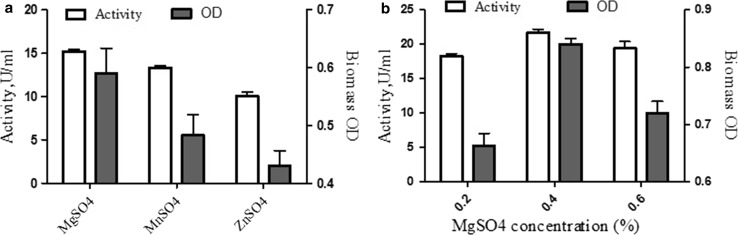
The effect of metal ions on the biosynthesis of enzyme production was screened by supplementing medium with 0.1 percent of metal sources such as MgSO4, MnSO4, and ZnSO4. The enzyme activity was improved with MgSO4 compared to MnSO4 and ZnSO4 and the results are displayed in Fig. 4a. In addition, the impact of MgSO4 was further investigated at different concentrations. Among the three ionic compounds examined, MgSO4 produced the highest activity of 27.12 U/ml with 0.4% of MgSO4 (Fig. 4b). The Mg2+ ion was found to enhance the release of lactase to the medium and this metal ion role as a channeling agent in membrane permeabilization has been reported (Karpen and Ruiz 2002). Similarly, Gopal et al.’s (2015) findings showed that the production of lactase was significantly improved with the supplementation of 10 mg of MgSO4 per gram of substrate. On the other hand, Kim and Rajagopal (2000) found the considerable effect of MnSO4 on the production of lactase through fermentation with L. crispatus but it is less efficient than MgSO4. In contrast, Ismail et al. (2010) observed that the addition of 0.02 M of MnCl2 to the medium had slight stimulatory influence on lactase production with L. acidophilus NRRL4495. They concluded that the addition of divalent ion sources such as Mg2+ and Mn2+ to growth medium may enhance the yields.
Fig. 4.
Effect of minerals on lactase production and biomass. a Different minerals, i.e., MgSO4, ZnSO4, MnSO4, and b MgSO4 concentrations in percent
Central composite design for medium optimization
After one factor at a time optimization study, activity of lactase was significantly improved as compared with basal medium. For further enhancing the lactase yield, the central composite design (CCD) was adopted for designing optimum concentration of nutrient components such as lactose, yeast extract, tryptophan, and MgSO4. The experimental results of CCD for enhancing the production of lactase are shown in Table 2.
Table 2.
The design data of CCD and lactase activity values
| Run no | Lactose (g/L) | Yeast extract (g/L) | Tryptophan (g/L) | MgSO4 (g/L) | Activity, U/ml | |
|---|---|---|---|---|---|---|
| Experimental | Predicted | |||||
| 1 | 5 | 5 | 0.6 | 6 | 28.31 | 26.17 |
| 2 | 20 | 5 | 0.6 | 6 | 27.46 | 29.10 |
| 3 | 5 | 5 | 0.2 | 2 | 25.18 | 24.11 |
