Abstract
Two distinct thioredoxin/thioredoxin reductase systems are present in the cytosol and the mitochondria of mammalian cells. Thioredoxins (Txn), the main substrates of thioredoxin reductases (Txnrd), are involved in numerous physiological processes, including cell-cell communication, redox metabolism, proliferation, and apoptosis. To investigate the individual contribution of mitochondrial (Txnrd2) and cytoplasmic (Txnrd1) thioredoxin reductases in vivo, we generated a mouse strain with a conditionally targeted deletion of Txnrd1. We show here that the ubiquitous Cre-mediated inactivation of Txnrd1 leads to early embryonic lethality. Homozygous mutant embryos display severe growth retardation and fail to turn. In accordance with the observed growth impairment in vivo, Txnrd1-deficient embryonic fibroblasts do not proliferate in vitro. In contrast, ex vivo-cultured embryonic Txnrd1-deficient cardiomyocytes are not affected, and mice with a heart-specific inactivation of Txnrd1 develop normally and appear healthy. Our results indicate that Txnrd1 plays an essential role during embryogenesis in most developing tissues except the heart.
Thioredoxins (Txn) are small redox-reactive proteins that regulate many cellular processes (3). Txn exert a cytokine-like influence on blood cells (29), modulate the activity of redox-regulated transcription factors such as NF-κB (34) and AP-1 (20), mediate peroxiredoxin antioxidant properties (35), and are putatively involved in DNA synthesis. The activity of Txn is controlled by thioredoxin reductases (Txnrd) together with NADPH as a cofactor. Three mammalian thioredoxin reductases are known, including a cytosolic (Txnrd1) (10), a mitochondrial (Txnrd2) (9), and a testis-specific (41) isoform. Thioredoxin reductases are homodimeric flavoproteins with two N- and C-terminally located interacting catalytic centers (12, 26). The C-terminal redox center contains a selenocysteine (Sec) residue which is part of a conserved Gly-Cys-Sec-Gly motif that is crucial for Txnrd function. Besides thioredoxins, thioredoxin reductases can also reduce other substrates, such as lipoic acid, NK-lysin, ascorbate, and ubiquinone (1, 31, 45).
Several gene targeting approaches with mice have been performed to investigate the participation of the Txn/Txnrd systems in development and adult physiology. The results revealed that cytosolic (Txn1) and mitochondrial (Txn2) thioredoxins are indispensable for embryonic development (24, 30). In Txn1−/− mutants, early embryonic death (by embryonic day 6.5 [E6.5]) is associated with a dramatically reduced proliferation of inner mass cells. Txn2-deficient embryos develop exencephaly, show markedly increased apoptosis, and die during midgestation around E10.5. Using a conditional targeting approach, we have shown recently that Txnrd2 is also essential for embryonic development (5). Txnrd2-null embryos die around E13.0 due to defects in hematopoiesis and heart development. The cardiac-specific deletion of Txnrd2 leads to fatal dilated cardiomyopathy and morphological abnormalities of cardiomyocytes. These results support and extend the observation that the heart-specific overexpression of dominant-negative Txn1 leads to oxidative stress and cardiac hypertrophy (46).
Txnrd1 is expressed in cells with a strong proliferative activity, such as neoplastic tissues and tumor cell lines (13, 23), and it has also been shown to be a c-Myc target in a human B-cell line (39), indicating a role in cell proliferation. The presence of several thioredoxin reductases obscures the contribution of the individual thioredoxin reductases to cellular growth and survival, and it is unknown to what extent Txnrd1 and Txnrd2 can complement each other. To study the impact of Txnrd1 on apoptosis and proliferation in vivo, we used cre/loxP technology to create mice with a floxed Txnrd1 allele. Ubiquitous Cre-mediated deletion resulted in early embryonic death around E10.5 due to severe growth retardation and widespread developmental abnormalities in most tissues, excluding the heart. This notion was further supported by the heart-specific deletion of Txnrd1, which resulted in viable mice with no obvious phenotypes. It thus appears that only Txnrd2 is crucial for heart development and function, whereas Txnrd1 activity is required in other developing tissues.
MATERIALS AND METHODS
Gene targeting, mouse breeding, and genotyping.
