Abstract
Scaffold proteins are believed to enhance specificity in cell signaling when different pathways share common components. The prototype scaffold Ste5 binds to multiple components of the Saccharomyces cerevisiae mating pheromone response pathway, thereby conducting the mating signal to the Fus3 mitogen-activated protein kinase (MAPK). Some of the kinases that Ste5 binds to, however, are also shared with other pathways. Thus, it has been presumed that Ste5 prevents its bound kinases from transgressing into other pathways and protects them from intrusions from those pathways. Here we found that Fus3MAPK required Ste5 scaffolding to receive legitimate signals from the mating pathway as well as misdirected signals leaking from other pathways. Furthermore, increasing the cellular concentration of active Ste5 enhanced the channeling of inappropriate stimuli to Fus3. This aberrant signal crossover resulted in the erroneous induction of cell cycle arrest and mating. In contrast to Fus3, the Kss1 MAPK did not require Ste5 scaffolding to receive either authentic or leaking signals. Furthermore, the Ste11 kinase, once activated via Ste5, was able to signal to Kss1 independently of Ste5 scaffolding. These results argue that Ste5 does not act as a barrier that actively prevents signal crossover to Fus3 and that Ste5 may not effectively sequester its activated kinases away from other pathways. Rather, we suggest that specificity in this network is promoted by the selective activation of Ste5 and the distinct requirements of the MAPKs for Ste5 scaffolding.
Different cellular signals can be transmitted by the same components yet produce distinct responses. In the yeast Saccharomyces cerevisiae, elements of the same mitogen-activated protein kinase (MAPK) cascade regulate the mating and filamentous invasive growth differentiation programs and the response to osmotic stress (26, 37, 40). Mating and haploid invasive growth use the same MAPKs, Fus3 and Kss1, the same MAPK kinase (MKK or MEK), Ste7, and the same MEK kinase (MEKK), Ste11. Ste11MEKK also activates the Pbs2 MEK and the Hog1 MAPK in response to osmotic stress (Fig. 1A). These three pathways react to different stimuli (pheromone, nutrient status, and osmotic stress, respectively) and regulate distinct endpoints (mating, invasive growth, and glycerol production, respectively). Both Fus3 and Kss1 are activated during mating, while Kss1 is preferentially activated during invasive growth, and Hog1 is activated only by stress (8, 12, 37, 41). These patterns make sense given the distinct, often antagonistic functions of these MAPKs.
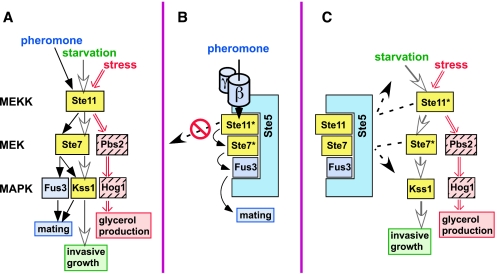
FIG. 1.
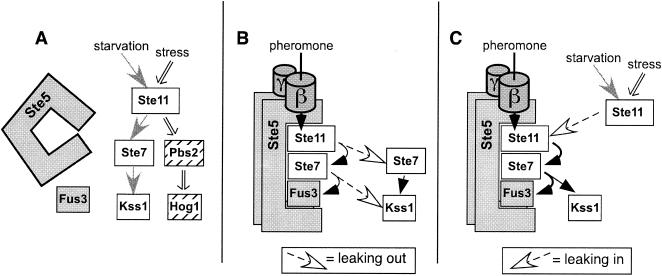
(A) Shared MAPK cascade components signal to three distinct endpoints. See the text for details. Shared components (Ste11MEKK, Ste7MEK, and Kss1MAPK) are shown in yellow. Nutrient limitation (starvation) is known to activate the invasive growth program, but whether this or some other signal is transmitted by the MAPK cascade is unknown. Whatever this signal is, it appears to be relayed by the Msb2 protein (12). (B and C) Models for how the Ste5 scaffold protein may promote signaling specificity. The active, phosphorylated Ste11 and Ste7 isoforms are indicated by an asterisk. (B) By the selective recognition of mating components, Ste5 enhances signaling within the mating pathway. Also, Ste5 may sequester its bound components and prevent them from straying into other pathways. (C) Ste5 may act as a sequestering barrier to protect mating components, particularly Fus3MAPK, from Ste11MEKK or Ste7MEK activated by other pathways.
How is selective MAPK activation achieved when each signal gets funneled through a common set of components? Scaffold proteins, such as yeast Ste5, are thought to provide some of the requisite specificity. Scaffolds are proteins that bind to several sequentially acting pathway members. For example, Ste5, a scaffold dedicated to the yeast mating pathway (21, 37, 40), binds to Ste11MEKK, Ste7MEK, Fus3MAPK, and, with somewhat lower affinity, Kss1MAPK (10, 25). Ste5 and other scaffolds have been proposed to enhance specificity in several ways (15, 17, 20, 28, 30, 35, 47, 49) (Fig. 1B and C). First, by selectively recognizing, concentrating and spatially orienting pathway-specific components, they are thought to promote within-pathway signal transmission (Fig. 1B). For instance, Ste5 is thought to promote Ste11MEKK activation of Ste7MEK but not of Pbs2MEK because Ste5 binds Ste7 but not Pbs2. This is a manifestation of the well-known role of molecular recognition in specificity (36); as such, it is not a unique property of scaffolds.
Scaffold proteins are also believed to promote specificity by additional mechanisms which exploit their defining ability to bind simultaneously to multiple components of the same pathway. In particular, it has been proposed that scaffolds may sequester their bound components into pathway-dedicated signaling particles, thereby physically isolating them from other pathways. In so doing, scaffolds may both prevent their bound compounds from straying into other pathways when active (Fig. 1B) and protect them from intrusions from other pathways when inactive (Fig. 1C). For instance, Ste5 may prevent pheromone-activated Ste11 from straying into the invasive growth and osmostress pathways and insulate Fus3 from Ste11 activated by these pathways.
Ste5's first function during mating is not as a scaffold but as an adapter. Following pheromone stimulation, Ste5 binds to the released βγ subunits of the pheromone receptor-coupled G protein, towing bound Ste11MEKK to the vicinity of the plasma membrane (38), where Ste11 gets phosphorylated and activated by the Ste20 protein kinase (46; reviewed in reference 3). The scaffold function of Ste5 then facilitates signal transmission from Ste11MEKK to Ste7MEK to Fus3MAPK (Fig. 1B) (15). The crucial role of both the adapter and scaffold functions of Ste5 in pheromone signal transmission makes it difficult to test whether Ste5 actively prevents signals from leaking out of the mating pathway (Fig. 1B). It is possible, however, to test whether Ste5 actively prevents signals from leaking into the mating pathway (Fig. 1C). Here we have done so, and performed additional experiments that provide insight into the influence of the Ste5 scaffold protein and the distinct scaffolding requirements of the Fus3 and Kss1 MAPKs on signaling specificity.
MATERIALS AND METHODS
Plasmid construction.
The plasmids used in this study are shown in Table 1. Reporter genes YEpU-FT1Z (used in Fig. 3A, 3B, and 4B) and YEpL-FT1Z (Fig. 3C), which contain the filamentation response element (FRE) (29) from the TEC1 gene driving lacZ, has been described (41). YEpT-FUS1Z (Fig. 3C) (5), YEpU-FUS1Z (Fig. 3B and 4B) (5), and YEpL-FUS1Z (Fig. 5B and Fig. S1 in the supplemental material) (2) are described where indicated. In these reporters, the promoter of the FUS1 gene drives lacZ; FUS1 is representative of a battery of genes induced by mating pheromone (39).
