Abstract
In the fission yeast Schizosaccharomyces pombe, meiosis normally takes place in diploid zygotes resulting from conjugation of haploid cells. In the present study, we report that the expression of a constitutively activated version of the pheromone-responsive mitogen-activated protein kinase kinase kinase (MAP3K) Byr2 can induce ectopic meiosis directly in haploid cells. We find that the Ste11 transcription factor becomes constitutively expressed in these cells and that the expression of pheromone-responsive genes no longer depends on nitrogen starvation. Epistasis analysis revealed that these conditions bypassed the requirement for the meiotic activator Mei3. Since Mei3 is normally needed for inactivation of the meiosis-repressing protein kinase Pat1, this finding suggests that the strong Byr2 signal causes inactivation of Pat1 by an alternative mechanism. Consistent with this possibility, we found that haploid meiosis was dramatically reduced when Ste11 was mutated to mimic phosphorylation by Pat1. The mutation of two putative MAPK sites in Ste11 also dramatically reduced the level of haploid meiosis, suggesting that Ste11 is a direct target of Spk1. Supporting this, we show that Spk1 can interact physically with Ste11 and also phosphorylate the transcription factor in vitro. Finally, we demonstrate that ste11 is required for pheromone-induced G1 arrest. Interestingly, when we mutated Ste11 in the sites for Pat1 and Spk1 phosphorylation simultaneously, the cells could still arrest in G1 in response to pheromone, suggesting the existence of yet a third bifurcation of the signaling pathway.
Eukaryotic cells integrate multiple signals from the environment when deciding whether to divide or undergo differentiation. In the fission yeast Schizosaccharomyces pombe, a sexual differentiation program is activated in response to nutritional deprivation (37, 64). Starvation, in particular of a nitrogen source, causes haploid cells to exhibit either M (minus) or P (plus) mating behavior, depending on which allele they express from their mat1 locus. Hereby a pheromone communication system that enables mating between the two cell types is established. The resulting diploid zygotes subsequently enter meiosis and sporulation, and this also depends on nitrogen starvation and pheromone signaling. Hence, the sexual differentiation process appears to be orchestrated by a complex integration of signals from the environment.
The high-mobility-group protein Ste11 constitutes a key transcription factor in the switch from mitotic to meiotic cell division. Ste11 activates numerous genes required for mating and meiosis, including the mat genes that control pheromone signaling and mei2, encoding an RNA-binding protein that triggers meiosis (6, 52, 58). During mitotic growth, transcription of the ste11+ gene is kept low due to the absence of starvation signals (52). Furthermore, the activity of the Ste11 protein is repressed by the Pat1 (or Ran1) protein kinase, a general inhibitor of sexual differentiation (4, 16, 17, 31, 40). Pat1 can phosphorylate Ste11 on Thr173 and Ser218 (27), and the 14-3-3 protein Rad24 binds Ste11 phosphorylated on these residues and inhibits its nuclear accumulation (22, 43). Since the ste11+ gene is autoregulated (25), this nuclear exclusion contributes to reducing ste11+ expression in vegetative cells.
Nitrogen limitation and pheromone signaling cause transcriptional induction of Ste11-controlled genes, and both of these signals are also required for the accumulation of Ste11 in the nucleus (43). Activation of Ste11 appears to involve stimulation of a positive feedback loop initiated by a gradual inhibition of the Pat1 protein kinase (4, 39). Thus, inactivation of a pat1 temperature-sensitive allele causes the induction of Ste11-controlled genes and mating in rich medium (4, 39, 41), and Ste11 is constantly nuclear in the absence of Pat1 (43). However, an activated mutant carrying alanine substitutions in the Pat1 phosphorylation sites of Ste11 still requires pheromone signaling in order to accumulate Ste11 in the nucleus, suggesting that Pat1 both directly and indirectly prevents nuclear accumulation of Ste11 (43).
The balance between the activities of Pat1 and Ste11 also regulates entry into meiosis. Nitrogen starvation rapidly induces Ste11-dependent expression of the mei2 gene (48, 52), but the Mei2 protein is kept inactive by Pat1-mediated phosphorylation until mating has occurred (57). Successful conjugation allows expression of the Mei3 protein, which serves as an inhibitory pseudosubstrate for Pat1 (27). Consequently, Pat1 can no longer phosphorylate Mei2, which then triggers meiosis in its unphosphorylated form (31, 57).
Since mei3 induction requires the expression of the two cell type-specific genes mat1-Pm and mat1-Mm, only diploid zygotes that are heterozygous for the mating type loci can enter meiosis (21, 32, 61). Furthermore, because these two genes are pheromone stimulated, entry into meiosis requires an intact pheromone communication pathway.
In order to transduce the mating pheromone signal, Schizosaccharomyces pombe employs a G protein-coupled receptor system and a mitogen-activated protein (MAP) kinase cascade, comprised of Byr2 (a MAP kinase kinase kinase [MAP3K]), Byr1 (a MAP2K), and Spk1 (a MAPK). This signaling pathway is essential for both conjugation and meiosis (34, 35, 51, 55, 56) and is stimulated by the Ras1 protein, a unique homolog of the mammalian Ras protein (36, 38). Like other MAP3Ks, Byr2 is presumably activated by the displacement of its N-terminal regulatory domain from the C-terminal kinase domain, and it has been demonstrated that Ras1 interacts with the regulatory domain directly (2).
The factor(s) activated by the Byr2-Byr1-Spk1 pathway has still to be identified, but Ste11 is an attractive candidate (1, 23, 42, 52). Thus, a heterologous promoter carrying eight copies of the TR box (to which Ste11 binds) is pheromone inducible (24). Hence, Ste11 appears to be directly responsible for the induction of transcription in response to both nitrogen starvation and pheromone signaling, and the pheromone MAPK pathway may actually participate in the transmission of the nutritional signal as well. Thus, byr2 and byr1 are required for the induction of the M-factor genes and the mam2 gene by both nitrogen starvation and pheromone signaling (23, 63).
In this study, we show that hyperactivation of the Byr2-Byr1-Spk1 pathway by the expression of a truncated version of Byr2 lacking its regulatory domain induces ectopic meiosis in haploid S. pombe cells. Quite unexpectedly, this induction of meiosis does not require Mei3. Hence, unregulated Byr2 activity can bypass the meiotic requirement for heterozygosity at the mat locus. Moreover, entry into meiosis becomes independent of nitrogen starvation, supporting the notion that the pheromone response pathway conveys the nutritional signal. Finally, the study of the hyperactivated Byr2 allele allowed us to obtain evidence in support of Ste11 being a direct target of Spk1 phosphorylation.
MATERIALS AND METHODS
Yeast strains, genetic procedures, and media.
The fission yeast strains used in this study are listed in Table 1. Standard genetic procedures were carried out as described previously (33). For physiological experiments, cells were grown in minimal sporulation liquid (MSL) (11). To induce sexual differentiation, cells with a density of 2.5 × 106 to 5 × 106 cells/ml were shifted to nitrogen-deficient MSL (MSL lacking arginine; MSL-N) and incubated for 3 to 8 h. To induce expression from the nmt promoter (3, 29), transformants were grown in MSL containing 6 μM thiamine, then shifted to fresh medium lacking thiamine, and grown for 14 h before starting induction of sexual differentiation. When indicated, chemically synthesized M-factor (made by Schafer-N, Copenhagen, Denmark) was added to a final concentration of 1 μg/ml. Strains were grown at 30°C unless otherwise indicated.
TABLE 1.
Fission yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Eg328 | h−smt0 ura4-D18 | Lab stock |
| Eg494 | h90 ste11 leu1 ade6-M210 | M. Yamamoto, JY7 |
| Eg545 | h+ Δmat2/3::LEU2 | Lab stock |
| Eg576 | h90 ade6-M216 leu1 ura4 mei2::ura4+ | M. Yamamoto, JZ127 |
| Eg578 | h90 ade6-M216 leu1 mam2 map3-74 | M. Yamamoto, JZ362 |
| Eg589 | h+ Δmat2/3::leu2−leu1 | Lab stock |
| Eg627 | h90 ras1::ura4+ura4-D18 leu1 ade6-M216 | C. Shimoda, JZ522 |
| Eg640 | h90 leu1 ura4-D18 | Lab stock |
| Eg709 | Δmat1::ura4+leu1 ura4-D18 | K. Ekwall, KE40 |
| Eg710 | h90 ade6 leu1 gpa1::ura4+ | M. Yamamoto |
| Eg768 | h− Δmat2/3::leu2−leu1 ura4-D18 | Lab stock |
| Eg930 | h90 Δbyr1 ura4-D18 leu1 | This study |
| Eg931 | h90 Δbyr2 ura4-D18 leu1 | This study |
| Eg934 | h90 Δspk1 ura4-D18 leu1 | This study |
| Eg950 | h− Δste11::LEU2 leu1 ura4-D18 | Lab stock |
| Eg1062 | h−mik1-6myc-His sup3-5 ura4-D18 leu1-32 ade6-704 | Lab stock |
| Eg1085 | h90 ste11::ura4+leu1-32 ura4-D18 | This study |
| Eg1092 | h90 ste11T173A, S218Aleu1-32 ura4-D18 | This study |
| Eg1093 | h90 ste11T173D, S218Dleu1-32 ura4-D18 | This study |
| Eg1125 | h+ Δmat2/3-leu2−leu1 ura4-D18 ste11::ura4+ | This study |
| Eg1207 | h90 ste11T305A, T317Aleu1-32 ura4-D18 | This study |
| Eg1226 | h− Δmat2/3::leu2−ste11T173D, S218D leu1-32 ura4-D18 | This study |
| Eg1259 | h− Δmat2/3::LEU2+ste11T305A, T317Aura4-D18 | This study |
| Eg1638 | h90 ste11T173D, S218D, T305A, T317A leu1-32 ura4-D18 | This study |
| Eg1645 | h90 ste11T305D, T317Dleu1-32 ura4-D18 | This study |
| Eg1646 | h90 ste11T173A, S218A, T305D, T317Dleu1-32 ura4-D18 | This study |
| Eg1671 | h− ste11T173D, S218D, T305A, T317A ura4-D18 | This study |
| Eg1672 | h+ste11T305D, T317D | This study |
| Eg1673 | h+ste11T173A, S218A, T305D, T317D | This study |
| Eg1730 | h90 mei3::ura4+leu1-32 ura4-D18 ade6-M210 | M. McLeod, spB203 |
Standard DNA manipulations were performed according to Sambrook et al. (45).
Disruption of byr2, byr1, and spk1.