An insert of cosmid clone MPMGc121Io7462Q2 (Deutsches Resourcenzentrum für Genomforschung GmbH, Berlin, Germany) (38), covering exons 12 to 15 of murine Txnrd1, was used for the generation of the targeting construct. An XmaI/EcoRV fragment representing the 3′ arm for homologous recombination was cloned between the XmaI and EcoRV sites of pBluescript SK(+) (Stratagene, La Jolla, Calif.), resulting in pCJ1. Subsequently, this arm was cloned as a SalI/BamHI fragment in the XhoI and BamHI sites of pPNT4 (4), a vector which already contained an frt-flanked neomycin cassette and one loxP site, to generate pCJ2. Several consecutive cloning steps were required to insert the 5′ arm, including the second loxP site and two characteristic restriction sites necessary for the confirmation of homologous recombination of the targeting construct into embryonic stem (ES) cells. Firstly, a HindIII/XmaI cosmid fragment including exon 15 was subcloned into pBluescript SK(+) (pCJ4). Secondly, a synthetic linker (created with the primers OligoloxPb [5′-CCGGGATAACTTCGTATAATGTATGCTATACGAAGTTATGAATTCGTTACCGATATCATTAAT-3′] and OligoloxPa [5′-CCGGATTAATGATATCGGTAACGAATTCATAACTTCGTATAGCATACATTATACGAAGTTATC-3′]) containing one loxP site and two characteristic restriction enzyme sites (EcoRV and AseI) was inserted into pCJ4 that had been digested with XmaI (pCJ4+L). Thirdly, a 4.5-kb XmaI fragment containing exon 15 was introduced into pCJ4+L (creating pCJ6). An XhoI/NotI fragment representing the modified 5′ arm was transferred into pCJ2 which had been linearized with SalI and NotI to obtain the targeting vector pCJ7. Gene targeting in E14 ES cells was performed as described previously (17, 18). Homologously recombined ES cell clones arose at a frequency of 1% when tested with both 3′ and 5′ external probes (see Fig. 1A and B). To simplify the genotyping of mice with floxed Txnrd1 alleles obtained after the backcrossing of chimeric mice with C57BL/6j mice, we used a primer pair (Txnrd1floxfor1 [5′-TCC ACC TCA CAG GAG TGA TCC C-3′] and Txnrd1floxrev1 [5′-TGC CTA AAG ATG AAC TCG CAG C-3′]) which detected wild-type (WT) (66 bp) and floxed (130 bp) Txnrd1 alleles in one PCR step. To discriminate between hemizygous and WT animals, we used one primer pair specific for the deleted Txnrd1 allele (320 bp) (Txnrd1wtfor2 [5′-GGT CTG AGC TAG CGT GAA GTG TTC C-3′] and PGKpromrev1 [5′-GGG CTG CTA AAG CGC ATG CTC CAG ACT GC-3′]) and one pair specific for the WT allele (290 bp) (Txnrd1wtf1 [5′CTA CCC CCA CAA GAA GGA GTA TAC G-3′] and Txnrd1flprev1 [5′-AGC CTG TAC GGA TAC CCT CAG C-3′]). Txnrd1 was deleted from cardiomyocytes by use of the MLC2a-Cre transgenic mouse line (44). Txnrd1+/− mice were first crossed with MLC2a-Cre mice, and subsequently, Txnrd1+/− MLC2a-Cre and Txnrd1loxP/loxP mice were mated to obtain Txnrd1loxP/− MLC2a-Cre mice.
FIG. 1.
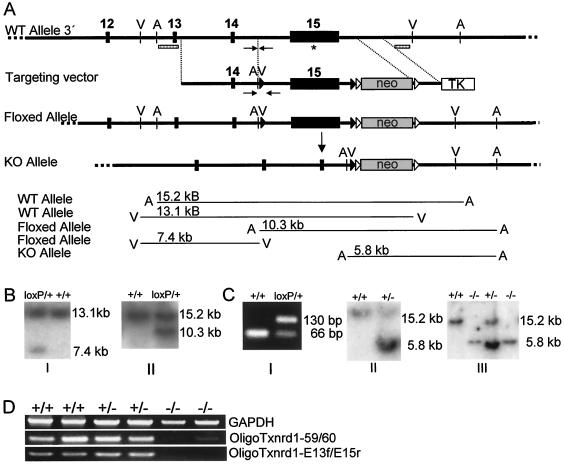
Gene targeting of Txnrd1. (A) Schematic drawing of mouse Txnrd1 3′ region including exons 12 to 15. The last exon harbors the coding region for the redox center located at the C terminus, including the Sec codon UGA (marked with an asterisk), the SECIS element, the AU-rich elements, and the endogenous transcription termination signal. The bottom panels depict the conditional gene targeting strategy to flank exon 15 with loxP sites (black triangles) (floxed). The neo cassette is flanked by frt sites (open triangles). TK, thymidine kinase. The Cre-mediated deletion of the floxed Txnrd1 allele led to the inactivation of Txnrd1. Expected DNA fragments after characteristic restriction digests (V, EcoRV; A, AseI) detected with corresponding 5′ and 3′ external probes (hatched rectangles) are drawn schematically. (B) Homologous recombination of the targeting construct in ES cells was verified by Southern blotting with both 3′ (I) and 5′ (II) external probes. (C) Germ line transmission of the conditional Txnrd1 allele was tested by PCR (the binding regions of the specific primers are marked with arrows in panel A). Panel I, the floxed Txnrd1 allele gives rise to a 64-bp-longer product than the WT allele; panel II, genotyping by Southern blotting of hemizygous KO mice obtained by breeding of floxed Txnrd1 mice with Cre deletion mice; panel III, genotyping of embryos isolated at E9.5 from hemizygous Txnrd1 intercrosses. (D) RT-PCR analysis with mRNAs isolated from E9.5 embryos of each genotype as templates. The primer pair Oligo Txnrd1-E13f and -E15r indicated the absence of exon 15 in Txnrd1 KO embryos. Very low levels of a truncated Txnrd1 message could be detected in Txnrd1 KO embryos by use of the primer pair Oligo Txnrd1-59 and -60 specific for the central region of Txnrd1.
Mice were kept under standard conditions with food and water provided ad libitum (Altromin GmbH, Lage, Germany). All animal experiments were performed in compliance with the German animal welfare law and have been approved by the institutional committee on animal experimentation and the government of Upper Bavaria.
RT-PCR analysis.
Total RNAs were isolated from embryos at gestational day 9.5 by the use of peqGOLDTriFast (peqLab, Erlangen, Germany). DNase-treated total RNAs (DNase I and RNase I free; Roche, Mannheim, Germany) were reverse transcribed by use of a reverse transcription (RT) system (Promega Corporation, Madison, Wis.). Primer pairs were designed to amplify products from exon 13 to exon 15 (Oligo Txnrd1-E13f, 5′-TTG GCC ATT GGA ATG GAC AGT CC-3′; Oligo Txnrd1-E15r, 5′-AGC ACC TTG AAT TGG CGC CTA GG-3′) and from exon 5 to exon 11 (Oligo Txnrd1-59, 5′-CGA AGA CAC AGT GAA GCA TGA CTG GG-3′; Oligo Txnrd1-60, 5′-TCC CCT CCA GGA TGT CAC CGA TGG CG-3′). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a control (GAPDH1, 5′-CTT CAT TGA CCT CAA CTA CAT GG-3′; GAPDH2, 5′-GCC TGC TTC ACC ACC TTC TTG-3′).
