Abstract
Biofuels are the promising sources which are produced by various microalgae or in the form of metabolic by-products from organic or food waste products. Microalgae have been widely reported for the production of biofuels since these have a high storage of lipids as triacylglycerides, which can mainly be converted into biofuels. Recently, products such as biodiesel, bioethanol and biogas have renewed the interest toward the microalgae. The proteomics alone will not pave the way toward finding an ideal alga which will fulfill the current energy demands, but a combined approach of proteomics, genomics and bioinformatics can be pivotal for a sustainable solution. The present review emphasizes various technologies currently involved in algal proteomics for the efficient production of biofuels.
Keywords: Microalgae, Biofuel, Proteomics, ITRAQ, Systems biology
Introduction
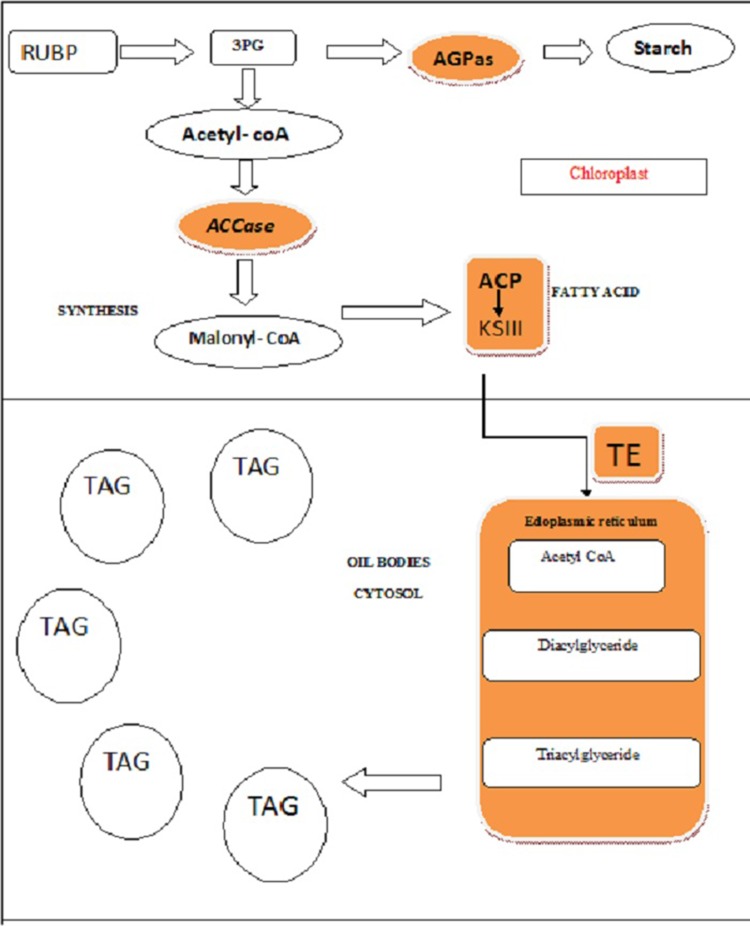
The molecular biology of microalgae is of great scientific attraction in recent years because of its potential for biofuel production. Researchers have shown their interest in microalgae from both the points of view, i.e., as public communities which can be used as a potent source toward biofuel potential as well as other high value compounds. Molecular and biochemical analyses show interesting differences in lipid metabolism between the microalgae and plants. These differences are mainly due to distinct acyl groups that are present in algal lipids. Plastids are also involved in the assembly of TAGs, which are the reasons for the glycerolipid metabolism in the subcellular organization (Hu et al. 2008). Thus, lipid metabolism in microalgae points to a new approach in such groups of organisms, which also deals with the analysis of lipid metabolism in plants in the field of genetic engineering (Fig. 1). Due to limited availability of conventional fossil fuels, the use of microalgae has attracted great attention of scientists in these days. Biodiesel production from microalgae and its utilization as fuel have become the topics of research in recent years and this has attracted the scientists toward exploring the microalgal cellular biology in detail (James et al. 2011). Fossil fuels are non-renewable source of energy, going to be limited by the time and its uses which has also influenced toward the global warming and pollutions. The concepts of alternative energy or sustainable energy are needed to overcome the twin problems of global temperature rise and air pollution. A few strains of microalgae have been identified as a source of biofuel production, since they store solar energy as chemical energy in various forms, which is used for the production of biodiesel, biomethane, and biohydrogen (Kruse et al. 2005; Chisti 2007; Schenk et al. 2008). Exploitation of algal diversity and its sustainable use for biodiesel production is the need of the hour to ensure that the energy crisis is alleviated. Microalgae have high-storage lipid content; such stored lipids can be converted into biodiesel. Research is needed on biodiesel production and exploring this at a commercial scale to screen out the existing strains of available species for the maximum yield.
Fig. 1.
Overview of metabolic pathway in microalgae for lipid synthesis
In the current review, we have elucidated the proteomic analysis and lipid profiling of algae and its relevance toward biofuel production. Without proteomics, studies of algal omics is incomplete as it gives an idea of the precise functional group in the state of static and dynamic physiological contexts. Particular or targeted proteomics studies on microalgae have not been reported yet. Proteomics has evolved as a better technique approach for understanding the complexity in biological system. This can be successfully exploited for obtaining algal oil and its biochemical conversion to biodiesel. The motivation behind this study is to attract awareness toward sustainable utilization of microalgae resources for obtaining indigenous strains toward exploring their possible role in the development of bioenergy production in the country.
