Abstract
Although both osteoblasts and adipocytes have a common origin, i.e., mesenchymal cells, the molecular mechanisms that define the direction of two different lineages are presently unknown. In this study, we investigated the role of a transcription factor, CCAAT/enhancer binding protein β (C/EBPβ), and its isoform in the regulation of balance between osteoblast and adipocyte differentiation. We found that C/EBPβ, which is induced along with osteoblast differentiation, promotes the differentiation of mesenchymal cells into an osteoblast lineage in cooperation with Runx2, an essential transcription factor for osteogenesis. Surprisingly, an isoform of C/EBPβ, liver-enriched inhibitory protein (LIP), which lacks the transcriptional activation domain, stimulates transcriptional activity and the osteogenic action of Runx2, although LIP inhibits adipogenesis in a dominant-negative fashion. Furthermore, LIP physically associates with Runx2 and binds to the C/EBP binding element present in the osteocalcin gene promoter. These data indicate that LIP functions as a coactivator for Runx2 and preferentially promotes the osteoblast differentiation of mesenchymal cells. Thus, identification of a novel role of the C/EBPβ isoform provides insight into the molecular basis of the regulation of osteoblast and adipocyte commitment.
Accelerated adipogenesis in bone marrow, namely fatty marrow, is often observed in patients with osteoporosis in association with bone loss (7). Because osteoblasts that play a central role in bone formation are derived from undifferentiated mesenchymal cells which have the capacity to differentiate into adipocytes (34), it is likely that balance between osteoblastogenesis and adipogenesis is critical to the maintenance of bone volume. Recent studies revealed that osteoblast differentiation and bone formation are regulated by critical transcription factors, Runx2 and Osterix (13, 44). Runx2 directly regulates osteoblast-specific genes, including osteocalcin, osteopontin, and type I collagen, through its specific DNA binding element, OSE2 (14), and ectopic expression of Runx2 induces osteoblast differentiation of mesenchymal cells (29). Mice deficient in Runx2 show no bone formation (24), and the inherited mutations of the Runx2 gene in humans cause cleidocranial dysplasia, characterized by severely impaired osteogenesis (27). Target disruption of the Osterix gene in mice results in abnormal skeletogenesis with a complete lack of osteogenesis (28). These findings indicate that Runx2 and Osterix are essential transcription factors for osteoblast differentiation. On the other hand, the program of adipocyte differentiation is sequentially regulated by CCAAT/enhancer binding protein (C/EBP) family members and peroxisome proliferator-activating protein gamma (PPARγ) (37). A recent study showed that PPARγ insufficiency enhances osteogenesis by stimulating osteoblast differentiation of bone marrow progenitors (2). Consistently, the introduction of PPARγ2 or activation of PPARγ by treatment with its ligand inhibits Runx2 expression and osteoblast differentiation (25). Moreover, impairment of Runx2 expression increases adipocyte differentiation (16, 23). These studies suggest that the relative expression level of Runx2 and PPARγ is involved in the regulation of balance between osteoblastogenesis and adipogenesis.
C/EBPβ, which belongs to the leucine zipper family of transcription factors, forms a homodimer or heterodimer complex with other C/EBP family members (11). Overexpression of C/EBPβ in mesenchymal cells induces PPARγ expression and adipocyte differentiation in cooperation with C/EBPδ (43). C/EBPβ-deficient mice manifest the reduction in adipogenesis (41). These biochemical and genetic studies indicate that C/EBPβ plays contributive roles in the early stages of adipogenesis by regulating transcription of the genes necessary for adipocytic differentiation, such as the PPARγ gene (11, 41). Interestingly, it has been described that C/EBPβ is expressed in the osteoblastic cell lineages and up-regulated during osteoblast differentiation (3, 17, 32, 35). Gutierrez et al. also showed that C/EBPβ and C/EBPδ regulate the osteocalcin gene promoter through physical interaction with Runx2 (17). Collectively, these studies suggest the importance of C/EBPβ during osteoblast differentiation. In contrast, it has been reported that overexpression of C/EBPβ stimulates the proliferation of a mouse osteoblast cell line, MC3T3-E1, but suppresses its osteoblast differentiation (21). Therefore, it remains unclear whether C/EBPβ is implicated in osteoblast differentiation of mesenchymal cells.
An isoform of C/EBPβ, liver-enriched inhibitory protein (LIP), which is translated from the same mRNA of C/EBPβ (12), has been shown to play a unique role in several tissues (4, 6, 15, 39). Since LIP lacks a transcriptional activation domain, it usually functions as a transcriptional repressor (12). Likewise, LIP markedly inhibits the transcriptional function of C/EBPβ, thereby suppressing adipocyte differentiation (8). Consistently, LIP expression is negatively regulated during the adipocyte differentiation of mesenchymal cells (42). Therefore, it is most likely that the ration of C/EBPβ-LIP is a critical factor for biological phenomena that are controlled by C/EBPβ.
To understand the molecular basis that regulates the balance between osteoblastogenesis and adipogenesis, we investigated the role of C/EBPβ and LIP in osteoblast differentiation and adipocyte differentiation. In the present study, we found that C/EBPβ promotes osteoblast differentiation of mesenchymal cells in Runx2-dependent and -independent mechanisms. Moreover, our results indicate that LIP functions as a coactivator for Runx2 and stimulates osteoblastic differentiation of mesenchymal cells. Thus, these findings provide support for the notion that LIP is an important key regulator that determines the commitment of osteoblasts and adipocytes.
MATERIALS AND METHODS
Cell and antibodies.
C3H10T1/2, C2C12, ST2, and Cos7 cells were purchased from the RIKEN gene bank and cultured in alpha-modified minimal essential medium containing 10% fetal bovine serum. Anti-Runx2 and anti-C/EBPβ antibodies were purchased from Santa Cruz Biotechnology and Oncogene Science. The anti-C/EBPβ antibody recognizes LIP as well as C/EBPβ.
Isolation of primary osteoblasts and mesenchymal cells.
The calvaria were isolated from 2- or 3-day-old neonatal mice and digested with 0.1% collagenase and 0.2% dispase for 7 min at 37°C, and then the cells collected by centrifugation were used as the primary mesenchymal cells. These cells contained few alkaline phosphatase (ALP)-positive cells. The digested calvaria were sequentially digested four times with 0.1% collagenase and 0.2% dispase for 7 min at 37°C, and the last three fractionated cells were collected and used as the primary osteoblasts.
Constructs and transfection.
C/EBPβ and C/EBPδ (41) cDNA were kindly given by Shizuo Akira. To generate the LIP construct, a PCR product which contains the C-terminal domain of C/EBPβ (amino acids 152 through 296) was synthesized and then subcloned into pcDNA3 (Invitrogen) tagged with a Flag or Myc epitope in the N terminus. Deletion mutants of LIP, LIP(Δ73-145), and LIP(Δ1-70) were generated by subcloning the corresponding PCR products into pcDNA3 tagged with a Myc epitope in the N terminus. The sequences of the constructs were confirmed by DNA sequence analysis. Runx2/Cbfa1 (22) and Cbfβ (5) cDNA were kindly given by Yoshiaki Ito and Paul Liu, respectively. Dominant-negative mutant Runx2 was constructed as previously described (29). The Flag-tagged Runx2 mutant constructs Runx2(Δ242-513) and Runx2(Δ1-207) were generated by subcloning the corresponding PCR products into pcDNA3 tagged with a Flag epitope in the N terminus. The sequences of the constructs were confirmed by DNA sequence analysis. Transfection of C3H10T1/2 cells was carried out using FuGENE6 (Roche) according to the manufacturer's protocol.