| 4 | 5 | 5 | 0.6 | 2 | 28.75 | 30.48 |
| 5 | 12.5 | 10 | 0.4 | 4 | 63.04 | 60.70 |
| 6 | 5 | 15 | 0.2 | 6 | 34.25 | 34.73 |
| 7 | 5 | 15 | 0.6 | 6 | 41.09 | 41.76 |
| 8 | 12.5 | 10 | 0.4 | 2 | 61.74 | 61.87 |
| 9 | 5 | 5 | 0.2 | 6 | 27.41 | 27.06 |
| 10 | 20 | 10 | 0.4 | 4 | 51.62 | 52.46 |
| 11 | 12.5 | 5 | 0.4 | 4 | 38.75 | 42.04 |
| 12 | 12.5 | 10 | 0.4 | 4 | 62.84 | 60.70 |
| 13 | 12.5 | 10 | 0.4 | 4 | 61.02 | 60.70 |
| 14 | 5 | 15 | 0.2 | 2 | 23.14 | 22.39 |
| 15 | 12.5 | 10 | 0.4 | 4 | 60.54 | 60.70 |
| 16 | 20 | 5 | 0.2 | 6 | 37.91 | 36.57 |
| 17 | 20 | 5 | 0.6 | 2 | 35.75 | 33.77 |
| 18 | 5 | 15 | 0.6 | 2 | 36.85 | 36.69 |
| 19 | 20 | 15 | 0.2 | 6 | 57.12 | 56.28 |
| 20 | 12.5 | 15 | 0.4 | 4 | 55.85 | 54.99 |
| 21 | 20 | 15 | 0.6 | 6 | 57.16 | 56.73 |
| 22 | 12.5 | 10 | 0.6 | 4 | 62.47 | 61.89 |
| 23 | 20 | 15 | 0.6 | 2 | 50.76 | 52.00 |
| 24 | 20 | 15 | 0.2 | 2 | 43.65 | 44.29 |
| 25 | 20 | 5 | 0.2 | 2 | 33.75 | 33.97 |
| 26 | 12.5 | 10 | 0.4 | 6 | 63.42 | 65.71 |
| 27 | 5 | 10 | 0.4 | 4 | 38.45 | 40.04 |
| 28 | 12.5 | 10 | 0.2 | 4 | 55.48 | 58.48 |
| 29 | 12.5 | 10 | 0.4 | 4 | 63.01 | 60.70 |
| 30 | 12.5 | 10 | 0.4 | 4 | 61.05 | 60.70 |
The response (Y) fitted with the second-order polynomial equation
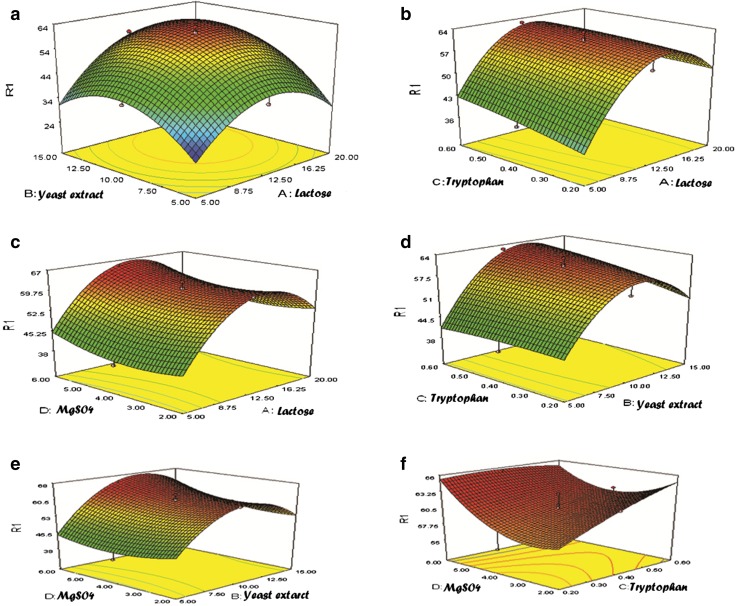
The model equation importance was statistically evaluated by the F test for the analysis of variance (ANOVA). The prob > F values (<0.0001) for the lactase production are lower than 0.05, which indicates that goodness of designed quadratic model was significant. Based on the observations from ANOVA table, it was found that the variables A, B, C, D, AB, AC, BC, BD, CD, A 2, B 2, and D 2 are significant model terms. The coefficient (R 2) value was found to be 0.98 which supports a high correlation between experimental and predicted values. The lower consistency of the experiment is generally indicated by the high value of the coefficient of variation (CV). In this case, low percent CV (4.55) represents the reliability of experiment performed. Adequate precision ratio greater than 4.00 is desirable. In the present study, the ratio is 29.12 which indicates an adequate signal (Table 3). The 3-D response surface curves were plotted to evaluate interactions of combinational medium variable effects on response and to find out optimal concentrations of nutrient sources for lactase production (Fig. 5a–f).