Physiology of microalgae
Algae are the oldest form of life at present, which come under the primitive plants thallophytes (Falkowski and Raven 1997). Thallophyta are the most primitive and the largest division of Cryptogams. The plant body of such plant kingdom is not differentiated into root, stem and leaves, due to which the whole plant body is termed as thallus. The thalloid plant body may be unicellular or multicellular. the physiological parameter includes lack of roots, reproductive leaves and lack of the covering around the cells of stems. Also, they contain chlorophyll, which is the pigment necessary for photosynthesis (Lee 1980) and whose primary function is to produce starch or carbohydrates. Simple physiology and habitat condition enable them to survive any environmental conditions and also ensure their survival for a long duration. These structures support the energy conversion, that is photosynthesis, without any development or change in their cellular composition (Falkowski and Raven 1997). Prokaryotic cells (cyanobacteria) generally do not have membrane-bound organelles and these are present in bacteria in comparison to algae which are eukaryotic. Different eukaryotic algae have been identified and placed in various groups, and are mainly recognized by the pigment, life cycle they follow, cellular structure and composition (Khan et al. 2009). Green algae (Chlorophyta), red algae (Rhodophyta) and diatoms (Bacillariophyta) are the algal types that have been identified. These microbes can be further grouped into autotrophs and heterotrophs. The autotrophic organisms require CO2, water and a light source for their development, since they are the producers of macromolecules such as carbohydrates; while the heterotrophic organisms are non-photosynthetic and so depend on the organic compounds along with the nutrients as an energy source rather than inorganic compounds. Some strains of microalgae are mixotrophic, that is, they can perform photosynthesis as well as they acquire exogenous organic nutrients (Lee 1980). Photosynthesis is necessary for the survival and growth of autotrophic microalgae, where adenosine triphosphate (ATP) is formed by the conversion of solar radiation and CO2 which is then absorbed by chloroplasts. O2 is used as the energy source at the cellular level, since O2 is used for respiration which in terms produces energy required for growth (Khan et al. 2009; Zilinskas Braun and Zilinskas Braun 1974).
Biofuel from microalgae
Microalgae have high-storage lipid content due to which it has gained importance among researchers, because neutral lipid, for example triacylglycerides, can be easily be converted to biodiesel (fatty acid methyl esters) through various methods such as supercritical process and microwave-assisted and ultrasound-assisted processes (Araujo et al. 2011). Biodiesel is a renewable source of energy which has the potential to replace non-renewable sources of energy. Supercritical processes have been identified as novel processes. Biofuel from algae has made the researcher identify the most suitable strain with a high storage of lipid content. There are few recent reports on various aspects of microalgal biofuel like metabolic engineering, systems biology and bioharvesting which have created attention recently (Banerjee et al. 2016a, b; Kumar et al. 2017). Macromolecules such as polysaccharides (sugars) and triacylglycerides (fats) which can also be obtained from the microalgae have made the researcher to show their interest on them as a potential feedstock, since microalgae grow on wetland. Besides these macromolecule, protein can also be extracted from algae, which can be used as a source of animal feed in the form of single cell protein (SCP), whereas some species of microalgae can also be used for the production of various commercially valuable compounds such as pigments and pharmaceuticals (Griffiths and Harrison 2009). Lipids are the stored products of many species of algae which are produced or stored in large amount, which may contribute more than 50% of its dry weight. Transesterification of such lipids, which are derived from the algae are very effective and a prospective source for biodiesel synthesis (Chaumont 1993).
The production of biofuel and its extraction from microalgae biomass are expensive and technologically challenging. Microalgae require light, CO2, water and inorganic salts to perform photosynthesis and grow (Banerjee et al. 2014). The temperature needed for their growth has to be strictly maintained. A temperature of 20–30 °C is required for the optimal growth and development of algae, whereas at 25–28 °C it attains its maximum growth (Banerjee et al. 2012). The cost for biodiesel production can be reduced by relying on freely available sunlight, instead of depending on the daily and seasonal natural light (Chisti 2007; Borowitzka 1999, 2005; Pulz 2001; Mallick 2002). Biomass obtained from the microalgae can be used as energy source by converting them through different ways, such as biochemical process, chemical reaction and direct combustion or through thermochemical conversion (Kumar et al. 2010). The different microalgal species that are used in biofuel production are shown in Table 1.
Table 1.
Microalgae used for the production of biofuel
| Algae species | Products | Culture technique | References |
|---|---|---|---|
| Botryococcus braunii | Triterpene oils | Photobioreactor | Courchesne et al. (2009) |
| Chlorella spp. | Carbohydrates, protein | Ponds, photobioreactor | Croft et al. (2007) |
| Chlamydomonas reinhardtii | Oils, carbohydrates, hydrogen and methane | Photobioreactor | Croft et al. (2006) |
| Dunaliella salina | β-Carotene | Brackish seawater ponds | Doebbe et al. (2007) |
| Nannochloropsis | Polyunsaturated fatty acids | Seawater ponds | Ermakova et al. (2013) |
| Ostreococcus tauri | Oils | Photobioreactor | Falkowski and Raven (1997) |
| Pavlova lutheri | Fatty acids, aquaculture feedstock | Photobioreactor | Forster et al. (2003) |
Proteomics
Proteomics by itself cannot be used for analysis because to get information about the gene expression and its regulation, proteomics has to be correlated with genomics and transcriptomics (Robinson et al. 2009). Since proteomics can be used for the analysis of protein expression along with their interactions so it can be further mediated with genomics and transcriptomics. The main idea behind proteomics is to analyze the post-translational modification and also to identify the different levels of proteins in submulti-protein complexes and their position in different types of cells and tissues. Genomics gives the genome sequence of the given microbes, which include coding as well as regulatory sequences. Transcripts [messenger RNA (mRNA)] too relate to genomics and proteomics, since the transcript gives information about gene expression, alternating splicing, RNA editing, polyadenylation site, etc. Transcript knowledge can be used for the development of algorithms and software. When the study on the functional gene is carried out in whole organisms, then such analysis is called phenomics, because in such case the phenotype of an organism is observed (Paigen and Eppig 2000). Another phenomics sequence analysis method is performed at different levels of different oligonucleotides sequences instated of different protein level. Metabolic pathways can also be understood through the study of proteomics. Comparative proteomics analysis is performed under different environmental conditions, which may be in the form of a fraction of a protein or the entire family of the one protein sample, can be compared to that of another sample or to that of the standard. Expressions of proteins and such differences are mainly by the different environmental stimuli, and which are analyzed, along with its metabolic pathway to which they are part of it can be analyzed through the proteomics analysis (Gasch et al. 2000).