Whole-mount in situ hybridization.
C57BL/6j embryos were dissected at E8.5, E9.5, and E10.5. Sense and antisense RNA probes were generated by the use of DIG RNA labeling mix (Roche Molecular Biochemicals) according to the manufacturer's instructions. Whole-mount in situ hybridization was performed as previously described (40). Embryos were stained with BM Purple AP substrate (Roche Molecular Biochemicals) and postfixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline. A Txnrd1-specific probe corresponding to exons 5 to 11 was generated from mouse testis cDNA by use of the primer pair OligoTxnrd1-59 and OligoTxnrd1-60. A probe specific for T (brachyury) was used as described previously (15).
In situ end-labeling method (ISEL).
The detection of apoptotic cells was performed with an ApopTag kit (S7100; Serologicals Corporation, Norcross, Ga.) on 4% PFA-fixed paraffin-embedded sections according to the manufacturer's instructions.
Immunohistochemistry.
An antibody against phospho-histone H3 (Ser10) (New England Biolabs GmbH, Frankfurt am Main, Germany) was used to detect mitotic chromosome condensations at a dilution of 1:50 on 4% PFA-fixed paraffin-embedded sections according to the manufacturer's instructions.
Immunoblotting.
After dissection, the cytosolic fraction of cardiac tissue was prepared as described previously (27). Extracts were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and analyzed by immunoblotting with anti-Txnrd1 (LF-PA0023; LabFrontier, Seoul, Korea). The protein concentration was quantified by the Bradford assay (Bio-Rad, Hercules, Calif.).
Isolation and culture of embryonic cells.
Embryos were isolated at E9.5 or E10.5. The head and the primitive heart were removed from the body trunk. The heart and the remaining body trunk were minced separately, trypsinized, treated with DNase, and cultivated under standard conditions in Dulbecco's modified Eagle's medium (Invitrogen GmbH, Karlsruhe, Germany) supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany), 2 mM glutamine, 100 U of penicillin-streptomycin/ml, and 0.1 mM β-mercaptoethanol.
RESULTS
Gene targeting of Txnrd1 in mice.
The murine Txnrd1 gene comprises 15 exons (Fig. 1A). The last exon encodes 22 amino acids, including the C-terminally located redox center consisting of Gly-Cys-Sec-Gly. Additionally, the 1.7-kb 3′ untranslated region contains the selenocysteine insertion sequence element (SECIS), which is essential for cotranslational Sec incorporation at the UGA codon (8), AU-rich mRNA instability elements, and the endogenous transcription termination signal. Since mutational and biochemical modification of the Sec codon UGA (11, 22, 48) as well as deletion of the SECIS element (7) results in the inactivation of Txnrd1 and, moreover, since alternative first exon usage may restrict gene targeting of the 5′ region of Txnrd1 (37, 42), we chose to flank exon 15 with loxP sites for Cre-mediated conditional inactivation of the Txnrd1 gene (Fig. 1A). Gene targeting in ES cells, the detection of germ line transmission, and the ubiquitous Cre-mediated deletion of the floxed (loxP) Txnrd1 allele are outlined in Fig. 1B and C. To confirm the excision of exon 15, we performed RT-PCR analysis with embryonic mRNAs isolated at E9.5. The primer pair Txnrd1-E13f and -E15r yielded no product with mRNAs from knockout (KO) embryos, whereas the primer pair Txnrd1-59 and -60 revealed low levels of truncated transcripts in Txnrd1 KO embryos (Fig. 1D).
Targeted disruption of Txnrd1 results in early embryonic lethality.
The intercross of hemizygous Txnrd1+/− mice, obtained by the mating of floxed Txnrd1 mice with ubiquitously expressing Cre deletion mice, never resulted in viable homozygous Txnrd1 KO mice. Of 79 pups, 32 (40.5%) were WT and 47 (59.5%) were hemizygous for Txnrd1. The genotyping of embryos dissected from hemizygous intercrosses on different days of gestation revealed that the expected Mendelian ratio was maintained up to E10.5, although the resorption of Txnrd1 KO embryos was frequently observed between gestational days 9.5 and 10.5 (Table 1).
TABLE 1.
Genotypes of embryos of Txnrd1+/− intercross on different gestational days
| Embryonic day | % of embryos with genotype (n)
|
||
|---|---|---|---|
| +/+ | +/− | −/− | |
| E10.0-E11.0 | 23 (9) | 59 (23) | 18 (7)a |
| E9.0-9.5 | 27 (19) | 49 (35) | 24 (17) |
| E7.0-8.5 | 25 (5) | 50 (15) | 25 (5) |
Five resorptions occurred.
Expression pattern of Txnrd1 in the mouse embryo.
To better understand the basis of the early null phenotype, we analyzed the expression profile of Txnrd1 at several developmental stages by performing whole-mount in situ hybridization. At E8.5, Txnrd1 was expressed throughout the entire embryo, with the exception of the primitive heart, with the highest levels detected in neuronal tissues such as the developing forebrain and rhombomeres (Fig. 2A). At E9.5, Txnrd1 expression was confined to the neural tube, the forebrain, branchial arches, somites, and the limb buds (Fig. 2B). At E10.5, Txnrd1 was present in developing somites, in the apical ectodermal ridge of the limb buds, in the first and second branchial arches, and in the lateral edges of the nasal pit. In short, this analysis revealed a complex and dynamic expression pattern of the Txnrd1 gene in early developmental stages. Note that no Txnrd1 expression was detected in the embryonic heart, in contrast to the high expression level of Txnrd2 in this region (5). The distinct expression patterns of Txnrd1 and Txnrd2 suggest particular roles for these redox systems during embryonic development.