To obtain a byr2 null allele, a 281-bp XbaI-XbaI fragment was removed from the byr2 open reading frame (ORF), giving pUS78 (51). A 1,708-bp HindIII-EcoRI Δbyr2 fragment from pUS78 was ligated into pDW227 (59), resulting in pIL143. This plasmid contains a Δbyr2 allele adjacent to the ura4+ gene, allowing disruption of the byr2 locus by the loop-in-loop-out gene replacement technique (47). In brief, first pIL143 was linearized with XhoI and transformed into Eg640. Second, stable ura+ loop-in transformants were selected. Third, stable ura−, 5-fluoroorotic acid-resistant, and sterile colonies were identified. To delete the byr1 gene, a 1,989-bp EcoRI-XbaI fragment comprising the byr1 gene was cloned into SphI-BamHI-digested pDW227. From this plasmid, a 734-bp fragment comprising the byr1 ORF was removed by BamHI and BclI digestion and religation. The resulting plasmid, pIL144, contains the Δbyr1 allele next to the ura4+ gene. pIL144 was linearized with SnaBI before transformation into Eg640 and screening for sterile colonies as described above. To delete the spk1 gene, a 1,860-bp fragment harboring spk1 was amplified by PCR using the oligonucleotides 5′-GCGCGGATCCCTCAAGGCATCTTTG and 5′-GCCGCATGCACATGACCTACTACTG and ligated into SphI-BamHI-digested pGEM3. The resulting vector, pUS128, was digested with BamHI and BclI followed by religation, thereby deleting a 904-bp fragment from the spk1 ORF. From this plasmid, a 740-bp SphI-BamHI fragment was cloned into pDW227, producing pIL145. pIL145 was linearized by partial digestion with HindIII before transformation into Eg640 and screening for sterile colonies as described above. To confirm disruption of the byr2, byr1, and spk1 loci, total DNA was isolated from each of the sterile strains and subjected to PCR analysis using the following oligonucleotides: for byr2, 5′-CGGCCCCGGGAAACGAACGGGTGGCTCAAC and 5′-GCGCGTCGACTAAGGGAGGCAGGTT; for byr1, 5′-GCGCCCGGGGTTTAAACGACGTCGAA and 5′-GCGGCTCGAGAAAAAAGCGGAAATAT; and for spk1, 5′-GCGCGGATCCCTCAAGGCATCTTTG and 5′-GCCGCATGCACATGACCTACTACTG.
Construction of nmt-byr2-ΔN and nmt-byr2.
In order to express the catalytic domain of Byr2 (amino acids 340 to 659) from the medium-strength nmt promoter (3), the plasmid pUS72 containing a genomic clone of the byr2 gene (51) was digested with NcoI, end filled with Klenow, and digested by SacI. The DNA fragment was ligated into SmaI-SacI-digested pREP42X and pREP41X (13) to produce pIL169 and pIL171, respectively. To express full-length Byr2 from the medium-strength nmt promoter, the N-terminal part of the byr2 ORF was amplified by PCR using the oligonucleotides 5′-GCGGCATATGGAATATTATACCTCGA and 5′-TTGAACTGCTCGAGTTTGCC and digested with NdeI and XhoI. Next, the PCR fragment was ligated with an XhoI-BamHI fragment from pUS72 comprising the residual part of the byr2 ORF and inserted into NdeI-BamHI-digested pREP41X, generating pIL164.
HA tagging of Ste11.
The ORF of ste11 was amplified by PCR using the oligonucleotides 5′-CCCGCTCGAGATGTCTGCTTCTTTAACAGCC and 5′-CGCGGATCCGAAAATAAATTAGAATTGGG. Reaction products were digested with XhoI and BamHI and ligated into XhoI-BglII-digested pSFL172 (14), producing a C-terminal triple hemagglutinin (HA)-tagged version of Ste11 expressed from the nmt1 promoter (pSK66). pSK66 was digested with XhoI and SacI and ligated into XhoI-SacI-digested pREP41X, producing pSK85.
Generation of ste11 mutants.
Mutagenesis of Ste11 to alter Thr305 and Thr317 was performed in the following way. First, a ste11 vector was constructed by moving a 3,600-bp BamHI-EcoRI fragment containing a genomic clone of ste11 into BamHI-EcoRI-digested pDW227. This plasmid was digested with EcoRI and SacI, end filled with Klenow, and religated, producing pON559. Mutagenesis of Thr305 and Thr317 to alanine was accomplished by using the QuikChange kit (Stratagene), pON559 as template, and the oligonucleotides 5′-GCTCGACCCTCAGCTCCCTCTTGCCCC and 5′-GGGGCAAGAGGGAGCTGAGGGTCGAGC for T305A and 5′-GCCAGTATATCACCTAAAGCCCCGAATACCGGT and 5′-ACCGGTATTCGGGGCTTTAGGTGATATACTGGC for T317A, thereby creating pST220. Mutagenesis of Thr305 and Thr317 to aspartate was accomplished by using the oligonucleotides 5′-GCTCGACCCTCAGATCCCTCTTGCCCC and 5′-GGGGCAAGAGGGATCTGAGGGTCGAGC for T305D and 5′-CCAGTATATCACCTAAAGACCCGAATACCGGTG and 5′-CACCGGTATTCGGGTCTTTAGGTGATATACTGG for T317D, thereby producing pSK143. Ste11 harboring T173D, S218D, T305A, and T317A mutations was created by inserting a 445-bp PstI-MfeI fragment from pSTE11.54 (27) into PstI-MfeI-digested pST220. Ste11 harboring T173A, S218A, T305D, and T317D mutations was created by inserting a 445-bp PstI-MfeI fragment from pSTE11.55 (27) into PstI-MfeI-digested pSK143.
The different ste11 alleles were integrated into the ste11 loci. A two-step sequential gene replacement was designed to obtain strains congenic with the wild-type strain except for the amino acid changes introduced into Ste11. First, the endogenous ste11 allele was replaced by a DNA fragment containing ste11 interrupted by the ura4+ gene. Cells with ste11 disruptions were selected as sterile ura+ cells. The resulting strain, Eg1085, was used for subsequent gene replacements using 3,500-bp EcoRV-BamHI fragments comprising the different ste11 alleles described above. ste11T305A, T317A, ste11T305D, T317D, ste11T173D, S218D, T305A, T317A, and ste11T173A, S218A, T305D, T317D cells were selected as ura− cells on plates containing 5-fluoroorotic acid (5). Genomic PCR combined with sequencing analysis was employed to confirm site-specific integration.
Construction of GST-Spk1, GST-Spk1K68R, and Spk1-Myc.
In order to express a glutathione S-transferase (GST)-tagged version of Spk1 from its own promoter, spk1 was initially amplified by PCR. First, a 900-bp fragment comprising the promoter region was amplified by using the oligonucleotides 5′-CCGCCTGCAGAACATAAAATTAACAACC and 5′-GCGCCATATGACTACAAATTGAAAACT and cloned into PstI-NdeI-digested pREP2 (30), giving pIL194. Second, a fragment containing the spk1 ORF was amplified by using the oligonucleotides 5′-GCGCCATATGGCGAGCGCTACTTCC and 5′-CGCCGGATCCAGCATAGTTGCCTTTG and ligated into NdeI-BamHI-digested pIL194, thereby producing pIL195. An NdeI fragment harboring GST was obtained from pREP41-GST-cdc2H (26) and was ligated into NdeI-digested pIL195, giving pIL201. The GST-spk1 fusion gene complements the spk1 null allele, and Western analysis of the protein encoded by the fusion gene revealed that full-length protein was produced.
To make a kinase-dead version of Spk1, Lys68 was replaced by arginine. Mutagenesis of Lys68 to arginine was accomplished by use of the QuikChange kit (Stratagene), pUS128 as template, and the oligonucleotides 5′-CCTAAAGGTGGCTGTTAGAAAAATACATCCTTTC and 5′-GAAAGGATGTATTTTTCTAACAGCCACCTTTAGG, thereby creating pSK162. An Eco47III-BglII fragment comprising the K68R substitution from pSK162 was ligated into Eco47III-BglII-digested pIL201, producing pSK161.
Spk1 was fused to the c-myc epitope tag as described below. The ORF of spk1 was amplified by PCR using pUS128 as template and the oligonucleotides 5′-GGGGTCGACTAATGGCGAGCGCTACT and 5′-GGCCTCGCGATATCTGAAATTTACTTCACG. The resulting PCR product was digested with SalI and NruI and ligated into SalI-NruI-digested pMNS36LEU (a kind gift from Sue Dorrington), producing a C-terminal myc-tagged version of Spk1 expressed from the nmt1 promoter (pIL180).
RNA preparation and Northern analysis.
Total RNA was extracted by using hot acidic phenol as described by Lyne et al. (28). Northern analysis was performed as described previously (39) using probes specific for mat1-Pm, mfm1, mei2, and ste11. Single-stranded RNA probes were transcribed from the pGEM3 vector (Promega) containing a probe-specific insert, either a 336-bp PCR fragment of the mei2 ORF or a 627-bp PstI-BclI fragment of the ste11 gene. Probes specific for mat1-Pm and mfm1 have been described previously (23, 39).
Recombinant Ste11 proteins.
To produce full-length Ste11 protein in Escherichia coli, GST and a His6 tag, respectively, were fused to the N and C termini of Ste11. To tag Ste11 C terminally with six histidines, ste11 was amplified by PCR using the oligonucleotides 5′-CATTGGTCAGCTGTGGAG and 5′-AATGAATTCCTAGATGATGATGATGATGATGTAAAAATAAATTAGAATTGGG. The resulting PCR fragment was digested with AvrII and EcoRI and ligated into AvrII-EcoRI-digested pGEX-Ste111-468 (24), producing pSK139. To create a vector expressing GST-Ste11T305A, T317A-His6, a 778-bp NdeI-AvrII fragment from pST220 was cloned into NdeI-AvrII-digested pSK139. The two plasmids were transformed into the DH5 E. coli strain. A two-step sequential purification protocol was designed to enrich for full-length Ste11 protein. First, Ste11 was affinity purified on glutathione-Sepharose (Amersham Biosciences). After purification on glutathione-Sepharose, the eluted recombinant Ste11 protein was further affinity purified by Ni-nitrilotriacetic acid (NTA)-agarose (QIAGEN) chromatography. This second purification step was utilized for the removal of C-terminally degraded Ste11 protein. Expression and purification on glutathione-Sepharose were described previously (24). Ni-NTA-agarose chromatography was performed according to the manufacturer's instructions. Purification and the size of recombinant proteins were tested by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie staining and Western analysis. Production of recombinant GST-Ste11 and GST-Ste11HMG fusion proteins have been described elsewhere (24).
Generation of anti-Ste11 monoclonal antibodies.
Production of monoclonal antibodies was performed according to Castrop et al. (7). In short, 6-week-old BALB/c mice were immunized by subcutaneous injection of 50 μg of GST-Ste11 fusion protein (24) in complete Freund's adjuvant on day 1, followed by a similar injection in incomplete Freund's adjuvant at day 14, followed by repeated intraperitoneal injections at weekly intervals of 50 μg of fusion protein in phosphate-buffered saline. When a high titer was obtained, as detected by enzyme-linked immunosorbent assay using GST-Ste11 as antigen, the spleen was isolated, and 108 splenocytes were fused to an equal number of Ag8 myeloma cells by using the standard polyethylene glycol protocol. Selection in hypoxanthine-aminopterin-thymidine was initiated directly after plating the cell suspension into 15 96-well plates. Supernatants were screened 10 to 14 days after the hybridoma fusion, and positive clones were repeatedly subcloned. Anti-Ste11 positive hybridomas were identified by enzyme-linked immunosorbent assay, according to Harlow and Lane (15), using GST-Ste11 and GST as antigens. Stable positive clones were grown in RPMI 1640 with 10% fetal calf serum or in serum-free and protein-free hybridoma medium (Hybri-Max; Sigma). Seven different clones were identified as producing high-affinity anti-Ste11 antibodies. In this study, clone 18 was used as the anti-Ste11 antibody source. This antibody binds to an epitope mapping between residues 96 and 113.
Protein extraction and Western analysis.
Protein extracts were made by trichloroacetic acid precipitation as described previously (12). Fifty micrograms of protein was loaded per lane, separated by using SDS-10% PAGE, and transferred to mixed cellulose ester membranes (Advantec). Ste11 was detected by using the monoclonal anti-Ste11 antibody, and α-tubulin was detected by using a monoclonal anti-Tat1 antibody (62). The secondary antibody was horseradish peroxidase-conjugated rabbit anti-mouse antibody (DakoCytomation) detected with enhanced chemiluminescence (Amersham Biosciences).
Protein extraction and kinase assays.
