Abstract
Studies ex vivo have shown that phosphoinositide 3-kinase (PI3K) activity is necessary but not sufficient for insulin-stimulated glucose uptake. Unexpectedly, mice lacking either of the PI3K regulatory subunits p85α or p85β exhibit increased insulin sensitivity. The insulin hypersensitivity is particularly unexpected in p85α−/− p55α−/− p50α−/− mice, where a decrease in p110α and p110β catalytic subunits was observed in insulin-sensitive tissues. These results raised the possibility that decreasing total PI3K available for stimulation by insulin might circumvent negative feedback loops that ultimately shut off insulin-dependent glucose uptake in vivo. Here we present results arguing against this explanation. We show that p110α+/− p110β+/− mice exhibit mild glucose intolerance and hyperinsulinemia in the fasted state. Unexpectedly, p110α+/− p110β+/− mice showed a ∼50% decrease in p85 expression in liver and muscle. Consistent with this in vivo observation, knockdown of p110 by RNA interference in mammalian cells resulted in loss of p85 proteins due to decreased protein stability. We propose that insulin sensitivity is regulated by a delicate balance between p85 and p110 subunits and that p85 subunits mediate a negative role in insulin signaling independent of their role as mediators of PI3K activation.
Type 2 diabetes is a serious public health problem that is growing rapidly in the developed world. The disease is characterized by insulin resistance of the peripheral organs and hyperglycemia. Initially, patients compensate by secreting elevated insulin, but over the course of the disease, the pancreatic beta cells fail to produce compensating levels of insulin. Based on ex vivo experiments, phosphoinositide 3-kinase (PI3K) is required for insulin-induced glucose uptake into muscle and fat and inhibition of glucose production in the liver (10, 30, 35, 38). Polymorphism in the human PI3K regulatory isoform p85α gene has been associated with increased risk for developing type 2 diabetes (5). However, the consequences of the polymorphism on the expression level or function of p85α have not yet been analyzed. Many of the metabolic effects of insulin require activation of the PI3K downstream target Akt. While constitutively active Akt induces translocation of the glucose transporter GLUT4 to the plasma membrane in adipocytes, dominant negative Akt inhibits it (13). Consistent with this, targeted disruption of Akt2 in the mouse resulted in insulin resistance and severe diabetes, and RNA interference (RNAi) for Akt2 inhibited insulin-dependent glucose uptake in 3T3L1 cells (12, 19, 28). An inactivating mutation in human AKT2 has been associated with severe insulin resistance and diabetes mellitus, demonstrating a pivotal role of AKT signaling in glucose homeostasis in humans (20).
Surprisingly, deletion of PI3K regulatory subunits in mice improves insulin signaling. Despite the necessity of PI3K activity for insulin-induced glucose uptake, genetic ablation of PI3K regulatory isoforms p85α or p85β resulted in improved insulin signaling (15, 33, 39, 42, 43). Based on studies with mice lacking the first exon of p85α (where p55α and p50α alternative splice forms are upregulated), Terauchi et al. hypothesized that the p55α and p50α isoforms were more efficient than p85α in mediating insulin responses (39). However, mice deficient in all p85α splice variants (p85α−/− p55α−/− p50α−/−) and p85β null mice also exhibited hypersensitivity to insulin (15, 43). More recently, mice lacking p55α and p50α isoforms but expressing p85α were also shown to have increased insulin sensitivity (11).
There is evidence in some cell lines that p85 isoforms are in excess over p110 isoforms and can compete with p85/p110 enzyme for binding to IRS proteins (41). Although this model can explain how excess p85 impairs insulin signaling in many situations, it cannot explain why p85α−/− p55α−/− p50α−/− mice display hypersensitivity to insulin despite a reduction in total PI3K and a 60% decrease of PI3K recruitment to IRS complexes after insulin treatment (15).
Recent studies point to a PI3K activity-dependent feedback inhibition of insulin signaling. The negative feedback loop results in an increase in IRS1 Ser-307 (Ser-312 in humans) phosphorylation, decreased kinase activity of the insulin receptor, decreased IR-IRS1 interaction, increased IRS1 cytosolic distribution, increased IRS1 degradation, and decreased association between IRS1 and SH2 domain-containing proteins, such as p85 (2, 22, 31, 47). Ser-307/Ser-312 phosphorylation is sensitive to inhibitors of the PI3K/Akt/aPKC/mTor pathway, as well as a c-jun kinase (JNK) small-molecule inhibitor and an IKK inhibitor (18, 22, 23, 31, 32). Obesity has been shown to result in insulin resistance (21). One mechanism by which obesity may cause insulin resistance is thought to occur via tumor necrosis factor (TNF)-induced, PI3K-dependent serine (Ser-307) phosphorylation on IRS proteins (27, 36, 45). Additionally, JNK has been implied in obesity-induced insulin resistance, as obese mice lacking JNK1 (JNK1−/−) exhibit decreased IRS1 Ser-307 phosphorylation and increased sensitivity to insulin in comparison to obese JNK1+/+ mice (26). In addition, inhibition of PI3K or the downstream kinase mTOR has been shown to prevent downregulation of insulin/IRS-1 signaling (34, 37). Furthermore, a recent study has shown that deletion of p70S6kinase in the mouse increases insulin sensitivity (44). These results suggest the existence of a strictly controlled, complex, negative feedback regulation of insulin signaling downstream of PI3K and raise the possibility that deletion of PI3K genes (regulatory or catalytic) might improve insulin signaling by preventing insulin-responsive tissues from reaching the threshold of PI3K response that activates a negative feedback loop.
To test the model that reduction in total PI3K activity in insulin-sensitive tissues can improve insulin-dependent glucose disposal in vivo, we examined mice heterozygous for PI3K p110α and/or p110β. Heterozygous loss of p110α or p110β alone had no effect on insulin sensitivity as judged by insulin tolerance tests or glucose tolerance tests. However, mice with double heterozygous loss of p110α and p110β were slightly glucose intolerant and exhibited hyperinsulinemia in the fasting state. Interestingly, not only were the PI3K catalytic isoforms decreased by ∼50% in tissues from p110α+/− p110β+/− mice, but also p85 protein levels decreased by a similar amount. Downregulation of p110α in cell lines using RNAi also resulted in a reduction of the p85 regulatory subunit, indicating a dynamic intracellular mechanism for control of p85/p110 ratios.
Thus, although heterozygous loss of p110α and p110β results in a similar reduction in total class Ia PI3K in muscle and liver to that observed in the p85α+/− p55α+/− p50α+/− mice, the effect on insulin-dependent glucose disposal is the opposite. These data argue against the model that loss of p85 isoforms improves insulin signaling by preventing a PI3K-dependent feedback shutoff of insulin signaling. The data imply that p85 has a negative role in insulin signaling independent of PI3K regulation and suggest that the ratio of p85 to p110 plays a critical role in setting in vivo insulin sensitivity.
MATERIAL AND METHODS
Mice.
Mice on a 129 background with heterozygous loss of p110α (p110α+/−) or p110α (+/−) were intercrossed to produce wild-type, single heterozygous, and double heterozygous mice. All animals were housed in microisolator cages on a 12-h light-12-h dark cycle and were fed a standard rodent chow (Purina) or put on a high-fat diet (Teklad) after they reached 2 months of age. Animal maintenance and experimental procedures were according to the National Institutes of Health guidelines and were approved by the Animal Care Use Committee of Harvard Medical School.
PCR.
Mice were genotyped by PCR of crude tail genomic DNA (digested using modified Gitschier's buffer). In order to genotype p110α+/− mice, we used primers PαF (5′TGG CAT CCT CAA ATG ATA GTA ACA 3′), PαR (5′GAA TAA AAT AAA GAG GAG GCA TCA TAG 3′), and PNEO (5′GAA GGC TCT TTA CTA TTG CTT TAT 3′). In order to genotype p110β mutant mice, we used primers PβF (5′GTA TTT GGA CCT GAT TTG ATG ATA G 3′), PβR (5′GTT TTT ATT TGA AAT TAA ATC ACA TCG C 3′), and PNEO (5′GAA GGC TCT TTA CTA TTG CTT TAT 3′).