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| YEpU-FT1Z | URA3 2μm FRE(Tec1)-lacZ | 41 |
| YEpL-FT1Z | LEU2 2μm FRE(Tec1)-lacZ | 41 |
| YEpT-FUS1Z | TRP1 2μm FUS1-lacZ | 5 |
| YEpU-FUS1Z | URA3 2μm FUS1-lacZ | 5 |
| YEpL-FUS1Z | LEU2 2μm FUS1-lacZ | 2 |
| YCpU | URA3 CEN | 2 |
| YCpH | HIS3 CEN | 2 |
| YCpT | TRP1 CEN | 2 |
| YCpU-STE5 | URA3 CEN STE5 | 22 |
| YCpU-STE5VASP | URA3 CEN STE5-V763AS861P | 22 |
| YCpU-STE5F14L | URA3 CEN STE5-F14L | 22 |
| YCpH-STE5 | HIS3 CEN STE5 | This study |
| YCpT3-STE7 | TRP1 CEN STE7 | 2 |
| YCpH-STE11-4 | HIS3 CEN STE11-T596I | 2 |
| YCpL-STE11-4 | LEU2 CEN STE11-T596I | This study |
| YGU-11ΔN | URA3 CEN GAL1-STE11 ΔN | 9 |
| YCpUG | URA3 CEN GAL1 | 23 |
| pCJ148/YCpUG-STE5-GST | URA3 CEN GAL1-STE5-GST | 23 |
| pCJ149/YCpUG-STE5-GST* | URA3 CEN GAL1-STE5-GST(C177AC180A) | 23 |
| YCpTG-STE5 | TRP1 CEN GAL1-STE5 | This study |
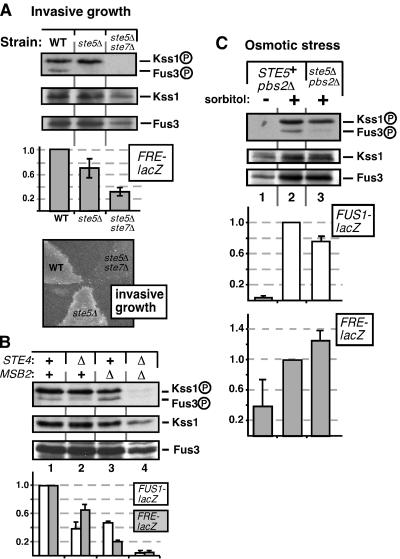
FIG. 3.
Differential requirements of the Fus3 and Kss1 MAPKs for Ste5 scaffolding during invasive growth and the osmotic stress response. (A) Fus3MAPK activation during invasive growth requires Ste5, but Kss1MAPK activation does not. Strains containing wild-type Ste5 (WT), lacking Ste5 (ste5Δ), or lacking Ste5 and Ste7 (ste5Δ ste7Δ) were grown on plates for 24 h and assayed for MAPK phosphorylation. FRE-lacZ reporter gene activity was also measured; standard error bars are shown (n = 7). Results were normalized by setting the mean β-galactosidase activity of the wild-type strain (1,200 U/min/mg of total yeast protein) to 1.0. Similar results were obtained when cells were grown in liquid prior to harvesting for analysis of FRE-lacZ expression (data not shown). Parallel plates were washed and scored for invasive growth. (B) Concurrent leaks. The invasive growth pathway contributes to the basal phosphorylation of Fus3, and the mating pathway contributes to the basal phosphorylation of Kss1. Strains containing wild-type Ste4 and/or Msb2 (+) or lacking Ste4 and/or Msb2 (Δ) were grown in liquid to mid-log phase and assayed for MAPK phosphorylation. Similar results were obtained when the cells were grown on plates for 24 h (data not shown). FUS1-lacZ and FRE-lacZ reporter gene activity was also measured; error bars represent standard deviations (n = 2). Note that the FUS1-lacZ expression being measured is the basal level (i.e., in the absence of pheromone stimulation). (C) Leaking from the osmostress pathway is preferentially channeled into Kss1, particularly when Ste5 is absent. Liquid cultures of strains containing wild-type Ste5 (STE5+ pbs2Δ) or not (ste5Δ pbs2Δ) were treated with 0.7 M sorbitol for 3 h after reaching mid-log phase. Lysates were probed for MAPK phosphorylation, and FUS1-lacZ or FRE-lacZ reporter gene activity was measured. The β-galactosidase activity for the reference strain (normalized to 1.0) was 1,200 U/min/mg of protein (FUS1-lacZ) and 1,150 U/min/mg of protein (FRE-lacZ). Error bars represent standard deviations (n = 2).
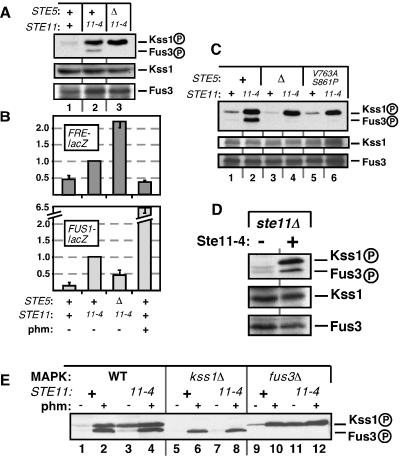
FIG. 4.
Influence of Ste5 on Ste11-4-generated signaling to Kss1MAPK and Fus3MAPK. + indicates the wild-type allele, Δ indicates a gene deletion, and 11-4 indicates strains containing Ste11-4. (A) Ste11-4-stimulated phosphorylation of Fus3 but not of Kss1 requires Ste5. Cells were grown on plates for 24 h before harvesting. The immunoblot in this figure was exposed for a shorter time than the immunoblot in Fig. 3A; this is why the equivalent lane 1's look different. (B) FRE-lacZ expression in the strains shown in A (upper panel) and FUS1-lacZ expression of strains grown in liquid and treated (+) or not (−) with 1 μM pheromone for 2 h (lower panel). Standard deviations are shown (n = 2 to 4). (C) Scaffolding-defective mutant of Ste5 (V763A S861P) does not support Ste11-4-stimulated Fus3 phosphorylation. (D) Removal of endogenous Ste11 has no effect on the inability of plasmid-expressed Ste11-4 to efficiently stimulate Fus3 phosphorylation. Cells were grown on plates for 24 h before harvesting. (E) Effect of Kss1 removal and pheromone stimulation on Fus3 phosphorylation in STE11-4 strains. Liquid-grown cells were treated or not for 15 min with 1 μM pheromone.
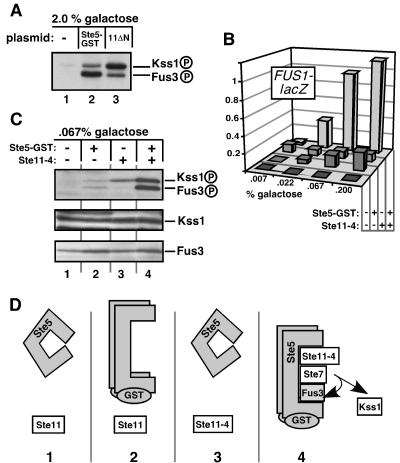
FIG. 5.
Active Ste5 guides signals carried by Ste11MEKK to Fus3MAPK during Ste11-4-generated signaling. Ste5-GST expression was regulated by galactose; the percent galactose used is shown. (A) MAPK phosphorylation by overexpressed and constitutively active Ste5 or Ste11. Ste11ΔN expression (lane 3) was also controlled by galactose levels. (B) FUS1-lacZ reporter gene expression in strains grown on plates containing 2% sucrose and the inducer galactose at increasing concentrations (0.007% to 0.2%). Strains containing Ste5-GST or Ste11-4 are indicated with a + in the appropriate row. (C) MAPK activation loop phosphorylation of the strains from B induced by 2% sucrose and 0.067% galactose. (D) Illustrative description and interpretation of the experiment in C. Panels 1 to 4 correspond to lanes 1 to 4 in C. Active Ste5 has no signal to channel (panel 2), and Ste11-4 needs active Ste5 to hyperactivate Fus3 (panel 3). Together, active Ste5 can channel the Ste11-4 signal to Fus3 (panel 4).