For kinase assays, extracts from 108 cells were made by using buffer L (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 50 mM NaF, 1 mM Na3VO4, 100 μg of Pefabloc SC/ml, 10 μg of aprotinin/ml, 2 μg of pepstatin/ml, 2 μg of leupeptin/ml) (50). Cells were disrupted by the use of glass beads and a FastPrep Instrument (Bio101) at settings of 6.5 for 3 s. After cell breakage, 100 μl of buffer L was added, and the soluble protein fraction was recovered by two centrifugations for 10 min at 20,000 × g. GST-Spk1 and GST-Spk1K68R were purified from the extracts by using glutathione-Sepharose chromatography as described previously (50). Purified GST-Spk1 or GST-Spk1K68R, attached to glutathione-Sepharose, was resuspended in 25 μl of buffer K (25 mM Tris-HCl [pH 7.2], 10 mM MgCl2, 0.1 mM EGTA, 1 mM dithiothreitol, 0.1 mM Na3VO4) containing 50 μM ATP, 2.5 μCi of [γ-32P]ATP, and 0.5 μg of affinity-purified GST-Ste11-His6 or GST-Ste11T305A, T317A-His6 and incubated at 25°C for 15 min. The reactions were stopped with 25 μl of 2× SDS sample buffer and denatured at 100°C for 5 min, and samples were run on an SDS-8% polyacrylamide gel. Phosphorylated proteins were detected and quantitated by use of a Storm (Molecular Dynamics).
GST pull-down assay and immunoprecipitations.
Five micrograms of purified recombinant GST, GST-Ste11, and GST-Ste11HMG (amino acids 1 to 113) (24) and 1 mg of protein extracts prepared from Eg934 transformed with pIL180 (pREP3X-Spk1-myc) or prepared from Eg1062 were incubated in buffer L at 4°C for 1 h with glutathione (GSH) beads (Amersham Biosciences). Beads were washed five times with 1 ml of lysis buffer, and GSH bead-bound proteins were finally analyzed by SDS-PAGE, followed by Coomassie staining or immunoblotting against rabbit anti-Myc antibodies (Research Diagnostics Inc.). Spk1-Myc was immunoprecipitated from 1 mg of protein extracts with 0.5 μg of rabbit anti-Myc antibodies by incubating at 4°C for 30 min. The formed complexes were collected with protein G-Sepharose (Amersham Biosciences) by incubating at 4°C for 30 min. The pellet was washed three times with buffer L and finally analyzed by SDS-PAGE followed by immunoblotting against rabbit anti-Myc antibodies and anti-HA (12CA5).
Mating and sporulation assay.
To monitor haploid sporulation, transformants were grown in MSL with 6 μM thiamine to a density of 5 × 106 cells/ml. Five-microliter aliquots of these exponentially grown cultures were spotted onto minimal sporulation agar (MSA) plates and MSA plates with thiamine and incubated for 48 h at 30°C before the number of asci was recorded. Haploid sporulation frequencies were calculated according to the equation: haploid sporulation frequency = number of haploid asci/total number of haploid entities. To determine mating and sporulation efficiency, homothallic strains were grown in MSL to a density of 5 × 106 cells/ml, then shifted to MSL without nitrogen source, and incubated at 30°C. Samples were withdrawn every hour, and the number of asci and zygotes was recorded. The efficiency of mating was calculated as the following ratio: (2 × number of asci and zygotes formed)/(total number of cells + 2 × number of asci and zygotes).
Flow cytometry and microscopy.
About 3 × 106 cells were harvested, fixed with 70% ice-cold ethanol, and processed for 4′,6′-diamidino-2-phenylindole (DAPI) staining and flow cytometry, as described previously (33, 46) with the following exception: prior to RNase treatment, the cells were incubated with 1 mg of pepsin/ml in 0.1 M HCl for 1 h at room temperature (E. Boye, personal communication). A Becton-Dickinson FACScan was used for flow cytometry. An Axiplan 2 (Carl Zeiss) was used for microscopy, and images were captured by use of a cooled CCD camera (MicroMax; Roper Scientific) and MetaMorph software (Universal Imaging).
Two-hybrid analysis.
The yeast two-hybrid assay was essentially performed as described previously (19). An NcoI-BamHI fragment from pSTE11.9 (27) carrying the ste11 ORF was fused in frame to the GAL4 activation domain in pGADT7 (ClonTech). To fuse spk1 to the GAL4 DNA binding domain, the ORF was amplified by PCR using the oligonucleotides 5′-GTCATATGGCGAGCGCTACTTC and 5′-ACGTCGACTTTGATGAGGAGAA and ligated into NdeI-SalI-digested pGBKT7 (ClonTech). To fuse Sty1 to the GAL4 DNA binding domain, the ORF was amplified by PCR using the oligonucleotides 5′-GTCATATGGCAGAATTTATTCG and 5′-GCGTCGACTTTTAAGGCTTTAT and inserted into NdeI-SalI-digested pGBKT7. Saccharomyces cerevisiae strain PJ69-4a (19) was transformed with various combinations of plasmids and assayed for β-galactosidase activity as described previously (44).
RESULTS
Unregulated Byr2 activity induces ectopic meiosis.
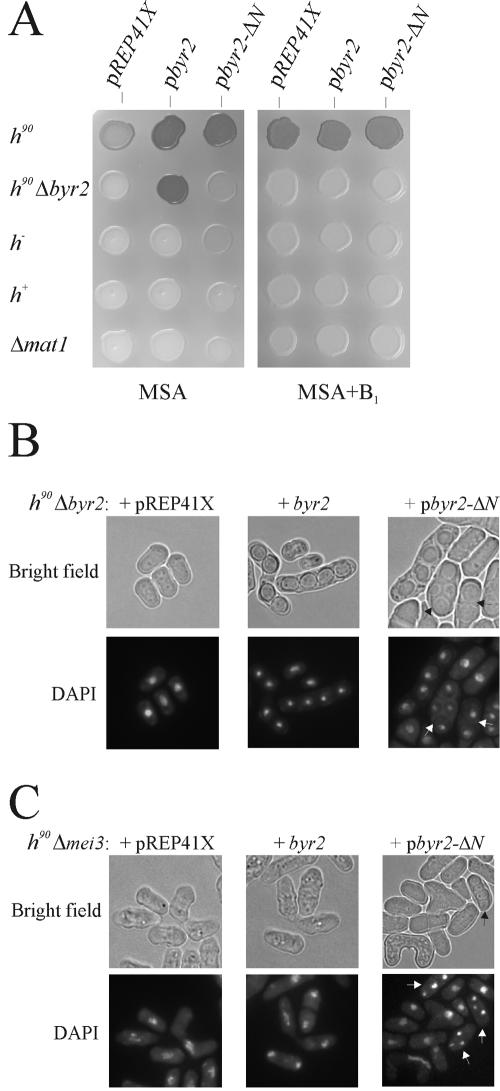
In order to activate the Byr2-Byr1-Spk1 pathway in the absence of pheromone signaling, we expressed a constitutively active version of Byr2. This was achieved by constructing a truncated version of Byr2 lacking the N-terminal regulatory domain, designated byr2-ΔN. To examine whether byr2-ΔN could complement a byr2 deletion, we expressed the mutant protein from a repressible promoter in homothallic h90 Δbyr2 haploid cells (see Materials and Methods). Under conditions when the promoter was induced, the expression of byr2-ΔN enabled the homothallic strain to go through meiosis, as revealed by iodine staining of plated cells (Fig. 1A). Unexpectedly, however, microscopic examination of the sporulating cells showed the presence of mainly azygotic asci (Fig. 1B). This result indicates that haploid cells had entered meiosis without preceding conjugation, suggesting that the requirement for zygotic expression of the mating type genes had been bypassed. In contrast, cells expressing the wild-type byr2 gene exclusively gave rise to zygotic asci, demonstrating that they, like wild-type cells, had undergone conjugation prior to entering meiosis (Fig. 1B). These findings indicate that the expression of uncontrolled Byr2 activity prevented conjugation and rather caused cells to enter meiosis directly. Presumably, this result reflects that dynamic changes in Byr2 activity are important for conjugation.
FIG. 1.
Expression of the catalytic domain of Byr2 results in ectopic meiosis. (A) Iodine staining of cells plated on MSA sporulation medium with or without 6 μM thiamine (B1). Eg640, Eg931, Eg768, Eg589, and Eg709 were transformed with the vectors pREP41X, pbyr2, expressing wild-type Byr2 from the medium-strength thiamine-repressible nmt1 promoter, and pbyr2-ΔN, expressing the catalytic domain of Byr2 from the medium-strength nmt1 promoter. The fact that cells transformed with pbyr2 or pbyr2-ΔN stain stronger than cells transformed with an empty vector reflects that Byr2 expression enhances sexual differentiation on MSA medium. (B) Microscopic examination of Eg931 transformed with vector pREP41X (left panel), pbyr2 (middle panel), and pbyr2-ΔN (right panel). Cells were plated on MSA medium at 30°C and fixed after 48 h of incubation. Nuclei were stained with DAPI. Bright-field images of the same cells are shown above. Arrowsindicate azygotic asci with two to four spores. (C) Deletion of the mei3 gene does not suppress Byr2-ΔN-induced ectopic meiosis. Shown is a microscopic examination of h90 Δmei3 cells (Eg1730) transformed with the vectors pREP41X (left panel), pbyr2 (middle panel), and pbyr2-ΔN (right panel). Cells were plated on MSA medium at 30°C and fixed after 48 h of incubation. Nuclei were stained with DAPI. Bright-field images of the same cells are shown above. Arrows indicate asci with spores.
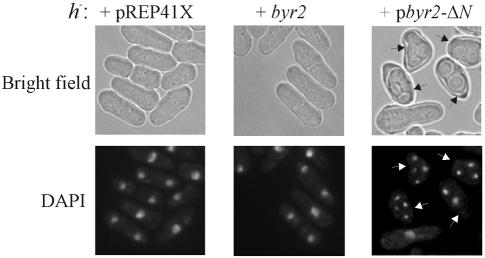
Derepression of the silent mating type loci mat2-P and mat3-M, which normally are tightly repressed, causes the induction of haploid meiosis (54). Hence, the haploid sporulation induced by byr2-ΔN could be due to derepression of the silent mating type loci in the h90 Δbyr2 strain. To examine this possibility, we expressed byr2-ΔN in h− and h+ strains with deletions of the mat2-P and mat3-M cassettes. In both cell types, the presence of byr2-ΔN caused a distorted morphology, and the cells were forced into meiosis (Fig. 1A and 2 and Table 2). This result clearly demonstrates that the requirement for zygotic expression of mating type genes for induction of meiosis was bypassed. Intriguingly, the level of haploid meiosis was approximately twofold higher in h− cells than in h+ cells. Either the M information promotes haploid meiosis or the P information impairs it. To distinguish between these two possibilities, byr2-ΔN was expressed in cells with mat1 deletions. The level of haploid meiosis in Δmat1 cells was equal to that found for h+ cells (Fig. 1A and Table 2). This observation implies that the M mating type somehow augments the haploid meiosis caused by unregulated Byr2 activity.
FIG. 2.
Expression of the catalytic domain of Byr2 induces haploid meiosis in the presence of nitrogen. h− cells (Eg768) transformed with vector pREP41X (left panel), pbyr2 (middle panel), and pbyr2-ΔN (right panel) were induced by the removal of thiamine. During growth, the cell concentration was kept below 107 cells/ml by diluting with fresh medium. After 20 h of induction, the cells were fixed and nuclei were stained with DAPI. Bright-field images of the same cells are shown above. Arrows indicate cells with two to four spores.