TaqMan real-time PCR.
CHO-IR cells were transiently transfected with bovine HA-p110α, mouse Flag-p85α, and either empty vector or short hairpin RNA (shRNA) for knockdown of expression of p110α. After 48 h total RNA was isolated and analyzed by real-time PCR. Quantities were normalized to 18S rRNA quantities measured in the same samples. The following primers were used to amplify mouse p85α: mp85a-539F (CCTTGTCCGGGAGAGCAGTA), mp85a-604R (TTGACTTCGCCGTCTACCACT), and mp85a-561T (CAGGGCTGCTATGCCTGCTCCG) coupled to tetramethyl carboxyrhodamine.
Tissue lysate preparation and Western blotting.
Mice were starved overnight, anesthetized with pentobarbital, and injected with 5 U of regular human insulin or phosphate-buffered saline into the inferior vena cava. After 5 min, liver and quadricepts muscles were removed and flash frozen in liquid nitrogen. Immunoprecipitation and immunoblot analysis of insulin-signaling molecules were performed on tissue homogenates extracted with buffer A containing 25 mM Tris-HCl (pH 7.4), 10 mM EDTA, 10 mM EGTA, 10 mM Na3VO4, 50 mM Na4P2O7, 100 mM NaF, 5 μg of leupeptin/ml, 5 μg of aprotinin/ml, 2 mM phenylmethylsulfonyl fluoride, and 1% (vol/vol) Nonidet P-40, as described before (8).
Antibodies.
Rabbit polyclonal anti-p85 antibody (anti-p85pan) was purchased from Upstate Biotechnology, Lake Placid, N.Y. Rabbit polyclonal anti-p110α antibody and anti-p110β antibody were purchased from Santa Cruz Biotechnology. Goat polyclonal anti-Akt antibody was purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-phospho-Akt antibodies recognizing phosphorylated Ser-473 or Thr-308 of Akt1 were purchased from Cell Signaling Technology, Beverly, Mass. Rabbit polyclonal anti-IR antibody, anti-IRS-1 antibody, and anti-IRS-2 antibody were generated (8). Mouse monoclonal antiphosphotyrosine antibody (4G10) was purchased from Upstate Biotechnology. Mouse monoclonal anti-HA11 antibody was purchased from Covance/Babco, rabbit polyclonal anti-FLAG antibody was purchased from Sigma, and anti-FLAG M2 agarose was purchased from Sigma.
PI3K activity assays.
The immunoprecipitates with anti-p110α, anti-p110β, anti-IRS-1, or anti-IRS-2 were washed twice with buffer A and twice with TNE (10 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA). The PI3K assay was performed by adding 20 μl of HEPES (100 mM; pH 7), 20 μl of lipids (67 μM phosphoinositide) (PI; Avanti Polar Lipids), 133 μM phosphoserine (PS; Avanti Polar Lipids) and 10 μl of ATP mix (70 mM HEPES [pH 7], 50 mM MgCl2, 0.5 mM ATP, [γ-32P]ATP [10 μCi/assay mixture]) to 50 μl of immunoprecipitate. The reactions were performed at room temperature and stopped after 20 min by adding 25 μl of HCl (5 M). The lipids were extracted with 160 μl of CHCl3-MeOH (1:1). The phosphorylated lipids were spotted on a thin-layer chromatography (TLC) and separated overnight with 1-propanol-2 M acetic acid (65:35). The radioactivity was visualized by using a PhosphorImager (Molecular Dynamics).
Akt activity assays.
Tissue homogenates were immunoprecipitated with anti-Akt polyclonal antibody that recognizes both Akt1 and Akt2, and an Akt kinase assay with cross-tide as a substrate was performed as described elsewhere (40). Briefly, the immunoprecipitates were washed and resuspended in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 1 mM dithiothreitol (DTT) to which 20 μM ATP, 5 μCi of [γ-32P]ATP, and 5 μg of cross-tide for AKT assay (Upstate Biotechnology) had been added. The reaction was performed at 30°C and after 20 min the reaction was stopped, and the aliquots were spotted onto P-81 paper, washed with 0.5% phosphoric acid, and counted for radioactivity.
High-fat diet.
Mice were fed a high-fat diet (containing 55% fat and 24% carbohydrates by calories; catalog no. TD 93075; Harlan Tekland, Madison, Wis.) beginning at the time of weaning (21 days of age).
Weight measurements.
Mice were weighed weekly between the ages of 2 and 6 months.
Glucose tolerance tests.
Overnight-starved mice were injected intraperitoneally with 2 g of glucose per kg of body weight at 9 a.m. Blood glucose levels were measured with a One Touch Basic glucose meter at 0, 15, 30, 60, 120, and 180 min after injection.
Insulin tolerance tests.
Mice were starved at 9:00 a.m. and injected intraperitoneally at 1:00 p.m. on the same day with regular human insulin at a dosage of 0.5 to 1.5 U per kg of body weight, dependent on age and gender of the animals. (In detail, female mice were injected with 0.5 U/kg at 2 months of age, 0.75 U/kg at 4 months of age, and 1 U/kg at 6 months of age. Male mice were injected with 0.75 U/kg at 2 months of age, 1 U/kg at 4 months of age, and 1.5 U/kg at 6 months of age.) Blood glucose levels were measured with a One Touch Basic glucose meter at 0, 15, 30, 45, 60, and 90 min after injection.
Insulin measurements.
Serum insulin levels were measured in duplicate by using enzyme-linked immunosorbent assay reagents from Crystal Chem Inc. (catalog no. 90060).
Statistical analysis.
Glucose and insulin tolerance tests were analyzed by repeated-measures analysis of variance. Weight growth curves, kinase assays, and glucose and insulin levels were analyzed by Student's t test.
RNA interference.
Oligonucleotides designed to produce shRNAs targeting p110 α were cloned into a retroviral vector that utilizes a U6 promoter (14). The oligonucleotides were cloned into the vector after digestion of the vector with ApaI (Klenow)/EcoRI. The oligonucleotide sequences were GGAGGAGAACCCTTATGTGACTTCTGAAGAGTCACATAAGGGTTCTCCTCCCTTTTTG (top strand; nucleotides in bold are those targeting p110α, human and bovine) and AATTCAAAAAGGGAGGAGAACCCTTATGTGACTCTTCAGAAGTCACATAAGGGTTCTCCTCC (bottom strand). Retroviral infection of HeLa cells was performed with the supernatants of 293T cells cotransfected with the p110α shRNA retroviral vector and PCL Ampho packaging vector (a generous gift from William Hahn). Analysis of protein expression in the infected HeLa cells was conducted after 7 days of selection with puromycin.
Transient transfections.
CHO cells were transfected with expression vectors for HA-p110α (bovine), Flag-p85α (mouse), Flag-p55α (mouse), HA-Δp85α (human), and either empty vector or vector expressing shRNA for targeting of human p110α. Two days later the cells were either lysed for Western blotting and immunoprecipitation or used for metabolic labeling. CHO cells were transfected with expression vectors for HA-Akt and increasing amounts of Flag-p85α in pCMV6 and decreasing amounts of empty vector pCMV6. The cells were allowed to recover for 3 h in Dulbecco's modified Eagle's medium (DMEM)-F12-10% fetal calf serum (FCS). The cells were starved overnight in DMEM-F12-0% FCS. The next day cells were stimulated with 20 nM IGF-1 for 5 min, lysed, and immunoprecipitated with 5 μl of anti-HA-11/500 μg of total cell lysate. The lysate and immunoprecipitate were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by immunoblotting with anti-p85pan, anti-AktT308P, and anti-HA-11 antibodies. All transfections were done according to the manufacturer's protocol (Lipofectamine Plus; Invitrogen).