Generation of adenovirus.
The recombinant adenovirus carrying a wild-type or mutant form of C/EBPβ or Runx2 was constructed by homologous recombination between the expression cosmid cassette (pAxCAwt) and the parental virus genome in 293 cells (RIKEN cell bank) by using an adenovirus construction kit (Takara) as previously described (19). The viruses showed no proliferative activity due to a lack of E1A-E1B (26). Titers of the viruses were determined by a modified point assay (26). Infection of C3H10T1/2, C2C12, or ST2 cells, primary osteoblasts, or mesenchymal cells with recombinant adenoviruses was performed by incubation with adenoviruses at a multiplicity of infection (MOI) of 50 except where specifically indicated.
Immunoprecipitation and Western blotting.
The cells were washed three times with ice-cold phosphate-buffered saline (PBS) and solubilized in lysis buffer (20 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM EGTA, 1.5 mM MgCl2, 10% glycerol, 1% Triton X-100, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.2 mM sodium orthovanadate) (19). The lysates were centrifuged for 15 min at 4°C at 16,000 × g and incubated with antibodies for 4 h at 4°C, followed by immunoprecipitation with protein A-Sepharose (Zymed) or protein G-agarose (Roche). Immunoprecipitates were washed five times with lysis buffer and boiled in sodium dodecyl sulfate (SDS) sample buffer, and supernatants were recovered as immunoprecipitate samples. These samples were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, immunoblotted with corresponding antibodies, and visualized with horseradish peroxidase coupled to protein A (KPL) or horseradish peroxidase coupled to anti-mouse immunoglobulin G (IgG) antibodies (Cappel) with an enhancement by use of ECL detection kits (Amersham).
Immunocytochemistry.
Cultured cells were washed three times with ice-cold PBS and fixed with 3.7% paraformaldehyde-PBS for 20 min. After a 20-min incubation with 0.1% Triton X-100-PBS, the cells were blocked with PBS containing 1% bovine serum albumin for 2 h, incubated with anti-C/EBPβ polyclonal antibodies in 1% bovine serum albumin-PBS, washed six times with PBS, and incubated with fluorescein isothiocyanate-conjugated affinity-purified anti-rabbit IgG antibodies (Jackson Laboratory). The cells were washed extensively with PBS and visualized with a fluorescence microscope (Zeiss).
Luciferase assay.
A total of 1.3 kb of the mouse osteocalcin gene promoter, which contains the OSE2 site (positions −137 to −131), was used as described previously (13). A mouse osteocalcin gene promoter construct which lacks a putative C/EBP binding element (positions −454 to −446) was generated by subcloning the corresponding PCR products. The osteocalcin gene promoter fused to firefly luciferase was cotransfected with the Runx2 and Cbfβ expression vector and a thymidine kinase (TK)-renilla luciferase construct (Promega) into C3H10T1/2 cells. Two days after transfection, the cells were lysed and luciferase activity was determined by using specific substrates in a luminometer (Promega) according to the manufacturer's protocol. Transfection efficiency was normalized by determining the activity of renilla luciferase.
Reverse transcriptase-PCR (RT-PCR).
After denaturation of total RNA at 70°C for 10 min, cDNA was synthesized with the oligo-dT primer and reverse transcriptase (Gibco). PCR amplification was performed by using the specific primers for mouse osteocalcin (sense primer, 5′-GACAAAGCCTTCATGTCCAAGC-3′; antisense primer, 5′-AAAGCCGAGCTGCCAGAGTTTG-3′), ALP (sense primer, 5′-GCTGATCATTCCCACGTTTT-3′; antisense primer, 5′-CTGGGCCTGGTAGTTGTTGT-3′), type IA collagen (sense primer, 5′-CCTGGTAAAGATGGTGCC-3′; antisense primer, 5′-CACCAGGTTCACCTTTCGCACC-3′), or Osterix (sense primer, 5′-GAAAGGAGGCACAAAGAAG-3′; antisense primer, 5′-CACCAAGGAGTAGGTGTGTT-3′). PCR products were loaded onto an agarose gel and stained with ethidium bromide. After the PCR products were subcloned into a TA cloning vector, DNA sequences of the PCR products were determined.
Oligonucleotide precipitation assay.
Cells were lysed in lysis buffer (10 mM HEPES [pH 7.9], 100 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, 0.5% NP-40, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 1 mM PMSF, 0.2 mM sodium orthovanadate). The lysates preincubated with streptavidin-agarose beads (Vector) were incubated with 1 mg of a biotinylated double-stranded oligonucleotide probe that contains three repeats of C/EBP binding element present in the osteocalcin gene promoter (sense primer, 5′-GATCGGACATTACTGAACACTACGGGACATTACTGAACACTCCCGGGACATTACTGAACACT-3′; antisense primer, 5′-GATCAGTGTTCAGTAATGTCCCGGGAGTGTTCAGTAATGTCCCGTAGTGTTCAGTAATGTCC-3′) and 10 mg of poly(dI-dC) · poly(dI-dC) for 16 h. DNA-bound proteins were collected with streptavidin-agarose beads for 1 h, washed with the lysis buffer, separated on an SDS-polyacrylamide gel, and identified by Western blotting.
Chromatin immunoprecipitation analysis.
C3H10T1/2 cells were cross-linked with 1% formaldehyde for 15 min at room temperature and washed three times with ice-cold PBS containing 1 mM PMSF and 1 μg of aprotinin/ml. The cells were collected by scraping and centrifugation and lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8], 1 mM PMSF, 1 μg of aprotinin/ml). The lysates were vortexed and sonicated with a Bioruptor (Cosmo Bio). The average length of DNA fragments ranged between 300 and 800 bp. The lysates were then clarified by centrifugation and diluted fivefold in chromatin immunoprecipitation buffer (15 mM Tris [pH 8], 1% Triton X-100, 0.01% SDS, 1 mM EDTA, 150 mM NaCl, 1 mM PMSF, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml). The samples were precleared using protein G-Sepharose beads for 1 h at 4°C. Next, 0.1, 0.5, and 1% of each sample were used for input control. The samples were then immunoprecipitated with anti-Flag antibody or control IgG, and the immune complexes were washed with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8], 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8], 500 mM NaCl), LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8]), and TE buffer (10 mM Tris-Hcl [pH 8], 1 mM EDTA) and eluted with elution buffer (50 mM Tris-HCl [pH 8], 10 mM EDTA, 1% SDS). Immunoprecipitated DNA was reverse cross-linked at 65°C for 12 h and purified using a PCR purification kit (QIAGEN). A total of 5 μl of the purified DNA was subjected to PCR amplification using the specific primer for the C/EBP binding element present in the osteocalcin gene (sense primer, 5′-TGCCCTACAACCGGATCTTA-3′; antisense primer, 5′-AAACTGGGCTCCAACTCTCA-3′).