TABLE 2.
Estimation of haploid meiosis in different strains
| Strain genotype | % Sporulationa
|
|
|---|---|---|
| pbyr2 | pbyr2-ΔN | |
| h90 Δbyr2 | 83c | 17 |
| h− | 0.0 | 12.2 |
| h+ | 0.0 | 5.8b |
| Δmat1 | 0.0 | 6.1b |
| h− Δste11 | 0.0 | 0.0b |
| h−ste11T173D, S218D | 0.0 | 1.2b |
| h−ste11T305A, T317A | 0.0 | 1.8b |
| h−ste11T173D, S218D, T305, T317A | 0.0 | 1.3b |
Cultures of Eg931, Eg768, Eg589, Eg709, Eg1085, Eg1226, Eg1259, and Eg1638 expressing either wild-type Byr2 (pbyr2) or the catalytic domain of Byr2 (pbyr2-ΔN) were grown for 24 h in MSL with thiamine and then spotted on MSA plates. Cells were incubated for 48 h at 30°C, and the percentage of sporulation was estimated by microscopic examination as described in Materials and Methods. Values represent means of results from three separate trials.
Sporulation is significantly reduced compared with that of wild-type h− cells (P < 0.05; tested by Student's t test).
This value represents normal zygotic sporulation.
Induction of haploid meiosis by byr2-ΔN requires the downstream Byr1 and Spk1 kinases.
We next determined the epistatic relationship of byr2-ΔN overexpression to a number of different mutants (Table 3). Six different homothallic strains with mutations in different components of the pheromone signaling pathway were transformed with vectors expressing either wild-type byr2 or byr2-ΔN. Cells lacking the pheromone receptors, mam2 and map3; the G protein α subunit, gpa1; and the Ras protein, ras1, all acting upstream of Byr2, entered haploid meiosis upon byr2-ΔN expression. In contrast, mutations in the downstream Byr1 and Spk1 kinases suppressed haploid meiosis. This observation demonstrates that the signal goes through Byr1 and Spk1 and suggests that the substrate specificity of Byr2-ΔN protein kinase has not been altered by removal of the regulatory domain. As a control, expression of the wild-type Byr2 protein did not induce haploid meiosis in any of the examined mutants (Table 3).
TABLE 3.
Epistatic analysis of haploid meiosis induced by unregulated Byr2
| Strain genotype | % Sporulationa
|
|
|---|---|---|
| pbyr2 | pbyr2-ΔN | |
| h90 Δmam2 Δmap3 | 0.0 | 9.7 |
| h90 Δgpa1 | 0.0 | 10.3 |
| h90 Δras1 | 0.0 | 8.1 |
| h90 Δbyr2 | 77b | 20 |
| h90 Δbyr1 | 0.0 | 0.0 |
| h90 Δspk1 | 0.0 | 0.0 |
| h90 Δmei2 | 0.0 | 0.0 |
| h90 Δste11 | 0.0 | 0.0 |
| h90 Δmei3 | 0.0 | 8.5 |
Cultures of Eg578, Eg710, Eg627, Eg931, Eg930, Eg934, Eg576, Eg1085, and Eg1730 expressing either wild-type Byr2 (pbyr2) or the catalytic domain of Byr2 (pbyr2-ΔN) were grown for 24 h in MSL with thiamine and then spotted on MSA plates. Cells were incubated for 48 h at 30°C, and the percentage of sporulation was estimated by microscopic examination as described in Materials and Methods. Values represent means of results from three separate trials.
This value represents normal zygotic sporulation.
ste11 and mei2, but not mei3, are essential for induction of haploid meiosis.
The fact that mat1-Pm and mat1-Mm are dispensable for Byr2-ΔN-induced haploid sporulation suggests that mei3 is not required. Indeed, cells lacking the Mei3 protein still entered meiosis and completed sporulation upon byr2-ΔN expression (Fig. 1C). This result demonstrates that haploid meiosis is activated by a Mei3-independent mechanism and hence is not a result of activation of unscheduled mei3 expression.
The mei2 gene is indispensable for meiosis (65), and its expression is dependent on the Ste11 transcription factor. We next examined whether these two critical gene functions were required for byr2-ΔN-induced haploid meiosis. ste11 and mei2 cells expressing byr2-ΔN did not enter meiosis (Table 3). This finding suggests that the expression of the catalytic domain of Byr2 activates Mei2 in a Ste11-dependent manner.
The pheromone-responsive genes mfm1 and mat1-Pm are constitutively induced by byr2-ΔN expression.
We noticed that even when cultured in rich liquid medium, cells became deformed and entered meiosis upon expression of byr2-ΔN (Fig. 2). Under the same conditions, control cells expressing wild-type Byr2 continued growth, and no asci were observed. This result indicates that unregulated Byr2 activates the downstream Byr1-Spk1 kinases even in the presence of nitrogen.
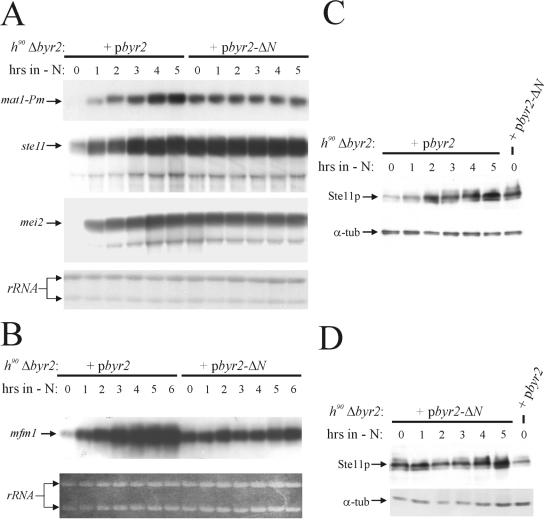
Transmission of the pheromone signal through the Byr2-Byr1-Spk1 kinase cascade induces the expression of a number of genes, among them mat1-Pm (1, 38). To test whether the signal generated by unregulated Byr2 activity mimics the pheromone signal, we monitored the mRNA levels of mat1-Pm in h90 Δbyr2 cells expressing byr2-ΔN and wild-type byr2. To induce expression of byr2-ΔN and wild-type byr2, thiamine was removed from the medium; 14 h after the induction, the cultures were shifted to medium without nitrogen source, and the level of the mat1-Pm transcript was determined by Northern analysis (Fig. 3). In agreement with previous observations, cells expressing wild-type byr2 displayed a gradual increase in the level of mat1-Pm mRNA during nitrogen starvation, and no transcript was present in nitrogen-rich medium (Fig. 3A). In contrast, in cells expressing the Byr2 catalytic domain, the level of mat1-Pm mRNA was constant during starvation, and mat1-Pm was highly transcribed even in the presence of nitrogen. Hence, byr2-ΔN mimics the pheromone signal and activates transcription of the pheromone-responsive mat1-Pm gene. However, the requirement for nitrogen starvation apparently is also bypassed. We performed a similar experiment monitoring the expression of mfm1. While the expression of mat1-Pm is absolutely dependent on pheromone signaling, the mfm1 gene is induced by nitrogen starvation and then further enhanced by pheromone signaling (23). Similar to mat1-Pm, we found that the mfm1 gene was constitutively transcribed in cells containing byr2-ΔN and that there was no requirement for nitrogen starvation (Fig. 3B). Curiously, the level of mfm1 transcript in cells expressing byr2-ΔN was somewhat lower than the level found in nitrogen-starved cells expressing wild-type byr2.
FIG. 3.
Ste11 is constitutively expressed in cells producing the catalytic domain of Byr2 and induces the transcription of pheromone-responsive genes and mei2 in the presence of nitrogen. h90 Δbyr2 cells (Eg931) were transformed with either pbyr2 (expressing wild-type Byr2) or pbyr2-ΔN (expressing the catalytic domain of Byr2). Transformants were cultured to 2 × 105 cells/ml in MSL with thiamine. After removal of the thiamine, the cells were incubated in MSL for 14 h before being shifted to MSL without a nitrogen source. Samples were withdrawn from the cultures every hour after the start of nitrogen starvation and subjected to Northern (A and B) and Western (C and D) analysis. (A) Northern analysis of mat1-Pm, ste11, and mei2 transcripts. The lower panel shows the RNA samples hybridized to 18S and 26S rRNAs. (B) Northern analysis of mfm1 transcript. The lower panel shows ethidium bromide staining of the loaded RNA samples. (C) Western analysis of Ste11 and α-tubulin proteins in cells expressing wild-type Byr2. Proteins were detected by mouse monoclonal antibodies against Ste11 and Tat1. (D) Western analysis of Ste11 and α-tubulin proteins in cells expressing the catalytic domain of Byr2. Proteins were detected by mouse monoclonal antibodies against Ste11 and Tat1.
Ste11 is constitutively expressed in cells with unregulated Byr2 activity.
To examine whether the constitutive expression of the mat1-Pm and mfm1 transcripts in cells harboring byr2-ΔN was caused by ectopic expression of ste11, we analyzed the samples described above for ste11 mRNA and Ste11 protein (Fig. 3A, C, and D). In the presence of nitrogen, cells expressing wild-type Byr2 expressed only small amounts of ste11 mRNA and Ste11 protein, and during nitrogen starvation the cells displayed a gradual increase in the level of the mRNA and protein. On the other hand, in cells expressing the Byr2 catalytic domain, the levels of ste11 mRNA and Ste11 protein were constant during starvation, and ste11 was highly expressed even in the presence of nitrogen. Thus, ste11 is ectopically expressed in cells with unregulated Byr2 kinase activity, enabling the cells to activate the Byr2-Byr1-Spk1 kinase cascade in the presence of nitrogen.
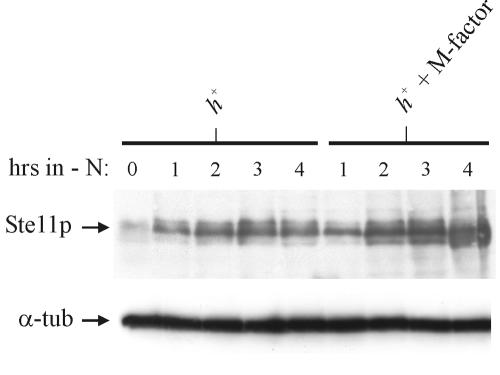
We next examined whether pheromone signaling augments the expression of the Ste11 protein in wild-type cells. As shown in Fig. 4 the level of Ste11 protein is higher in h+ cells starved for nitrogen and treated with M-factor pheromone than in untreated nitrogen-starved h+ cells. Thus, also in wild-type cells does pheromone signaling contribute to the expression the Ste11 protein. Since pheromone signaling allows accumulation of Ste11 in the nucleus (43), the increase in the Ste11 level is likely caused by autostimulation of the ste11 gene (25).
FIG. 4.
The Ste11 protein is regulated by the pheromone pathway in wild-type cells. Shown is a Western analysis of the expression of Ste11 in h+ with and without M-factor treatment. Cells were grown to a density of 5 × 106 cells/ml in MSL and then shifted to MSL-N or MSL-N with 1 μg of M-factor/ml. Samples were withdrawn from the cultures every hour after the start of nitrogen starvation and subjected to Western analysis using mouse monoclonal antibodies against Ste11 (upper panel) and Tat1 (lower panel).
One of the direct targets of the Ste11 transcription factor is the meiotic inducer mei2. Massive expression of mei2 causes ectopic meiosis (58). Therefore, we next examined whether the haploid meiosis observed in cells expressing byr2-ΔN could be due to overexpression of mei2 as a result of unregulated Ste11 activity. Although the mei2 gene is constitutively expressed in byr2-ΔN cells, the overall level of mei2 transcript is not higher than that found for nitrogen-starved wild-type cells (Fig. 3A). Thus, the haploid meiosis induced by the catalytic domain of Byr2 is not due to overexpression of mei2, although this experiment cannot exclude the possibility that the level of Mei2 protein is abnormally high.