Metabolic labeling experiments.
CHO cells were transfected with expression vectors for HA-p110α (bovine) or Flag-p85α (mouse) in the presence or absence of an expression vector for shRNAi targeting p110α. Two days after transfection, CHO cells were incubated for 1.5 h with methionine-cysteine-free medium containing 0.1 mCi of methionine-cysteine (Easy-Tag Express 35S; NEG 772; NEN)/ml and then chased for the indicated times in regular growth medium (DMEM-F12-10% FCS). The cells were lysed and immunoprecipitated with 50 μl of anti-Flag (M2) agarose beads/500 μg of total cell lysate or with 250 μl of anti-Flag (M2) agarose beads/5,000 μg of total cell lysate, eluted with 0.5 mg of Flag peptide/ml for 1 h at room temperature, and subsequently immunoprecipitated with 25 μl of anti-HA-11 antibody and 100 μl of protein A/G mix/5,000 μg of total cell lysate in a 3-ml total volume. The immunoprecipitate was separated by SDS-PAGE. The gel was dried, and the radiolabeled proteins were visualized by phosphorimager and autoradiography.
RESULTS
p110α+/− p110β+/− mice are slightly glucose intolerant.
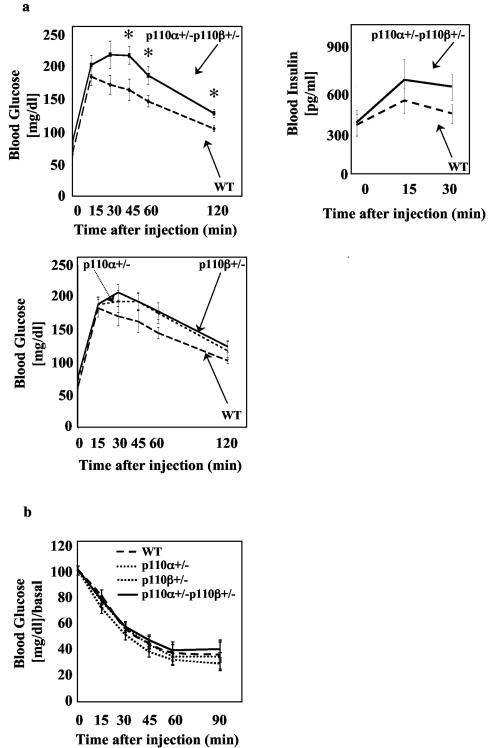
Since PI3K activity has been shown to be necessary for insulin-induced glucose uptake in vitro (10, 25), it was surprising that targeted disruption of any PI3K p85 regulatory isoform caused improved insulin sensitivity in mice even when total PI3K activity in tissues was reduced (11, 15, 39, 43). In order to understand if the increased insulin sensitivity was due to decreased PI3K available for downstream signaling, we examined mice in which catalytic subunits of PI3K were deleted. Since homozygous loss of either p110 isoform in mice is embryonic lethal (6, 7), mice with heterozygous loss of the isoforms were analyzed. At early ages, comparison between the different genotypes did not reveal any significant differences in blood glucose clearance during glucose or insulin tolerance tests (ATT or ITT) (data not shown). However, at 6 months of age, the male p110α+/− p110β+/− mice showed a mild but statistically significant glucose intolerance (Fig. 1a, upper left) and a small (but not statistically significant) elevation in insulin during the GTT (Fig. 1a, upper right). No differences between p110α+/− p110β+/− and wild-type mice in glucose clearance during an insulin tolerance test were observed (Fig. 1b). Mice heterozygous for only p110α or only p110β were not statistically different from wild-type mice in the GTT or ITT but showed reproducibly an intermediate phenotype between p110α+/− p110β+/− and wild-type mice in glucose clearance during the GTT that did not achieve statistical significance (Fig. 1a, lower).
FIG. 1.
p110α+/− p110β+/− mice are mildly glucose intolerant. (a) Blood glucose levels (p110α+/− p110β+/−, n = 13; wild type [WT], n = 13; p110α+/−, n = 11; p110β+/−, n = 21) and blood insulin levels (p110α+/− p110β+/−, n = 10; WT, n = 5) were measured in 6-month-old male mice subjected to glucose tolerance tests as described in Materials and Methods. (b) Insulin tolerance test (p110α+/− p110β+/−, n = 10; WT, n = 5; p110α+/−, n = 8; p110β+/−, n = 12) on 6-month-old male mice. Results are means ± standard errors of the means of the blood glucose (GTT) or blood glucose/basal blood glucose ratio (ITT). *, P < 0.05, compared to WT.
p110α+/− p110β+/− mice exhibit mild hyperinsulinemia in the fasted state.
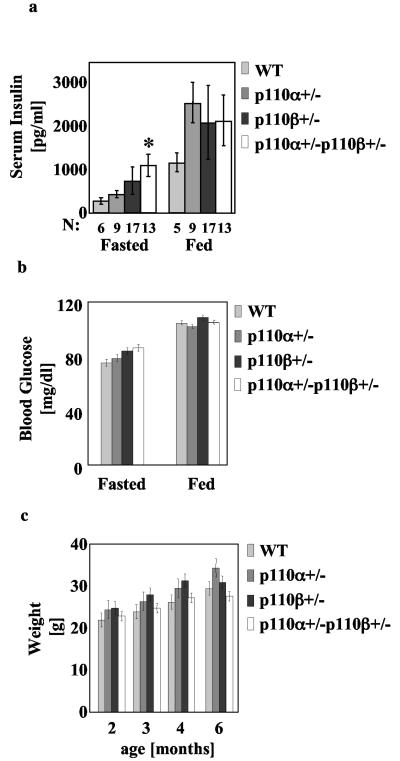
We assessed the blood insulin and glucose concentration in the fasted (after overnight food withdrawal) and fed (at 11 p.m.) state of male wild-type, p110α+/−, p110β+/−, and p110α+/− p110β+/− mice at 6 months of age. The p110α+/− p110β+/− but not the p110α+/− or p110β+/− mice exhibited significantly elevated insulin in the fasted state (Fig. 2a). In the fed state, the heterozygous mice exhibited normal insulin levels in comparison to wild-type mice (Fig. 2a). Also, the fasted and fed glucose levels of the heterozygous mice were not significantly different from those of wild-type mice (Fig. 2b). Since abnormalities in glucose homeostasis are often reflected in overall weight differences, we periodically weighed wild-type, p110α+/−, p110β+/−, and p110α+/− p110β+/− mice from weaning up to 6 months of age. The p110α+/−, p110β+/−, and p110α+/− p110β+/− mice exhibited a similar weight to wild-type mice (Fig. 2c).
FIG. 2.
Mild hyperinsulinemia in p110α+/− p110β+/− mice. (a) Plasma insulin concentrations in 6-month-old fasting and fed male mice. Results are means ± standard deviations of the means. *, P < 0.05, compared to wild type (WT). (b) Weights for male mice with the indicated genotypes at different ages. The data express the means ± standard deviations of the means (WT, n = 6; p110α+/−, n = 8; p110β+/−, n = 15; p110α+/− p110β+/−, n = 12). P values were calculated by using the Student t test. There was no statistically significant difference between genotypes. (c) Blood glucose levels of 6-month-old male fasting or fed mice with the indicated genotypes. The data express the means ± standard deviations of the means (WT, n = 6; p110α+/−, n = 8; p110β+/−, n = 15; p110α+/− p110β+/−, n = 12). P values were calculated by using the Student t test. There was no statistically significant difference between genotypes.
Reduced PI3K catalytic and regulatory subunits in insulin-sensitive tissues of p110α+/− p110β+/− mice.