Plasmids YCpU, YCpH, and YCpT are low-copy-number (centromere-containing) yeast expression plasmids described in more detail elsewhere (2). The YCpU-based plasmids expressing STE5 alleles from the STE5 promoter are described elsewhere (22). YCpH-STE5 contains the STE5 promoter and open reading frame on a SacI-BamHI fragment inserted into YCpH. YCpT3-STE7, a low-copy-number (CEN) plasmid containing the STE7 promoter and open reading frame, is described elsewhere (2). YCpH-STE11-4 (2) contains the STE11-4 allele (45) driven by its own (STE11) promoter in YCpH. YCpUG (22) consists of the GAL1,10 promoter in YCpU. YCpUG-STE5-GST (also known as pCJ148) (23) contains STE5-GST driven by the GAL1 promoter. YCpUG-STE5-GSTC177AC180A (also known as pCJ149) (23) contains STE5(C177A C180A)-GST driven by the GAL1 promoter. YGU-11ΔN (9) contains the STE11ΔN allele driven by the GAL1 promoter.
Yeast strain construction.
The yeast strains used in this study are shown in Table 2. Strain LFY105 (MATa ste5Δ, Σ1278b lineage) was transformed with a low-copy-number plasmid expressing wild-type Ste5 from its own promoter, the corresponding empty vector (5Δ), the adapter-defective mutant Ste5F514L (F514L), or the scaffolding-defective mutant Ste5V763A S861P (VASP).
TABLE 2.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| JCY100 | MATaSTE+; Σ1278b lineage | 11 |
| JCY107 | JCY100 ste7Δ::ura3 | 11 |
| JCY120 | JCY100 fus3Δ::TRP1 | 11 |
| LFY105 | JCY100 ste5Δ::LEU2 | 41 |
| LFY157 | JCY100 ste5Δ::LEU2 ste7Δ::ura3 | This work |
| YDV500 | JCY100 ste11Δ | D. Voora and J. Thorner |
| YPH499 | MATaSTE+; S288C lineage | 5 |
| YLB105 | YPH499 ste5Δ::LYS2 ste7Δ::ADE2 | 2 |
| YLB257 | YPH499 ste5Δ::LYS2 ste7Δ::ADE2 pbs2Δ::URA3 | This work |
| YDM600 | YPH499 kss1Δ::hisG | 5 |
| YDM200 | YPH499 fus3Δ::LEU2 | 5 |
| BYB87 | YPH499 ste4Δ::TRP1 | C. Inouye and J. Thorner |
| DC17 | MATα his1 | J. B. Hicks |
| SY3089 | MATaura3-52; Σ1278b lineage | 13 |
| SY3927 | SY3089 ste4Δ FUS1-HIS3 FUS1-lacZ | 12 |
| SY3941 | SY3089 msb2Δ | 12 |
| SY3930 | SY3927 msb2Δ::KAN | 12 |
The strain shown in Fig. 3A, LFY157 (MATa ste5Δ::LEU2 ste7Δ::ura3 Σ1278b lineage) was derived from JCY107 (ste7Δ::ura3) with a 3.5-kb ste5Δ::LEU2 allele described previously (41). The resulting strain was cotransformed with YEpU-FT1Z and either YCpT3-STE7 (ste5Δ) or YCpT (5Δ7Δ). The MATa Σ1278b lineage strains used in Fig. 3B have been described (12, 13); they were transformed with either YEpU-FUS1Z or YEpU-FT1Z. YLB257 (MATa ste5Δ::LYS2 ste7Δ::ADE2 pbs2Δ::URA3), used in Fig. 3C, was derived from YLB105 (MATa ste5Δ::LYS2 ste7Δ::ADE2; S288C lineage) with a 4.1-kb pbs2Δ::URA3 allele (7). The resulting strain was cotransformed with wild-type STE7 and YEpT-FUS1Z plus either YCpH-STE5 (STE5) or YCpH (ste5Δ).
In Fig. 4A to C, LFY105 was transformed with either “empty” YCpU (Δ) or YCpU-STE5 (+) and cotransformed with YEpU-FT1Z. The resulting strains were then further transformed with either YCpH (+) or YCpH-STE11-4 (11-4). Transformation with plasmid YCpU-STE5-V763AS861P (22) was used to generate the indicated strains in Fig. 4C. Figure 4D strains were derived by transforming YDV500 (MATa ste11Δ) with either YCpH (−) or YCpH-STE11-4 (+). Strain YPH499 (MATa STE+; S288C lineage) and the isogenic derivatives YDM600 (kss1Δ::hisG) and YDM200 (fus3Δ::LEU2) used in Fig. 4E were transformed with YCpH-STE11-4 (11-4) or empty vector (+), as described above.
In Fig. 5A, JCY100 was transformed with either YCpUG (−), YCpUG-STE5-GST (Ste5-GST), or YGU-STE11ΔN (11ΔN). In Fig. 5B and 5C, YPH499 was transformed with YEpL-FUS1Z, and either YCpUG (−) or YCpUG-STE5-GST (+). The resulting strains were then further transformed with either YCpH (−) or YCpH-STE11-4 (+).
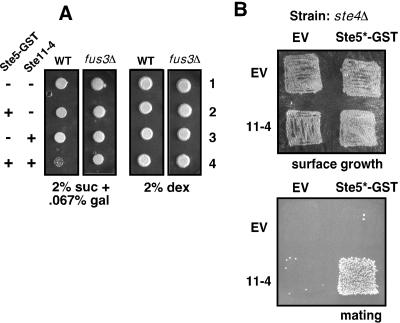
In Fig. 6A, strains JCY100 and its isogenic derivative JCY120 (fus3Δ::TRP1) (11) were transformed with YCpUG (−) or YCpUG-STE5-GST (+). The resulting strains were then further transformed with either YCpH (−) or YCpH-STE11-4 (+). In Fig. 6B, strain BYB87 (MATa ste4Δ) was transformed with either YCpUG (EV) or YCpUG-STE5(C177A C180A)-GST (STE5*-GST) and then either YCpH (EV) or YCpH-STE11-4 (Ste11-4). Strain DC17 (MATα his1) was used as the α tester strain for the mating assay.
FIG. 6.
Ste5-promoted signal crossover results in miscued induction of cell cycle arrest and mating. (A) Cell cycle arrest in strains with active Ste5 and Ste11-4. Wild-type (WT) and fus3Δ strains were transformed with Ste5-GST and/or Ste11-4 or the corresponding empty vector as indicated, and low-level Ste5-GST expression was induced or not by growth on plates containing 2% sucrose and 0.067% galactose or 2% dextrose, respectively. Equal numbers of cells (roughly 30,000) were spotted and grown for 6 days. (B) Mating in strains with active Ste5 and Ste11-4. MATa strains lacking Ste4 and containing Ste5-GSTC177A,C180A (Ste5*-GST) and/or Ste11-4 or the corresponding empty vector (EV) were grown on a plate containing 2% sucrose and 0.067% galactose for 24 h to induce low-level Ste5-GST (surface growth) and then incubated with a MATα strain and transferred to 2% dextrose plates selective for diploids (mating). Ste5-GSTC177A,C180A is a mutant of Ste5-GST that is active even in the absence of Ste4 (23).
Media and growth conditions.
The growth temperature for all experiments was 30°C. Yeast were grown in standard media with rotary shaking to mid-exponential phase (A595 of ≈0.8). Pheromone (Fig. 2A and 4E) or sorbitol (Fig. 3C) was added directly to growing cultures with no additional manipulation. Alternatively, ≈2 optical densities of yeast cells were plated as a lawn and grown for 24 h on plates selective for plasmid maintenance prior to harvesting (Fig. 3A, 4A, and 4C). To overexpress genes driven by the GAL1 promoter (Fig. 5A), S. cerevisiae cells were grown in standard media (with dextrose) to mid-exponential phase, washed with water, and resuspended in media containing 2% galactose and 0.2% sucrose. The cultures were harvested 3 h later. For low-level GAL1-driven expression in Fig. 5B and 5C, yeast cells were grown as a lawn on plates containing 2% sucrose and 0.007% to 0.2% galactose for 24 h before harvesting. In Fig. 6A, cells were spotted on 2% sucrose and 0.067% galactose plates or plates with 2.0% dextrose. For Fig. 6B, cells were grown on plates containing 2% sucrose and 0.067% galactose for 24 h before initiating the mating assay.