Determination of ALP activity.
ALP activity was determined as described previously (31). C3H10T1/2, C2C12, or ST2 cells were washed with PBS and lysed with 0.05% Triton X-100 solution. The ALP activities of the lysates were determined using p-nitrophenol-phosphate as a substrate. Protein contents of the lysates were measured by use of the Bradford protein assay reagent (Bio-Rad). For cytochemical analysis, cells were washed with PBS, fixed with 3.7% formaldehyde, and stained with a mixture of 330 μg of Nitro Blue Tetrazolium/ml, 165 μg of bromochloroindolyl phosphate/ml, 100 mM NaCl, 5 mM MgCl2, and 100 mM Tris (pH 9.5).
Oil red O staining.
C3H10T1/2 cells were washed with PBS and fixed with 10% formalin for 20 min. After the cells were washed twice with PBS and once with 60% isopropyl alcohol, they were stained with Oil red O solution (Sigma). The area of the cells stained with Oil red O was measured with an Image Pro Plus analyzer (Palmerton, Inc.).
Determination of osteocalcin production.
Osteocalcin production in the culture media was determined by using a mouse osteocalcin enzyme immunoassay kit (Biomedical Technologies, Inc.) according to the manufacturer's protocol.
Statistical analysis.
All data were analyzed by analysis of variance, followed by a paired t test. Values are given as means ± standard deviations.
RESULTS
Promotion of osteoblast differentiation by C/EBPβ.
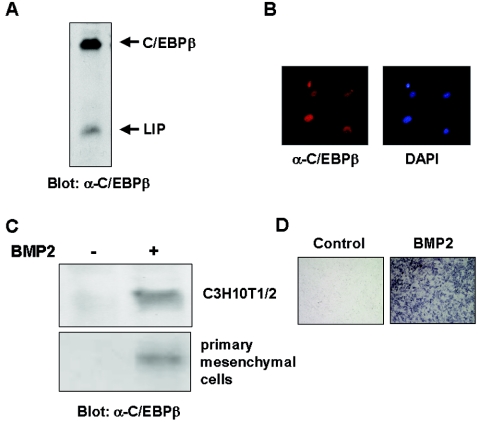
To examine whether C/EBPβ is involved in the regulation of osteoblast differentiation, we first determined the expression of C/EBPβ in mouse primary osteoblasts by Western blot analysis. As shown in Fig. 1A, primary osteoblasts constitutively expressed C/EBPβ. Immunocytochemical analysis using the anti-C/EBPβ antibody showed nuclear localization of C/EBPβ in primary osteoblasts (Fig. 1B). To examine whether C/EBPβ expression is associated with osteoblast differentiation, we determined the C/EBPβ expression in a multipotent mesenchymal cell line, C3H10T1/2. C3H10T1/2 cells faintly expressed C/EBPβ in an undifferentiated fashion (Fig. 1C). In contrast, when treated with bone morphogenetic protein 2 (BMP2), the cells showed distinct expression of C/EBPβ (Fig. 1C) and induction of ALP activity (Fig. 1D), which is a well-known phenotypic marker for an osteoblast lineage (30). In addition, BMP2 also induced C/EBPβ expression in primary mesenchymal cells (Fig. 1C). These results, consistent with those of previous studies (3, 35), demonstrated that C/EBPβ expression is induced along with osteoblast differentiation of mesenchymal cells and suggested that C/EBPβ is involved in the regulation of osteoblast differentiation.
FIG. 1.
C/EBPβ expression associated with osteoblastogenesis. Expression of C/EBPβ and its isoform, LIP, in mouse primary osteoblasts as determined by immunoblotting analysis (A) and immunochytochemical analysis (B) is shown. C/EBPβ expression (C) and ALP activity (D) of C3H10T1/2 or primary mesenchymal cells cultured with or without BMP2 for 7 days are also shown.
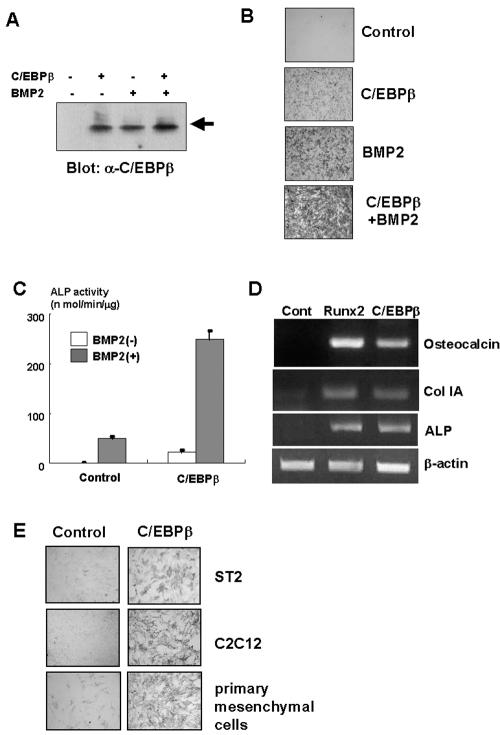
To investigate the role of C/EBPβ in the regulation of osteoblast differentiation of mesenchymal cells, we examined the effects of C/EBPβ overexpression on osteoblast differentiation by using an adenovirus system (Fig. 2A), which is one of the most powerful and efficient gene delivery systems (19, 20, 26, 29). As shown in Fig. 2B and C, ALP activity was induced in C3H10T1/2 cells overexpressing C/EBPβ. Of interest, this effect of C/EBPβ was markedly enhanced in the presence of BMP2 (Fig. 2B and C). Furthermore, overexpression of C/EBPβ induced osteocalcin, type IA collagen, and ALP expression in C3H10T1/2 cells (Fig. 2D). Consistent with these data, overexpression of C/EBPβ markedly increased ALP activity in C2C12, ST2, and primary mesenchymal cells isolated from mouse calvaria (Fig. 2E). These data indicate that C/EBPβ promotes the differentiation of mesenchymal cells toward an osteoblast lineage.
FIG. 2.
C/EBPβ promotes osteoblast differentiation of mesenchymal cells. (A through C) C3H10T1/2 cells were infected with control or C/EBPβ adenovirus at an MOI of 40 and incubated with BMP2 for 7 days. The cells were determined by immunoblotting (A), ALP staining (B), or ALP activity (C). (D) C3H10T1/2 cells were infected with control, Runx2, or C/EBPβ adenovirus at an MOI of 40 and cultured for 7 days. Osteocalcin, type IA collagen (Col IA), or ALP expression in the cells was assessed by RT-PCR. (E) ST2, C2C12, and mouse primary mesenchymal cells were infected with control or C/EBPβ adenovirus at an MOI of 40 and incubated for 7 days. The cells were subjected to ALP staining.