Proteomics of eukaryotic algae and cyanobacteria
A wide range of algae are described in terms of genomics and transcriptomics, but the information in terms of proteomics is not adequate. Chlamydomonas reinhardtii is a eukaryotic algae, whose proteomic analysis has been done primarily and taken as a model organism in various research activities. Full characterization of algal mitochondria was the first proteomic analysis performed in algae (Lis et al. 2003). It was analyzed with the help of blue native-polyacrylamide gel electrophoresis (BN-PAGE) and further analyzed with a 2D SDS-PAGE. Similarly, 2D gel electrophoresis combined with immunoblotting (2D-IS) and mass spectrometry was used for the examination of the thylakoid membrane extracted from wild-type along with mutant strains of C. reinhardtii (Hippler et al. 2001). They included separation and classification of transmembrane-spanning and antenna complex proteins in their method. The antenna complex was further characterized, in which gene tagging signifies differentially processed amino-terminus in antenna proteins and potential post-translational phosphoregulation. The results obtained from these studies gave an intuition into various differential processing and post-translational modifications observed in many algal systems (Stauber et al. 2003).
Algae have gained its importance in the area of energy such as production of algae-based biofuels where a number of proteins can be identified through proteomic analyses along with microalgal lipid accumulation. Lipid droplets from C. reinhardtii were extracted using the solvent extraction method and then examined with GeLC/MS/MS method to inspect lipid droplet-associated proteins (Moellering and Benning 2010). More than 250 proteins in a lipid droplet-enriched fraction have been recognized, along with major lipid droplet protein (MLDP), which is predominantly found in green algae. RNAi was predominantly used in examining the functionality of these proteins, its involvement in size of the lipid droplet, and examining its capability as a marker for amassing lipid droplets and triacylglyceride (TAG). Upon examination of several proteins accompanying lipid biosynthesis and protein pathway, their direct relation with lipid droplets are found and hence it is proved that besides the direct role of lipidogenesis, these lipid droplets also have an indirect role in carbon flux. Isoelectric focusing combined with 2D SDS-PAGE and fluorescent staining was employed in C. reinhardtii, in which the carbon-concentrating mechanism (CCM) was studied by Baba and colleagues (Baba et al. 2011). Proteomics has gained much of its importance due to the development of highly sophisticated instrument. Nowaday’s proteomics is on the way to inspect light variation tolerance strategy in algae, which is crucial for the deployment of algae in open pond systems where the light intensity fluctuate considerably.
Similar to algal proteomics analysis, cyanobacterial proteomics studies are also applied to Synechocystis PCC 6803 as a model organism which is mainly engrossed on its organelle configuration and stress reactions. The external membrane of PCC 6803 was sequestered and characterized by using two-dimensional gel isoelectric focusing by Huang and colleagues (2002). They identified 49 different proteins that correspond to 29 gene products. All sequestered proteins contained amino-terminus signal peptides, though the function of 40% proteins remained unknown. A collection of porins and transporters were also recognized, along with several membrane-associated proteases. Similarly, the plasma membrane of PCC 6803 was inspected by Pisareva et al. (2007) who identified 51 proteins from a purified plasma membrane fraction using 1D SDS/PAGE MALDI-TOF MS (Pisareva et al. 2007). Most of the proteins observed were new, with representation from chemotaxis, metalloprotease, and secretion ontologies. Moreover, a substantial portion is expected to be integral, trans-membrane helix-containing proteins.
Technologies of proteomics
Proteomics mainly deals with large-scale experiment. Such type of experiment or analysis requires specialized tools, which are developed for the particular type of experiment or the design of the experiment. Three aspects of proteomics technologies have been identified. One is mass spectrophotometer, which is highly sensitive, where the endogenous protein mixtures can be identified along with its analysis. Array-based proteomics is also one kind of proteomic technologies, which mainly relates to cDNA microarray and oligonucleotide chips (Gahoi et al. 2015). A third area of proteomic analysis deals with the structure and imaging of single proteins, or protein with multi-subunit are experimented in a large scale such as its 3D shape, localization, metabolism and physiological parameters.
Mass spectrophotometer
Proteomics through the mass spectrophotometer (MS) has improved the analysis of a number of proteins, which means a number of proteins can be identified in a single experiment (George et al. 2015). The analysis of protein through MS mainly depends on the beakdown of protein sample into its constituents with the use of specific enzyme such as sequence protease. The protein as a whole itself is of high molecular weight which enables the analysis, as the protein cannot be eluted from the gel. For example, C. reinhardtii has been subjected to proteomics studies, in which around 240 proteins have been identified by providing heat shock. Synechocystis 6803 went through the compositional analysis of membrane protein complexes in different growth conditions (Herranen et al. 2004).
Array-based proteomics
Proteomic array-based analysis can also be used for the identification of a large set of proteins or multiprotein complexes (Liu et al. 2015). In this array based on analysis, purified ligands are separated individually. It may be from protein, peptide or carbohydrates or from small molecules such as antibodies or antigens. These are placed on a plane surface where they can be used for analysis of protein and its expression at the level of protein profiling. Different types of protein microarray formats have been identified, such as tissue array, reverse-phase array, capture arrays and lectin arrays, which are gaining importance in recent years. These tools are used in various fields of analysis such as protein–protein interaction studies, immunological profiling, biomarker discoveries and vaccine development. Such tools are delivering meaningful biological insights into modern biology. Nostoc flagelliforme was studied for its diurnal changes (Liang et al. 2013) and Anabaena 7120 for the effect of deletion of specific heterocyst proteins Flv1B and Flv3B (Ermakova et al. 2013).
Next-generation proteomic tools
Conventional tools such 2D gel electrophoresis, which is mainly used for the separation of a large number of protein along with the MALDI-TOF-MS-based analysis were quite popular in the early period of proteomics; however they are time consuming and lack throughput and quantitative ability. The development of nano-liquid chromatography-based analysis with the high resolution of mass spectrometry has improved the method of quantitative analysis of the protein or any other biological sample. Furthermore, a technique like isobaric tags for relative and absolute quantification (iTRAQ) has the ability to identify the biomarker such as the tissue biomarker, serum biomarker and drug resistance marker. Stable isotope labeling by amino acids in cell culture (SILAC) is another type of mass spectrometry-based quantitative approach, which is more efficient toward the cell culture system and has become a highly acceptable tool for quantitative biology.