Ste11 is a substrate for the Spk1 kinase.
Although the genetic experiments described above demonstrate that the Byr2-Byr1-Spk1 kinase cascade regulates the Ste11 transcription factor, they do not distinguish direct regulations from indirect ones. The activity of transcription factors is often controlled by phosphorylation, and the Pat1 kinase directly regulates the activity of Ste11 by phosphorylation of Thr173 and Ser218 (27, 43). Western analysis using large 22- by 20-cm polyacrylamide gels revealed that Ste11 migrates as at least six closely spaced bands, which upon calf intestinal phosphatase treatment can be shifted to a single fast migrating band of approximately 50 kDa (unpublished observations). Thus, Ste11 is probably phosphorylated at multiple sites and may be a target for kinases other than Pat1. Interestingly, two PXS/TP motifs for MAP kinase phosphorylation are found in the Ste11 sequence at positions Thr305 and Thr317. Therefore, we next addressed whether the link between the Byr2-Byr1-Spk1 pathway and Ste11 was direct.
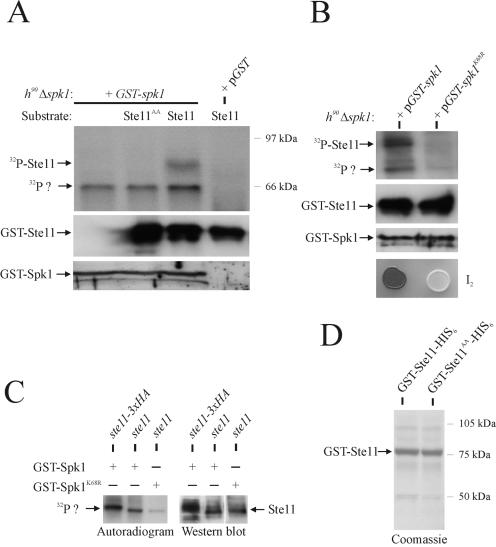
First, we used an in vitro kinase assay to determine whether the Spk1 MAP kinase was capable of phosphorylating Ste11. Nitrogen-starved cells containing a GST-Spk1 fusion were subjected to GSH-Sepharose chromatography. Kinase assays were performed with the affinity-purified GST-Spk1 fusion protein by using E. coli-expressed GST-Ste11-His6 as substrate (Fig. 5A and D). GST-Ste11-His6 became phosphorylated in this assay, and importantly, this phosphorylation was absolutely dependent on the presence of Spk1 (Fig. 5A, lanes 3 and 4). To determine whether the two putative MAP kinase sites were important for Ste11 to function as an Spk1 substrate, a mutant Ste11 allele was created. Site-specific mutagenesis was used to convert Thr305 and Thr317 to nonphosphorylatable alanine residues. The protein encoded by the mutant allele, Ste11T305A, T317A, was produced in E. coli as a GST and His6 fusion protein and used in the kinase assay described above (Fig. 5A). Purified GST-Ste11T305A, T317A-His6 fusion protein was not phosphorylated in this assay, demonstrating that Thr305 and Thr317 are crucial for phosphorylation of Ste11 by Spk1 in vitro.
FIG. 5.
Ste11 is a substrate for the Spk1 MAP kinase. (A) In vitro kinase assay. h90 Δspk1 cells (Eg934) expressing GST fused to Spk1 (pGST-spk1) and unfused GST (pGST) were nitrogen starved for 3 h. Protein extracts were prepared and subjected to GSH-Sepharose chromatography. The purified proteins were tested for kinase activity by using GST-Ste11-His6 and GST-Ste11T305A, T317A-His6 as substrates. Kinase assays were carried out for 15 min at 30°C, and the samples were separated by SDS-PAGE, followed by autoradiography (upper panel) or immunoblotting against anti-Ste11 antibodies (middle panel). The lower panel shows a Western analysis of the purified GST-Spk1 protein added to the kinase reactions; a polyclonal antibody against GST was used. The upper arrow (32P-Ste11) represents the position of recombinant GST-Ste11-His6. The lower arrow (32P?) indicates unidentified phosphorylated protein. (B) In vitro kinase assay. h90 Δspk1 cells (Eg934) expressing GST fused to wild-type Spk1 (pGST-spk1) or kinase-dead Spk1 (pGST-spk1K68R) were processed as described above, and purified proteins were tested for kinase activity by using GST-Ste11-His6 as substrate. Samples were separated by SDS-PAGE, followed by autoradiography (upper panel) or immunoblotting against anti-Ste11 antibodies (second panel from top). The third panel from the top shows a Western analysis of the purified GST-Spk1 proteins added to the kinase reactions; a polyclonal antibody against GST was used. The lower panel shows iodine staining (I2) of the two strains grown on MSA plates for 48 h at 30°C. (C) In vitro kinase assay. h90 Δspk1 cells (Eg934) expressing GST fused to wild-type Spk1 (GST-Spk1) or kinase-dead Spk1 (GST-Spk1K68R) were transformed with vectors harboring ste11 or ste11 tagged C-terminally with a triple HA epitope (ste11-3xHA). Cells were processed as described above, and purified proteins were tested for kinase activity without adding recombinant Ste11. Samples were separated by SDS-PAGE, followed by autoradiography (left panel). Extracts from the same cells were analyzed for the expression of Ste11 by immunoblotting against Ste11 (right panel). (D) SDS-PAGE and Coomassie brilliant blue staining of GST-Ste11-His6 and GST-Ste11T305A, T317A-His6 affinity purified by GSH-Sepharose and Ni2+-NTA-agarose chromatography. Arrow indicates full-length recombinant Ste11 protein.
These observations indicate that Spk1 or another kinase whose activity is dependent on Spk1 copurifies with and phosphorylates Ste11. To discriminate between these two possibilities, we next made a kinase-dead version of GST-Spk1 and analyzed whether a Ste11-phosphorylating kinase copurifies with Spk1. A kinase-dead version of Spk1 was made by substituting a conserved lysine at position 68, in the ATP-binding domain, with an arginine. Unlike wild-type GST-Spk1, GST-Spk1K68R does not complement the mating defect of h90 Δspk1 cells, although the mutant protein is stably expressed (Fig. 5B). Nitrogen-starved cells containing the GST-Spk1K68R fusion were subjected to GSH-Sepharose chromatography and analyzed for kinase activity with GST-Ste11-His6 as substrate. Importantly, no phosphorylation of the recombinant Ste11 could be observed with these cells, indicating that the observed Ste11 phosphorylation is due to Spk1 and not a kinase that copurifies with GST-Spk1.
Curiously, a 60-kDa protein copurifies with S. pombe-expressed GST-Spk1 and becomes phosphorylated in the kinase assay (Fig. 5A, lanes 1 to 3). This phosphorylated protein could represent endogenous Ste11 (60 kDa) or, alternatively, autophosphorylated GST-Spk1, which has a predicted molecular weight of 67.7 kDa. To distinguish between these two possibilities, we coexpressed GST-Spk1 with, respectively, untagged Ste11 and Ste11 tagged with a triple HA epitope in nitrogen-starved cells. The cells were subjected to GSH-Sepharose chromatography and analyzed for kinase activity without the addition of recombinant Ste11 protein (Fig. 5C). Interestingly, the unidentified phosphorylated protein that copurifies with GST-Spk1 migrates more slowly in cells expressing Ste11-triple HA than in cells expressing untagged Ste11, implying that this protein represents endogenous Ste11.
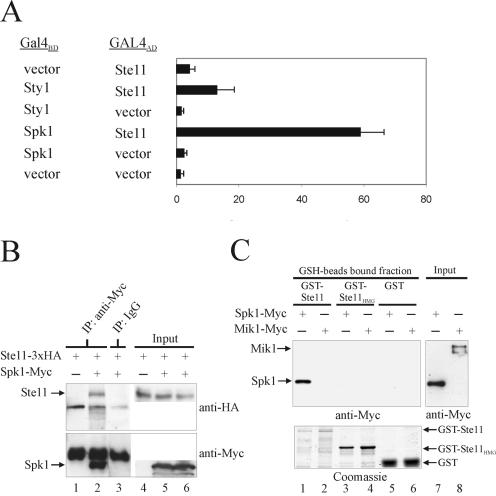
These observations strongly suggest that Ste11 is a target for Spk1 phosphorylation. To examine whether Ste11 and Spk1 physically associate, we next made use of the yeast two-hybrid assay. Ste11 was fused to the GAL4 activation domain, and the S. cerevisiae strain PJ69-4a (19) was transformed with a vector carrying this fusion protein together with either a vector expressing Spk1 fused to the GAL4 DNA binding domain (GAL4BD) or a vector expressing a GAL4BD-Sty1 fusion. We included sty1 in the analysis, since it encodes another S. pombe MAP kinase involved in the sexual differentiation process (20, 49, 60). Sty1 is a part of the Wis1-Sty1 pathway, the activity of which is important for transcription of the ste11 gene, and the Ste11 transcription factor could be a target of Sty1. In the two-hybrid assay, the combination of Ste11 and Spk1 sustained growth on selective medium without adenine and histidine, demonstrating that transcription of the GAL2-ADE2 and GAL1-HIS3 fusion genes was activated (data not shown). Ste11 combined with Sty1 did not allow growth on selective medium. To confirm these observations, the ability to activate transcription of the GAL7-lacZ fusion gene was monitored by β-galactosidase assays. The combination of Ste11 and Spk1 clearly displayed higher activity than Ste11 combined with Sty1 and various controls (Fig. 6A).
FIG. 6.
Spk1 and Ste11 physically interact in vivo and in vitro. (A) Yeast two-hybrid analysis of the interaction between Ste11 and the MAP kinases Spk1 and Sty1. The GAL4 DNA binding domain (GAL4BD) fused to either Spk1 or Sty1 was coexpressed with the GAL4 activation domain (GAL4AD) in the reporter S. cerevisiae strain PJ69-4a. PJ69-4a permits detection of interaction through transcriptional activation of the GAL1-HIS3, GAL2-ADE2, and GAL7-lacZ fusion genes. Transformants were grown to a density of 107 cells/ml and assayed for by β-galactosidase activity. Activity is expressed in Miller units and represents the mean value of results from three separate trials. Each error bar represents a standard deviation. (B) h90 Δspk1 cells (Eg934) expressing triple HA-tagged Ste11 (Ste11-3xHA) from the nmt1 promoter were cotransformed with pREB4X (lanes 1 and 4) or pREB4X-spk1-myc (lanes 2, 3, 5, and 6) and starved for nitrogen for 3 h. Cell extracts containing equal amounts of protein were immunoprecipitated (IP) with rabbit anti-myc antibodies (lanes 1 and 2) or control rabbit immunoglobulin G (lane 3). Immunoprecipitates (lanes 1 to 3) and total extracts (Input; lanes 4 to 6) were run on SDS-PAGE and immunoblotted against HA and myc epitopes. Arrows indicate the positions of Ste11 and Spk1. Other bands on the gel represent the heavy chain of immunoglobulin G. (C) GST pull-down assay. Approximately 5 μg of purified GST, GST-Ste11HMG (HMG box of Ste11), or GST-Ste11 (full-length Ste11) was mixed with GSH beads and cell extracts prepared from h90 Δspk1 cells (Eg934) expressing pREP3X-spk1-myc or h− mik1-6myc-His (Eg1062) cells, the latter serving as a negative control. The amount of total myc-tagged protein (Input; lanes 7 and 8) and bead-bound myc-tagged protein (lanes 1 to 6) was analyzed by immunoblotting against anti-myc antibody. Coomassie staining of the purified GST, GST-Ste11HMG, and GST-Ste11 proteins used in the assay is shown in the lower panel (lanes 1 to 6).