FIG. 2.
Fus3MAPK activation during mating requires Ste5 scaffolding, but Kss1MAPK activation does not. (A) Strains containing wild-type Ste5 (WT), lacking Ste5 (ste5Δ), or containing the adapter-defective mutant Ste5F514L (F514L) or the scaffolding-defective mutant Ste5V763A S861P (VASP) were treated (+) or not (−) with 1 μM α-factor mating pheromone (phm) for 15 min. Activation loop phosphorylation was detected by immunoblotting with a phosphorylation state-specific antibody (αpTEpY). Kss1-P and Fus3-P indicate the dually phosphorylated, activated species. Total protein levels were determined with anti-Kss1 (αKss1) and anti-Fus3 (αFus3) antisera. (B) Schematic interpretation of the results; see the text for details.
Biochemical methods.
Yeast cultures were harvested for biochemical analysis and extract preparation as described (41). The electrophoresis, immunoblotting, and immunostaining were done as described (4). The phosphorylated MAPKs were detected as described with an antibody raised against the dually phosphorylated activation loop of ERK2 under conditions in which it cross-reacted with both phosphorylated Kss1 and phosphorylated Fus3 (41). Despite a recent claim to the contrary (1), we have no evidence that leads us to believe that this antibody recognizes phosphorylated Kss1 with substantially greater avidity than phosphorylated Fus3. Total MAPK protein levels were determined by stripping and reprobing with anti-Kss1 or anti-Fus3 antiserum. Reporter gene assays were performed as described from liquid cultures (11) or from plates (41).
Bioassays.
Reporter gene assays were performed as described (4, 6), with either cell extracts (Fig. 3A and 3C) (4) or semipermeablized cells (Fig. 3B, 4B, and 5B) (6). Invasive growth (Fig. 3A) was scored as described (40). Cell cycle arrest (Fig. 6A) was assayed by spotting 10 μl of standard medium containing equal numbers of cells onto plates with the appropriate carbon source and then incubating for 6 days. The mating assay (Fig. 6B) was performed as described (22), except that after yeast cells were grown on plates containing 2% sucrose and 0.067% galactose for 24 h, they were replica plated onto a rich plate with the mating tester strain for 4 h and then replica plated onto 2% dextrose plates selective for diploids and grown overnight before being photographed.
RESULTS
Kss1MAPK and Fus3MAPK have different requirements for Ste5 scaffolding during mating.
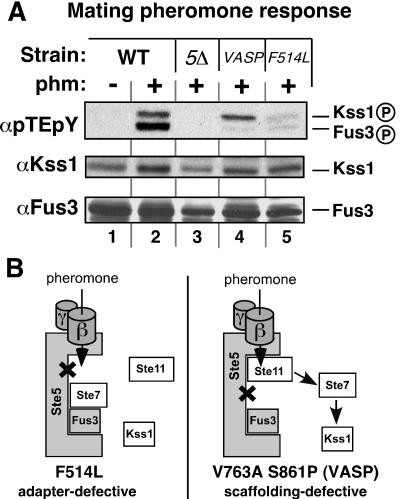
Because Ste5 functions as an adapter in Ste11MEKK activation, removal of Ste5 would be expected to block pheromone-stimulated Ste11 activation, eliminating the possibility of leaking into other MAPK pathways. Consistent with this expectation, removal of Ste5 prevented the pheromone-stimulated activation of both Kss1MAPK and Fus3MAPK, as assessed by staining with an antibody specific for the activated (dually phosphorylated) isoforms of both MAPKs (Fig. 2A). In addition, in cells in which Ste5 was replaced with Ste5F514L, a mutant that is specifically compromised for Ste11 binding (22), pheromone signaling to both Fus3 and Kss1 was substantially decreased (Fig. 2A, lane 5). This makes sense, since this mutant should be defective in the adapter function of Ste5 and therefore unable to promote Ste11 activation.
To specifically inactivate the scaffold function of Ste5 while leaving its adapter function intact, we replaced Ste5 with the Ste5V763A S861P mutant, which cannot bind to Ste7MEK but still binds to other ligands (22). When this was done, Fus3 activation by pheromone was abolished, but Kss1 activation was only slightly reduced (Fig. 2A, lane 4). Thus, Fus3 requires Ste5 scaffolding to receive the mating signal, as expected based on other results (22). Surprisingly, however, Kss1 does not require Ste5 scaffolding to receive the mating signal.
A schematic interpretation of these results is shown in Fig. 2B. In cells containing the adapter-defective Ste5, Ste11 cannot be activated, so neither Fus3 nor Kss1 is activated. In cells containing the scaffolding-defective Ste5, Ste11 can be activated and can apparently activate non-Ste5-bound Ste7, which then activates Kss1 but is unable to activate Fus3. Hence, these results suggest that active Ste11 may dissociate from Ste5 with a half-time that is relatively fast compared to the duration of pheromone stimulation (and in this case the stimulus duration was only 15 min). Alternatively, scaffold-bound Ste11 may be able to phosphorylate free Ste7. In either case, the results do not support the model shown in Fig. 1B; rather, they suggest that Ste5-activated Ste11 is not restricted to acting on Ste5-bound Ste7, and may be able to leak out into other pathways.
In the absence of Ste5, leaking from the invasive growth pathway to Fus3MAPK decreases.
A prediction of the sequestering barrier model of scaffold function (Fig. 1C) is that removing a scaffold might enhance signal crossover, or leaking, between pathways. That is, if a pathway-dedicated scaffold blocks leaking into its pathway, then removing the scaffold may increase such leaking. Specifically, if Ste5 protects Fus3MAPK from Ste7MEK or Ste11MEKK activated by the invasive growth pathway or from Ste11 activated by the osmostress pathway, then removing Ste5 would be predicted to increase Fus3 activation by these inappropriate stimuli.
The filamentation-invasion pathway signals via the Ste11-to- Ste7-to-Kss1 MAPK cascade to drive the expression of genes under the control of filamentation response elements (FREs) (11, 29, 31). Accordingly, a portion of the cellular pool of Kss1MAPK was phosphorylated in cells undergoing invasive growth (Fig. 3A, lane 1), as previously reported (41). Fus3MAPK activation presumably needs to be minimized in these circumstances, since Fus3 positively regulates mating-specific functions and inhibits invasive growth (11, 31, 40, 41). Consistent with this expectation, although some Fus3 activation was detected, it was minimal. Surprisingly, however, in invasively growing cells lacking Ste5, phosphorylated Fus3 was no longer detectable (Fig. 3A, lane 2). This result is the opposite of what would be expected if Ste5 were simply acting as a passive physical barrier to prevent invasive growth signals from activating Fus3. In contrast, when Ste5 was absent, Kss1 phosphorylation was still robust, and Kss1-dependent, FRE-driven gene expression was only slightly reduced (Fig. 3A). Consistent with this, such cells could grow invasively (Fig. 3A), as previously reported (40).
The source of the minimal Fus3 phosphorylation seen in Fig. 3A could have been leaking from the invasive growth pathway, or it could have been (Ste5-dependent) basal signaling from the mating pathway, or a combination of both. Similarly, some of the Kss1 phosphorylation seen in Fig. 3A could have come from basal signaling from the mating pathway (31, 41); this would explain why FRE-driven expression decreased slightly when Ste5 was removed (Fig. 3A). In order to clarify the source of the Fus3 and Kss1 phosphorylation in cells undergoing invasive growth, we examined strains lacking key, pathway-dedicated upstream components of the mating and invasive growth pathways.
First we examined a strain lacking Ste4, the β subunit of the pheromone receptor-coupled G protein. In this strain, spontaneous activation of the G protein is impossible; thus, basal signal flux through the mating pathway should diminish (18, 44). Indeed, phosphorylation of both Kss1 and Fus3 was decreased in cells lacking Ste4Gβ, though Kss1 phosphorylation was still clearly evident and Fus3 phosphorylation was still detectable (Fig. 3B, lane 2 versus lane 1). Furthermore, both FRE-driven expression and basal expression of the mating-specific gene FUS1 were decreased (Fig. 3B), consistent with the decrease in Kss1 and Fus3/Kss1 phosphorylation, respectively.