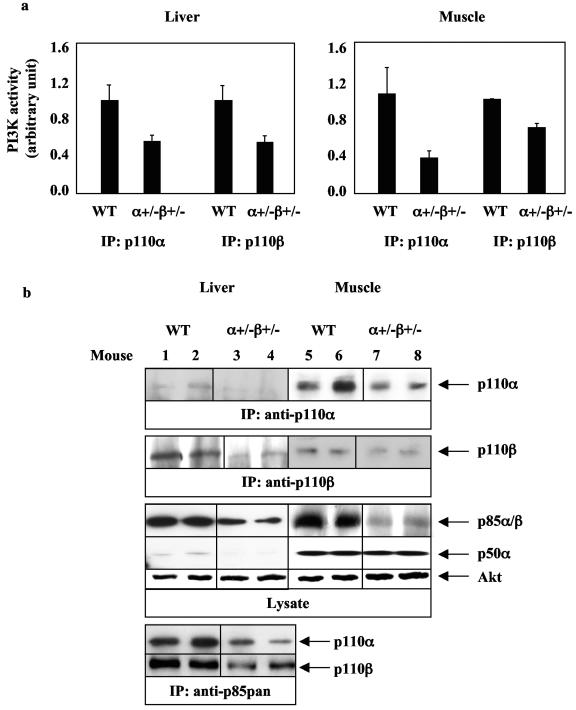
We analyzed protein levels and activation states of enzymes in the insulin signaling pathway in liver and muscle of wild-type and p110α+/− p110β+/− mice. As expected, double heterozygous loss of p110α and p110β (p110α+/− p110β+/−) resulted in a ∼50% reduction of PI3K activities associated with anti-p110α or anti-p110β immunoprecipitates from liver (Fig. 3a, left) and muscle (Fig. 3a, right) as well as a reduction of both p110α and p110β protein levels (Fig. 3b). In order to determine alterations in p85 protein levels, whole liver and muscle lysates were analyzed by Western blot analysis using an anti-p85pan antibody that recognizes all p85 isoforms. In both liver and muscle of p110α+/− p110β+/− mice, p85 protein levels were unexpectedly decreased to ∼50% of levels in wild-type tissue. The smaller p85 isoform, p50α, was also reduced in liver but not in muscle of p110α+/− p110β+/− mice in comparison to wild-type mice (Fig. 3b). It was difficult to visualize the p110α protein from anti-p110α immunoprecipitations in p110α+/− p110β+/− livers. Therefore, we performed an additional anti-p85 immunoprecipitation and anti-p110α or anti-p110β blotting. This approach also revealed approximately twice as much p110α and p110β in wild-type versus p110α+/− p110β+/− livers, consistent with our observation of twice as much PI3K activity.
FIG. 3.
p110α+/− p110β+/− mice have decreased p110 and p85 levels in liver and muscle. (a) PI3K assays in liver (for anti-p110α immunoprecipitation [IP]: wild type [WT], n = 3; p110α+/− p110β+/−, n = 3; for anti-p110β IP: WT, n = 3; p110α+/− p110β+/−, n = 6) and skeletal muscle (for anti-p110β and anti-p110α IP: WT, n = 3; p110α+/− p110β+/−, n = 3) from 4-month-old WT and p110α+/− p110β+/− mice. Results are means ± standard errors of the means and represent one of two experiments. (b) p110α, p110β, and p85 protein levels in liver (left) and skeletal muscle (right) from 4-month-old mice. IP was performed on 2 mg of muscle or 2 mg of liver from WT or p110α+/− p110β+/− mice. A representative experiment is shown. Each lane represents a single mouse.
Reduced PI3K activity associated with IRS proteins in insulin-sensitive tissues of p110α+/− p110β+/− mice upon insulin stimulation.
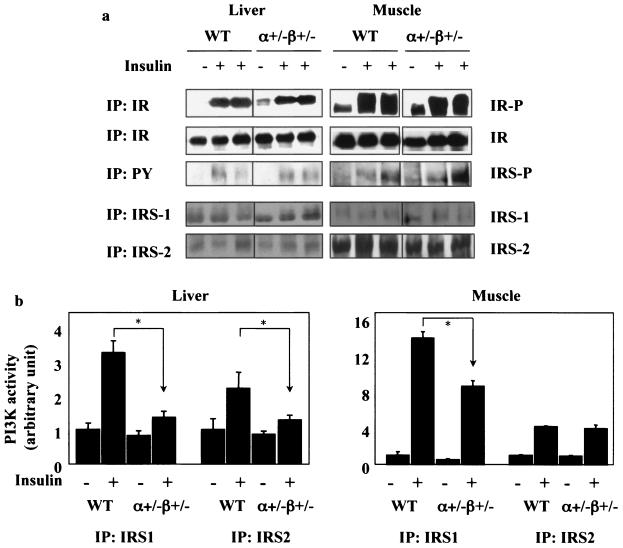
No difference was observed between wild-type and p110α+/− p110β+/− mice in insulin receptor autophosphorylation. Although there was variability in the amount of insulin receptor and IRS protein phosphorylation from mouse to mouse in response to insulin injection, we saw no significant difference between wild-type and p110α+/− p110β+/− mice in the tyrosine phosphorylation of IRS1 and IRS2 (Fig. 4a). However, insulin-induced interaction between IRS1 and PI3K was reduced to ∼42% in liver and ∼61% in muscle of p110α+/− p110β+/− mice in comparison to levels in wild-type mice (Fig. 4b). Insulin-stimulated association between IRS2 and PI3K was normal in muscle (∼95%) but reduced to ∼65% in liver of p110α+/− p110β+/− mice (Fig. 4b).
FIG. 4.
p110α+/− p110β+/− mice have decreased IRS- associated PI3K activity in liver and muscle. (a) Phosphorylation of insulin receptor (upper) and insulin receptor substrates (lower) in liver (left) and skeletal muscle (right) from 4-month-old wild-type (WT) and p110α+/− p110β+/− mice injected with saline or 5 U of insulin. Each lane represents a single mouse. (b) PI3K activity associated with IRS1 or IRS2 in liver (WT, n = 2 [−insulin] or n = 3 [+insulin]; p110α+/− p110β+/−, n = 4 [−insulin] or n = 4 [+insulin]) and skeletal muscle (WT, n = 2 [−insulin] or n = 3 [+insulin]; p110α+/− p110β+/−, n = 3 [−insulin] or n = 4 [+insulin]) from 4-month-old WT and p110α+/− p110β+/− mice injected with saline or 5 U of insulin. Results are means ± standard errors of the means and represent one of two experiments. *, P < 0.05 compared to WT. The samples were chosen for similar levels of phosphorylation of the insulin receptor from a total of 18 samples.
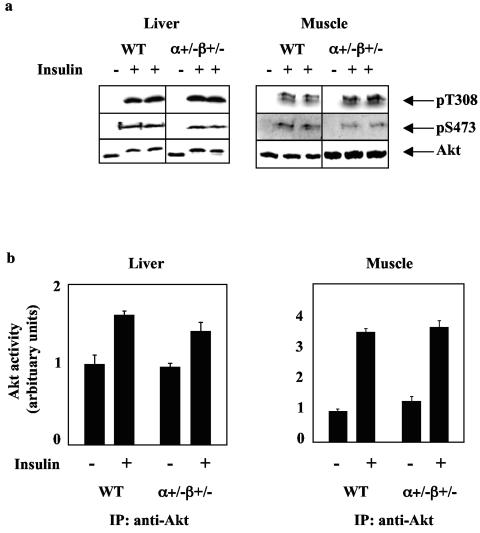
Preserved insulin-dependent Akt activation in muscle and liver of p110α+/− p110β+/− mice.