FIG. 2.
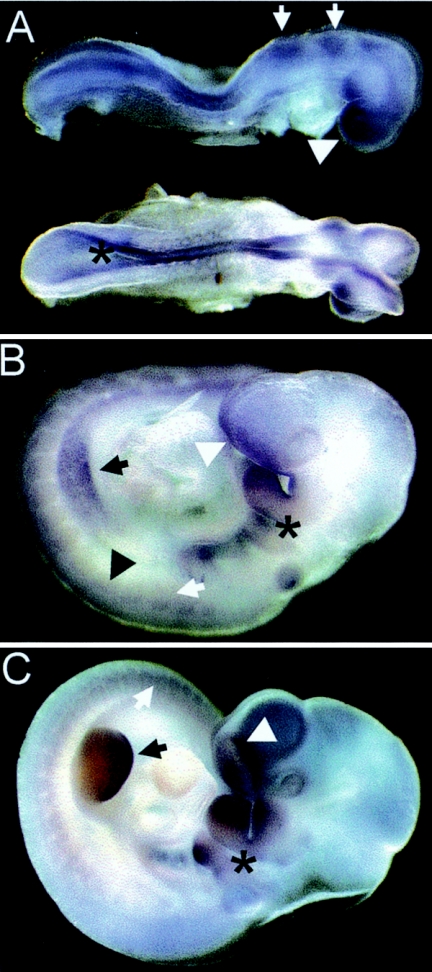
Expression pattern of Txnrd1 during embryonic development. (A) At E8.5, Txnrd1 was widely expressed in all areas, with the exception of the primitive heart. The strongest expression was in the forebrain (arrowhead), the rhombomeres (arrows), and the neural ectoderm (asterisk). (B) At E9.5, Txnrd1 was highly expressed in the forebrain (white arrowhead), the branchial arches (asterisk), the limb buds (black arrow), neuronal tissues (black arrowhead), and somites (white arrow). (C) At E10.5, intense expression was detected in the nasal pit (white arrowhead), the branchial arches (asterisk), the distal end of the limb buds (black arrow), neuronal tissues, and somites (white arrow).
Growth and developmental retardation of Txnrd1 KO embryos.
We observed a clear size reduction of the decidua of KO embryos between gestational days E8.5 and E10.5 compared to those of WT embryos (Fig. 3A). After the removal of maternal and extraembryonic tissues, the size difference between WT and KO embryos was more pronounced (Fig. 3B and C). Despite the facts that the expression of the gastrulation and midline marker brachyury was normal (Fig. 3D) and that no signs of resorption could be observed until E9.5 (Table 1), the development of Txnrd1-deficient embryos was highly abnormal and retarded (Fig. 3C and D). The anterior-posterior axis of Txnrd1 mutant embryos at E10.5 was markedly shortened, and somitogenesis was impaired. Interestingly, distinct regions within Txnrd1-deficient embryos were differently affected. For example, in E10.5 KO embryos, the head region resembled that of wild-type embryos at E9.0, whereas tissues caudal to the heart resembled the morphology at developmental stage E8.5 or earlier (Fig. 3C and D). At all stages, the primitive heart was the only organ that appeared to be unaffected by the Txnrd1 deficiency (Fig. 3C and D), and cardiac contractility was observed in freshly prepared KO embryos (data not shown). In accordance with the developmental retardation, we never observed proper turning of Txnrd1 KO embryos. Also, in contrast to the case for WT embryos, primary vascular plexus formation in the yolk sacs of Txnrd1-deficient embryos was delayed, but blood vessel formation appeared unaffected (data not shown).
FIG. 3.
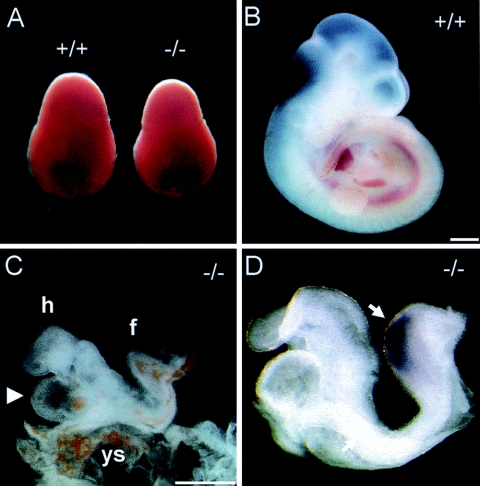
Phenotypic analysis of Txnrd1-deficient embryos. (A) Txnrd1 KO embryos (−/−) at E8.5 showed a smaller decidua than their wild-type siblings (+/+). (B and C) WT (B) and Txnrd1 KO (C) siblings dissected at E10.5. The obvious growth retardation of the KO embryo was manifested in a less developed head region and a fold in the short embryonic trunk. The heart appeared normal but oversized relative to the reduced overall body size (arrowhead). (D) Txnrd1 KO embryos dissected at E9.5. The expression of brachyury (arrow) in the axial midline and the posterior mesoderm indicated that gastrulation is not affected in Txnrd1 KO embryos. Abbreviations: f, fold; h, embryonic heart; ys, yolk sac. Bars = 500 μm.