Functional and physical interaction of C/EBPβ with Runx2 during osteoblastogenesis.
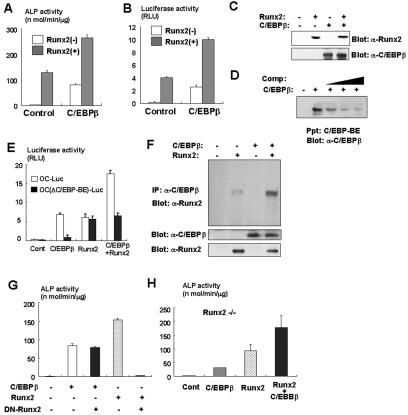
Because we observed that treatment with BMP2 enhances C/EBPβ-induced osteoblast differentiation (Fig. 2B and C), it is possible that C/EBPβ regulates osteoblast differentiation in cooperation with BMP2 signaling. In this context, Runx2 has been shown to play an important role in the BMP2-regulated osteoblast differentiation of mesenchymal cells (29) and to physically associate with C/EBPβ and C/EBPδ (17). We therefore examined whether C/EBPβ functionally collaborates with Runx2 in the regulation of osteoblast differentiation. As previously reported (14, 29), overexpression of Runx2 induces ALP activity (Fig. 3A) and osteocalcin, type IA collagen, and ALP expression in C3H10T1/2 cells (Fig. 2D). When C/EBPβ and Runx2 were cooverexpressed in C3H10T1/2 cells, ALP activity was significantly increased (Fig. 3A). These data show that C/EBPβ and Runx2 functionally collaborate in promotion of osteoblast differentiation of mesenchymal cells. To verify the cooperative role of C/EBPβ and Runx2, we next performed a reporter assay using the osteocalcin gene promoter, which is directly regulated by Runx2 though the specific DNA binding element OSE2 (14). As described previously (17), overexpression of C/EBPβ significantly increases osteocalcin gene promoter activity in the absence of Runx2 (Fig. 3B) and further enhances its activity in the presence of Runx2 (Fig. 3B). We have confirmed that C/EBPβ overexpression does not induce Runx2 expression in C3H10T1/2 cells (Fig. 3C). Since there is a consensus sequence of C/EBP binding element in the osteocalcin gene promoter, we determined whether C/EBPβ binds to the element using a biotinylated oligonucleotide probe. As shown in Fig. 3D, C/EBPβ binds to the element. The deletion of the element suppressed C/EBPβ-induced osteocalcin gene promoter activity, whereas the deletion did not affect Runx2-induced promoter activity (Fig. 3E). These data indicate that C/EBPβ regulates the osteocalcin gene promoter through association with its DNA binding element. Furthermore, a coimmunoprecipitation experiment confirmed that C/EBPβ physically associates with Runx2 (Fig. 3F), as shown previously (17). Together with the cooperative effects of C/EBPβ and Runx2 on ALP activity, these results indicate that C/EBPβ controls osteoblast differentiation of mesenchymal cells through a physical association with Runx2.
FIG. 3.
C/EBPβ regulates osteoblastogenesis in a Runx2-dependent and -independent fashion. (A) C3H10T1/2 cells were infected with control or C/EBPβ adenovirus with or without Runx2 adenovirus and cultured for 7 days. ALP activity in the cells was measured. (B) Osteocalcin gene promoter fused to the luciferase reporter construct and TK-renilla reporter constructs were transfected into C3H10T1/2 cells together with pcDNA3 (control), the C/EBPβ expression vector, the Runx2 expression vector, or both vectors. Luciferase activity of the cell lysates was measured in relative light units (RLU). (C) The lysates of C3H10T1/2 cells infected with control or C/EBPβ adenovirus together with or without Runx2 adenovirus were examined by immunoblotting with anti-Runx2 or C/EBPβ antibody. (D) The lysates of C3H10T1/2 cells infected with control or C/EBPβ adenovirus were incubated with a biotinylated probe containing the C/EBP binding element (BE) in osteocalcin gene promoter in the presence or absence of unbiotinylated probe (Comp). Associated protein with biotinylated probe was determined by immunoblotting with anti-C/EBPβ antibody. Ppt, precipitation. (E) Luciferase reporter construct-fused osteocalcin gene promoter containing osteocalcin luciferase (OC-Luc) or lacking the C/EBP binding element [OC(ΔC/EBP-BE)-Luc] and the TK-renilla reporter construct were transfected into C3H10T1/2 cells together with pcDNA3 (control), the C/EBPβ expression vector, the Runx2 expression vector, or both vectors. Luciferase activity of the cell lysates was measured. (F) C3H10T1/2 cells were infected with the control, C/EBPβ adenovirus, Runx2 adenovirus, or both adenovirus strains. The cell lysates were immunoprecipitated (IP) with anti-C/EBPβ antibody, and immunoprecipitates were determined by immunoblotting with anti-Runx2 antibody. (G) C3T10T1/2 cells were infected with adenoviruses as indicated in the text and cultured for 7 days. ALP activity in the cells was measured. (H) Mesenchymal cells isolated from Runx2-deficient mice were infected with adenoviruses as indicated in the text and cultured for 7 days. ALP activity of the cells was measured.
Runx2-independent osteoblast differentiation by C/EBPβ.
Komori et al. described that ALP-positive immature osteoblast-like cells are present in Runx2-deficient cells (24). This finding suggests that a certain transcription factor(s) might be involved in the regulation of osteoblast differentiation in the absence of Runx2. Because we observed that C/EBPβ overexpression induces ALP activity in the absence of BMP2 or Runx2 (Fig. 2B and C and 3A), we asked whether C/EBPβ has the ability to promote the differentiation of mesenchymal cells into osteoblastic cells in an Runx2-independent manner. To test this, we determined the effects of dominant-negative Runx2 (29) on ALP activity induced by C/EBPβ. As shown in Fig. 3G, overexpression of dominant-negative Runx2 did not affect ALP activity induced by C/EBPβ. These data suggested that C/EBPβ can promote osteoblast differentiation independently, at least in part, of Runx2. To further confirm the Runx2-independent action of C/EBPβ, we determined the effects of C/EBPβ on mesenchymal cells isolated from Runx2-deficient mice (24). In Runx2-deficient mesenchymal cells, C/EBPβ could induce ALP activity even in the absence of BMP2 and Runx2 (Fig. 3H). This effect of C/EBPβ was enhanced by the introduction of Runx2 (Fig. 3H). Collectively, these results indicated that C/EBPβ regulates osteoblast differentiation in both Runx2-dependent and -independent mechanisms.
Regulation of osteoblast and adipocyte commitment by LIP.