Quantification methods
MRM (multiple reaction monitoring) and SRM (selected reaction monitoring) are the tools which have overcome the triple–quadrupole based mass spectrometers and have made a high impact in the area of proteomics in the case of biomarker identification. MRM-based quantification has been used extensively and has gained much importance in the proteomics area, since these tools have replaced some of the expensive methods of quantification such as antibody-based analysis like western blotting and ELISA. MRM performed on QTOFs and Orbit raps are called pseudo-MRM or high-resolution MRM (HR-MRM), which can also be called parallel reaction monitoring (PRM). In comparison, between MRM and PRM, the specific fragment ions obtained during acquisition are not possible through PRM.
Post-translation modification
Post-translation modifications are the mechanism used for the cellular processes along with the cellular control. Protein activity is curbed by the use of covalent modifiers like phosphate groups or ubiquitin moieties or by proteolytic cleavage. Protein turnover, localization, activity, or binding interactions can be affected through PTMs. Phosphorylation, acetylation, methylation, glycosylation, ubiquitination, and lipid modifications are some of the most common PTMs which can affect the cellular processes, since they directly affect the protein structure resulting in the change in the function of the given protein. Since, such modifications result in the variation in the molecular mass of the amino acid, these are quite important for being studied in detail. Mass spectrometry with its sensitivity, high mass accuracy and ability to deal with complex mixtures are some of the options for describing post-translational modifications. A broad description of protein stability in the chloroplast stroma of algae such as Chlamydomonas, their PTMs and the connection between the two was developed. It included 2D gel electrophoresis for the sequestration of the soluble stromal proteins, their classification and the characterization of the PTMs by MS combined with bioinformatics treatment of the data.
Future needs for the integration of “omics” in system biology
Complete, consistent and in-depth knowledge of the biological world by analyzing the behavior and interaction between its individual components and the surroundings is the main objective of systems biology (Andersen 2004). Systems biology includes many steps which follow the basic structure and formulation of the system and is the basic step in systems biology. Elements of system analysis, such as gene network, protein interaction and metabolic pathways, form the basic step in systems biology. This information will help us to understand and develop the basic model or interpretation of their behavior. A recent study has already reported the role of various microbial interactions and their studies using a systems biology approach (Singh and Shukla 2015). Based on the information gained about the various elements of the system, different experiments can be performed at different levels. There are also different types of analysis through which the whole system can be analyzed and understood, but before such analysis can be done, an understanding of certain parameters or elements of the system is required. Moreover, when the system is affected by genetic or environmental stimuli, high output measurement tools are necessary for their corresponding responses. These data can be compared with the different levels of hierarchy in the organization and can be compared with each other and also with the recently developed system. Such model has to be designed according to the data which are experimentally observed and are similar to the model’s predictions.
Algae genome projects should be increased in number with a view to understanding more in detail of biological organization, especially microalgae. Microarray tool has found to be good for a broad experimental analysis in algal studies. A lesser amount of research have been done so far in the analysis of the whole proteome or metabolome or larger parts of the algae. Therefore, initiation of computer databases should be done to store the information obtained through the experiment in a proper and systematic way.
Genome-scale metabolic reconstruction
Such modeling is used to study the metabolism involved in the reconstruction, pathway analysis and other applications thereof. This is employed for the metabolic engineering at the organism’s level, optimization or novel design of metabolic pathways and computational analysis for improving the quality of the strain. In the reconstruction process, enzymes and reactions are categorized from metabolic pathways and this is improved further in entire selected network. It is evident from literature that mathematical models embrace the metabolic information (Rudd 2000; Forster et al. 2003; Thiele and Palsson 2010; Prigent et al. 2014). The network topology has a significant role in the finding of missing gene when the coding gene is responsible for the biosynthesis of amino acid. The investigation of missing genes or annotations is a challenging task which is achieved through genome-scale metabolic reconstructions.
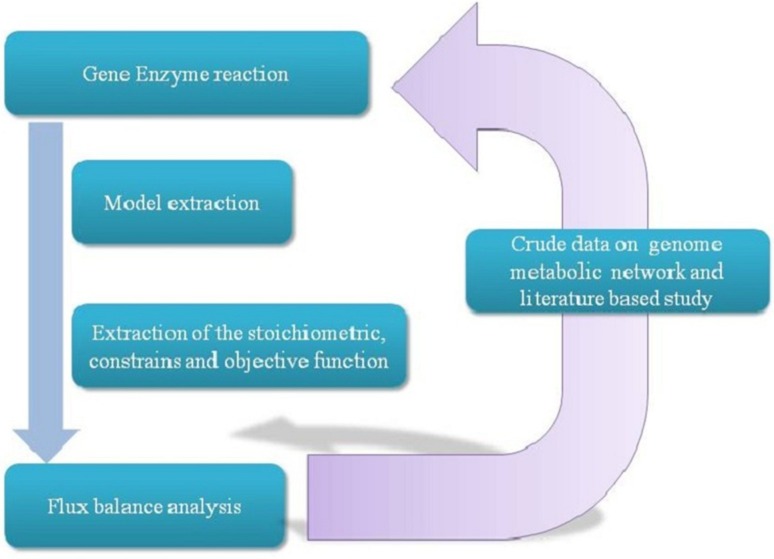
A report published by Oberhardt et al. (2009) pointed out that the range of typical genome-scale reformation of eukaryotes includes on average 1000 genes, 1200 metabolites and 1500 reactions (Oberhardt et al. 2009). Genome-scale metabolic models provide a platform to perform high-throughput computational analysis and also greatly influence the systems biology field. In silico reconstructions and successive study of genome-scale metabolic models through constraint-based modeling facilitate the identification of target gene which are to be manipulated (Lee et al. 2011; Jang et al. 2012). Such models are consistently tested in the perspective of metabolic engineering of the target microorganism (Blazeck and Alper 2010). The algae systems biology could be useful for studying the gene and metabolic network, and Chlamydomonas reconstructions represent a significant accomplishment in this aspect. Flux balance analysis demonstrates the prospective of various genome-scale models to look at composite, classified networks and for understanding the various hypotheses in this area (Walker et al. 2005). Genome-scale modeling of algae and flux balance analysis is a prevailing method for analyzing insights into the genomic and metabolomic level of an organism (Fig. 2).
Fig. 2.