The developmental delay of embryonic structures became particularly evident when we compared serial sections of heterozygous (phenotypically wild type) and homozygous Txnrd1 KO siblings at E9.5 and E10.5 with sections of corresponding WT embryos (C57BL/6j) at E7.5 and E8.5 (Fig. 4A to M). The positioning and size of the E9.5 KO embryos (Fig. 4B, E, and H) resembled those of E7.5 WT embryos (Fig. 4C, F, and I) rather than those of their heterozygous siblings (Fig. 4A, D, and G). At E10.5, the Txnrd1-deficient embryos failed to turn and showed a defect in neural tube closure with an overall structure comparable to that of E8.5 WT embryos (Fig. 4, compare panels L and M). In contrast, the layering and size of trophoblast giant cells at the proximal pole of the decidua seemed to be unaffected by the Txnrd1 deficiency, and these structures were comparable in heterozygous and homozygous siblings of the same age (Fig. 4G and H).
FIG. 4.
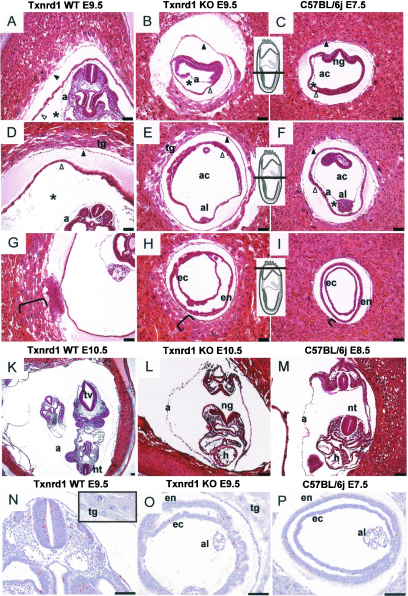
Histological analysis of paraffin-embedded Txnrd1-deficient and wild-type embryos. (A to I) Serial sections of hemizygous Txnrd1 (A, D, and G) and homozygous Txnrd1 KO (B, E, and H) siblings at E9.5 as well as corresponding wild-type embryos at E7.5 (C, F, and I). Inserts indicate section levels. The brackets in panels G, H, and I indicate the layering of trophoblast giant cells. Whereas the homozygous KO embryo had retarded growth and resembled E7.5 wild-type embryos, extraembryonic tissues seemed unaffected by the Txnrd1 deficiency. Asterisks indicate the exocoelomic cavity, black arrowheads mark the Reichert's membrane, and white arrowheads point to the yolk sac. (K to L) Cross sections of hemizygous Txnrd1 (K) and homozygous Txnrd1 KO (L) siblings at E10.5 compared to a wild-type embryo at E8.5 (M). Note that the open neuralgroove of the homozygous KO embryo (L) is opposed to the caudal neuropore, indicating that turning had not occurred. (N to P) Mitotically active cells were detected by phospho-histone H3 (PH3) immunostaining (brown color). Hemizygous embryos at E9.5 (N) displayed significantly more mitotically active cells than Txnrd1 KO embryos of the same age (O), whereas the number of PH3-positive cells at E9.5 in Txnrd1 KO embryos was similar to that in WT embryos at E7.5 (P). Sections A to M were stained with hematoxylin and eosin, and sections N to P were counterstained with hematoxylin after immunodetection. Bars = 1,000 μm. Abbreviations: a, amnion; ac, amniotic cavity; al, allantois; ec, ectoderm; en, endoderm; h, embryonic heart; ng, neural groove; nt, neural tube; tg, trophoblast giant cells; tv, telencephalic vesicle.
Taken together, our results suggest that the Txnrd1 deficiency critically affects most aspects of early embryonic development, sparing only the heart and a few extraembryonic structures.
Proliferation defects in Txnrd1 KO embryos.
To investigate whether increased apoptosis or reduced proliferation was responsible for the obvious growth retardation of Txnrd1−/− embryos, we performed ISEL immunostaining of serial sections of paraffin-embedded embryos. In contrast to the case for WT sections at E10.5, only a few ISEL-positive cells could be detected in sections of Txnrd1-deficient embryos of the same age (data not shown). This was in line with the findings for WT sections at E8.5, indicating that apoptosis levels were comparable in WT and KO embryos. In contrast, cell proliferation in Txnrd1 knockout embryos was markedly reduced compared to that in their wild-type siblings. Using phospho-histone H3 (PH3) as a marker (6, 43), we observed clear differences in the numbers and densities of PH3-positive cells between Txnrd1 KO and wild-type siblings at E9.5 (Fig. 4N and O) as well as E10.5 (data not shown). The number of proliferating cells in KO embryos was clearly reduced and comparable to the number of PH3-positive cells in WT embryos at earlier stages.
To further investigate the effect of Txnrd1 deficiency on cell proliferation, we isolated mouse embryonic fibroblast (MEFs) from WT and KO cells. Whereas the cultivation of MEFs isolated from hemizygous Txnrd1 embryos at E8.5 and E9.5 was always successful, the establishment of proliferating MEFs from homozygous knockout embryos invariably failed (data not shown). Although some cells attached to the culture dish after seeding, they were not capable of proliferating. Notably, the isolation and cultivation of spontaneously contracting cardiomyocytes from E10.5 Txnrd1 KO embryos were possible (data not shown).
Taken together, the results show that Txnrd1 knockout embryos display general growth retardation with the exception of cardiomyocytes, most likely due to a global proliferation defect, and reach a developmental stage comparable to E8.5 wild-type littermates when dissected at gestational day E10.5.
No phenotype after heart-specific elimination of Txnrd1.