Next, we examined a strain lacking Msb2, a transmembrane protein that has recently been shown to be a component of the invasive growth pathway and to reside upstream of the MAPK cascade (12). In this strain, Kss1 phosphorylation but not Fus3 phosphorylation was noticeably reduced, and both FUS1 and (especially) FRE-driven expression were decreased (Fig. 3B, lane 3). Finally, in a strain lacking both Ste4Gb and Msb2, both Kss1 and Fus3 phosphorylation was absent and both FUS1 and FRE-driven expression was dramatically reduced (Fig. 3B, lane 4).
These observations lead to the following conclusions. Ste4-dependent signals appear to be preferentially channeled to Fus3, and Msb2-dependent signals are preferentially channeled to Kss1. However, in ste4Δ cells, some Fus3 phosphorylation is still detectable, and this is dependent upon Msb2. Thus, the invasive growth pathway does leak into the mating pathway to weakly activate Fus3. Conversely, in msb2Δ cells, some Kss1 phosphorylation is still detectable, and this is dependent upon Ste4. Thus, some of the Kss1 phosphorylation seen in invasively growing cells is attributable to basal signaling from the mating pathway. Removal of Ste4 or Ste5 apparently has two opposing effects on Kss1 phosphorylation and invasive growth that seem to essentially cancel each other out: a decrease in Kss1 phosphorylation due to reduced basal signaling through the mating pathway, and an increase in Kss1 phosphorylation and invasive growth due to reduced inhibition by Fus3-dependent mechanisms (41).
Despite these complications, it is evident that in invasively growing cells, the removal of Ste5 did not increase Fus3 activation (as would be predicted by the sequestering barrier model); instead, Ste5 removal decreased Fus3 activation.
In the absence of Ste5, leaking from the osmostress pathway to Fus3MAPK decreases.
In wild-type cells, osmotic stress signals activate Hog1MAPK and do not leak into either the mating or invasive growth pathway (14, 37). However, a leak can be created by removing or inactivating the kinase activity of Hog1 or Pbs2MEK (14, 19, 34). In such cells, presumably due to the lack of a negative-feedback loop that normally limits the extent of Ste11MEKK activation, osmotic stress increases the expression of mating genes such as FUS1 (19, 34) as well as FRE-driven invasive growth genes (14) and stimulates Fus3MAPK (19) and Kss1MAPK (14) phosphorylation. As determined by monitoring gene expression, the osmostress-to-mating leak occurs in cells containing Ste5 and is not increased in the absence of Ste5 (34), suggesting that Ste5 does not play a strong role in preventing this leak.
To more precisely determine the role of Ste5 in modulating signal crossover from the osmostress pathway and to ascertain if the MAPKs were differentially affected by such crossover, strains lacking Pbs2 and either lacking or containing Ste5 were constructed and treated with sorbitol to induce osmotic stress. This treatment resulted in potent Kss1 phosphorylation in cells containing Ste5 (Fig. 3C, lane 1 versus lane 2). Fus3 was also phosphorylated in these circumstances, but not nearly to the same extent as Kss1 (Fig. 3C, lane 1 versus lane 2). Strikingly, removal of Ste5 almost completely eliminated this low-level signal crossover to Fus3 but had less of an effect on signal crossover to Kss1 (Fig. 3C, lane 3). Consistent with the phosphorylation data, mating gene expression (as reported by FUS1-lacZ) decreased in the absence of Ste5, while filamentation gene expression (FRE-lacZ) did not (Fig. 3C). These results indicate that Ste5 enhances rather than lessens misdirected osmotic stress signaling to Fus3.
Fus3MAPK activation by Ste11-4 is weak and dependent on Ste5 scaffolding.
To bypass the adapter function of Ste5 and study its scaffold function in isolation, we used a constitutively active version of Ste11MEKK, Ste11-4, which has a point mutation in its kinase domain that renders it independent of Ste5 adapter function (45). The constitutive activity of this mutant (expressed at nearly endogenous levels under the control of its own promoter on a single-copy plasmid) mimics the sustained signaling presumed to occur during filamentation and invasion; indeed, in haploid cells containing Ste11-4, FRE-driven expression was increased (Fig. 4B), as previously reported for diploids (31). Moreover, both Kss1MAPK and Fus3MAPK phosphorylation levels were increased (Fig. 4A, lane 2 versus lane 1, also Fig. 4C, lane 2 versus lane 1), but not to the same degree: the increase in the level of Kss1 phosphorylation was consistently and substantially greater than that of Fus3.
Initially, the weak activation of the mating pathway seemed surprising, since the Ste11-4 allele was isolated in a screen for hyperactivators of FUS1, a mating gene (45). Ste11-4 did in fact drive FUS1 expression, but it did so inefficiently relative to pheromone stimulation (Fig. 4B). Strikingly, when Ste5 was absent from cells containing Ste11-4, phosphorylated Fus3 disappeared and FUS1 expression decreased, but Kss1 phosphorylation levels remained high and FRE-driven expression increased (Fig. 4A, lane 2 versus lane 3, and Fig. 4B). Activation of Fus3 required the scaffolding function of Ste5, because the scaffolding-defective mutant of Ste5 (Ste5V763A S861P) was unable to channel the signal from Ste11-4 to Fus3 (Fig. 4C).
A modest decrease in FUS1 expression levels when Ste5 was removed from cells containing Ste11-4 was observed previously (45). The above results indicate that this modest change in gene expression is the net result of dramatic and opposing changes in the phosphorylation levels of the two MAPKs.
The relative insensitivity of Fus3 to Ste11-4-promoted activation was not caused by a decrease in Fus3 protein levels (Fig. 4A and 4C), nor was it attributable to competition between the plasmid-expressed Ste11-4 and endogenous Ste11 protein, because it also occurred when Ste11-4 was expressed in an ste11Δ background (Fig. 4D). In addition, there was no restoration of Ste11-4-promoted Fus3 activation in cells lacking Kss1 (Fig. 4E, lane 7); thus, Kss1 was not blocking Fus3 activation. Yet another possibility was that exposure to chronic stimulation was somehow desensitizing Fus3. However, Fus3 was capable of being activated, because it was rapidly phosphorylated following the addition of mating pheromone (Fig. 4E, lanes 4 and 8). Finally, it was unlikely that Ste5 was sequestering Fus3 into a complex from which Ste11-4 was excluded because if this were true, Fus3 activation should have increased (and not decreased) when Ste5 was removed. Ste11-4-promoted activation of Kss1 still occurred in cells lacking Fus3 (Fig. 4E, lane 11).
Although the observation that Ste11-4 signaling to Kss1 increased in the absence of Ste5 is consistent with the idea that Ste5 sequesters limiting amounts of Ste11 and/or Ste7 away from the invasive growth pathway, this model does not explain why Fus3 phosphorylation decreased in the absence of Ste5. Hence, we favor an alternative explanation for the increase in Ste11-4 signaling to Kss1 in the absence of Ste5, that it is a result of the loss of the Fus3-dependent feedback circuit documented previously (41).
To summarize, as in the previous experiments, Kss1 phosphorylation did not require Ste5 scaffolding, whereas Fus3 phosphorylation was strongly dependent on Ste5 even when Fus3 was receiving a misdirected, leaking signal.
Active Ste5 signals preferentially to Fus3.
Ste5 is thought to exist in an inactive conformation in resting cells and to adopt an active, open conformation by binding to the pheromone receptor-coupled G protein during mating (15, 23, 43, 48). However, a low level of active Ste5 is present even in the absence of pheromone due to a low level of spontaneous activation of the G protein (44). We speculated that this low level of active Ste5 was responsible for the small amount of signal crossover to Fus3MAPK that we observed (e.g., in Fig. 3A, 3C, 4A, and 4C). In other words, perhaps signal crossover was limited because a component required for crossover (active Ste5) was limiting.