Next, the interaction of endogenous Spk1 and Ste11 was confirmed by immunoprecipitation. As shown in Fig. 6B, Ste11 coimmunoprecipitates with Spk1. To confirm the binding specificity between Ste11 and Spk1 and to examine the interaction further, we tested the ability of myc-tagged Spk1 obtained from cell extracts to bind full-length Ste11 and the HMG box of Ste11 fused to GST. Whereas GST-Ste11 efficiently interacted with Spk1-myc, no interaction with GST-Ste11HMG or GST was observed (Fig. 6C). As a control, myc-tagged Mik1 failed to bind the three recombinant GST proteins. These data, together with the previous results, strongly support that Ste11 is a direct downstream target of the Spk1 kinase.
Mutations of the Spk1 sites in Ste11 impair haploid meiosis but not normal meiosis.
Taken together, the above experiments demonstrate that threonines 305 and 317 are critical for the phosphorylation of Ste11 by Spk1 in vitro. If Ste11 is a direct target of the Byr2-Byr1-Spk1 pathway, one would expect that mutations in Thr305 and Thr317 affect conjugation and sporulation. To investigate this possibility, we constructed a homothallic h90 strain in which the endogenous ste11 gene was replaced with the ste11T305A, T317A allele (for details, see Materials and Methods). This ste11 allele carrying nonphosphorylatable alanines instead of threonines at residues 305 and 317 is predicted to represent an attenuated form of the transcription factor. We monitored the ability of the h90 ste11T305A, T317A cells to conjugate and sporulate and found that they entered meiosis with same frequency as wild-type h90 cells (Table 4).
TABLE 4.
Effect of mutations in ste11 on mating and meiosis
| Strain genotype | % Sporulationa
|
|||||
|---|---|---|---|---|---|---|
| 0 h | 2 h | 4 h | 6 h | 8 h | 19 h | |
| h90 | 0.0 | 0.0 | 0.9 | 5.3 | 19.8 | 51.9 |
| h90 ste11T173A, S218A | 0.0 | 0.0 | 2.4 | 41.7b | 59.1b | 81.1b |
| h90 ste11T173D, S218D | 0.0 | 0.0 | 0.0 | 1.6 | 4.0b | 25.0b |
| h90 ste11T305A, T317A | 0.0 | 0.0 | 1.7 | 8.2 | 24.0 | 55.4 |
| h90 ste11T173D, S218D, T305A, T317A | 0.0 | 0.0 | 0.7 | 2.4 | 6.8b | 53.3 |
| h90 ste11T305D, T317D | 2.4b | 5.9b | 21.3b | 34.6b | 47.5b | 82.3b |
| h90 ste11T173A, S218A, T305D, T317D | 3.1b | 11.3b | 25.4b | 39.2b | 50.9b | 84.9b |
Cultures of Eg640, Eg1207, Eg1093, Eg1092, Eg1638, Eg1645, and Eg1646 were grown in MSL to a density of 2.5 × 106 cells/ml and then shifted to MSL without nitrogen. Samples were taken at the indicated times, and the percentage of sporulation was estimated by microscopic examination as described in Materials and Methods. Values represent means of results from three separate trials.
Sporulation is significantly different from that of wild-type h90 cells (P < 0.05; tested by Student's t test).
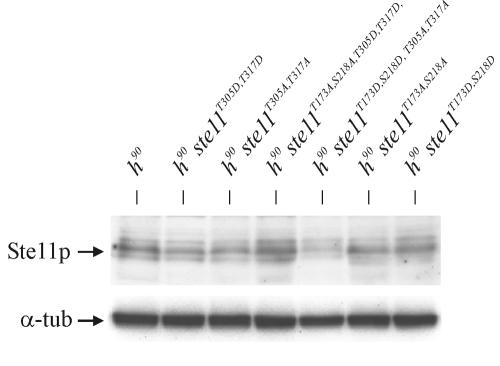
Next, we wished to investigate whether the ste11T305A, T317A allele could impair the haploid meiosis induced by unregulated Byr2 activity. To this end, a heterothallic h− ste11T305A, T317A strain was transformed with a vector expressing byr2-ΔN, and the frequency of haploid meiosis was monitored. We found that the level of byr2-ΔN-induced meiosis was significantly reduced in h− ste11T305A, T317A cells compared to wild-type h− cells (Table 2). This result supports the idea that Ste11 is a direct target of Spk1. The Ste11T305A, T317A mutant protein was expressed to a level similar to the wild-type Ste11 protein, ruling out the possibility that the suppression of haploid meiosis seen in h− ste11T305A, T317A cells was the result of an unstable Ste11 mutant protein (Fig. 7).
FIG. 7.
The mutant Ste11 proteins are stably expressed. Shown is a Western analysis of wild-type Ste11 and mutant Ste11 proteins. h90 strains Eg640, Eg1645, Eg1207, Eg1646, Eg1638, Eg1092, and Eg1093 were grown to a density of 5 × 106 cells/ml in MSL, then shifted to MSL without nitrogen, and incubated for 3 h before sampling. Extracts were subjected to Western analysis using mouse monoclonal antibodies against Ste11 (upper panel) and Tat1 (lower panel).
Mutations of the Pat1 sites in Ste11 impair haploid meiosis.
It has been demonstrated that Pat1 directly represses Ste11 activity by phosphorylation of Thr173 and Ser218 (27, 43), and substitution of Thr173 and Ser218 with aspartic residues, mimicking constitutive Pat1 phosphorylation, did indeed reduce the ability of Ste11 to promote sexual differentiation (Table 4).
To examine whether mutations of the Pat1 phosphorylation sites in Ste11 could suppress byr2-ΔN-induced meiosis, we con-structed an h− ste11T173D, S218D strain. Interestingly, in an h− ste11T173D, S218D strain, haploid meiosis was suppressed to the same level as in an h− ste11T305A, T317A strain (Table 2). Neither of the two Ste11 alleles inhibits haploid meiosis completely. Therefore, we combined the four point mutations in a composite allele, Ste11T173D, S218D, T305A, T317A, to see wheth-er haploid meiosis was totally suppressed. However, h− ste11T173D, S218D, T305A, T317A cells entered haploid meiosis with the same frequency as h− ste11T173D, S218D and h− ste11T305A, T317A cells, showing that there was no additive effect of combining the mutations in the Pat1 and Spk1 phosphorylation sites (Table 2).
These observations suggest that the pheromone signal is transduced to Ste11 by two pathways: a direct one where Spk1 activates Ste11 by phosphorylating threonines 305 and 317 and an indirect one where Spk1 impairs Pat1 kinase activity, thereby relieving its inhibitory phosphorylation of Ste11 at residues 173 and 218.
Cells carrying mutations in Ste11 that mimic constant MAPK phosphorylation enter meiosis in the presence of nitrogen.
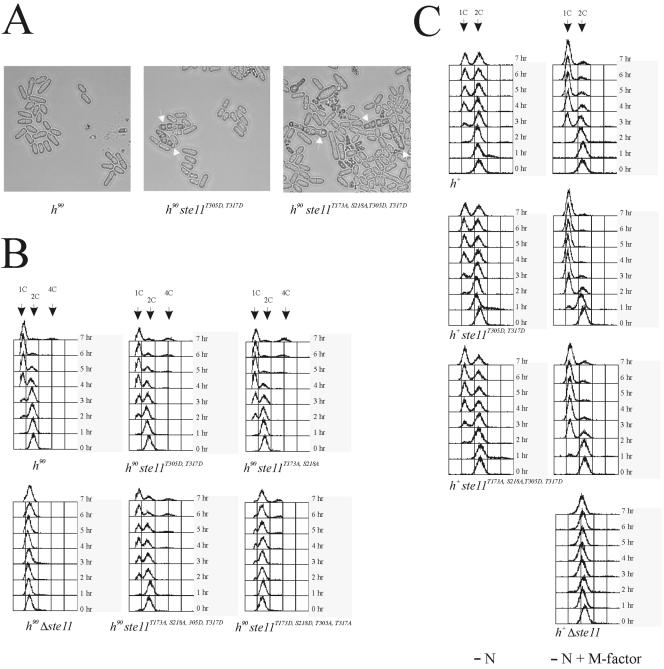
The ste11T305A, T317A allele is predicted to encode an attenuated version of the Ste11 transcription factor. However, as described above, this allele has no obvious effect on mating and meiosis in wild-type cells. Maybe the activity of Ste11 is controlled by several redundant means. We therefore asked whether it was possible to make a hyperactive allele of Ste11 by exchanging threonines 305 and 317 with aspartates. Interestingly, homothallic h90 cells carrying a ste11T305D, T317D allele mated and sporulated, although at low frequency, when grown in minimal medium with nitrogen (Fig. 8A and Table 4). Normally, nitrogen represses sexual differentiation, and wild-type cells grown under the same conditions did not mate (Fig. 8A and Table 4). The effect of Ste11T305D, T317D on conjugation and sporulation was also measured in nitrogen-starved liquid cultures. This experiment revealed that Ste11T305D, T317D augments both the onset and the extent of sexual differentiation (Table 4). Hence, the T305D and T317D substitutions mimic the effects of a constantly phosphorylated and activated Ste11 protein.
FIG. 8.
Ste11T305D, T317D cells are derepressed for mating and arrest faster in G1 in response to pheromone signaling. (A) h90 strains Eg640, Eg1645, and Eg1646 were grown to a density of 5 × 106 cells/ml in MSL and examined by microscopy. Arrows indicate asci. (B) h90 strains Eg640, Eg1645, Eg1092, Eg1085, Eg1646, and Eg1638 were grown to a density of 5 × 106 cells/ml in MSL and then shifted to MSL without nitrogen. Samples were taken every hour until 7 h, fixed in 70% ethanol, and processed for flow cytometry. (C) h+ strains Eg545, Eg1672, Eg1673, and Eg1125 were grown to a density of 5 × 106 cells/ml in MSL and then shifted to MSL-N or MSL-N with 1 μg of M-factor/ml. Samples were taken every hour until 7 h, fixed in 70% ethanol, and processed for flow cytometry.
Ste11 is required for arresting cells in G1.
An important aspect of sexual differentiation is the ability to arrest in G1 in response to nitrogen limitation and pheromone signaling (9, 10, 18). Since Ste11 is required for the expression of many genes necessary for pheromone communication, we investigated whether the transcription factor contributes to this G1 arrest. Homothallic strains were transferred to nitrogen-free medium, and the DNA content of the cell population was monitored by using flow cytometry (Fig. 8B). In wild-type strains, G1 cells start accumulating 2 to 3 h following nutritional shift. In contrast, cells with ste11 deletions exhibited mostly a C2 DNA content even after 7 h of incubation in nitrogen-free medium. After 16 h of starvation, G1 cells can be observed for a Δste11 strain (data not shown). Hence, Δste11 cells are severely impaired in their ability to arrest in G1 in response to nitrogen limitation. Interestingly, Ste11T305D, T317D-expressing cells arrest somewhat faster in G1 than wild-type cells, confirming that the T305D and T317D substitutions mimic an activated Ste11 protein.