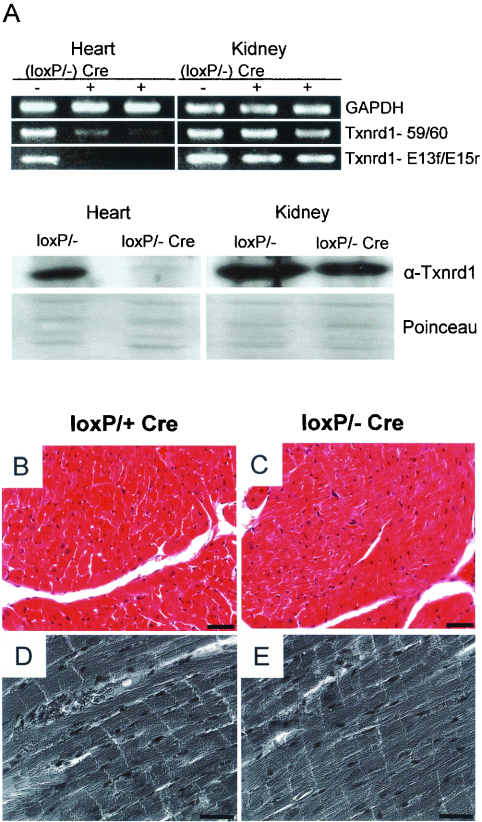
Thioredoxin has recently attracted considerable interest in cardiac research (47). The heart-specific overexpression of dominant-negative Txn1 was shown to be associated with increased oxidative stress and cardiac hypertrophy in mice, whereas the overexpression of wild-type Txn1 was beneficial (46). Yet the expression studies of Txnrd1 by in situ hybridization described above and analyses of the Txnrd1 null phenotype did not provide evidence for the suspected role of Txnrd1 in the heart. There are two possibilities to explain this discrepancy. Firstly, Txnrd1 might contribute to proper heart functioning at later stages of development or eventually only after birth, when an entirely self-sustaining circulatory system must be maintained by the heart. The second possibility is that Txnrd2, and not Txnrd1, plays a decisive role in proper heart development and function. To discriminate between these possibilities, we specifically deleted Txnrd1 from the heart. To this end, we first crossed Txnrd1+/− mice with MLC2a-Cre-expressing mice (44). The resulting Txnrd1+/− MLC2a-Cre mice were subsequently crossed with Txnrd1loxP/loxP mice to obtain Txnrd1loxP/− MLC2a-Cre mice. The efficiency of the heart-specific deletion of Txnrd1 was monitored by RT-PCR analysis and Western blotting (Fig. 5A) and appeared to be similarly effective as the MLC2a-Cre-mediated elimination of Txnrd2. Heart-specific Txnrd1 KO mice were viable and fully fertile. The preparation of cardiac tissue did not show any signs of cardiac insufficiency (data not shown). Similarly, a histological examination did not reveal any differences in heart shape or in the cellular morphology of cardiomyocytes between WT and heart-specific Txnrd1 KO mice (Fig. 5B to E). These results indicate that Txnrd1 is dispensable for embryonic heart development. We conclude that Txnrd2 rather than Txnrd1 plays the decisive role in heart development.
FIG. 5.
Histological examination of cardiac tissues of heart-specific Txnrd1 KO mice. (A) Semiquantitative RT-PCRs revealed reduced levels of Txnrd1 transcripts in the hearts of heart-specific Txnrd1 KO mice (loxP/− Cre+) compared to those in the hearts of control animals (loxP/− Cre−). Note that Txnrd1 mRNA levels were comparable in other tissues, such as the kidneys, showing the specificity of the MLC2a-Cre line. In the bottom panels, immunoblotting reveals the high efficacy of deletion of the targeted Txnrd1 allele in the hearts of Txnrd1loxP/− Mlc2a-Cre mice, but not in the kidneys. Provided that the antibody used was highly specific, the faint band migrating at the position of Txnrd1 was most likely due to cell and tissue heterogeneity within the heart and to an incomplete deletion of the targeted allele by Cre recombinase in vivo. Serial sections of control (B and D) and Txnrd1-deficient (C and E) hearts show no differences in nuclear and cellular morphology between WT and KO cells by hematoxylin and eosin staining (B and C) and phase-contrast imaging (D and E) of cardiomyocytes. Bars = 25 μm (B and C) and 20 μm (D and E).
DISCUSSION
The Txnrd1 knockout approach described here sheds light on as yet unresolved questions regarding a putative functional redundancy between Txnrd1 and Txnrd2 and whether (or how closely) the phenotype of Txnrd1−/− mice mimics the phenotype of Txn1−/− mice. With this study and our recent work (5), we provide evidence that Txnrd1 and Txnrd2 fulfill nonredundant functions to a large extent. Both enzymes play distinct and essential roles in embryonic development and in cell and organ functions and cannot substitute for each other. A ubiquitous inactivation of Txnrd1 in mice was associated with remarkable embryonic growth and developmental retardation, a failure of embryonic turning, an impairment of somitogenesis, and early embryonic death between E9.5 and E10.5. Txnrd1−/− embryos reached a developmental stage resembling E8.5 wild-type embryos. A severe impairment of proliferation was noted for Txnrd1−/− embryos by use of a phospho-histone 3-specific antibody, whereas no obvious increase in the number of apoptotic cells was observed. Remarkably, the heart appeared to develop normally and was spared from dramatic growth and developmental retardation. The development of extraembryonic tissues also appeared unaffected in Txnrd1−/− embryos.
Only the last of 15 exons of the Txnrd1 gene was deleted in our gene targeting approach. This exon includes the selenocysteine-containing active site and the SECIS element required for selenocysteine incorporation. The strategy of deleting only a small part of the gene bears the risk that a truncated protein will be synthesized from the targeted allele and that the truncated protein may impose a gain-of-function rather than a loss-of-function phenotype. Given the facts that small amounts of truncated transcripts were indeed detected and that selenium-compromised Txnrd1 was shown to induce apoptosis (2), it was particularly important to test whether a truncated protein that might be responsible for the phenotype observed was made from the targeted allele. A Western blot analysis of heart tissue obtained from Txnrd1loxP/− MLC2a-Cre mice (and Txnrd1loxP/− mice as controls) revealed that the deletion of the Txnrd1 gene in the heart was indeed very effective and apparently did not lead to the formation of a truncated protein (Fig. 5A). The notion that our targeting strategy led to a loss- and not to a gain-of-function phenotype is corroborated by the fact that hemizygous Txnrd1+/− and Txnrd1loxP/− mice were normal and did not show any evidence of increased cell death in vivo.