In vivo Akt activation in response to insulin injection was monitored using anti-phospho-threonine 308 antibody and anti-phospho-serine 473 antibody and by protein kinase activity in anti-Akt immunoprecipitates from liver and muscle. Despite the reduced total PI3K protein and anti-IRS-associated PI3K activity (Fig. 4), there did not appear to be a significantly reduced Akt activation in liver or muscle of the p110α+/− p110β+/− mice at the insulin dose used in these experiments (Fig. 5). This result is consistent with the normal insulin tolerance test observed for these mice (Fig. 1).
FIG. 5.
p110α+/− p110β+/− mice have normal insulin-induced Akt activation in liver and muscle. (a) Akt phosphorylation at threonine 308 (pT308) and serine 473 (pS473) and total Akt protein levels in liver and skeletal muscle from 4-month-old wild-type (WT) and p110α+/− p110β+/− mice injected with saline or insulin. (b) Akt activity assays in liver (WT, n = 3 [−insulin] or n = 3 [+insulin]; p110α+/− p110β+/−, n = 3 [−insulin] or n = 6 [+insulin]) and skeletal muscle (WT, n = 2 [−insulin] and n = 3 [+Ins]; p110α+/− p110β+/−, n = 3 [−insulin] or n = 4 [+insulin]) from 4-month-old WT and p110α+/− p110β+/− mice injected with saline or insulin. Results are means ± standard errors of the means and represent one of two experiments. *, P < 0.05 compared to WT. The samples were chosen for similar levels of phosphorylation of the insulin receptor from a total of 18 samples.
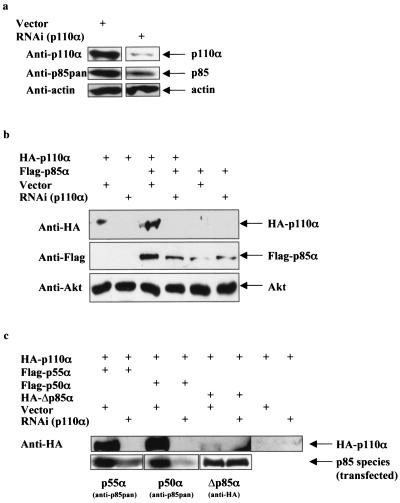
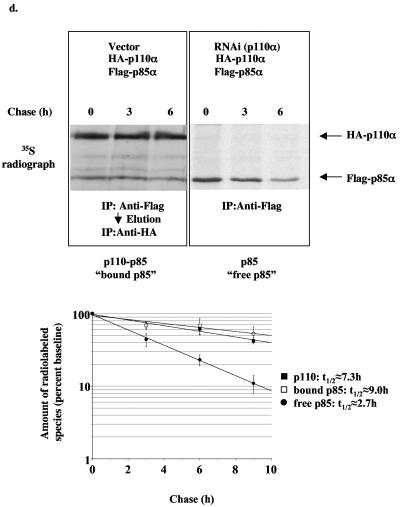
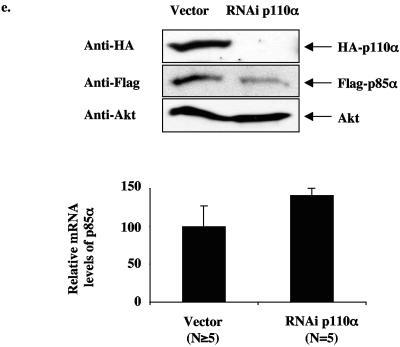
Reduction of the PI3K catalytic subunit results in decreased protein stability of the PI3K regulatory subunit.
The reduction in p85 regulatory subunits in the liver and muscle of the p110α+/− p110β+/− mice was unexpected and led us to further explore the effect of reducing p110 isoforms on p85 protein levels. We were able to dramatically reduce p110α in HeLa cells by RNAi, and this resulted in a significant reduction in endogenous p85 proteins (Fig. 6a). In addition, coexpression of p110α with p85α in CHO-IR cells resulted in more p85α protein than in cells expressing p85α alone, and this effect was blocked by p110α RNAi (Fig. 6b). Furthermore, knockdown of p110α decreased both the protein levels of the smaller p85α isoforms (p55α and p50α) lacking the N-terminal domains (SH3, proline-rich regions, and the Rho-GAP domain) but had no effect on the protein levels of a mutant form of p85α lacking the p110-binding site (Δp85α) (Fig. 6c). Consistent with previous observations that p85 binding to p110α stabilizes p110α transient overexpression of p85α, p55α or p50α increased the p110α protein levels whereas overexpression of Δp85α did not (Fig. 6b and c). To directly test whether protein stability of p85α is altered upon binding to p110, we examined the half-life of p85α complexed with p110α versus free p85α in metabolically labeled CHO-IR cells. Transiently overexpressed free p85α turned over with a half-life of approximately 2.7 h, whereas p85α complexed with p110α had a half-life of approximately 9.0 h (Fig. 6d). In addition, p110α turned over at approximately the same rate (7.3 h) as the subset of p85 bound to p110α. This result is consistent with our previous observation that p85 and p110 form an essentially irreversible complex in vivo (9, 16). The mRNA levels of p85α were not decreased upon knockdown of p110α (Fig. 6e). Thus, the results indicate that the subset of p85 bound to p110α turns over in concert with p110α, while free p85 turns over approximately 2.5 to 3 times faster, explaining why loss of p110 results in loss of p85.
FIG. 6.
Loss of p110 protein by RNAi causes instability of the p85 protein. (a) Endogenous p110α and p85 protein levels in HeLa cells infected with either empty vector or shRNA to knock down expression of p110α and selected for 7 days with puromycin. (b) p110α and p85α protein levels in CHO cells transiently transfected with bovine HA-p110α or mouse Flag-p85α and either empty vector or shRNA to knock down expression of p110α. (c) p110α, p55α, p50α, and Δp85α protein levels in CHO cells transiently transfected with bovine HA-p110α, mouse Flag-p55α, mouse Flag-p50α, or human HA-Δp85α (p85α lacking the inter-SH2 domain that binds p110) and either empty vector or shRNA to knock down expression of p110α. (d) Pulse-chase experiment of CHO cells transiently transfected with bovine HA-p110α, mouse Flag-p85α, and either empty vector or shRNA for knockdown expression of p110α. Graph shows the amount of radiolabeled p110, p85 bound to p110, and free p85 relative to baseline (t = 0 h). Data express means ± standard errors of the means (p110, n = 5; bound p85, n = 3; free p85, n = 5). (e) Protein and relative mRNA levels of mouse Flag-p85α from CHO cells transiently transfected with bovine HA-p110α, mouse Flag-p85α, and either empty vector or shRNA to knock down expression of p110α. Relative mRNA levels were determined by real-time PCR and normalized to the expression level of 18S rRNA. Results are means of relative mRNA levels ± standard deviations of the means.
Free p85 blocks IGF-1-induced Akt activation.
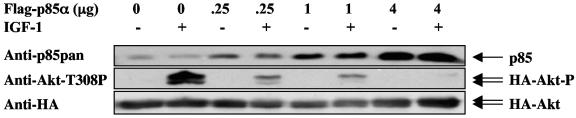
The effect of excess free p85 on IGF-1-mediated Akt activation was investigated by transient overexpression of p85 in CHO cells. Similar to insulin, IGF-1 causes activation of the PI3K-Akt pathway by tyrosine phosphorylation of IRS proteins mostly through the IGF receptor and to a minor degree through the insulin receptor. A twofold overexpression of p85 over endogenous p85 caused a dramatic decrease in IGF-1-induced Akt activation (Fig. 7). The IGF-1-induced Akt activation was completely abolished by higher levels of p85.
FIG. 7.
Overexpression of p85 results in decreased insulin sensitivity. HA-Akt and increasing amounts of p85α were cotransfected into CHO cells. The cells were starved overnight,and one set of cells was stimulated with 20 nM IGF-1 for 5 min. The cells were lysed and HA-Akt was immunoprecipitated and blotted with anti-AktT308P antisera.