Recently, it was reported that Ste11 is activated when T173 and S218, the two amino acid residues phosphorylated by Pat1, are replaced by alanines (43). The phenotype of h90 ste11T173A, S218A cells is similar to that found for h90 ste11T305D, T317D cells. Thus, in cells expressing Ste11T173A, S218A, the onset and extent of sexual differentiation are enhanced and the cells tend to arrest faster in G1 when they are nitrogen starved (43) (Fig. 8B and Table 4). However, unlike the ste11T305D, T317D allele, the T173A and S218A substitutions do not allow mating and sporulation in the presence of nitrogen (Table 4). To see whether there was an additive effect on combining these activating point mutations, we next created a ste11T173A, S218A, T305D, T317D allele. However, the phenotype of cells carrying this ste11 allele did not differ significantly from the phenotypes we had observed for cells carrying only T305D and T317D substitutions (Fig. 8 and Table 4). This result is strikingly reminiscent of what we found when we combined the inactivating point mutations T173D and S218D with T305A and T317A (see above).
In the experiments described above, we used homothallic strains, which make it impossible to separate the effects of nitrogen starvation from pheromone signaling. In order to discriminate between the effects of nitrogen starvation and pheromone signaling, we did another series of flow cytometry analyses using heterothallic h+ cells. Exponentially growing heterothallic strains were shifted to nitrogen-depleted medium and grown in the presence or absence of M-factor pheromone. When cells were only nitrogen starved, there was no significant difference between wild-type h+ cells and h+ ste11T305D, T317D cells in their ability to arrest in G1 (Fig. 8C). In contrast, when treated with M-factor, h+ ste11T305D, T317D cells arrest more rapidly than wild-type cells. After 2 h of incubation, most of the h+ ste11T305D, T317D cells have a G1 DNA content, whereas a substantial fraction of wild-type h+ cells have a C2 DNA content (Fig. 8C). We next compared h+ cells expressing the ste11T173A, S218A, T305D, T317D allele with h+ cells expressing the ste11T305D, T317D allele. h+ ste11T173A, S218A, T305D, T317D cells respond to nitrogen starvation and pheromone signaling with the same kinetics as h+ ste11T305D, T317D cells. However, when cells were nitrogen starved in the absence of M-factor, the T173A, S218A, T305D, and T317D substitutions seem to augment the G1 arrest slightly. Thus, after 7 h of nitrogen starvation, approximately 75% of the h+ ste11T173A, S218A, T305D, T317D cells are in G1, whereas the number for h+ ste11T305D, T317Dand wild-type cells is only 50%.
In conclusion, these results confirm that ste11T173A, S218A and ste11T305D, T317D represent activated alleles of ste11. However, they also imply that pheromone signaling activates Ste11 by at least one additional mechanism besides Spk1 phosphorylation and relief of inhibitory Pat1 phosphorylation.
DISCUSSION
In this report, we have demonstrated that the activation of the pheromone-responsive signaling cascade is sufficient for the activation of meiosis in S. pombe. Thus, the expression of a constitutive allele of the MAP3K Byr2 (byr2-ΔN) induced haploid cells to directly enter meiosis and sporulation. Furthermore, the normal requirement of nitrogen starvation for meiotic entry was bypassed. Curiously, mei3+ was not required for the byr2-ΔN-induced meiosis. This observation challenges our present understanding of the regulation of meiosis, where Mei3 is required for inactivation of the Pat1 protein kinase in order to relieve its inhibitory phosphorylation of Mei2 (22, 31, 32, 57). Normally, only diploid cells enter meiosis, because induction of the mei3 gene requires coexpression of the mat1-Pm and mat1-Mm genes expressed from the mat1 locus (32, 61). Our finding implies that pheromone signaling can cause Mei3-independent inactivation of Pat1, as previously anticipated (39), although we cannot rule out the alternative explanation that Mei2 can be activated in a Pat1-independent manner.
The Byr2 pathway activates Ste11 in response to both nitrogen starvation and pheromone signaling.
Despite the fact that the pheromone-responsive Byr2-Byr1-Spk1 pathway has been known for more than a decade, no direct targets of Spk1 have been identified so far. The HMG box transcription factor Ste11 is a likely candidate, as most genes induced by pheromone appear to be controlled by Ste11 (1, 23, 42, 52). Moreover, eight copies of the Ste11 binding site confer pheromone-induced transcription to a heterologous S. cerevisiae promoter (24). However, investigations of Ste11 as a target for the Byr2-Byr1-Spk1 pathway have been complicated by the fact that Ste11 itself is required for the expression of many components of the pathway, including the receptors and spk1 (53). In addition, most Ste11-controlled genes seem to be induced to various extents both by nitrogen starvation and by pheromone signaling. Curiously, Δbyr2 and Δbyr1 mutants are also defective in the induction of mam2 and the three mfm genes in response to nitrogen starvation (23, 63). We found that the expression of byr2-ΔN in fact bypasses the requirement for starvation, since the transcription of mat1-Pm and mfm1 now takes place in rich medium. Taken together, these observations suggest that the Byr2-Byr1-Spk1 pathway conveys both the nitrogen starvation and the pheromone signals and that Ste11 is an important target for both signals.
Consistent with this, we show that the mutation of two putative sites for MAPK phosphorylation in Ste11 impairs byr2-ΔN-induced meiosis. Thus, replacing threonines 305 and 317 with alanines caused an approximately 10-fold reduction in haploid meiosis. This result provides the first direct evidence for Ste11 being a downstream target of Spk1. Further supporting this idea, we found that Spk1 interacts physically with Ste11 and that Spk1 can phosphorylate Ste11 in vitro. This phosphorylation relies on the two putative MAPK sites in Ste11, as the exchange of threonines 305 and 317 with alanines completely abolishes in vitro phosphorylation. Furthermore, a kinase-dead version of Spk1 could no longer phosphorylate Ste11.
Under normal conditions, the Spk1 site mutations had no detectable physiological consequences, i.e., h90 ste11T305A, T317Acells conjugated and sporulated as wild-type cells. However, we found that changing threonines 305 and 317 into aspartates, mimicking constant Spk1 phosphorylation, resulted in an activated Ste11 protein that permitted conjugation and sporulation in rich medium, albeit at low frequencies. Taken together, these observations strongly suggest that Spk1 directly phosphorylates Ste11 to promote its activity.
Pat1 function and pheromone signaling.
Sexual differentiation appears to be accompanied by a gradual down-regulation of Pat1 activity (4, 39). First, a Mei3-independent partial inactivation of Pat1 occurs in response to nitrogen starvation, which activates Ste11 in a positive feedback loop. This enables the expression of genes required for the establishment of pheromone communication. Upon successful cell conjugation, Pat1 becomes completely inactivated by Mei3, allowing the activation of Mei2 and meiosis. Since pheromone-controlled genes become induced upon inactivation of a pat1 temperature-sensitive allele (39, 61), we speculate that unregulated Byr2-Byr1-Spk1 activity may directly inactivate Pat1 to a level adequate for Mei2 activation, and indeed, the Pat1 sequence harbors five potential sites for MAPK phosphorylation (data not shown). Under normal conditions, however, the output from pheromone signaling may never reach such high levels, and Pat1 is inactivated only to a level that allows conjugation but not meiosis.
Presumably, unregulated Byr2 activity causes constitutive activation of Ste11, an interpretation supported by the high level of the protein irrespective of growth conditions (Fig. 3). Pat1 inhibits Ste11 by phosphorylation of the residues T173 and S218, and this creates binding sites for the 14-3-3 protein Rad24, which prevents nuclear accumulation of Ste11 and hence its transcriptional activity (22, 27, 43). Therefore, the constitutive activation of Ste11 is likely to reflect increased nuclear accumulation and subsequent autostimulation of the ste11 gene (25). Consistent with byr2-ΔN causing inactivation of Pat1, we found that the mutagenesis of T173 and S218 into aspartates, mimicking constant Pat1 phosphorylation, impaired haploid sporulation triggered by unregulated Byr2 activity.
Neither ste11T305A, T317A nor ste11T173D, S218D completely block haploid meiosis, and we found no additive effects of combining them in the same Ste11 molecule. This result suggests that Ste11 activation by the Byr2-Byr1-Spk1 pathway involves both dephosphorylation of the Pat1 sites (T173 and S218) and phosphorylation of the Spk1 sites (T305 and T317). This is consistent with the observation that ste11T173A, S218A cells still require a pheromone signal in order to accumulate Ste11 in the nucleus (43).
Ste11 is required for G1 arrest.
Both nitrogen depletion and pheromone signaling cause cell cycle arrest in G1 (9, 10, 18), and we have presented evidence that ste11 cells are defective in G1 arrest, consistent with Ste11 being a target for the Byr2-Byr1-Spk1 pathway conveying both these signals. In support of this model, we also found that the two activated ste11 alleles, ste11T173A, S218A and ste11T305D, T317D, accelerated the arrest upon nitrogen starvation and pheromone signaling. Furthermore, we did not observe synergy when combining the Spk1 and Pat1 site mutations in the same allele, again arguing that activation of Ste11 requires both activation of Spk1 and inactivation of Pat1.
Intriguingly, this activated Ste11 mutant required pheromone signaling for acceleration of the G1 arrest. This result means that ste11T173A, S218A, T305D, T317D cells are still capable of conveying a pheromone signal to promote G1 arrest. These observations imply that the pheromone signal can be transduced by a third mechanism that involves neither the Pat1 nor the Spk1 phosphorylation sites in Ste11. Cells containing a loss-of-function pat1 allele have Ste11 constantly in the nucleus irrespective of nutritional starvation and pheromone signaling, whereas wild-type cells expressing Ste11T173A, S218A still require pheromone signaling for nuclear accumulation of the transcription factor (43). We therefore speculate that this third mechanism of activating Ste11 also involves the inactivation of Pat1 (Fig. 9). This mechanism is reminiscent of pheromone-induced transcription in S. cerevisiae, where the downstream transcription factor of the pheromone signaling pathway, Ste12, is regulated by several means. Thus, Ste12 activity is controlled directly by MAP kinase phosphorylation and indirectly through the inhibitors Dig1 and Dig2 (8).
FIG. 9.
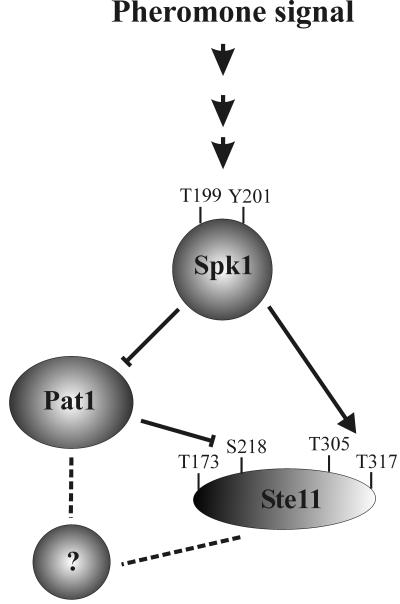
Model for regulation of Ste11 by pheromone signaling. Activation and inhibition are indicated by arrows and crossing bars, respectively. Dotted lines indicate pathways that may be indirect and can either be activating or inhibitory. In mitotically growing cells, Ste11 is kept in an inactive form by Pat1-mediated phosphorylation of threonine 173 and serine 218. In nitrogen-starved cells, pheromone signaling activates Spk1, which then activates Ste11 via three different pathways, a direct one where Spk1 activates Ste11 by phosphorylating threonines 305 and 317 and two indirect pathways that both involve the Pat1 kinase. Upon pheromone signaling, Pat1 is inactivated to a level that inhibits its phosphorylation of Ste11 at residues T173 and S218. Additionally, Pat1 probably regulates an unknown factor that controls the activity of Ste11.