The Txnrd1−/− phenotype described here clearly differs from that of Txnrd2−/− embryos (5). Txnrd2−/− embryos were smaller and pale compared to their wild-type siblings and died of severe anemia and improper heart development at E13.5. The importance of Txnrd2 for heart development and function was emphasized by the heart-specific deletion of Txnrd2 leading to congestive dilated cardiomyopathy and perinatal death, whereas mice with a heart-specific elimination of Txnrd1 developed normally and appeared healthy. The importance of thioredoxin for proper heart function has recently been noticed by Yamamoto et al. (46). They showed that the heart-specific overexpression of a dominant-negative mutant of Txn1 in mice results in increased oxidative stress and cardiac hypertrophy. Since dominant-negative Txn1 most likely also interferes with Txn2 function, the transgenic approach did not determine which of the thioredoxin/thioredoxin reductase systems is responsible for the observed phenotype. Thus, our data clearly indicate a pivotal role for Txnrd2 in proper heart function but do not exclude a potential contribution of Txn1. If Txn1, in addition to Txn2, is also of critical importance in the heart, then Txn1 should be a substrate of Txnrd2.
The time of embryonic death and the appearance of Txnrd1 KO embryos closely reflected the expression pattern. At E8.5, Txnrd1 was highly expressed in the developing neural tissues, i.e., the neural tube, rhombomeres, and forebrain, and at E9.5 and E10.5, expression was confined to branchial arches, somites, and the apicoectodermal ridge of the limb buds, but the primitive heart was largely excluded. Notably, Jurado and colleagues showed that Txnrd1 expression peaks around E10.5 (19). The discrepancy in the expression patterns of Txnrd1 and Txnrd2 in the heart, the different subcellular localization of Txnrd1 and Txnrd2, and potential differences in the enzymes themselves suggest the nonredundancy of both thioredoxin reductases (28).
Our analysis of the phenotypes of Txnrd1−/− and Txnrd2−/− embryos has clearly revealed the nonredundant functions of Txnrd1 and Txnrd2, but we could not assess to what extent both thioredoxin/thioredoxin reductase systems might functionally overlap. Additional information was provided by comparing the phenotypes of Txn1−/− to Txnrd1−/− and of Txn2−/− to Txnrd2−/− embryos. Mice deficient in Txn1 show a severely perturbed proliferation of the inner cell mass and embryonic death at around E6.5, which is much earlier than that for Txnrd1−/− embryos (E10.5). Likewise, mice lacking the Txn2 gene die at E10.5 from exencephaly and massive apoptosis, which is significantly earlier than the death of Txnrd2−/− embryos (E13). The fact that Txn1−/− and Txn2−/− mice die much earlier than mice lacking the respective thioredoxin reductases may suggest a partial functional overlap of Txnrd1 and Txnrd2. Alternatively, other pivotal functions of thioredoxins that do not require thioredoxin reductase activity might exist. A C-terminally truncated form of Txn (referred to as Txn80) exerts a potent mitogenic activity on resting human peripheral mononuclear cells (32). This truncated form lacks reductase activity and tends to form homodimers, unlike full-length Txn. It may be possible to discriminate between these possibilities by generating mice with a Txn1 gene that is mutated in the catalytic cysteine residues Cys 32 and Cys 35.
It is remarkable, but not surprising, that the phenotypes of both Txn1−/− and Txnrd1−/− embryos are dominated by a defect in cell proliferation. This supports the notion that Txn1 is the main substrate of Txnrd1 and strongly argues that Txnrd1−/− embryos die from compromised Txn1 activity. An important role of the Txn1/Txnrd1 system in cell proliferation is also suggested by the facts that the Txn/Txnrd system is abundantly expressed in tumors and tumor cell lines (23, 33, 36), that Txnrd1 is a direct target gene of the proto-oncogene c-myc (39), and that it was impossible to establish primary mouse embryonic fibroblast cultures of Txnrd1−/− embryos. It is of crucial importance now to identify the molecular targets of Txn1 in cell proliferation. Due to the pleiotropic activity of Txn1 and its interaction with many different target molecules, it will remain a very difficult task to define the actions of Txn1 and Txnrd1 in diverse biological processes at a biochemical level. Several reports indicate that the Txn/Txnrd1 system—either directly or through substrates such as Ref-1 and peroxiredoxins—participates in the regulation of transcription, replication, and DNA repair (3, 16). Whether the mammalian Txn/Txnrd system provides reducing equivalents for the synthesis of deoxyribonucleotides by ribonucleotide reductase, as shown for Escherichia coli (21), Saccharomyces cerevisiae (25), and Xenopus laevis (14), is still being debated.
Its association with proliferation makes Txnrd1 an interesting drug target for cancer therapy in the future. Our data stress the importance of inhibiting Txnrd1 without affecting Txnrd2 activity. We predict that drugs lacking specificity for Txnrd1 would most likely also impair the activity of Txnrd2, eventually leading to serious cardiac side effects. Since the catalytic centers of Txnrd1 and Txnrd2 are virtually identical, it will be extremely difficult to design chemical drugs with specificity for Txnrd1. Txnrd1 may therefore be an ideal target for inhibition by RNA interference.