DISCUSSION
Extensive studies have shown that PI3K activity is required for insulin-induced glucose uptake (10, 25, 35). It was therefore surprising that loss of any p85 isoform resulted in increased insulin signaling even under conditions where total PI3K levels were reduced in insulin-sensitive tissues (11, 15, 33, 39, 43). We were interested to know whether reduction of PI3K by deletion of p110 subunits might also improve insulin sensitivity. We show here that combined heterozygous loss of p110α and p110β results in a ∼50% decrease of class Ia PI3K without improved insulin signaling. Indeed, mutant mice have a mild glucose intolerance and exhibit mild hyperinsulinemia in the fasting state. Elevated insulin levels point to insulin resistance similar to the early stages of diabetes, in which pancreatic β cells secrete more insulin in order to compensate for insulin resistance in the peripheral tissues. We therefore conclude that reduction in total PI3K activity does not improve insulin-dependent glucose disposal and that increased insulin sensitivity observed in p85α−/− p55α−/− p50α−/− mice is due to elimination of a negative role played by p85 isoforms rather than due to a general effect of reducing PI3K signaling.
The mechanism by which deletion of p85 causes increased insulin sensitivity in mice is still not clear but could involve multiple components. Insulin-dependent Akt activation is enhanced in both p85α+/− p55α+/− p50α+/− mice and p85β−/− mice without increased PI3K activity associated with IRS proteins (33, 43). These findings raise the possibility that p85 isoforms unbound to p110 might activate a PIP3 phosphatase or an Akt phosphatase and thereby contribute to a shutoff of Akt signaling (24). Another possibility is that unbound p85 contributes to JNK activation. JNK activity causes phosphorylation on serine 307 of IRS-1, a site implicated in inhibition of insulin signaling (1, 26). In p85α−/− p55α−/− p50α−/− brown adipocytes, insulin-dependent JNK activation is impaired in comparison to brown adipocytes from wild-type mice and the defect can be rescued by reintroduction of p85 isoforms as well as by a p85 mutant (delta-p85) that lacks the binding site for p110 (42). Finally, unbound p85 can compete for p85/p110 complexes for binding to IRS proteins and thus limit signaling.
We found that heterozygous loss of either p110α or p110β alone does not lead to significant abnormalities in the GTT or ITT, while loss of both proteins causes a significant impairment in the GTT. These results suggest that p110α and p110β both contribute to insulin signaling in vivo. Asano et al. suggested that p110β specifically mediates insulin-induced GLUT4 translocation: microinjection of inhibitory antibodies against p110β blocked insulin-induced translocation of GLUT4 to the plasma membrane in 3T3-L1 adipocytes, whereas inhibitory antibodies against p110α had only a minor effect (3). The difference between our results and those of Asano et al. may be explained by differences in expression levels of p110 isoforms in insulin-sensitive tissues in vivo versus expression levels in 3T3-L1 adipocytes. Mice with combined heterozygous loss of p110α and p110β have a mild glucose intolerance, especially at the later time points of the GTT, and exhibit mild hyperinsulinemia in the fasting state. These findings point to a defect in liver function of the mutant mice.
Previous studies have shown that monomeric p110 is thermally unstable, while heterodimerization with p85 causes stabilization of p110 protein (17, 46). We show here that the stabilization of p110 by p85 isoforms requires the direct association between the two proteins, since all splice variants of p85α (p85α, p55α, and p50α) can augment p110α protein levels, whereas the Δp85α mutant which is lacking the p110 interaction domain cannot. The studies presented here further indicate that p85 protein stability is positively regulated by the p110 protein level both in vivo and in cells in culture. Also, this mechanism seems to be dependent on the direct association between p110 and p85, since Δp85α protein levels are not affected by knockdown of p110. The decrease of p85 upon loss of p110 could be a compensatory mechanism to ensure that a system with decreased PI3K activity is not additionally hampered by the resulting excess of monomeric p85. These results indicate that the ratio of p85 to p110 is tightly controlled in the cell. The results suggest that free p85 is preferentially degraded compared to p85 complexed with p110. However, the mechanism for this discriminatory degradation is not yet clear.
Figure 8 presents a pictorial representation of how deletion of p85 versus p110 subunits can have opposing effects on insulin sensitivity. Our previous studies have shown that p85 can exist in vivo, not only as a dimer complexed to p110 (PI3K) but also in an unbound form (41). While the p110-p85 complex acts as a mediator in insulin signaling by producing PI-3,4,5-P3 for Akt activation, free p85 provides a negative signal, as shown in Fig. 7, where a twofold overexpression of p85 over endogenous p85 potently blocked IGF-1-induced Akt activation. The ratio between p110-p85 and p85 might therefore define the normal state of insulin sensitivity in a wild-type setting. In the case of a mild reduction of total p85 levels in mice without loss of p110 proteins (e.g., p85α+/− or p85β−/− mice), increased insulin sensitivity is observed (33, 43). These findings argue that a negative regulator of insulin sensitivity is lost. Since wild-type p110 can only exist as a dimer bound to p85, but p110 levels are not reduced in p85α+/− or p85β−/− mice, only the free p85 pool can be reduced in these mutant mice. Therefore, we argue that free p85 is a negative regulator in insulin signaling and that the pool of free p85 (which has a shorter half-life and is not protected by p110) is preferably decreased in p85α+/− or p85β−/− mice. This is in agreement with the findings of Ueki et al. (41), showing that in p85α+/− and p85α−/− mouse embryonic fibroblasts the ratio between p110-p85 and free p85 is shifted towards p110-p85. The shorter half-life for free p85 that we report here explains why free p85 is preferentially lost in the p85 knockout mice.
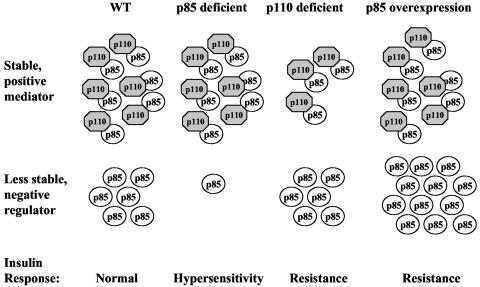
FIG. 8.
Model. Insulin sensitivity is tightly regulated by the ratio of the positive mediator, p110-p85, and the negative regulator, unbound p85. A given ratio between p110-p85 and p85 might define the normal state of insulin sensitivity in a wild-type setting. In p85α+/− or p85β−/− mice, free p85 is decreased preferentially. Therefore, the balance between p110-p85 and free p85 is shifted towards the positive mediator, p110-p85, causing increased insulin sensitivity in the mutant mice. In contrast, in p110α+/− p110β+/− mice the p110-p85 pool is preferentially decreased over free p85. Hence, the balance is shifted towards the negative regulator, free p85, resulting in decreased insulin sensitivity. Overexpression of p85 causes a substantial increase in free p85 and therefore shifts the balance towards the negative regulator, resulting in decreased insulin sensitivity.
In the case of p110α+/− p110β+/− mice, we observe decreased insulin sensitivity. This finding now argues that the balance is shifted toward the negative regulator, meaning that preferably p110-p85 complexes are decreased over free p85. We suggest that upon reduction of p110 the p85 pool normally bound to p110 is less stable and therefore more rapidly degraded and hence preferentially reduced, while the pool of free p85 is not affected upon loss of p110. The proposed model fits best the present data but requires further investigation. Importantly, evidence is emerging that under physiological or pathological conditions levels of p85 are altered in insulin-sensitive tissues. Treatment of female mice with placental growth hormone to mimic pregnancy-induced insulin resistance resulted in an increase in p85 proteins but not p110 proteins in skeletal muscle (4). In addition, p85 levels decrease in skeletal muscle of postpartum women in parallel with increased insulin sensitivity (29).