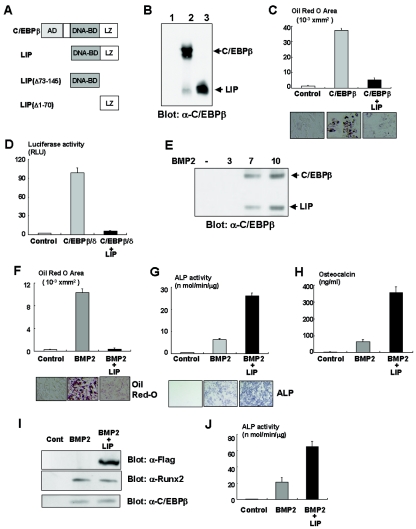
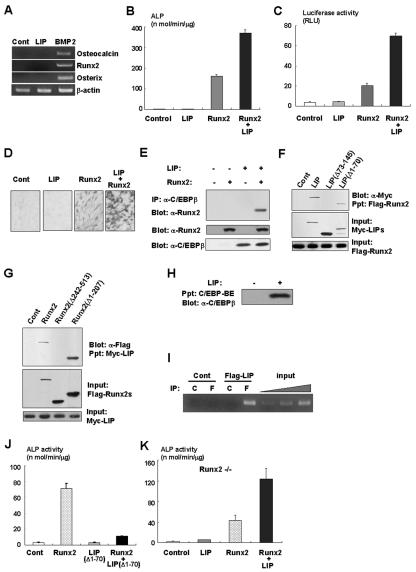
It has been shown that a naturally occurring isoform of C/EBPβ, LIP, exhibits a dominant-negative effect on adipogenesis due to a lack of the transcriptional activation domain (18) (Fig. 4A). We observed that overexpression of LIP inhibits C/EBPβ-induced adipocytic differentiation in C3H10T1/2 cells (Fig. 4B and C). Moreover, LIP overexpression also inhibits the PPARγ gene promoter activity transactivated by C/EBPβ and C/EBPδ (Fig. 4D). These data support the notion that LIP inhibits adipogenesis in a dominant-negative fashion. Notably, we found that LIP was expressed in primary osteoblasts (Fig. 1A) and induced in C3H10T1/2 cells in response to BMP2 treatment (Fig. 4E), suggesting that LIP plays a role in osteoblast differentiation. To define whether LIP affects osteoblast differentiation as well as adipocytic differentiation of mesenchymal cells, we examined the effects of LIP overexpression on osteoblast differentiation of C3H10T1/2 cells in the presence of BMP2 that concomitantly induced osteoblastic and adipocytic differentiation of C3H10T1/2 cells (1, 19). As expected, overexpression of LIP markedly inhibited adipogenesis induced by BMP2 (Fig. 4F). However, surprisingly, LIP overexpression enhanced the osteogenic action of BMP2 on osteoblast differentiation (Fig. 4G and H) without affecting Runx2 expression (Fig. 4I). Consistently, LIP increased BMP2-induced ALP activity in C2C12 cells (Fig. 4J). We found that overexpression of LIP did not induce osteocalcin, Runx2, and Osterix expression (Fig. 5A), indicating that LIP alone is unable to promote osteoblast differentiation. Since C/EBPβ physically and functionally associates with Runx2 (Fig. 3F) (17), we next examined whether LIP has physical and functional interactions with Runx2. Although the effect of overexpression of LIP alone had no effects on ALP activity (Fig. 5B), ALP activity was dramatically increased when LIP was cooverexpressed with Runx2 (Fig. 5B). Furthermore, overexpression of LIP together with Runx2 also transactivated the osteocalcin gene promoter activity more than Runx2 alone (Fig. 5C). In addition, LIP also stimulated Runx2-dependent ALP activity in C2C12 cells (Fig. 5D). Coimmunoprecipitation experiments showed the physical interaction between LIP and Runx2 (Fig. 5E). To gain further insight into the molecular interaction between LIP and Runx2, we examined the association by using the deletion mutants of LIP or Runx2. As shown in Fig. 5F, the leucine zipper domain of LIP is responsible for the binding to Runx2. Figure 5G indicates that Runx2 associates with LIP through the C-terminal portion containing the transcriptional activation domain. To address whether LIP can directly bind to the osteocalcin gene, we performed an oligonucleotide precipitation assay and a chromatin immunoprecipitation experiment. As shown in Fig. 5H and I, LIP has the capacity to bind to the osteocalcin gene promoter. These data suggest that LIP functions as a coactivator for Runx2 during osteoblastogenesis. To explore the importance of LIP as a coactivator for Runx2, we examined the effects of a mutant of LIP, namely, LIP(Δ1-70), which lacks both transcriptional activation and DNA binding domains (Fig. 4A). Because the mutant retains the ability to associate with Runx2 (Fig. 5F) and C3H10T1/2 cells slightly express C/EBPβ (Fig. 1C), overexpression of the mutant profoundly inhibited Runx2-induced ALP activity (Fig. 5J) by inhibiting the interaction of C/EBPβ with Runx2. These data suggest that LIP functions as a coactivator for Runx2 during osteoblast differentiation. To further understand the relationship between LIP and Runx2 during osteoblast differentiation, we next evaluated the effects of LIP on osteoblast differentiation by using Runx2-deficient mesenchymal cells. Consistent with the results shown in Fig. 5B and C, overexpression of LIP failed to induce osteoblast differentiation of Runx2-deficient mesenchymal cells (Fig. 5K), whereas overexpression of Runx2 or C/EBPβ was able to promote osteoblast differentiation of Runx2-deficient mesenchymal cells (Fig. 3H). In contrast, when Runx2 was introduced into Runx2-deficient mesenchymal cells, LIP overexpression stimulated osteoblast differentiation (Fig. 5K). These data indicate that LIP, which lacks transcriptional activity, requires Runx2 to induce osteoblast differentiation of mesenchymal cells.
FIG. 4.
LIP inhibits adipocyte differentiation in a dominant-negative fashion. (A) Schematic structure of C/EBPβ, LIP, and its deletion mutants. AD, activation domain; BD, binding domain; LZ, leucine zipper. (B) C3H10T1/2 cells infected with control, C/EBPβ, or LIP adenovirus were analyzed by immunoblotting with anti-C/EBPβ antibody. (C) C3H10T1/2 cells were infected with adenoviruses as indicated in Materials and Methods, incubated for 7 days, and then stained with Oil red O. Oil red O-stained area was measured as described in Materials and Methods. (D) PPARγ gene promoter fused to the luciferase reporter construct and TK-renilla reporter constructs was transfected into C3H10T1/2 cells together with expression vectors as indicated. Luciferase activity of the cell lysates was measured. (E) C3H10T1/2 cells were incubated with BMP2 as indicated, and expression of LIP and C/EBPβ was determined by immunoblotting. (F) C3H10T1/2 cells infected with control or Flag-tagged LIP adenovirus were incubated with BMP2 for 7 days and stained with Oil red O. The Oil red O-stained area was measured. (G and H) C3H10T1/2 cells infected with control or Flag-tagged LIP adenovirus were incubated with BMP2 for 7 days and ALP activity (G) and osteocalcin production (H) were determined. (I) C3H10T1/2 cells infected with control or Flag-tagged LIP adenovirus were incubated with BMP2 for 7 days, and the cell lysates were examined by immunoblotting with anti-Flag, Runx2, or C/EBPβ antibody. (J) C2C12 cells infected with control or Flag-tagged LIP adenovirus were incubated with BMP2 for 7 days, and ALP activity was determined.