Table 3.
ANOVA analysis of model and medium components
| Source | Sum of squares | df | Mean square | F value | p value |
|---|---|---|---|---|---|
| Prob > F | |||||
| Model | 5707.57 | 14 | 407.68 | 92.08 | <0.0001 |
| A—lactose | 693.78 | 1 | 693.78 | 156.70 | <0.0001 |
| B—yeast extract | 755.31 | 1 | 755.31 | 170.59 | <0.0001 |
| C—tryptophan | 52.39 | 1 | 52.39 | 11.83 | 0.0036 |
| D—MgSO4 | 66.36 | 1 | 66.36 | 14.99 | 0.0015 |
| AB | 144.84 | 1 | 144.84 | 32.71 | <0.0001 |
| AC | 43.30 | 1 | 43.30 | 9.78 | 0.0069 |
| AD | 0.12 | 1 | 0.12 | 0.03 | 0.8701 |
| BC | 62.73 | 1 | 62.73 | 14.17 | 0.0019 |
| BD | 88.17 | 1 | 88.17 | 19.91 | 0.0005 |
| CD | 52.78 | 1 | 52.78 | 11.92 | 0.0036 |
| A 2 | 541.39 | 1 | 541.39 | 122.28 | <0.0001 |
| B 2 | 385.02 | 1 | 385.02 | 86.96 | <0.0001 |
| C 2 | 0.69 | 1 | 0.69 | 0.16 | 0.699 |
| D 2 | 24.73 | 1 | 24.73 | 5.59 | 0.032 |
| Residual | 66.41 | 15 | 4.43 | ||
| Lack of fit | 59.65 | 10 | 5.97 | 4.41 | 0.0575 |
| Pure error | 6.76 | 5 | 1.35 | ||
| Cor total | 5773.99 | 29 |
R-squared 0.98
Adj R-squared 0.97
Pred R-squared 0.94
Adeq precision 29.12
C.V. % 4.55
Fig. 5.
a–f Response surface plot showing the combinational effects of multiple variables on lactase production
Validation of RSM model
The RSM model is validated by conducting an experiment at best-predicted solution for production of lactase. Under optimized conditions, the enzyme activity reached 63.54 U/ml from B. subtilis strain VUVD001, which is almost near to the RSM predicted value (Table 4). The maximum lactase activity previously reported was 18.6 U/ml obtained in submerged fermentation with Bifidobacterium longum CCRC 15708. The process was run at optimum conditions of pH 6.5, temperature 37 °C and incubation time 16 h in liquid medium containing 4% lactose, 3.5% yeast extract, 0.3% K2HPO4, 0.1% KH2PO4, 0.05% MgSO4·7H2O, and 0.03% l-cysteine (Hsu et al. 2007). Jokar and Karbassi in 2009 designed a new medium by factorial method. This medium provided the maximum lactase activity of 4.924 U/ml at optimum conditions of 3% yeast extract, 1.5% whey powder, and 2% wheat steep liquor. Compared to these results, our newly designed medium produced an improved activity of 12.9-folds and it is economically feasible. Similarly, Lee et al. (2013) obtained the highest activity of 1.17 U/ml through submerged fermentation with Bacillus sp. LX-1 with the optimized medium consisting of 16.5 g/L galactose, 6.2 g/L peptone, and 0.0018 g/L. It seems that in the study by Lee et al., the effect of galactose and MnSO4 has not much influence on the production. But in our experimental studies, we got 54-fold higher lactase production than Lee et al.’s report. The reason for improved activity may be the addition of tryptophan. Similarly, Prasad et al. (2013) reported the highest intracellular lactase activity of 6.80 U/ml and 7.7 U/ml by culturing Bifidobacterium animalis spp. lactis Bb12 and Lactobacillus delbrueckii spp. bulgaricus ATCC11842 on whey. Both microbes were grown in protein-free whey medium containing yeast extract (3.0 g/L), peptone (5.0 g/L), and glucose (10.0 g/L) for 18 h, at 37 °C for B. animalis ssp. lactis Bb12 and at 45 °C for L. delbrueckii ssp. bulgaricus ATCC11842, respectively. Compared to the Prasad et al. report, our strain is an extracellular producer and the lactase activity was found to be 9.3- and 8.4-folds higher. In the fermentation process, they used multiple nitrogen sources which increase the overall cost of the process but in the present study, a single nitrogen source was used which may reduce the cost of the process. Based on these findings, it was concluded that the tryptophan supplement and design of medium with four nutritional components is a novel attempt for improving the production of lactase yield through submerged fermentation using B. subtilis VUVD001.