In conclusion, our observations suggest that high signal intensity of the pheromone response can activate meiosis by a new Mei3-independent pathway in S. pombe. This outcome is consistent with a model where gradual inactivation of the Pat1 kinase orchestrates the various steps of sexual differentiation by gradually activating Ste11 functions.
After the manuscript was submitted for publication, Yamamoto et al. (66) reported that the expression of a constitutively active version of Byr1 causes haploid cells to enter meiosis directly and independently of Mei3. Interestingly, these authors found that meiosis under these conditions is accompanied by normal telomere clustering, as opposed to meiosis triggered by Pat1 inactivation.
Acknowledgments
We thank M. McLeod, K. Ekwall, and M. Yamamoto for plasmids and strains; Hanne Jørgensen and Karin Holm for expert technical assistance; and Richard Egel for comments on the manuscript.
This work was supported by the Danish Natural Science Research Council, the Novo Nordisk Foundation, and NorFA.
REFERENCES
- 1.Aono, T., H. Yanai, F. Miki, J. Davey, and C. Shimoda. 1994. Mating pheromone-induced expression of the mat1-Pm gene of Schizosaccharomyces pombe: identification of signalling components and characterization of upstream controlling elements. Yeast 10:757-770. [DOI] [PubMed] [Google Scholar]
- 2.Barr, M. M., H. Tu, L. Van Aelst, and M. Wigler. 1996. Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol. Cell. Biol. 16:5597-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 4.Beach, D., L. Rodgers, and J. Gould. 1985. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr. Genet. 10:297-311. [DOI] [PubMed] [Google Scholar]
- 5.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 6.Bresch, C., G. Muller, and R. Egel. 1968. Genes involved in meiosis and sporulation of a yeast. Mol. Gen. Genet. 102:301-306. [DOI] [PubMed] [Google Scholar]
- 7.Castrop, J., D. van Wichen, M. Koomans-Bitter, M. van de Wetering, R. de Weger, J. van Dongen, and H. Clevers. 1995. The human TCF-1 gene encodes a nuclear DNA-binding protein uniquely expressed in normal and neoplastic T-lineage lymphocytes. Blood 86:3050-3059. [PubMed] [Google Scholar]
- 8.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831-2848. [DOI] [PubMed] [Google Scholar]
- 9.Davey, J., and O. Nielsen. 1994. Mutations in cyr1 and pat1 reveal pheromone-induced G1 arrest in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 26:105-112. [DOI] [PubMed] [Google Scholar]
- 10.Egel, R., and M. Egel-Mitani. 1974. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 88:127-134. [DOI] [PubMed] [Google Scholar]
- 11.Egel, R., M. Willer, S. Kjaerulff, J. Davey, and O. Nielsen. 1994. Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast 10:1347-1354. [DOI] [PubMed] [Google Scholar]
- 12.Foiani, M., F. Marini, D. Gamba, G. Lucchini, and P. Plevani. 1994. The B subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsburg, S. L., and D. A. Sherman. 1997. General purpose tagging vectors for fission yeast. Gene 191:191-195. [DOI] [PubMed] [Google Scholar]
- 15.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 16.Iino, Y., and M. Yamamoto. 1985. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol. Gen. Genet. 198:416-421. [DOI] [PubMed] [Google Scholar]
- 17.Iino, Y., and M. Yamamoto. 1985. Negative control for the initiation of meiosis in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 82:2447-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai, Y., and M. Yamamoto. 1994. The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev. 8:328-338. [DOI] [PubMed] [Google Scholar]
- 19.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, T., Jr., K. Okazaki, H. Murakami, S. Stettler, P. A. Fantes, and H. Okayama. 1996. Stress signal, mediated by a Hog1-like MAP kinase, controls sexual development in fission yeast. FEBS Lett. 378:207-212. [DOI] [PubMed] [Google Scholar]
- 21.Kelly, M., J. Burke, M. Smith, A. Klar, and D. Beach. 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitamura, K., S. Katayama, S. Dhut, M. Sato, Y. Watanabe, M. Yamamoto, and T. Toda. 2001. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev. Cell 1:389-399. [DOI] [PubMed] [Google Scholar]
- 23.Kjærulff, S., J. Davey, and O. Nielsen. 1994. Analysis of the structural genes encoding M-factor in the fission yeast Schizosaccharomyces pombe: identification of a third gene, mfm3. Mol. Cell. Biol. 14:3895-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjærulff, S., D. Dooijes, H. Clevers, and O. Nielsen. 1997. Cell differentiation by interaction of two HMG-box proteins: Mat1-Mc activates M cell-specific genes in S. pombe by recruiting the ubiquitous transcription factor Ste11 to weak binding sites. EMBO J. 16:4021-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunitomo, H., T. Higuchi, Y. Iino, and M. Yamamoto. 2000. A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11(+) gene, which encodes a pivotal transcription factor for sexual development. Mol. Biol. Cell 11:3205-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leroy, D., V. Baldin, and B. Ducommun. 1994. Characterization of an active GST-human Cdc2 fusion protein kinase expressed in the fission yeast Schizosaccharomyces pombe: a new approach to the study of cell cycle control proteins. Yeast 10:1631-1638. [DOI] [PubMed] [Google Scholar]
- 27.Li, P., and M. McLeod. 1996. Molecular mimicry in development: identification of ste11+ as a substrate and mei3+ as a pseudosubstrate inhibitor of ran1+ kinase. Cell 87:869-880. [DOI] [PubMed] [Google Scholar]
- 28.Lyne, R., G. Burns, J. Mata, C. J. Penkett, G. Rustici, D. Chen, C. Langford, D. Vetrie, and J. Bahler. 2003. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maundrell, K. 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265:10857-10864. [PubMed] [Google Scholar]
- 30.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 31.McLeod, M., and D. Beach. 1988. A specific inhibitor of the ran1+ protein kinase regulates entry into meiosis in Schizosaccharomyces pombe. Nature 332:509-514. [DOI] [PubMed] [Google Scholar]
- 32.McLeod, M., M. Stein, and D. Beach. 1987. The product of the mei3+ gene, expressed under control of the mating-type locus, induces meiosis and sporulation in fission yeast. EMBO J. 6:729-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 34.Nadin-Davis, S. A., and A. Nasim. 1988. A gene which encodes a predicted protein kinase can restore some functions of the ras gene in fission yeast. EMBO J. 7:985-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nadin-Davis, S. A., and A. Nasim. 1990. Schizosaccharomyces pombe ras1 and byr1 are functionally related genes of the ste family that affect starvation-induced transcription of mating-type genes. Mol. Cell. Biol. 10:549-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadin-Davis, S. A., R. C. Yang, S. A. Narang, and A. Nasim. 1986. The cloning and characterization of a RAS gene from Schizosaccharomyces pombe. J. Mol. Evol. 23:41-51. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen, O. 2004. Mating-type control and differentiation, p. 281-296. In R. Egel (ed.), The molecular biology of Schizosaccharomyces pombe. Springer, Berlin, Germany.
- 38.Nielsen, O., J. Davey, and R. Egel. 1992. The ras1 function of Schizosaccharomyces pombe mediates pheromone-induced transcription. EMBO J. 11:1391-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen, O., and R. Egel. 1990. The pat1 protein kinase controls transcription of the mating-type genes in fission yeast. EMBO J. 9:1401-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nurse, P. 1985. Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell-proliferation and sporulation. Mol. Gen. Genet. 198:497-502. [Google Scholar]
- 41.Okazaki, N., K. Okazaki, Y. Watanabe, M. Kato-Hayashi, M. Yamamoto, and H. Okayama. 1998. Novel factor highly conserved among eukaryotes controls sexual development in fission yeast. Mol. Cell. Biol. 18:887-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen, J., D. Weilguny, R. Egel, and O. Nielsen. 1995. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol. Cell. Biol. 15:3697-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin, J., W. Kang, B. Leung, and M. McLeod. 2003. Ste11p, a high-mobility-group box DNA-binding protein, undergoes pheromone- and nutrient-regulated nuclear-cytoplasmic shuttling. Mol. Cell. Biol. 23:3253-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose, M., and D. Botstein. 1983. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 101:167-180. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Sazer, S., and S. W. Sherwood. 1990. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97:509-516. [DOI] [PubMed] [Google Scholar]
- 47.Scherer, S., and R. W. Davis. 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. USA 76:4951-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimoda, C., M. Uehira, M. Kishida, H. Fujioka, Y. Iino, Y. Watanabe, and M. Yamamoto. 1987. Cloning and analysis of transcription of the mei2 gene responsible for initiation of meiosis in the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 169:93-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10:2276-2288. [DOI] [PubMed] [Google Scholar]
- 50.Shiozaki, K., and P. Russell. 1997. Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol. 283:506-520. [DOI] [PubMed] [Google Scholar]
- 51.Styrkarsdottir, U., R. Egel, and O. Nielsen. 1992. Functional conservation between Schizosaccharomyces pombe ste8 and Saccharomyces cerevisiae STE11 protein kinases in yeast signal transduction. Mol. Gen. Genet. 235:122-130. [DOI] [PubMed] [Google Scholar]
- 52.Sugimoto, A., Y. Iino, T. Maeda, Y. Watanabe, and M. Yamamoto. 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5:1990-1999. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka, K., J. Davey, Y. Imai, and M. Yamamoto. 1993. Schizosaccharomyces pombe map3+ encodes the putative M-factor receptor. Mol. Cell. Biol. 13:80-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thon, G., and A. J. Klar. 1992. The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131:287-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toda, T., M. Shimanuki, and M. Yanagida. 1991. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 5:60-73. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Y., H. P. Xu, M. Riggs, L. Rodgers, and M. Wigler. 1991. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol. Cell. Biol. 11:3554-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe, Y., S. Shinozaki-Yabana, Y. Chikashige, Y. Hiraoka, and M. Yamamoto. 1997. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386:187-190. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe, Y., and M. Yamamoto. 1994. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78:487-498. [DOI] [PubMed] [Google Scholar]
- 59.Weilguny, D., M. Praetorius, A. Carr, R. Egel, and O. Nielsen. 1991. New vectors in fission yeast: application for cloning the his2 gene. Gene 99:47-54. [DOI] [PubMed] [Google Scholar]
- 60.Wilkinson, M. G., M. Samuels, T. Takeda, W. M. Toone, J. C. Shieh, T. Toda, J. B. Millar, and N. Jones. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10:2289-2301. [DOI] [PubMed] [Google Scholar]
- 61.Willer, M., L. Hoffmann, U. Styrkarsdottir, R. Egel, J. Davey, and O. Nielsen. 1995. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol. Cell. Biol. 15:4964-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woods, A., T. Sherwin, R. Sasse, T. H. MacRae, A. J. Baines, and K. Gull. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491-500. [DOI] [PubMed] [Google Scholar]
- 63.Xu, H. P., M. White, S. Marcus, and M. Wigler. 1994. Concerted action of RAS and G proteins in the sexual response pathways of Schizosaccharomyces pombe. Mol. Cell. Biol. 14:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto, M. 2004. Initiation of meiosis, p. 297-309. In R. Egel (ed.), The molecular biology of Schizosaccharomyces pombe. Springer, Berlin, Germany.
- 65.Yamamoto, M. 1996. Regulation of meiosis in fission yeast. Cell Struct. Funct. 21:431-436. [DOI] [PubMed] [Google Scholar]
- 66.Yamamoto, T. G., Y. Chikashige, F. Ozoe, M. Kawamukai, and Y. Hiraoka. 2004. Activation of the pheromone-responsive MAP kinase drives haploid cells to undergo ectopic meiosis with normal telomere clustering and sister chromatid segregation in fission yeast. J. Cell Sci. 117:3875-3886. [DOI] [PubMed] [Google Scholar]