FIG. 5.
LIP enhances osteogenic action of Runx2 through its physical association. (A) Total RNA of C3H10T1/2 cells infected with control (Cont) or Flag-LIP adenovirus or incubated with BMP2 was determined by RT-PCR analysis. (B) C3H10T1/2 cells were infected with adenoviruses as indicated in the text and incubated for 7 days. ALP activity of the cells was measured. (C) Osteocalcin gene promoters fused to luciferase reporter construct and TK-renilla reporter constructs were transfected into C3H10T1/2 cells together with pcDNA3 (control), LIP expression vector, Runx2 expression vector, or both vectors. Luciferase activity of the cell lysates was measured. (D) C2C12 cells infected with adenovirus as indicated in the text were cultured for 7 days and then examined by ALP staining. (E) C3H10T1/2 cells were infected with control, LIP, or Runx2 adenovirus or both LIP and Runx2 adenovirus. The cell lysates were immunoprecipitated with anti-C/EBPβ antibody, and immunoprecipitates (IP) were determined by immunoblotting with anti-Runx2 antibody. (F) The lysates of Cos7 cells were transfected with Myc-tagged LIP constructs as indicated and incubated with Flag-tagged Runx2 protein immobilized on protein G-agarose beads. Precipitated proteins (Ppt) were examined by immunoblotting with anti-Myc antibody. (G) The lysates of Cos7 cells were transfected with Flag-tagged Runx2 constructs as indicated and incubated with Myc-tagged LIP protein immobilized on protein G-agarose beads. Precipitated proteins were examined by immunoblotting with anti-Flag antibody. (H) The lysates of C3H10T1/2 cells infected with control or LIP adenovirus were incubated with biotinylated probe containing C/EBP binding element (BE) in the osteocalcin gene. Associated protein with biotinylated probe was determined by immunoblotting with anti-C/EBPβ antibody. (I) The lysates of C3H10T1/2 cells infected with control or Flag-tagged LIP adenovirus were subjected to chromatin immunoprecipitation analysis. C, immunoprecipitation with control mouse IgG; F, immunoprecipitation with anti-Flag antibody. (J) C3H10T1/2 cells were infected with adenoviruses as indicated in the text. Seven days after culture, ALP activity was examined. (K) Runx2-deficient mesenchymal cells were infected with control, either LIP adenovirus or Runx2 adenovirus, or both adenoviruses and then cultured for 7 days. ALP activity was determined.
DISCUSSION
Several clinical and experimental studies suggest that the differentiation program of mesenchymal cells in bone marrow is controlled by various hormones, growth factors, and cytokines through specific transcription factors (7, 13, 40, 44). Consistent with clinical evidence (7), estrogen preferentially stimulates the commitment of mesenchymal cells into osteoblasts (10, 33). Similarly, sonic hedgehog seems to be one of the cytokines that promote osteoblast differentiation and suppress adipocytic differentiation (38). In addition, the involvement of several transcription factors in regulation of the balance between osteoblastogenesis and adipogenesis has been described. Several studies suggest the reciprocal roles of Runx2 and PPARγ during osteoblast and adipocyte differentiation (2, 16, 23). Recently, Cheng et al. (9) and Ichida et al. (20) indicated that a homeobox gene, the Msx2 gene, stimulates the commitment of mesenchymal cells into an osteoblast lineage in association with inhibition of adipogenesis. Because transgenic mice overexpressing the ΔFosB transcription factor showed increases in bone formation and decreases in adipogenesis (36), it is likely that AP-1 transcription factors are also involved in the commitment of bone marrow mesenchymal cells. However, to date, investigation of the transcriptional events involved in the commitment of mesenchymal cells has not been accomplished. In the present study, we have shown that LIP preferentially stimulates osteoblast differentiation of undifferentiated mesenchymal cells but inhibits adipocyte differentiation. To support this, we found that expression of LIP is up-regulated during osteoblast differentiation. In contrast, Tang et al. demonstrated that LIP expression is down-regulated during adipocyte differentiation of 3T3-L1 cells (42). Previous studies (18, 45) and our results also indicate that LIP blocks the adipogenic function of C/EBPβ in a dominant-negative mechanism. Thus, LIP is one of several key transcription factors that define the balance between osteoblastogenesis and adipogenesis in bone marrow.
Since LIP is an isoform of C/EBPβ that lacks a transcriptional activation domain, it how LIP stimulates osteoblastogenesis is an interesting question. We have demonstrated that LIP physically associates with Runx2 and binds to the DNA binding element present in the osteocalcin gene promoter which is one of the targets for Runx2. These biochemical experiments indicate that LIP functions as a coactivator for Runx2. Indeed, LIP enhances the transcriptional activity of Runx2 and its osteogenic action. Consistently, unlike C/EBPβ, LIP was not able to promote osteoblast differentiation in Runx2-deficient cells. Collectively, LIP has dual functional roles in the regulation of osteoblastogenesis and adipogenesis.
The C/EBPβ gene has only one exon, and both C/EBPβ and LIP are transcribed from the common mRNA (12). Calkhoven et al. showed the molecular regulatory mechanisms of expression of C/EBPβ isoforms in an adipogenic cell line, 3T3-L1, during adipogenesis (8). However, the regulation of LIP expression during osteoblast differentiation is presently unknown. Because we found that BMP2, one of most powerful osteogenic cytokines (44), induces LIP expression, investigation of the role of BMP2 signaling in LIP expression may solve this issue.
Large bodies of evidence indicate that Runx2 is an essential transcription factor for bone formation (13, 44). However, it has been suggested that other transcriptional regulators are cooperatively involved in the osteogenic action of Runx2 because the transcriptional activity of Runx2 itself is relatively weak (22). As described previously (17), we have also observed that C/EBPβ physically interacts with Runx2. In addition, we showed that C/EBPβ induces ALP activity and osteogenic markers as type IA collagen and osteocalcin. Furthermore, a mutant of C/EBPβ, which lacks a DNA binding domain but retains the activity of binding to Runx2, markedly inhibited the osteogenic action of Runx2 presumably by competing with endogenous C/EBPβ or its related family. Together with the results from a previous study (17), we show that C/EBPβ is an important transcriptional partner for Runx2 during osteogenesis. In conflict with our data, an earlier study reported that osteoblast differentiation was inhibited in MC3T3-E1 cells stably transfected with C/EBPβ (21). Although we cannot completely exclude the possibility that this discrepancy is due to a difference in the experimental model or culture condition, we believe that C/EBPβ positively regulates osteoblastogenesis based on the following reasons: (i) overexpression of C/EBPβ by using an adenovirus system, which is one of the most powerful, efficient, and reproducible technologies to introduce the exogenous genes into the variety type of cells without making a clonal difference, promoted osteoblast differentiation of ST2, C2C12, and primary mesenchymal cells as well as C3H10T1/2 cells; (ii) these effects of C/EBPβ adenovirus are dose dependent (data not shown); (iii) C/EBPβ physically interacts with Runx2 and enhances its transcriptional activity and osteogenic action; and (iv) in our hands, we did not observe the stimulative effects of C/EBPβ on the proliferation of osteoblasts. Generation and analyses of appropriate transgenic mice or knockout mice as described below solve this issue.