In summary, we report that double heterozygous loss of PI3K catalytic subunits p110α and p110β results in mice with slight glucose intolerance and with hyperinsulinemia in the fasting state. Besides the ∼50% decrease in PI3K catalytic isoforms, also p85 protein levels were reduced in p110α+/− p110β+/− mice, indicating a surprising role of p110 subunits in controlling p85 levels. Given that genetic ablation of p85 isoforms leads to increased insulin sensitivity and glucose tolerance, we conclude that p85 plays a negative role in insulin signaling independent of its role in mediating activation of the p110 catalytic subunit. Therefore, pharmacological intervention into p85 expression in insulin-sensitive tissues might improve insulin responses in diabetic patients.
Acknowledgments
We thank Robert Nussbaum for providing the p110α+/− and p110β+/− mice, William Hahn for providing the sequences for knocking down p110α, Ji Luo for the bovine HA-p110α construct, and John M. Watt, Nicole Logsdon, Nikki Madson, and Monica Kosmatka for their help in breeding and genotyping the p110α+/− p110β+/− mice.
This work was supported by GM41890 and PO1-CA89021 (to L.C.C.) and DK 55545 (to C.R.K.). S.M.B. was supported by a scholarship from Boehringer Ingelheim Fonds.
REFERENCES
- 1.Aguirre, V., T. Uchida, L. Yenush, R. Davis, and M. F. White. 2000. The c-Jun NH2-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser(307). J. Biol. Chem. 275:9047-9054. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre, V., E. D. Werner, J. Giraud, Y. H. Lee, S. E. Shoelson, and M. F. White. 2002. Phosphorylation of Ser307 in insulin receptor substrate-1 blocks interactions with the insulin receptor and inhibits insulin action. J. Biol. Chem. 277:1531-1537. [DOI] [PubMed] [Google Scholar]
- 3.Asano, T., A. Kanda, H. Katagiri, M. Nawano, T. Ogihara, K. Inukai, M. Anai, Y. Fukushima, Y. Yazaki, M. Kikuchi, R. Hooshmand-Rad, C. H. Heldin, Y. Oka, and M. Funaki. 2000. p110α is up-regulated during differentiation of 3T3-L1 cells and contributes to the highly insulin-responsive glucose transport activity. J. Biol. Chem. 275:17671-17676. [DOI] [PubMed] [Google Scholar]
- 4.Barbour, L. A., J. Shao, L. Qiao, W. Leitner, M. Anderson, J. E. Friedman, and B. Draznin. 2004. Human placental growth hormone increases expression of the p85 regulatory unit of phosphatidylinositol 3-kinase and triggers severe insulin resistance in skeletal muscle. Endocrinology 145:1144-1150. [DOI] [PubMed] [Google Scholar]
- 5.Barroso, I., J. Luan, R. P. Middelberg, A. H. Harding, P. W. Franks, R. W. Jakes, D. Clayton, A. J. Schafer, S. O'Rahilly, and N. J. Wareham. 2003. Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action. PLoS Biol. 1:E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi, L., I. Okabe, D. J. Bernard, and R. L. Nussbaum. 2002. Early embryonic lethality in mice deficient in the p110β catalytic subunit of PI 3-kinase. Mamm. Genome 13:169-172. [DOI] [PubMed] [Google Scholar]
- 7.Bi, L., I. Okabe, D. J. Bernard, A. Wynshaw-Boris, and R. L. Nussbaum. 1999. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110α subunit of phosphoinositide 3-kinase. J. Biol. Chem. 274:10963-10968. [DOI] [PubMed] [Google Scholar]
- 8.Bruning, J. C., J. Winnay, S. Bonner-Weir, S. I. Taylor, D. Accili, and C. R. Kahn. 1997. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell 88:561-572. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, C. L., B. C. Duckworth, K. R. Auger, B. Cohen, B. S. Schaffhausen, and L. C. Cantley. 1990. Purification and characterization of phosphoinositide 3-kinase from rat liver. J. Biol. Chem. 265:19704-19711. [PubMed] [Google Scholar]
- 10.Cheatham, B., C. J. Vlahos, L. Cheatham, L. Wang, J. Blenis, and C. R. Kahn. 1994. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol. Cell. Biol. 14:4902-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, D., F. Mauvais-Jarvis, M. Bluher, S. J. Fisher, A. Jozsi, L. J. Goodyear, K. Ueki, and C. R. Kahn. 2004. p50α/p55α phosphoinositide 3-kinase knockout mice exhibit enhanced insulin sensitivity. Mol. Cell. Biol. 24:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, H., J. Mu, J. K. Kim, J. L. Thorvaldsen, Q. Chu, E. B. Crenshaw III, K. H. Kaestner, M. S. Bartolomei, G. I. Shulman, and M. J. Birnbaum. 2001. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science 292:1728-1731. [DOI] [PubMed] [Google Scholar]
- 13.Cong, L. N., H. Chen, Y. Li, L. Zhou, M. A. McGibbon, S. I. Taylor, and M. J. Quon. 1997. Physiological role of Akt in insulin-stimulated translocation of GLUT4 in transfected rat adipose cells. Mol. Endocrinol. 11:1881-1890. [DOI] [PubMed] [Google Scholar]
- 14.Devroe, E., and P. A. Silver. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fruman, D. A., F. Mauvais-Jarvis, D. A. Pollard, C. M. Yballe, D. Brazil, R. T. Bronson, C. R. Kahn, and L. C. Cantley. 2000. Hypoglycaemia, liver necrosis and perinatal death in mice lacking all isoforms of phosphoinositide 3-kinase p85 alpha. Nat. Genet. 26:379-382. [DOI] [PubMed] [Google Scholar]
- 16.Fruman, D. A., R. E. Meyers, and L. C. Cantley. 1998. Phosphoinositide kinases. Annu. Rev. Biochem. 67:481-507. [DOI] [PubMed] [Google Scholar]
- 17.Fruman, D. A., S. B. Snapper, C. M. Yballe, L. Davidson, J. Y. Yu, F. W. Alt, and L. C. Cantley. 1999. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85α. Science 283:393-397. [DOI] [PubMed] [Google Scholar]
- 18.Gao, Z., D. Hwang, F. Bataille, M. Lefevre, D. York, M. J. Quon, and J. Ye. 2002. Serine phosphorylation of insulin receptor substrate 1 by inhibitor kappa B kinase complex. J. Biol. Chem. 277:48115-48121. [DOI] [PubMed] [Google Scholar]
- 19.Garofalo, R. S., S. J. Orena, K. Rafidi, A. J. Torchia, J. L. Stock, A. L. Hildebrandt, T. Coskran, S. C. Black, D. J. Brees, J. R. Wicks, J. D. McNeish, and K. G. Coleman. 2003. Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB beta. J. Clin. Investig. 112:197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George, S., J. J. Rochford, C. Wolfrum, S. L. Gray, S. Schinner, J. C. Wilson, M. A. Soos, P. R. Murgatroyd, R. M. Williams, C. L. Acerini, D. B. Dunger, D. Barford, A. M. Umpleby, N. J. Wareham, H. A. Davies, A. J. Schafer, M. Stoffel, S. O'Rahilly, and I. Barroso. 2004. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science 304:1325-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein, B. J. 2002. Insulin resistance as the core defect in type 2 diabetes mellitus. Am. J. Cardiol. 90:3G-10G. [DOI] [PubMed] [Google Scholar]
- 22.Greene, M. W., H. Sakaue, L. Wang, D. R. Alessi, and R. A. Roth. 2003. Modulation of insulin-stimulated degradation of human insulin receptor substrate-1 by serine 312 phosphorylation. J. Biol. Chem. 278:8199-8211. [DOI] [PubMed] [Google Scholar]
- 23.Gual, P., T. Gremeaux, T. Gonzalez, Y. Le Marchand-Brustel, and J. F. Tanti. 2003. MAP kinases and mTOR mediate insulin-induced phosphorylation of insulin receptor substrate-1 on serine residues 307, 612 and 632. Diabetologia 46:1532-1542. [DOI] [PubMed] [Google Scholar]