The fusion of glutathione S-transferase (GST) to the C terminus of otherwise wild-type Ste5 creates a constitutively active Ste5 mutant by inducing Ste5 to adopt an active conformation (22, 48). This mutant signals in the absence of pheromone but still requires Ste4Gβ, indicating that it spontaneously associates with Gβγ to form a membrane-associated signaling complex. When Ste5-GST was greatly overproduced (by expressing it from the inducible GAL1 promoter under fully inducing conditions) in otherwise normal, unstimulated cells, both Kss1 and Fus3 were hyperphosphorylated (Fig. 5A). Notably, the signal from active Ste5 was preferentially channeled into Fus3 (Fig. 5A), perhaps because Ste5 binds to Fus3 with a higher affinity than it does to Kss1 (25). These results suggest that Ste5 can channel signal to both Fus3 and Kss1 but shows a preference for Fus3.
In contrast, when another constitutively active allele of Ste11MEKK (Ste11ΔN) (9) was greatly overproduced from the GAL1 promoter, Kss1 was preferentially hyperphosphorylated (Fig. 5A), recapitulating the results obtained with the Ste11-4 mutant or osmostress-activated Ste11 (Fig. 3 and 4). The Ste11ΔN mutant lacks the Ste5-binding domain of Ste11 (24) and thus is unlikely to signal to Fus3 and Kss1 via Ste5. Accordingly, the absence of Ste5 did not diminish Fus3 activation by overproduced Ste11ΔN (data not shown). Thus, the requirement for Ste5-mediated scaffolding in Fus3 phosphorylation can be partially bypassed by overexpressing active Ste11.
Active Ste5 is responsible for signal crossover to the mating pathway.
If signal crossover was constrained because the amount of active Ste5 was limiting, then increasing the amount of active Ste5 should increase crossover. To test this proposition, we sought to express the Ste5-GST protein at a level that would not, by itself, substantially activate Fus3; this was achieved by growing cells containing the Ste5-GST expression plasmid in a low concentration (0.067%) of the inducer galactose. This treatment resulted in a modest induction of FUS1 expression (Fig. 5B) and Kss1 and Fus3 phosphorylation (Fig. 5C, compare lane 2 to lane 1). Cells expressing only the Ste11-4 allele also exhibited modest FUS1 expression (Fig. 5B) and preferential phosphorylation of Kss1 (Fig. 5C, lane 3), as seen above (Fig. 4). Strikingly, cells expressing Ste11-4 in conjunction with Ste5-GST displayed greatly increased FUS1 expression (Fig. 5B) and Fus3 phosphorylation (Fig. 5C, lane 4). Kss1 phosphorylation was also increased by this combination (Fig. 5C, lane 4), consistent with the ability of Ste5 to guide some signal to Kss1 (Fig. 3).
Our interpretation of these results is illustrated in Fig. 5D. Only when both Ste5 and Ste11 were active was signal effectively channeled to Fus3. Efficient channeling required active Ste5 protein, because wild-type Ste5, when expressed at a low level comparable to Ste5-GST, was unable to channel the Ste11-4-generated signal to Fus3 (Fig. S1). Hence, active Ste5 allowed the otherwise inefficient Ste11-4-generated signal to further activate Kss1 and to potently activate Fus3, leading to increased signal crossover.
Increased signal crossover changes the developmental option chosen by the cell.
Presumably the reason why S. cerevisiae and other organisms have evolved multiple mechanisms to limit signal crossover is because such crossovers would have a negative impact on their fitness. To begin to explore this issue, we investigated some of the functional consequences of the increased leaking that we observed when the cellular concentration of active Ste5 was increased. Two dramatic consequences were readily apparent.
First, whereas otherwise wild-type cells containing the Ste11-4 mutant exhibited increased invasiveness (data not shown), cells containing both Ste11-4 and low levels of Ste5-GST went into cell cycle arrest, as indicated by a lack of growth under inducing conditions (Fig. 6A). This arrest was partially suppressed in the absence of Fus3MAPK, consistent with the hypothesis that the aberrant activation of Fus3 was (at least partially) responsible for the arrest.
Second, Ste5-promoted signal crossover was able to induce the mating program. To demonstrate this, Ste5-GSTC177A C180A, a mutant of Ste5-GST that is active even in the absence of Ste4 (23), was expressed or not in an ste4Δ strain, with or without coexpression of Ste11-4. Ste5-GSTC177A C180A expressed at a low levels did not stimulate mating on its own (Fig. 6B), although when expressed at higher levels it is capable of doing so (23). Similarly, cells containing only Ste11-4 did not mate efficiently. In contrast, cells containing both Ste11-4 and Ste5-GSTC177A C180Awere able to mate efficiently with a partner of the opposite mating type even though they lacked Ste4Gβ and thus were unable to effectively receive the pheromone signal from that mating partner (Fig. 6B). Hence, aberrant signal crossover caused by Ste5's being active at an inappropriate time resulted in a miscued induction of cell cycle arrest and mating.
DISCUSSION
Scaffold proteins have been thought to enhance signaling specificity by multiple mechanisms. Many of these ideas have been difficult to test, however, although progress has been made recently (20, 35). Here, we examined the role of a prototype scaffold protein, S. cerevisiae Ste5, in maintaining specificity between MAPK pathways that share components. In particular, we dissected the requirement for Ste5 in signal transmission to the Kss1 and Fus3 MAPKs and investigated the hypothesis that Ste5 actively prevents signal crossover, or leaking, into the mating pheromone response pathway from other pathways. We obtained three main findings, which prompt a reevaluation of the influence of Ste5 on some aspects of signaling specificity.
First, we tested the following simple hypothesis: if the Ste5 scaffold protein acts as a passive physical barrier to protect the Fus3 MAPK from misdirected signals and thereby prevents leaking into the mating pathway, then removing Ste5 should increase such leaking. In contrast to this prediction, we found that signal crossover from the invasive growth and osmostress pathways to Fus3 was not increased in the absence of Ste5; in fact, it was diminished (Fig. 3 and 4). Therefore, we conclude that Ste5 does not act as a sequestering barrier that isolates the mating pathway from intrusions from other pathways (i.e., it does not act as shown in Fig. 1C).
Second, we found that rather than preventing leaking into the mating pathway, active Ste5 actually promotes such leaking. Increasing the cellular concentration of active Ste5 resulted in increased signal crossover into the mating pathway, as measured by Fus3MAPK and Kss1MAPK activation, mating gene expression, and the downstream endpoints of mating and cell cycle arrest (Fig. 5 and 6). This aberrant signal crossover caused the cells to undergo Fus3-promoted cell cycle arrest and to attempt to mate under conditions that would normally encourage invasive growth (Fig. 6). These results suggest that when Ste5 is active, it directs some of the signal from activated Ste11MEKK to Fus3 and does not distinguish which pathway the activated Ste11 came from. The idea that constitutively active scaffolds may promote enhanced signal crossover is potentially relevant to the pathogenesis of cancer and other diseases.
Distinct requirements of MAPKs for scaffolding.
Our third main finding was a dramatic difference in the dependency of the MAPKs Fus3 and Kss1 on Ste5 scaffolding. Whereas both MAPKs required the adapter function of Ste5 during mating, Ste5 scaffolding was not required for Kss1 activation by legitimate signals from the mating or invasive growth pathways or for Kss1 activation via signal crossover from the osmostress pathway. In marked contrast, Fus3 phosphorylation was strongly dependent on Ste5 whether Fus3 was receiving a bona fide mating signal or a misdirected signal leaking from the invasive growth or osmostress pathway (Fig. 2, 3, and 4).
Kss1 activation during mating required Ste5's adapter function (i.e., its ability to bind Ste11MEKK) but not its scaffold function (i.e., its ability to bind Ste7MEK). The adapter function requirement is consistent with the generally accepted proposal that Ste11 is activated at the membrane while bound to Ste5. Apparently, however, Ste5 did not prevent bound, active Ste11 from leaking out and encountering nonscaffolded Ste7. Hence, this experiment (Fig. 2A, lane 4) provides some evidence against the model shown in Fig. 1B. A caveat to this interpretation is that it is based upon analysis of a mutant of Ste5 that cannot bind Ste7; perhaps Ste7 binding to Ste5 reduces Ste11 dissociation from Ste5.