The method for genome-scale model recreation. Metabolic features including relations between genes, enzymes and various reactions were recovered from the genomic metabolic records, which were then stored and curated
Metabolic engineering in algae
Genetic manipulation remains limited to a few select algal laboratory models (e.g., C. reinhardtii, Volvox carteri, and the diatom Phaeodactylum tricornutum); the increasing interest in the area of algal biofuels will likely lead to the development of techniques in other organisms and the establishment of new model systems. Algal transgenics have been previously reviewed (Neupert et al. 2008); however, the ‘molecular toolkit’ has since expanded because of recent seminal studies. Significant advances include: (a) the efficient expression of transgenes (Croft et al. 2007); (b) a novel mechanism for gene regulation in algae using riboswitches (Shao and Bock 2008); (c) inducible nuclear promoters and luciferase reporter genes (Surzycki et al. 2007; Bogen et al. 2013), and (d) inducible chloroplast gene expression (Anarat-Cappillino and Sattely 2014). Molecular toolkit or genetic tool information too are mandatory along with metabolites and metabolic pathways. The sequence analysis will include the information on metabolomics which opens the area for the metabolic flux analysis along with the development of metabolic networks (Kliebenstein 2014; Courchesne et al. 2009). Algae having a single cell which are easily cultivable can be focused toward metabolic engineering in relation to their metabolites (Hallmann and Sumper 1996). Moreover, these algae perform photosynthesis which helps in understanding the carbon assimilation with relation to its flux; this will boost the research toward bioenergy production. In a recent report, it is deciphered that an algae Volvox carteri has been genetically transformed (Doebbe et al. 2007). Similarly, trophic conversion of C. reinhardtii and P. tricornutum was also reported which is very good step toward establishing single gene change (Zaslavskaia et al. 2001; Mussgnug et al. 2007). Moreover, RNAi technology can also be used to downregulate the expression of light-harvesting antenna complexes (LHC) proteins in C. reinhardtii (Leon Banares et al. 2004; Mayfield and Franklin 2005). Thus, engineered light-harvesting strains have higher photodamage resistivity and augmented light penetration capacity. Along with this, the time span from early transformation to production level is also less compared to other mammalian-based platforms (Blatti et al. 2013). It is well established that algae is an attractive source for the production of diverse metabolites and other similar proteins. In several studies, C reinhardtii has been reported to be a good source for the production of fatty acids, biohydrogen, etc. (Paterson et al. 2009; Arruda 2012; Wu and Birch 2007). Other biofuel-related resources and their applications are presented in Table 2.
Table 2.
Other biofuel feedstocks with their applications
| Resource | Applications | References |
|---|---|---|
| Sorghum | Ideal plant for both proteomics and genomics research in cereals High tolerances to environment stress |
Liu et al. (2015) |
| Sugarcane | Highly demanded for ethanol production Abundant proteins are found which may be target for further manipulation for increased sugar content |
Mallick (2002) |
| Maize | Contains a large amount of soluble sugars and have greater biomass Proteomics of maize with respect to growth and development and stress response serve as base for using the tools and technique for stalk proteome analysis |
Matsuo et al. (2005) |
| Sugar beet | Protein presents which expression was significantly and reproducibly altered under salt stress condition | Mayfield and Franklin (2005) |
| Jatropha | Rich source of protein which contains toxic phorbol esters and anti-nutritional factors; studies are being made to detoxify make it fit for animal diet | Moellering and Benning (2010) |
Algae–bacterial interaction
Symbiotic associations of algae with bacteria were reported by various psychologists. The production of vitamins, fixation of nitrogen or the liberation of CO2 and minerals are few things which are complementary for algal growth (Wakeel et al. 2011; Kumar et al. 2013; Croft et al. 2006). Ectocarpus fasciculatus (Goecke et al. 2010), Monostroma (Wahl et al. 2012) and Ulva (Pedersen 1968) are few examples which provide evidence that algae are somehow dependent on the factors provided by bacteria. It has been reported that almost half of the algae species are probably dependent on the growth factors released by the bacteria (Matsuo et al. 2005). To understand the algae–bacterial association and evolution there is need to explore the existing tools and develop new powerful molecular biology tools.
The falling costs of sequencing and availability of potent analysis technologies has limited the algal genomics studies. If possible, each component of the holobiont is sequenced separately and then metagenomic or metatranscriptomic studies are advisable to analyze the non-cultivable organisms. The residual reads are then combined to represent such group of microorganisms or further components of the holobiont. Finally, it can be established that cautious observations of the genomic dataset is very crucial for better understanding and proper conclusions. There are certain examples of in silico studies and riboswitch engineering which could also be helpful in studying algae–microbe interaction (Pedersen 1973; Matsuo et al. 2005).
Conclusion
The interest in microalgal lipids has grown in recent years, owing to their high growth rate and productivity with less possibility of pathogenicity. Recent technologies have shown the progresses and the manipulation of one or more genes to system-based approaches. Moreover, recent applications of gene editing, novel platform designs for proteins and computational modeling can also be helpful toward improvising the production (Cock et al. 2010; Banerjee et al. 2016c, d; Gupta and Shukla 2015, 2016a, b; Shukla and Karthik 2015). Table 3 highlights the latest tools used in proteomics study and their application.
Table 3.
Different proteomics tools and their applications
| Sl. no. | Proteomics tools | Applications |
|---|---|---|
| 1 | One and 2D gel electrophoresis | To determine the comparative mass of protein and its isoelectric point |
| 2 | X-ray crystallography and nuclear magnetic resonance | To describe the 3D arrangement of peptides and proteins |
| 3 | Mass spectrometry | To identify the proteins by peptide mass fingerprinting |
| 4 | Affinity chromatography, yeast two hybrid techniques, fluorescence resonance energy transfer (FRET), and surface plasmon resonance (SPR) | To identify protein–protein and protein–DNA binding reactions |
| 5 | X-ray tomography | To identify the site of various proteins or protein complexes in an intact cell |
Genome sequencing and a suit of omics technologies which include genomics, transcriptomics, proteomics, metabolomics, and lipidomics are some of the recent technologies which have a high impact on the progresses and the manipulation of the microalgae. If these tools are used together with the view of transformation and molecular genetics toolboxes for particular algal strains, it can provide ample opportunities for researchers to redesign or construct new algal metabolism methods for the production of oils or any other chemical molecules which are useful for industrial applications. The various applications of genomic models have proved relevant in the development and hypothesis-based researches in algal metabolic engineering. Omics approaches can be used to characterize diverse biomolecules such as DNA, RNA, protein, and other relevant metabolic entities from one source sample of interest. Such modern experimentation tools balance the research gaps and offers better understanding of proteomic-based approaches in microalgae. Finally, enhanced perception of the microalgae, along with combinatory approaches of omics, will be helpful to decipher newer areas in microalgal biotechnology.