- 24.Gupta, N., A. M. Scharenberg, D. A. Fruman, L. C. Cantley, J. P. Kinet, and E. O. Long. 1999. The SH2 domain-containing inositol 5′-phosphatase (SHIP) recruits the p85 subunit of phosphoinositide 3-kinase during FcgammaRIIb1-mediated inhibition of B cell receptor signaling. J. Biol. Chem. 274:7489-7494. [DOI] [PubMed] [Google Scholar]
- 25.Hara, K., K. Yonezawa, H. Sakaue, A. Ando, K. Kotani, T. Kitamura, Y. Kitamura, H. Ueda, L. Stephens, T. R. Jackson, et al. 1994. 1-Phosphatidylinositol 3-kinase activity is required for insulin-stimulated glucose transport but not for RAS activation in CHO cells. Proc. Natl. Acad. Sci. USA 91:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirosumi, J., G. Tuncman, L. Chang, C. Z. Gorgun, K. T. Uysal, K. Maeda, M. Karin, and G. S. Hotamisligil. 2002. A central role for JNK in obesity and insulin resistance. Nature 420:333-336. [DOI] [PubMed] [Google Scholar]
- 27.Hotamisligil, G. S., N. S. Shargill, and B. M. Spiegelman. 1993. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 259:87-91. [DOI] [PubMed] [Google Scholar]
- 28.Katome, T., T. Obata, R. Matsushima, N. Masuyama, L. C. Cantley, Y. Gotoh, K. Kishi, H. Shiota, and Y. Ebina. 2003. Use of RNA interference-mediated gene silencing and adenoviral overexpression to elucidate the roles of AKT/protein kinase B isoforms in insulin actions. J. Biol. Chem. 278:28312-28323. [DOI] [PubMed] [Google Scholar]
- 29.Kirwan, J. P., A. Varastehpour, M. Jing, L. Presley, J. Shao, J. E. Friedman, and P. M. Catalano. 2004. Reversal of insulin resistance postpartum is linked to enhanced skeletal muscle insulin signaling. J. Clin. Endocrinol. Metab. 89:4678-4684. [DOI] [PubMed] [Google Scholar]
- 30.Kotani, K., A. J. Carozzi, H. Sakaue, K. Hara, L. J. Robinson, S. F. Clark, K. Yonezawa, D. E. James, and M. Kasuga. 1995. Requirement for phosphoinositide 3-kinase in insulin-stimulated GLUT4 translocation in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 209:343-348. [DOI] [PubMed] [Google Scholar]
- 31.Li, J., K. DeFea, and R. A. Roth. 1999. Modulation of insulin receptor substrate-1 tyrosine phosphorylation by an Akt/phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 274:9351-9356. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y. F., K. Paz, A. Herschkovitz, A. Alt, T. Tennenbaum, S. R. Sampson, M. Ohba, T. Kuroki, D. LeRoith, and Y. Zick. 2001. Insulin stimulates PKCζ-mediated phosphorylation of insulin receptor substrate-1 (IRS-1). A self-attenuated mechanism to negatively regulate the function of IRS proteins. J. Biol. Chem. 276:14459-14465. [DOI] [PubMed] [Google Scholar]
- 33.Mauvais-Jarvis, F., K. Ueki, D. A. Fruman, M. F. Hirshman, K. Sakamoto, L. J. Goodyear, M. Iannacone, D. Accili, L. C. Cantley, and C. R. Kahn. 2002. Reduced expression of the murine p85α subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes. J. Clin. Investig. 109:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connor, J. C., and G. G. Freund. 2003. Vanadate and rapamycin synergistically enhance insulin-stimulated glucose uptake. Metabolism 52:666-674. [DOI] [PubMed] [Google Scholar]
- 35.Okada, T., Y. Kawano, T. Sakakibara, O. Hazeki, and M. Ui. 1994. Essential role of phosphatidylinositol 3-kinase in insulin-induced glucose transport and antilipolysis in rat adipocytes. Studies with a selective inhibitor wortmannin. J. Biol. Chem. 269:3568-3573. [PubMed] [Google Scholar]
- 36.Ozes, O. N., H. Akca, L. D. Mayo, J. A. Gustin, T. Maehama, J. E. Dixon, and D. B. Donner. 2001. A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc. Natl. Acad. Sci. USA 98:4640-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pirola, L., S. Bonnafous, A. M. Johnston, C. Chaussade, F. Portis, and E. Van Obberghen. 2003. Phosphoinositide 3-kinase-mediated reduction of insulin receptor substrate-1/2 protein expression via different mechanisms contributes to the insulin-induced desensitization of its signaling pathways in L6 muscle cells. J. Biol. Chem. 278:15641-15651. [DOI] [PubMed] [Google Scholar]
- 38.Quon, M. J., H. Chen, B. L. Ing, M.-L. Liu, M. J. Zarnowski, K. Yonezawa, K. M., S. W. Cushman, and S. I. Taylor. 1995. Roles of 1-phosphatidylinositol 3-kinase and ras in regulating translocation of GLUT 4 in transfected rat adipose cells. Mol. Cell. Biol. 15:5403-5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terauchi, Y., Y. Tsuji, S. Satoh, H. Minoura, K. Murakami, A. Okuno, K. Inukai, T. Asano, Y. Kaburagi, K. Ueki, H. Nakajima, T. Hanafusa, Y. Matsuzawa, H. Sekihara, Y. Yin, J. C. Barrett, H. Oda, T. Ishikawa, Y. Akanuma, I. Komuro, M. Suzuki, K. Yamamura, T. Kodama, H. Suzuki, T. Kadowaki, et al. 1999. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat. Genet. 21:230-235. [DOI] [PubMed] [Google Scholar]
- 40.Ueki, K., P. Algenstaedt, F. Mauvais-Jarvis, and C. R. Kahn. 2000. Positive and negative regulation of phosphoinositide 3-kinase-dependent signaling pathways by three different gene products of the p85α regulatory subunit. Mol. Cell. Biol. 20:8035-8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueki, K., D. A. Fruman, S. M. Brachmann, Y. H. Tseng, L. C. Cantley, and C. R. Kahn. 2002. Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signaling and survival. Mol. Cell. Biol. 22:965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueki, K., D. A. Fruman, C. M. Yballe, M. Fasshauer, J. Klein, T. Asano, L. C. Cantley, and C. R. Kahn. 2003. Positive and negative roles of p85α and p85β regulatory subunits of phosphoinositide 3-kinase in insulin signaling. J. Biol. Chem. 278:48453-48466. [DOI] [PubMed] [Google Scholar]
- 43.Ueki, K., C. M. Yballe, S. M. Brachmann, D. Vicent, J. M. Watt, C. R. Kahn, and L. C. Cantley. 2002. Increased insulin sensitivity in mice lacking p85β subunit of phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA 99:419-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Um, S. H., F. Frigerio, M. Watanabe, F. Picard, M. Joaquin, M. Sticker, S. Fumagalli, P. R. Allegrini, S. C. Kozma, J. Auwerx, and G. Thomas. 2004. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431:200-205. [DOI] [PubMed] [Google Scholar]
- 45.Uysal, K. T., S. M. Wiesbrock, M. W. Marino, and G. S. Hotamisligil. 1997. Protection from obesity-induced insulin resistance in mice lacking TNF-alpha function. Nature 389:610-614. [DOI] [PubMed] [Google Scholar]
- 46.Yu, J., Y. Zhang, J. McIlroy, T. Rordorf-Nikolic, G. A. Orr, and J. M. Backer. 1998. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110α catalytic subunit by the p85 regulatory subunit. Mol. Cell. Biol. 18:1379-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zick, Y. 2001. Insulin resistance: a phosphorylation-based uncoupling of insulin signaling. Trends Cell Biol. 11:437-441. [DOI] [PubMed] [Google Scholar]