The mechanism underlying the strikingly different Ste5 dependencies of Fus3 versus Kss1 remains to be determined. These two MAPKs show greater than 50% sequence identity, are activated by the same MEK (Ste7), and have some key substrates in common (e.g., the Ste12, Dig1 and Dig2 transcription factors) (8). In vitro, Fus3 activation clearly requires Ste7 but does not require Ste5 (16, 33), although a differential enhancement of Fus3 versus Kss1 activation by purified Ste5 protein has been reported (8). The requirement for Ste5 in Fus3 activation is evidently intimately associated with the scaffolding function of Ste5, since the Ste5V763A S861P mutant, which cannot bind to Ste7, did not support Fus3 activation. Perhaps Ste5 causes a conformational change in Fus3 that is required for its phosphorylation by cobound Ste7. Presumably, Kss1 is exempt from these requirements or has them provided by another protein.
Possibly, Kss1 requires a different scaffold for its activation. If so, then our results suggest that this putative scaffold must be active most of the time (including during mating and osmostress) and that it does not protect Kss1 from signal crossover from the osmostress pathway (this work) or the mating pathway (41). Thus, this putative scaffold apparently does not contribute to specificity. The simpler model, which we favor, is that Kss1 activation is largely scaffold independent.
Model of how leaking into the mating pathway is prevented.
Apparently, the Ste5 scaffold acts not as an insulant or barrier but as a conduit or sluice that, when open, undiscerningly guides signals from shared upstream components towards Fus3MAPK. Despite this seeming handicap, signal crossover into the mating pathway is normally limited. A model for how this may be achieved is presented in Fig. 7. This model is based upon the idea that Ste5 is in an inactive, closed conformation in nonmating cells and converts to an active, open, membrane-localized configuration in response to pheromone stimulation (15, 23, 43, 48). We suggest that since Fus3 requires Ste5 scaffolding to receive potentially leaky signals and Ste5 is inactive in nonmating cells, Fus3 ignores such signals most of the time.
FIG. 7.
Model for specific signaling by the selective activation of the Ste5 scaffold protein combined with MAPKs with different scaffold requirements. (A) Fus3MAPK requires active Ste5 in order to effectively receive any signals. Hence, in nonmating cells, Fus3 mostly ignores signals from the invasive growth and osmostress pathways because Ste5 is inactive. Kss1MAPK uses an unknown scaffold or is scaffold independent. Hog1MAPK may use Pbs2MEK as a scaffold (37, 50). When inactive, Ste5 is presumably unable to organize Ste11MEKK, Ste7MEK, and Fus3 into a signaling-competent configuration. Here we have pictured this as a lack of Ste5-kinase association (48). (B and C) In mating cells, activated Ste5 relays the pheromone signal via activated Ste11 to Fus3. However, activated Ste11 can apparently also leak out and transmit signals in a scaffold-independent fashion (B). Furthermore, active Ste5 can also accept Ste11 activated by the other pathways and channel those signals to Fus3, promoting leaking (C).
In this view, the scaffold dependency of Fus3 and the fact that Ste5 is capable of effectively transmitting signals to Fus3 only during mating collaborate to insulate Fus3 from signal crossover. The Ste5-Fus3 interaction somehow creates the equivalent of a molecular AND gate. Fus3 activation requires the coincidence of two separable events, Ste11MEKK activation and Ste5 activation (Fig. 5). Both events occur during mating, but just Ste11 is activated during invasive growth or osmotic stress.
In contrast to Fus3, Hog1MAPK and Kss1MAPK are independent of Ste5 scaffolding and so can receive signals when Ste5 is inactive, although Kss1 still requires the adapter function of Ste5 during mating. During invasive growth, Kss1 is activated to a lesser extent than it is during mating (L. J. Flatauer, unpublished observations); Kss1 is also less effective than Fus3 at driving mating gene expression (39) or causing cell cycle arrest (8). Hence, invasive growth signals do not significantly stimulate mating via Kss1.
Comparison to other results.
Our experiments indicate that Ste5 does not act via sequestration to prevent signals from leaking into the mating pathway. Furthermore, they suggest that Ste5 may also not be very effective at preventing signals from leaking out of the mating pathway. That stable kinase-scaffold associations have the potential to limit leaking out of the scaffolded pathway was demonstrated by Harris et al. (20), who showed that Ste11MEKK, covalently attached to Ste5, stimulated mating but not stress responses whereas Ste11, covalently attached to Pbs2MEK, stimulated stress responses but not mating. Evidence consistent with the idea that selective recognition of Ste7MEK is one important factor in limiting leaking out of the mating pathway was provided by Park et al. (35), who found that covalently attaching Pbs2MEK to Ste5 created a mating-to-osmostress leak.
Pbs2MEK and Sho1 have been proposed to act together to scaffold the osmostress pathway (37, 50). However, leaking out of this pathway occurs in cells in which Sho1 and Pbs2 are both present (19, 34) and functional for signal transmission (42). Hence, the Sho1/Pbs2 scaffold apparently does not effectively sequester stress-activated Ste11 and restrain it from straying into other pathways, at least not under the conditions studied.
Other mechanisms act to restrict signal crossover in certain directions. MAPK-dependent mechanisms limit leaking from the osmostress pathway (14, 19, 34) and from mating into invasive growth (41). The mechanisms that prevent signals from leaking into the Hog1MAPK pathway are unknown at present. Possibly, in analogy to the model we have proposed here, Hog1 or Pbs2 requires activation by stress in order to receive signals from Ste11.
At first consideration, our finding that pheromone-activated Ste11 was apparently able to dissociate from Ste5 and phosphorylate Ste7 is inconsistent with the results of the diverter scaffold experiment of Park et al. (35): if pheromone-activated Ste11 readily dissociates from Ste5, then why did Park et al. find it necessary to attach Pbs2 to Ste5 in order to get pheromone-activated Ste11 to phosphorylate Pbs2? Perhaps membrane localization of Pbs2 activates it somehow, making it competent to be phosphorylated by Ste11. Further work will be required to clarify this issue.
Two other groups have also recently found evidence of differential MAPK activation. Maleri et al. found that constitutively active Ste7 selectively activates Kss1 (32); this finding is consistent with our proposal that Ste5 must be activated by upstream components of the mating pathway in order to transmit signals to Fus3. Andersson et al. (1) observed the selective, scaffold-independent activation of Kss1 in response to several manipulations, such as expression of Ste11-4. In contrast to our findings, however, these authors reported that Ste4 and Ste5 are needed to activate Kss1 during invasive growth. The difference between their results and ours seems to be attributable to their use of the noninvasive S228C lineage for most of their experiments. The S288C lineage has acquired multiple mutations during the process of repeated selection in the laboratory for nonclumpy cells that formed round colonies (27). Perhaps one or more of these mutations decreased the ability of Kss1 to be activated in a scaffold-independent fashion (1). Like Andersson et al., we too have observed that in S228C-derived strains, Kss1 activation during mating is more dependent on the scaffold function of Ste5 (data not shown). Furthermore, we found a strong requirement for Ste4 in Kss1 activation during invasive growth in a Σ1278b-derived strain that lacked the Msb2 protein (Fig. 3B). Hence, one possibility is that the S228C lineage may be defective in a component that signals (in an Ste5-independent fashion) from Msb2 to Kss1.
Conclusions.
Here we have shown the Ste5 scaffold protein does not act as an insulant or barrier to block signal crossover into the S. cerevisiae mating pathway. Instead of preventing misdirected signals from activating Fus3MAPK, Ste5 actually helps misdirected signals to activate Fus3. Leaking into the mating pathway is limited under normal circumstances because Fus3 requires active Ste5 to receive any signals and Ste5 is activated only during mating. Kss1MAPK does not require Ste5 scaffolding to receive signals, even during mating, and so can function in two different pathways, one which uses Ste5 and one which does not. If generalized, these observations imply that scaffold proteins promote specificity by directing signal flow in certain directions at certain times but do not actively prevent signal crossover by sequestration.