Table 4.
RSM optimized medium variables for lactase activity
| A—lactose (g/L) | B—yeast extract (g/L) | C—tryptophan (g/L) | D—MgSO4 (g/L) | Activity, U/ml | |
|---|---|---|---|---|---|
| Predicted (U/ml) | Experimental (U/ml) | ||||
| 14.01 | 10.30 | 0.43 | 5.32 | 64.49 | 63.54 |
Conclusion
The present study was undertaken to optimize the media components and enhance the production of lactase by RSM using B. subtilis strainVUVD001. The desired quantities of medium components established by RSM for the highest production of lactase were lactose, 14.01 g/L; yeast extract, 10.30 g/L; tryptophan, 0.43 g/L; and MgSO4, 5.32 g/L.
Acknowledgements
The authors acknowledge Vignan’s Foundation for Science, Technology and Research University, Vadlamudi, Guntur, Andhra Pradesh, India for providing necessary facilities to carry out this work.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
References
- Akcan N. High level production of extracellular galactosidase from Bacillus licheniformis ATCC 12759 in submerged fermentation. Afr J Microbiol Res. 2011;5:4615–4621. [Google Scholar]
- Alazzeh AY, Ibrahım SA, Song D, Shahbazi A, Abu Ghazaleh AA. Carbohydrate and protein sources influence the induction of α- and β-galactosidases in Lactobacillus reuteri. Food Chem. 2009;4:654–659. doi: 10.1016/j.foodchem.2009.04.065. [DOI] [Google Scholar]
- Amid A, Ismail N, Yusof F, Salleh HM. Expression, purification and characterization of a recombinant stem bromelain from Ananas comosus. Process Biochem. 2011;46:2232–2239. doi: 10.1016/j.procbio.2011.08.018. [DOI] [Google Scholar]
- Anisha GS, Prema P. Production of galactosidase by a novel actinomycete Streptomyces griseoloalbus and its application in soymilk hydrolysis. World J Microbiol Biotechnol. 2006;23:859–864. doi: 10.1007/s11274-006-9310-6. [DOI] [Google Scholar]
- Batra N, Singh J, Banerjee UC, Patnaik P, Sobti RC. Production and characterization of a thermostable b-galactosidase from Bacillus coagulans RCS3. Biotechnol Appl Biochem. 2002;36:1–6. doi: 10.1042/BA20010091. [DOI] [PubMed] [Google Scholar]
- Devi DM, Mukesh Kumar DJ, Thisiddhuraj J, Naganarayani K, Immaculate NRA, Kalaichelvan PT. Concomitant production of amylase and galactosidase by native Bacillus sp. MNJ23 isolated from dairy effluent, American-Eurasian. J Agric Environ Sci. 2012;12:579–587. [Google Scholar]
- Gajdhane SB, Bhagwat PK, Dandge PB. Response surface methodology-based optimization of production media and purification of a-galactosidase in solid-state fermentation by Fusarium moniliforme NCIM 1099. 3 Biotech. 2016;6:1–14. doi: 10.1007/s13205-016-0575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg SK, Jain A. Fermentative production of 2, 3-butanediol: a review. Bioresour Technol. 1995;51:103–109. doi: 10.1016/0960-8524(94)00136-O. [DOI] [Google Scholar]
- Garman J, Coolbear T, Smart J. The effect of cations on the hydrolysis of lactose and the transferase reactions catalysed by β-galactosidase from six strains of lactic acid bacteria. Appl Microbiol Biotechnol. 1996;46:22–27. doi: 10.1007/s002530050778. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Pulicherla KK, Rekha VPB, Kumarraja P, Rao KRSS. Cold active b-galactosidase from Thalassospira sp. 3SC-21 to use in milk lactose hydrolysis: a novel source from deep waters of Bay-of-Bengal. World J Microbiol Biotechnol. 2012;28:2859–2869. doi: 10.1007/s11274-012-1097-z. [DOI] [PubMed] [Google Scholar]