Acknowledgements
The authors acknowledge the support from the University Grants Commission (UGC), New Delhi, India (Grant No. 42-457/2013 (SR). CB greatly acknowledges the Department of Science and Technology (DST) Govt. of India for providing financial support as well as project grant from INSPIRE Faculty award scheme (DST/INSPIRE/04/2014/002322).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Vishal Anand and Chiranjib Banerjee contributed equally.
References
- Anarat-Cappillino G, Sattely ES. The chemical logic of plant natural product biosynthesis. Curr Opin Plant Biol. 2014;19:51–58. doi: 10.1016/j.pbi.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RA. Biology and systematics of heterokont and haptophyte algae. Am J Bot. 2004;91:1508–1522. doi: 10.3732/ajb.91.10.1508. [DOI] [PubMed] [Google Scholar]
- Araujo GS, Matos LJBL, Goncalves LRB, Fernandes FAN, Farias VRL. Bioprospecting for oil producing microalgal strains: evaluation of oil and biomass production for ten microalgal strains. Bioresour Technol. 2011;102:5248–5250. doi: 10.1016/j.biortech.2011.01.089. [DOI] [PubMed] [Google Scholar]
- Arruda P. Genetically modified sugarcane for bioenergy generation. Curr Opin Biotechnol. 2012;23:315–322. doi: 10.1016/j.copbio.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Baba M, Suzuki I, Shiraiwa Y. Proteomic analysis of high-CO(2)-inducible extracellular proteins in the unicellular green alga, Chlamydomonas reinhardtii. Plant Cell Physiol. 2011;52:1302–1314. doi: 10.1093/pcp/pcr078. [DOI] [PubMed] [Google Scholar]
- Banerjee C, Bandopadhyay R, Shukla P. A simple novel agar diffusion method for isolation of indigenous microalgae Chlamydomonas sp. CRP7 and Chlorella sp. CB4 from operational swampy top soil. Indian J Microbiol. 2012;52:710–712. doi: 10.1007/s12088-012-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee C, Ghosh S, Sen G, Mishra S, Shukla P, Bandopadhyay R. Study of algal biomass harvesting through cationic cassia gum, a natural plant based biopolymer. Bioresour Technol. 2014;151:6–11. doi: 10.1016/j.biortech.2013.10.035. [DOI] [PubMed] [Google Scholar]
- Banerjee C, Dubey KK, Shukla P. Metabolic engineering of microalgal based biofuel production: prospects and challenges. Front Microbiol. 2016;7:432. doi: 10.3389/fmicb.2016.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee C, Dubey KK, Shukla P. Metabolic engineering of microalgal based biofuel production: prospects and challenges. Front Microbiol. 2016;7(432):1–8. doi: 10.3389/fmicb.2016.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee C, Singh PK, Shukla P. Microalgal bioengineering for sustainable energy development: recent transgenesis and metabolic engineering strategies. Biotechnol J. 2016;11(3):303–314. doi: 10.1002/biot.201500284. [DOI] [PubMed] [Google Scholar]
- Banerjee C, Singh PK, Shukla P. Microalgal bioengineering for sustainable energy development: recent transgenesis and metabolic engineering strategies. Biotechnol J. 2016;11:303–314. doi: 10.1002/biot.201500284. [DOI] [PubMed] [Google Scholar]
- Blatti JL, Michaud J, Burkart MD. Engineering fatty acid biosynthesis in microalgae for sustainable biodiesel. Curr Opin Chem Biol. 2013;17:496–505. doi: 10.1016/j.cbpa.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Blazeck J, Alper H. Systems metabolic engineering: genome scale models and beyond. Biotechnol J. 2010;5:647–659. doi: 10.1002/biot.200900247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogen C, Al-Dilaimi A, Albersmeier A, Wichmann J, Grundmann M, Rupp O, et al. Reconstruction of the lipid metabolism for the microalga Monoraphidium neglectum from its genome sequence reveals characteristics suitable for biofuel production. BMC Genom. 2013;14:926. doi: 10.1186/1471-2164-14-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowitzka MA. Commercial production of microalgae: ponds, tanks, tubes and fermenters. J Biotechnol. 1999;70:313–321. doi: 10.1016/S0168-1656(99)00083-8. [DOI] [Google Scholar]
- Borowitzka MA. Culturing microalgae in outdoor ponds. In: Andersen RA, editor. Algal culturing techniques. Burlington: Elsevier Academic Press; 2005. pp. 205–218. [Google Scholar]
- Chaumont D. Biotechnology of algal biomass production: a review of systems for outdoor mass culture. J Appl Phycol. 1993;5:593–604. doi: 10.1007/BF02184638. [DOI] [Google Scholar]
- Chisti Y. Biodiesel from microalgae. Biotech Adv. 2007;25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouze P. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Courchesne NM, Parisien A, Wang B, Lan CQ. Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J Biotechnol. 2009;141:31–41. doi: 10.1016/j.jbiotec.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Croft MT, Warren MJ, Smith AG. Algae need their vitamins. Eukaryot Cell. 2006;5:1175. doi: 10.1128/EC.00097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebbe A, Rupprecht J, Beckmann J, Mussgnug JH, Hallman A, Hankamer B, Kruse O. Functional integration of the HUP1 hexose symporter gene into the genome of C. reinhardtii: impacts on biological H2 production. J Biotechnol. 2007;131:27–33. doi: 10.1016/j.jbiotec.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Ermakova M, Battchikova N, Allahverdiyeva Y, Aro EM. Novel heterocyst-specific flavodiiron proteins in Anabaena sp. PCC 7120. FEBS Lett. 2013;587:82–87. doi: 10.1016/j.febslet.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Raven JA. Aquatic photosynthesis. London: Blackwater Science; 1997. p. 375. [Google Scholar]
- Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahoi N, Ray S, Srivastava S. Array-based proteomic approaches to study signal transduction pathways: prospects, merits and challenges. Proteomics. 2015;15:218–231. doi: 10.1002/pmic.201400261. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George PM, Mlynash M, Adams CM, Kuo CJ, Albers GW, Olivot JM. Novel TIA biomarkers identified by mass spectrometry-based proteomics. Int J Stroke. 2015;10:1204–1211. doi: 10.1111/ijs.12293. [DOI] [PubMed] [Google Scholar]
- Goecke F, Labes A, Wiese J, Imhoff J. Chemical interactions between marine macroalgae and bacteria. Mar Ecol Prog Ser. 2010;409:267. doi: 10.3354/meps08607. [DOI] [Google Scholar]
- Griffiths MJ, Harrison STL. Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J Appl Phycol. 2009;21:493–507. doi: 10.1007/s10811-008-9392-7. [DOI] [Google Scholar]
- Gupta SK, Shukla P. Advanced technologies for improved expression of recombinant proteins in bacteria: perspectives and applications. Crit Rev Biotechnol. 2015;36(6):1089–1098. doi: 10.3109/07388551.2015.1084264. [DOI] [PubMed] [Google Scholar]
- Gupta S, Shukla P. Gene editing for cell engineering: trends and applications. Crit Rev Biotechnol. 2016;18:1–13. doi: 10.1080/07388551.2016.1214557. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Shukla P. Bacterial platform technology for recombinant antibody fragment production: a review. Crit Rev Microbiol. 2016;43:31–42. doi: 10.3109/1040841X.2016.1150959. [DOI] [PubMed] [Google Scholar]
- Hallmann A, Sumper M. The Chlorella hexose/H+ symporter is a useful selectable and biochemical reagent when expressed in Volvox. Proc Natl Acad Sci USA. 1996;93:669–673. doi: 10.1073/pnas.93.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranen M, Battchikova N, Zhang P, Graf A, Sirpio S, Paakkarinen V, Aro EM. Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol. 2004;134:470–481. doi: 10.1104/pp.103.032326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippler M, Klein J, Fink A, Allinger T, Hoerth P. Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 2001;28:595–606. doi: 10.1046/j.1365-313X.2001.01175.x. [DOI] [PubMed] [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 2008;54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- Huang F, Parmryd I, Nilsson F, Persson AL, Pakrasi HB, et al. Proteomics of Synechocystis sp. strain PCC 6803: identification of plasma membrane proteins. Mol Cell Proteomics. 2002;1:956–966. doi: 10.1074/mcp.M200043-MCP200. [DOI] [PubMed] [Google Scholar]
- James GO, Hocart CH, Hillier W, Chen H, Kordbacheh F, Price GD, Djordjevic MA. Fatty acid profiling of Chlamydomonas reinhardtii under nitrogen deprivation. Bioresour Technol. 2011;102:3343–3351. doi: 10.1016/j.biortech.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Jang YS, Lee J, Malaviya A, do Seung Y, Cho JH, Lee SY. Butanol production from renewable biomass: rediscovery of metabolic pathways and metabolic engineering. Biotechnol J. 2012;7:186–198. doi: 10.1002/biot.201100059. [DOI] [PubMed] [Google Scholar]
- Khan SA, Rashmi Hussain MZ, Prasad S, Banerjee UC. Prospects of biodiesel production from microalgae in India. Renew Sust Energ Rev. 2009;13:2361–2372. doi: 10.1016/j.rser.2009.04.005. [DOI] [Google Scholar]
- Kliebenstein DJ. Synthetic biology of metabolism: using natural variation to reverse engineer systems. Curr Opin Plant Biol. 2014;19:20–26. doi: 10.1016/j.pbi.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Kruse O, Rupprecht J, Bader K, Thomas-Hall S, Schenk PM, Finazzi G, Hankamer B. Improved photobiological H2 production in engineered green algal cells. J Biol Chem. 2005;280:34170–34177. doi: 10.1074/jbc.M503840200. [DOI] [PubMed] [Google Scholar]
- Kumar A, Yuan X, Sahu AK, Dewulf J, Ergas SJ, Van Langenhove H. A hollow fiber membrane photo-bioreactor for CO2 sequestration from combustion gas coupled with wastewater treatment: a process engineering approach. J Chem Technol Biotechnol. 2010;85:387–394. doi: 10.1002/jctb.2332. [DOI] [Google Scholar]
- Kumar GRK, Bapat VA, Johnson TS. Phorbol esters and other toxic constituents of Jatropha curcas L. In: Carels N, editor. Jatropha, challenges for a new energy crop. NY: Springer Science; 2013. pp. 441–460. [Google Scholar]
- Kumar R, Biswas K, Singh PK, Singh PK, Elumalai S, Shukla P, Pabbi S. Lipid production and molecular dynamics simulation for regulation of accD gene in cyanobacteria under different N and P regimes. Biotechnol Biofuels. 2017;10:94. doi: 10.1186/s13068-017-0776-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RE. Phycology. New York: Cambridge University Press; 1980. [Google Scholar]
- Lee JW, Kim TY, Jang YS, Choi S, Lee SY. Systems metabolic engineering for chemicals and materials. Trends Biotechnol. 2011;29:370–378. doi: 10.1016/j.tibtech.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Leon Banares R, Gonzalez Ballester D, Galvan A, Fernandez E. Transgenic microalgae as green cell factories. Trends Biotechnol. 2004;22:45–52. doi: 10.1016/j.tibtech.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Liang W, Wang L, Zhang Y, Lei X, Yang J, You X, Cheng C-L, Zhou Y, Wei C. Comparative proteomic and physiological analysis of diurnal changes in Nostoc flagelliforme. J Appl Phycol. 2013;25:1709–1721. doi: 10.1007/s10811-013-0032-5. [DOI] [Google Scholar]
- Liu H, Li L, Voss C, Wang F, Liu J, Li SS. A comprehensive immunoreceptor phosphotyrosine-based signaling network revealed by reciprocal protein-peptide array screening. Mol Cell Proteomics. 2015;14:1846–1858. doi: 10.1074/mcp.M115.047951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick K. Biotechnological potential of immobilized algae for wastewater N, P and metal removal: a review. Biometals. 2002;15:377–390. doi: 10.1023/A:1020238520948. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Imagawa H, Nishizawa M, Shizuri Y. Isolation of an algal morphogenesis inducer from a marine bacterium. Science. 2005;307:1598. doi: 10.1126/science.1105486. [DOI] [PubMed] [Google Scholar]