Interestingly, our results, using a dominant-negative Runx2 mutant or Runx2-deficient mesenchymal cells, indicate that C/EBPβ itself can induce osteoblast differentiation. Thus, C/EBPβ regulates osteoblast development in Runx2-dependent and -independent mechanisms. However, bone abnormality in C/EBPβ-deficient mice has not yet been reported (41). We have observed that C/EBPα and C/EBPδ are able to promote osteoblast differentiation of mesenchymal cells (R. Nishimura, K. Hata, and M. Ueda, unpublished data). Gutierrez et al. reported that C/EBPδ as well as C/EBPβ activates the rat osteocalcin gene promoter (17). It is therefore likely that other C/EBP family members would compensate for C/EBPβ deficiency during osteoblast differentiation. Since we have presently been generating transgenic mice in which the pan type of dominant-negative C/EBP mutant is specifically expressed in an osteoblast lineage, the analyses of the mouse model may allow further dissection of the roles of C/EBPβ in bone development in vivo.
In conclusion, the identification of dual functional roles of LIP in osteoblast and adipocyte differentiation provides a novel insight into understanding the molecular mechanism of the commitment of mesenchymal stem cells in bone marrow and may allow us to develop new therapeutic agents for bone disease such as osteoporosis.
Acknowledgments
We thank Shizuo Akira (Osaka University) for C/EBPβ and C/EBPδ cDNA.
This work was supported in part by Ministry of Education, Science, Sports and Culture Grants-in-Aid for Scientific Research A 11307041(T.Y.), B 15390560 (R.N.), and C 10671739 (R.N.) Support was also received from a Grant-in-Aid for Scientific Research on Priority Areas B 12137205 (T.Y) and from the Senri Life Science Foundation (R.N.) and The 21st Century COE Program (T.Y. and R.N.).
REFERENCES
- 1.Ahrens, M., T. Ankenbauer, D. Schroder, A. Hollnagel, H. Mayer, and G. Gross. 1993. Expression of human bone morphogenetic proteins-2 or -4 in murine mesenchymal progenitor C3H10T1/2 cells induces differentiation into distinct mesenchymal cell lineages. DNA Cell Biol. 12:871-880. [DOI] [PubMed] [Google Scholar]
- 2.Akune, T., S. Ohba, S. Kamekura, M. Yamaguchi, U. I. Chung, N. Kubota, Y. Terauchi, Y. Harada, Y. Azuma, K. Nakamura, T. Kadowaki, and H. Kawaguchi. 2004. PPARgamma insufficiency enhances osteogenesis through osteoblast formation from bone marrow progenitors. J. Clin. Investig. 113:846-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachner, D., M. Ahrens, D. Schroder, A. Hoffmann, J. Lauber, N. Betat, P. Steinert, L. Flohe, and G. Gross. 1998. Bmp-2 downstream targets in mesenchymal development identified by subtractive cloning from recombinant mesenchymal progenitors (C3H10T1/2). Dev. Dyn. 213:398-411. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, B. R., N. A. Timchenko, and C. A. Zahnow. 2004. Epidermal growth factor receptor stimulation activates the RNA binding protein CUG-BP1 and increases expression of C/EBPβ-LIP in mammary epithelial cells. Mol. Cell. Biol. 24:3682-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blake, T., N. Adya, C. H. Kim, A. C. Oates, L. Zon, A. Chitnis, B. M. Weinstein, and P. P. Liu. 2000. Zebrafish homolog of the leukemia gene CBFB: its expression during embryogenesis and its relationship to scl and gata-1 in hematopoiesis. Blood 96:4178-4184. [PubMed] [Google Scholar]
- 6.Buck, M., H. Turler, and M. Chojkier. 1994. LAP (NF-IL-6), a tissue-specific transcriptional activator, is an inhibitor of hepatoma cell proliferation. EMBO J. 13:851-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkhardt, R., G. Kettner, W. Bohm, M. Schmidmeier, R. Schlag, B. Frisch, B. Mallmann, W. Eisenmenger, and T. Gilg. 1987. Changes in trabecular bone, hematopoiesis and bone marrow vessels in aplastic anemia, primary osteoporosis, and old age: a comparative histomorphometric study. Bone 8:157-164. [DOI] [PubMed] [Google Scholar]
- 8.Calkhoven, C. F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 14:1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, S. L., J. S. Shao, N. Charlton-Kachigian, A. P. Loewy, and D. A. Towler. 2003. MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J. Biol. Chem. 278:45969-45977. [DOI] [PubMed] [Google Scholar]
- 10.Dang, Z., and C. W. Lowik. 2004. The balance between concurrent activation of ERs and PPARs determines daidzein-induced osteogenesis and adipogenesis. J. Bone Miner. Res. 19:853-861. [DOI] [PubMed] [Google Scholar]
- 11.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273:30057-30060. [DOI] [PubMed] [Google Scholar]
- 12.Descombes, P., and U. Schibler. 1991. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67:569-579. [DOI] [PubMed] [Google Scholar]
- 13.Ducy, P., T. Schinke, and G. Karsenty. 2000. The osteoblast: a sophisticated fibroblast under central surveillance. Science 289:1501-1504. [DOI] [PubMed] [Google Scholar]
- 14.Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. [DOI] [PubMed] [Google Scholar]
- 15.Gheorghiu, I., C. Deschenes, M. Blais, F. Boudreau, N. Rivard, and C. Asselin. 2001. Role of specific CCAAT/enhancer-binding protein isoforms in intestinal epithelial cells. J. Biol. Chem. 276:44331-44337. [DOI] [PubMed] [Google Scholar]
- 16.Gori, F., T. Thomas, K. C. Hicok, T. C. Spelsberg, and B. L. Riggs. 1999. Differentiation of human marrow stromal precursor cells: bone morphogenetic protein-2 increases OSF2/CBFA1, enhances osteoblast commitment, and inhibits late adipocyte maturation. J. Bone Miner. Res. 14:1522-1535. [DOI] [PubMed] [Google Scholar]
- 17.Gutierrez, S., A. Javed, D. K. Tennant, M. van Rees, M. Montecino, G. S. Stein, J. L. Stein, and J. B. Lian. 2002. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277:1316-1323. [DOI] [PubMed] [Google Scholar]
- 18.Hamm, J. K., B. H. Park, and S. R. Farmer. 2001. A role for C/EBPbeta in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J. Biol. Chem. 276:18464-18471. [DOI] [PubMed] [Google Scholar]