Supplementary Material
Acknowledgments
We thank G. Boguslawski, B. Cairns, G. Fink, G. Sprague, Jr., and J. Thorner for generous gifts of reagents.
This work was supported by Public Health Service Training Grant GM07311 (to L.J.F.) and by research grants GM60366 and GM69013 from the National Institute of General Medical Science.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andersson, J., D. M. Simpson, M. Qi, Y. Wang, and E. A. Elion. 2004. Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J. 23:2564-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, A. J., L. J. Flatauer, K. Matsukuma, J. Thorner, and L. Bardwell. 2001. A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276:10374-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, L. 2005. A walk-through of the yeast mating pheromone response pathway. Peptides 26:339-350. [DOI] [PMC free article] [PubMed]
- 4.Bardwell, L., J. G. Cook, E. C. Chang, B. R. Cairns, and J. Thorner. 1996. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16:3637-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell, L., J. G. Cook, J. X. Zhu-Shimoni, D. Voora, and J. Thorner. 1998. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. USA 95:15400-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel, P. L., and S. Fields. 1995. Analyzing protein-protein interactions using the two-hybrid system. Methods Enzymol. 254:241-263. [DOI] [PubMed] [Google Scholar]
- 7.Boguslawski, G. 1992. PBS2, a yeast gene encoding a putative protein kinase, interacts with the RAS2 pathway and affects the osmotic sensitivity of Saccharomyces cerevisiae. J. Gen. Microbiol. 138:2425-2432. [DOI] [PubMed] [Google Scholar]
- 8.Breitkreutz, A., L. Boucher, and M. Tyers. 2001. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol. 11:1266-1271. [DOI] [PubMed] [Google Scholar]
- 9.Cairns, B. R., S. W. Ramer, and R. D. Kornberg. 1992. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev. 6:1305-1318. [DOI] [PubMed] [Google Scholar]
- 10.Choi, K.-Y., B. Satterberg, D. M. Lyons, and E. A. Elion. 1994. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78:499-512. [DOI] [PubMed] [Google Scholar]
- 11.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous growth signaling pathway. Nature 390:85-88. [DOI] [PubMed] [Google Scholar]
- 12.Cullen, P. J., W. Sabbagh, Jr., E. Graham, M. M. Irick, E. K. van Olden, C. Neal, J. Delrow, L. Bardwell, and G. F. Sprague, Jr. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18:1695-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davenport, K. D., K. E. Williams, B. D. Ullmann, and M. C. Gustin. 1999. Activation of the Saccharomyces cerevisiae filamentation/invasion pathway by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153:1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elion, E. A. 2001. The Ste5p scaffold. J. Cell Sci. 114:3967-3978. [DOI] [PubMed] [Google Scholar]
- 16.Errede, B., A. Gartner, Z. Zhou, K. Naysmith, and G. Ammerer. 1993. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature 362:261-264. [DOI] [PubMed] [Google Scholar]
- 17.Ferrell, J. E., Jr., and K. A. Cimprich. 2003. Enforced proximity in the function of a famous scaffold. Mol. Cell 11:289-291. [DOI] [PubMed] [Google Scholar]
- 18.Hagen, D. C., G. McCaffrey, and G. F. Sprague, Jr. 1991. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11:2952-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, J. P., V. Cherkasova, E. Elion, M. C. Gustin, and E. Winter. 1996. The osmoregulatory pathway represses mating pathway activity in Saccharomyces cerevisiae: isolation of a FUS3 mutant that is insensitive to the repression mechanism. Mol. Cell. Biol. 16:6715-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, K., R. E. Lamson, B. Nelson, T. R. Hughes, M. J. Marton, C. J. Roberts, C. Boone, and P. M. Pryciak. 2001. Role of scaffolds in MAP kinase pathway specificity revealed by custom design of pathway-dedicated signaling proteins. Curr. Biol. 11:1815-1824. [PubMed] [Google Scholar]
- 21.Hartwell, L. H. 1980. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J. Cell Biol. 85:811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inouye, C., N. Dhillon, T. Durfee, P. C. Zambryski, and J. Thorner. 1997. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: application of a differential interaction trap assay for examining protein-protein interactions. Genetics 147:479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inouye, C., N. Dhillon, and J. Thorner. 1997. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science 278:103-106. [DOI] [PubMed] [Google Scholar]
- 24.Jansen, G., F. Buhring, C. P. Hollenberg, and M. Ramezani Rad. 2001. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:102-117. [DOI] [PubMed] [Google Scholar]
- 25.Kusari, A. B., D. M. Molina, W. Sabbagh, Jr., C. S. Lau, and L. Bardwell. 2004. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1. J. Cell Biol. 164:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, H., C. A. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741-1744. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H., C. A. Styles, and G. R. Fink. 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144:967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodish, H., A. Berk, P. Matsudaira, C. A. Kaiser, M. Krieger, M. P. Scott, S. L. Zipursky, and J. Darnell. 2003. Molecular cell biology, 5th ed. W. H. Freeman, New York, N.Y.
- 29.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 30.Madhani, H. D., and G. R. Fink. 1998. The riddle of MAP kinase signaling specificity. Trends Genet. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 31.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 32.Maleri, S., Q. Ge, E. A. Hackett, Y. Wang, H. G. Dohlman, and B. Errede. 2004. Persistent activation by constitutive Ste7 promotes Kss1-mediated invasive growth but fails to support Fus3-dependent mating in yeast. Mol. Cell. Biol. 24:9221-9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neiman, A. M., and I. Herskowitz. 1994. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homolog STE7 by STE11. Proc. Natl. Acad. Sci. USA 91:3398-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, S. H., A. Zarrinpar, and W. A. Lim. 2003. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299:1061-1064. [DOI] [PubMed] [Google Scholar]
- 36.Pawson, T., and P. Nash. 2000. Protein-protein interactions define specificity in signal transduction. Genes Dev. 14:1027-1047. [PubMed] [Google Scholar]
- 37.Posas, F., and H. Saito. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702-1705. [DOI] [PubMed] [Google Scholar]
- 38.Pryciak, P. M., and F. A. Huntress. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 12:2684-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 40.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 41.Sabbagh, W., Jr., L. J. Flatauer, A. J. Bardwell, and L. Bardwell. 2001. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuller, C., J. L. Brewster, M. R. Alexander, M. C. Gustin, and H. Ruis. 1994. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 13:4382-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sette, C., C. J. Inouye, S. L. Stroschein, P. J. Iaquinta, and J. Thorner. 2000. Mutational analysis suggests that activation of the yeast pheromone response mitogen-activated protein kinase pathway involves conformational changes in the Ste5 scaffold protein. Mol. Biol. Cell 11:4033-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siekhaus, D. E., and D. G. Drubin. 2003. Spontaneous receptor-independent heterotrimeric G-protein signalling in an RGS mutant. Nat. Cell Biol. 5:231-235. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson, B. J., N. Rhodes, B. Errede, and G. F. Sprague, Jr. 1992. Constitutive mutants of the protein kinase STE11 activate the yeast pheremone response pathway in the absence of the G protein. Genes Dev. 6:1293-1304. [DOI] [PubMed] [Google Scholar]
- 46.van Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 47.van Drogen, F., and M. Peter. 2002. MAP kinase cascades: scaffolding signal specificity. Curr. Biol. 12:R53-R55. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Y., and E. A. Elion. 2003. Nuclear export and plasma membrane recruitment of the Ste5 scaffold are coordinated with oligomerization and association with signal transduction components. Mol. Biol. Cell 14:2543-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitmarsh, A. J., and R. J. Davis. 1998. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 23:481-485. [DOI] [PubMed] [Google Scholar]
- 50.Zarrinpar, A., R. P. Bhattacharyya, M. P. Nittler, and W. A. Lim. 2004. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell 14:825-832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.