- Mayfield SP, Franklin SE. Expression of human antibodies in eukaryotic micro-algae. Vaccine. 2005;23:1828–1832. doi: 10.1016/j.vaccine.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Moellering ER, Benning C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell. 2010;9:97–106. doi: 10.1128/EC.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussgnug JH, Thomas-Hall S, Rupprecht J, Foo A, Klassen V, McDowal IA, et al. Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnol J. 2007;5:802–814. doi: 10.1111/j.1467-7652.2007.00285.x. [DOI] [PubMed] [Google Scholar]
- Neupert J, Karcher D, Bock R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2008;57:1140–1150. doi: 10.1111/j.1365-313X.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Eppig JT. A mouse phenome project. Mamm Genome. 2000;11:715–717. doi: 10.1007/s003350010152. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Pedersen M. Ectocarpus fasciculatus: marine brown alga requiring kinetin. Nature. 1968;218:776. doi: 10.1038/218776a0. [DOI] [Google Scholar]
- Pedersen M. Identification of a cytokinin, 6-(3 methyl-2- butenylamino) purine in sea water and the effect of cytokinins on brown algae. Physiol Plantarum. 1973;28:101. doi: 10.1111/j.1399-3054.1973.tb01158.x. [DOI] [Google Scholar]
- Pisareva T, Shumskaya M, Maddalo G, Ilag L, Norling B. Proteomics of Synechocystis sp. PCC 6803. Identification of novel integral plasma membrane proteins. FEBS J. 2007;274:791–804. doi: 10.1111/j.1742-4658.2006.05624.x. [DOI] [PubMed] [Google Scholar]
- Prigent S, Collet G, Dittami SM, Delage L, de Corny FE, Dameron O, et al. The genomescale metabolic network of Ectocarpus siliculosus (EctoGEM): a resource to study brown algal physiology and beyond. Plant J. 2014;80:367–381. doi: 10.1111/tpj.12627. [DOI] [PubMed] [Google Scholar]
- Pulz O. Photobioreactors: production systems for phototrophic microorganisms. Appl Microbiol Biotechnol. 2001;57:287–293. doi: 10.1007/s002530100702. [DOI] [PubMed] [Google Scholar]
- Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: proteins associated with invasion and infection of the mammalian host. Mol Cell Proteomics. 2009;8:1891–1907. doi: 10.1074/mcp.M900045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd KE. EcoGene: a genome sequence database for Escherichia coli K-12. Nucl Acids Res. 2000;28:60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Thomas-Hall SR, Stephens E, Marx U, Mussgnug JH, Posten C, Kruse O, Hankamer B. Second generation biofuels: high-efficiency microalgae for biodiesel production. Bioenerg Res. 2008;1:20–43. doi: 10.1007/s12155-008-9008-8. [DOI] [Google Scholar]
- Shao N, Bock R. A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr Genet. 2008;53:381–388. doi: 10.1007/s00294-008-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P, Karthik MVK (2015) Computational approaches in Chlamydomonas reinhardtii for effectual bio-hydrogen production. Springer, New York
- Singh PK, Shukla P. Systems biology as an approach for deciphering microbial interactions. Brief Funct Genom. 2015;14:166–168. doi: 10.1093/bfgp/elu023. [DOI] [PubMed] [Google Scholar]
- Stauber EJ, Fink A, Markert C, Kruse O, Johanningmeier U, et al. Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot Cell. 2003;2:978–994. doi: 10.1128/EC.2.5.978-994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surzycki R, Cournac L, Peltier G, Rochaix JD. Potential for hydrogen production with inducible chloroplast gene expression in Chlamydomonas. Proc Natl Acad Sci USA. 2007;104:17548–17553. doi: 10.1073/pnas.0704205104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I, Palsson BO. Reconstruction annotation jamborees: a community approach to systems biology. Mol Syst Biol. 2010;6:361. doi: 10.1038/msb.2010.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lis R, Atteia A, Mendoza-Hernandez G, Gonzalez-Halphen D. Identification of novel mitochondrial protein components of Chlamydomonas reinhardtii. A proteomic approach. Plant Physiol. 2003;132:318–330. doi: 10.1104/pp.102.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M, Goecke F, Labes A, Dobretsov S, Weinberger F. The second skin: ecological role of epibiotic biofilms on marine organisms. Front Microbiol. 2012;3:292. doi: 10.3389/fmicb.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeel A, Asif AR, Pitann B, Schubert S. Proteome analysis of sugar beet (Beta vulgaris L.) elucidates constitutive adaptation during the first phase of salt stress. J Plant Physiol. 2011;168:519–526. doi: 10.1016/j.jplph.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Walker TL, Collet C, Purton S. Algal transgenics in the genomic era. J Phycol. 2005;41:1077–1093. doi: 10.1111/j.1529-8817.2005.00133.x. [DOI] [Google Scholar]
- Wu L, Birch RG. Doubled sugar content in sugarcane plants modified to produce a sucrose isomer. Plant Biotechnol J. 2007;5:109–117. doi: 10.1111/j.1467-7652.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- Zaslavskaia LA, Lippmeier JC, Shih C, Ehehardt D, Grossman AR, Apt KE. Trophic conversion of an obligate photoautotrophic organism through metabolic engineering. Science. 2001;292:2073–2075. doi: 10.1126/science.160015. [DOI] [PubMed] [Google Scholar]
- Zilinskas Braun G, Zilinskas Braun B. Light absorption, emission and photosynthesis. In: Stewart WDP, editor. Algal physiology and biochemistry. Oxford: Oxford Blackwell Scientific Publications; 1974. [Google Scholar]