- Goodman RE, Pederson DM. β-Galactosidase from Bacillus sterothermophilus. Can J Microbiol. 1975;22:817–825. doi: 10.1139/m76-118. [DOI] [PubMed] [Google Scholar]
- Gopal G, Raol Raol BV, Prajapati Vimal S, Bhavsar NH. Utilization of agro-industrial waste for b-galactosidase production under solid state fermentation using halotolerant Aspergillus tubingensis GR1 isolate. 3 Biotech. 2015;5:411–421. doi: 10.1007/s13205-014-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B. Microbial α-amylases: a biotechnological perspective. Process Biochem. 2003;38:1599–1616. doi: 10.1016/S0032-9592(03)00053-0. [DOI] [Google Scholar]
- He T, Priebe MG, Zhong Y, Huang C, Harmsen HJ, Raangs GC, Antoine JM, Welling GW, Vonk RJ. Effects of yogurt and bifidobacteria supplementation on the colonic microbiota in lactose intolerant subjects. J Appl Microbiol. 2008;104:595–604. doi: 10.1111/j.1365-2672.2007.03579.x. [DOI] [PubMed] [Google Scholar]
- Hsu CA, Yu RC, Chou SL. Cultural condition affecting the growth and production of β-galactosidase by Bifidobacterium longum CCRC 15708 in a jar fermenter. Int J Food Microbiol. 2007;116:186–189. doi: 10.1016/j.ijfoodmicro.2006.12.034. [DOI] [PubMed] [Google Scholar]
- Ichikawa Y, Look GC, Wong CH. Enzyme-catalyzed oligosaccharide synthesis. Anal Biochem. 1992;202:215–238. doi: 10.1016/0003-2697(92)90099-S. [DOI] [PubMed] [Google Scholar]
- Ismail SAA, El-Mohamady Y, Helmy WA, Abou-Romia R, Hashem AM. Cultural condition affecting the growth and production of β-galactosidase by Lactobacillus acidophilus NRRL 4495. Aust J Basic Appl Scib. 2010;4:5051–5058. [Google Scholar]
- Jokar A, Karbassi A. Determination of proper conditions for the production of crude beta-galactosidase using Lactobacillus delbrueckii ssp. bulgaricus. J Agric Sci Technol. 2009;11:301–308. [Google Scholar]
- Jurado E, Camacho F, Luzón G, Vicaria JM. Kinetic models of activity for β-galactosidases influence of pH, ionic concentration and temperature. Enzyme Microb Technol. 2004;34:33–40. doi: 10.1016/j.enzmictec.2003.07.004. [DOI] [Google Scholar]
- Karasova P, Spiwok V, Mala S, Kralova B, Russell NJ. Betagalactosidase activity in psychrophic microorganisms and their potential use in food industry. Czech J Food Sci. 2002;20:43–47. [Google Scholar]
- Karpen JW, Ruiz ML. Ion channels: does each subunit do something on its own. Trends Biochem Sci. 2002;27:402–409. doi: 10.1016/S0968-0004(02)02124-2. [DOI] [PubMed] [Google Scholar]
- Kim JW, Rajagopal SN. Isolation and characterization of b-galactosidase from Lactobacillus crispatus. Folia Microbiol. 2000;45:29–34. doi: 10.1007/BF02817446. [DOI] [PubMed] [Google Scholar]
- Lee J, Park I, Cho S, Kim S, Choi Y, Cho J. Optimization of medium composition for-galactosidase production by Antarctic bacterial isolate, Bacillus sp. LX-1 using response surface methodology. Afr J Microbiol Res. 2013;7:3494–3500. [Google Scholar]