- 19.Hata, K., R. Nishimura, F. Ikeda, K. Yamashita, T. Matsubara, T. Nokubi, and T. Yoneda. 2002. Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator-activating receptor gamma during bone morphogenetic protein 2-induced adipogenesis. Mol. Biol. Cell 14:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichida, F., R. Nishimura, K. Hata, T. Matsubara, F. Ikeda, K. Hisada, H. Yatani, X. Cao, T. Komori, A. Yamaguchi, and T. Yoneda. 2004. Reciprocal roles of MSX2 in regulation of osteoblast and adipocyte differentiation. J. Biol. Chem. 279:34015-34022. [DOI] [PubMed] [Google Scholar]
- 21.Iyer, V. V., T. B. Kadakia, L. R. McCabe, and R. C. Schwartz. 2004. CCAAT/enhancer-binding protein-beta has a role in osteoblast proliferation and differentiation. Exp. Cell Res. 295:128-137. [DOI] [PubMed] [Google Scholar]
- 22.Kanno, T., Y. Kanno, L.-F. Chen, E. Ogawa, W.-Y. Kim, and Y. Ito. 1998. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol. Cell. Biol. 18:2444-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi, H., Y. Gao, C. Ueta, A. Yamaguchi, and T. Komori. 2000. Multilineage differentiation of Cbfa1-deficient calvarial cells in vitro. Biochem. Biophys. Res. Commun. 273:630-636. [DOI] [PubMed] [Google Scholar]
- 24.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y. H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764. [DOI] [PubMed] [Google Scholar]
- 25.Lecka-Czernik, B., I. Gubrij, E. J. Moerman, O. Kajkenova, D. A. Lipschitz, S. C. Manolagas, and R. L. Jilka. 1999. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARgamma2. J. Cell. Biochem. 74:357-371. [PubMed] [Google Scholar]
- 26.Miyake, S., M. Makimura, Y. Kanegae, S. Harada, Y. Sato, K. Takamori, C. Tokuda, and I. Saito. 1996. Efficient generation of recombinant adenoviruses using adenovirus DNA-terminal protein complex and a cosmid bearing the full-length virus genome. Proc. Natl. Acad. Sci. USA 93:1320-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mundlos, S., F. Otto, C. Mundlos, J. B. Mulliken, A. S. Aylsworth, S. Albright, D. Lindhout, W. G. Cole, W. Henn, J. H. Knoll, M. J. Owen, R. Mertelsmann, B. U. Zabel, and B. R. Olsen. 1997. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89:773-779. [DOI] [PubMed] [Google Scholar]
- 28.Nakashima, K., X. Zhou, G. Kunkel, Z. Zhang, J. M. Deng, R. R. Behringer, and B. de Crombrugghe. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108:17-29. [DOI] [PubMed] [Google Scholar]
- 29.Nishimura, R., K. Hata, S. E. Harris, F. Ikeda, and T. Yoneda. 2002. Core-binding factor alpha 1 (Cbfa1) induces osteoblastic differentiation of C2C12 cells without interactions with Smad1 and Smad5. Bone 31:303-312. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura, R., K. Hata, F. Ikeda, T. Matsubara, K. Yamashita, F. Ichida, and T. Yoneda. 2003. The role of Smads in BMP signaling. Front. Biosci. 8:s275-s284. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura, R., Y. Kato, D. Chen, S. E. Harris, G. R. Mundy, and T. Yoneda. 1998. Smad5 and DPC4 are key molecules in mediating BMP-2-induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J. Biol. Chem. 273:1872-1879. [DOI] [PubMed] [Google Scholar]
- 32.Ogasawara, A., T. Arakawa, T. Kaneda, T. Takuma, T. Sato, H. Kaneko, M. Kumegawa, and Y. Hakeda. 2001. Fluid shear stress-induced cyclooxygenase-2 expression is mediated by C/EBP beta, cAMP-response element-binding protein, and AP-1 in osteoblastic MC3T3-E1 cells. J. Biol. Chem. 276:7048-7054. [DOI] [PubMed] [Google Scholar]
- 33.Okazaki, R., D. Inoue, M. Shibata, M. Saika, S. Kido, H. Ooka, H. Tomiyama, Y. Sakamoto, and T. Matsumoto. 2002. Estrogen promotes early osteoblast differentiation and inhibits adipocyte differentiation in mouse bone marrow stromal cell lines that express estrogen receptor (ER) alpha or beta. Endocrinology 143:2349-2356. [DOI] [PubMed] [Google Scholar]
- 34.Owen, M. 1988. Marrow stromal stem cells. J. Cell Sci. Suppl. 10:63-76. [DOI] [PubMed] [Google Scholar]
- 35.Pereira, R. C., A. M. Delany, and E. Canalis. 2002. Effects of cortisol and bone morphogenetic protein-2 on stromal cell differentiation: correlation with CCAAT-enhancer binding protein expression. Bone 30:685-691. [DOI] [PubMed] [Google Scholar]
- 36.Sabatakos, G., N. A. Sims, J. Chen, K. Aoki, M. B. Kelz, M. Amling, Y. Bouali, K. Mukhopadhyay, K. Ford, E. J. Nestler, and R. Baron. 2000. Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat. Med. 6:985-990. [DOI] [PubMed] [Google Scholar]
- 37.Spiegelman, B. M., and J. S. Flier. 2001. Obesity and the regulation of energy balance. Cell 104:531-543. [DOI] [PubMed] [Google Scholar]
- 38.Spinella-Jaegle, S., G. Rawadi, S. Kawai, S. Gallea, C. Faucheu, P. Mollat, B. Courtois, B. Bergaud, V. Ramez, A. M. Blanchet, G. Adelmant, R. Baron, and S. Roman-Roman. 2001. Sonic hedgehog increases the commitment of pluripotent mesenchymal cells into the osteoblastic lineage and abolishes adipocytic differentiation. J. Cell Sci. 114:2085-2094. [DOI] [PubMed] [Google Scholar]
- 39.Su, W. C., H. Y. Chou, C. J. Chang, Y. M. Lee, W. H. Chen, K. H. Huang, M. Y. Lee, and S. C. Lee. 2003. Differential activation of a C/EBP beta isoform by a novel redox switch may confer the lipopolysaccharide-inducible expression of interleukin-6 gene. J. Biol. Chem. 278:51150-51158. [DOI] [PubMed] [Google Scholar]
- 40.Suzawa, M., I. Takada, J. Yanagisawa, F. Ohtake, S. Ogawa, T. Yamauchi, T. Kadowaki, Y. Takeuchi, H. Shibuya, Y. Gotoh, K. Matsumoto, and S. Kato. 2003. Cytokines suppress adipogenesis and PPAR-gamma function through the TAK1/TAB1/NIK cascade. Nat. Cell Biol. 5:224-230. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, T., N. Yoshida, T. Kishimoto, and S. Akira. 1997. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 16:7432-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang, Q. Q., T. C. Otto, and M. D. Lane. 2003. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc. Natl. Acad. Sci. USA 100:850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, Z., Y. Xie, N. L. Bucher, and S. R. Farmer. 1995. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 9:2350-2363. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi, A., T. Komori, and T. Suda. 2000. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocr. Rev. 21:393-411. [DOI] [PubMed] [Google Scholar]
- 45.Yeh, W. C., Z. Cao, M. Classon, and S. L. McKnight. 1995. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 9:168-181. [DOI] [PubMed] [Google Scholar]