- Liu C, Ruan H, Shen H, Chen Q, Zhou B, Li Y, He G. Optimization of the fermentation medium for galactosidase production from Aspergillus foetidus ZU-G1 using response surface methodology. J Food Sci. 2007;72:120–125. doi: 10.1111/j.1750-3841.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- Picard C, Fioramonti J, Francois A, Robinson T, Neant F, Matuchansky C. Bifidobacteria as probiotic agents—physiological effects and clinical benefits. Aliment Pharmacol Ther. 2005;22:495–512. doi: 10.1111/j.1365-2036.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- Prasad L, Ghosh N, Sherkat BC. Extraction and characterisation of β-galactosidase produced by Bifidobacterium animalis spp. lactis Bb12 and Lactobacillus delbrueckii spp. bulgaricus ATCC 11842 grown in whey. Int Food Res J. 2013;20:487–494. [Google Scholar]
- Pulicherla KK, Kumar PS, Manideep K, Rekha VP, Ghosh M, Sambasiva Rao KR. Statistical approach for the enhanced production of cold-active β-galactosidase from Thalassospira frigidphilosprofundus a novel marine psychrophile from deep waters of Bay of Bengal. Prep Biochem Biotechnol. 2013;43(8):766–780. doi: 10.1080/10826068.2013.773341. [DOI] [PubMed] [Google Scholar]
- Santos A, Ladero M, Garcia-Ochoa F. Kinetic modeling of lactose hydrolysis by a galactosidase from Kluyveromyces fragilis. Enzyme Microb Technol. 1998;22:558–567. doi: 10.1016/S0141-0229(97)00236-6. [DOI] [PubMed] [Google Scholar]
- Schneider ALS, Merkle R, Carvalho-jonas MF, Jonas R, Furlan S. Oxygen transfer on β-D-galactosidase production by Kluyveromyces marxianus using sugar cane molasses as carbon source. Biotechnol Lett. 2001;23:547–550. doi: 10.1023/A:1010338904870. [DOI] [Google Scholar]
- Shaikh SA, Khire JM, Khan MI. Production of galactosidase from thermophilic fungus Rhizomucor sp. J Ind Microbiol Biotechnol. 1997;17:239–245. doi: 10.1038/sj.jim.2900452. [DOI] [Google Scholar]
- Sharma S, Singh P. Isolation and characterization of Galactosidase enzyme producing microbe and optimization of its enzyme activity under different culture condition. Int J Curr Microbiol App Sci. 2014;3:148–155. [Google Scholar]
- Somkuti GA, Dominiecki ME, Steýnberg DH. Permeabilization of Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus with ethanol. Curr Microbiol. 1998;36:202–206. doi: 10.1007/s002849900294. [DOI] [PubMed] [Google Scholar]
- Sriphannam W, Unban K, Ashida H, Kenji Y, Khanongnuch C. Medium component improvement for β-galactosidase production by a probiotic strain Lactobacillus fermentum CM33. Afr J Biotechnol. 2012;11:11242–11251. [Google Scholar]
- Swift RJ, Karandikar A, Griffen AM, Punt PJ, Cees AMJJ, Hondel VD, Robson GD, Trinci APJ, Weibe MG. The effect of organic nitrogen sources on recombinant glucoamylase production by Aspergillus niger in chemostat culture. Fungal Gen Biol. 2000;31:125–133. doi: 10.1006/fgbi.2000.1241. [DOI] [PubMed] [Google Scholar]
- Vinderola CG, Reinheimer JA. Lactic acid starter and probiotic bacteria a comparative in vitro study of probiotic characteristics and biological barrier resistance. J Food Res Int. 2003;36:895–904. doi: 10.1016/S0963-9969(03)00098-X. [DOI] [Google Scholar]