Acknowledgments
This work was supported by the DFG-Priority Programs SPP1087 (to M.B., M.C., and G.W.B.) and SPP1069 (to A.K.H.), by a fellowship from Alexander von Humboldt-Stiftung to S. Moreno, and by Fonds der Chemischen Industrie.
We are most grateful to K. R. Chien for providing the MLC2a-Cre mice. We thank S. Lippl and members of the blastocyst injection unit for their excellent technical assistance and animal keepers for their support. We are grateful to A. Banjac, A. Seiler, W. Schmahl, and U. Heinzmann for fruitful discussions and to V. Mostert and R. Chapman for critically reading the manuscript.
REFERENCES
- 1.Andersson, M., A. Holmgren, and G. Spyrou. 1996. NK-lysin, a disulfide-containing effector peptide of T-lymphocytes, is reduced and inactivated by human thioredoxin reductase. Implication for a protective mechanism against NK-lysin cytotoxicity. J. Biol. Chem. 271:10116-10120. [DOI] [PubMed] [Google Scholar]
- 2.Anestal, K., and E. S. Arner. 2003. Rapid induction of cell death by selenium-compromised thioredoxin reductase 1 but not by the fully active enzyme containing selenocysteine. J. Biol. Chem. 278:15966-15972. [DOI] [PubMed] [Google Scholar]
- 3.Arner, E. S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267:6102-6109. [DOI] [PubMed] [Google Scholar]
- 4.Conrad, M., M. Brielmeier, W. Wurst, and G. W. Bornkamm. 2003. Optimized vector for conditional gene targeting in mouse embryonic stem cells. BioTechniques 34:1136-1138, 1140. [DOI] [PubMed] [Google Scholar]
- 5.Conrad, M., C. Jakupoglu, S. G. Moreno, S. Lippl, A. Banjac, M. Schneider, H. Beck, A. K. Hatzopoulos, U. Just, F. Sinowatz, W. Schmahl, K. R. Chien, W. Wurst, G. W. Bornkamm, and M. Brielmeier. 2004. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol. Cell. Biol. 24:9414-9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crosio, C., G. M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C. D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujiwara, N., T. Fujii, J. Fujii, and N. Taniguchi. 1999. Functional expression of rat thioredoxin reductase: selenocysteine insertion sequence element is essential for the active enzyme. Biochem. J. 340:439-444. [PMC free article] [PubMed] [Google Scholar]
- 8.Gasdaska, J. R., J. W. Harney, P. Y. Gasdaska, G. Powis, and M. J. Berry. 1999. Regulation of human thioredoxin reductase expression and activity by 3′-untranslated region selenocysteine insertion sequence and mRNA instability elements. J. Biol. Chem. 274:25379-25385. [DOI] [PubMed] [Google Scholar]
- 9.Gasdaska, P. Y., M. M. Berggren, M. J. Berry, and G. Powis. 1999. Cloning, sequencing and functional expression of a novel human thioredoxin reductase. FEBS Lett. 442:105-111. [DOI] [PubMed] [Google Scholar]
- 10.Gladyshev, V. N., K. T. Jeang, and T. C. Stadtman. 1996. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc. Natl. Acad. Sci. USA 93:6146-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorlatov, S. N., and T. C. Stadtman. 1998. Human thioredoxin reductase from HeLa cells: selective alkylation of selenocysteine in the protein inhibits enzyme activity and reduction with NADPH influences affinity to heparin. Proc. Natl. Acad. Sci. USA 95:8520-8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gromer, S., J. Wissing, D. Behne, K. Ashman, R. H. Schirmer, L. Flohe, and K. Becker. 1998. A hypothesis on the catalytic mechanism of the selenoenzyme thioredoxin reductase. Biochem. J. 332:591-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han, H., D. J. Bearss, L. W. Browne, R. Calaluce, R. B. Nagle, and D. D. Von Hoff. 2002. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 62:2890-2896. [PubMed] [Google Scholar]
- 14.Hartman, H., M. Wu, B. B. Buchanan, and J. C. Gerhart. 1993. Spinach thioredoxin m inhibits DNA synthesis in fertilized Xenopus eggs. Proc. Natl. Acad. Sci. USA 90:2271-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrmann, B. G. 1991. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development 113:913-917. [DOI] [PubMed] [Google Scholar]
- 16.Hirota, K., H. Nakamura, H. Masutani, and J. Yodoi. 2002. Thioredoxin superfamily and thioredoxin-inducing agents. Ann. N. Y. Acad. Sci. 957:189-199. [DOI] [PubMed] [Google Scholar]
- 17.Hogan, B., C. F. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Joyner, S. 1999. Gene targeting—a practical approach, 2nd ed. Institute of Biomolecular Medicine, NYU Medical Centre, New York, N.Y.
- 19.Jurado, J., M. J. Prieto-Alamo, J. Madrid-Risquez, and C. Pueyo. 2003. Absolute gene expression patterns of thioredoxin and glutaredoxin redox systems in mouse. J. Biol. Chem. 278:45546-45554. [DOI] [PubMed] [Google Scholar]
- 20.Karimpour, S., J. Lou, L. L. Lin, L. M. Rene, L. Lagunas, X. Ma, S. Karra, C. M. Bradbury, S. Markovina, P. C. Goswami, D. R. Spitz, K. Hirota, D. V. Kalvakolanu, J. Yodoi, and D. Gius. 2002. Thioredoxin reductase regulates AP-1 activity as well as thioredoxin nuclear localization via active cysteines in response to ionizing radiation. Oncogene 21:6317-6327. [DOI] [PubMed] [Google Scholar]
- 21.Laurent, T. C., E. C. Moore, and P. Reichard. 1964. Enzymatic synthesis of Deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J. Biol. Chem. 239:3436-3444. [PubMed] [Google Scholar]
- 22.Lee, S. R., S. Bar-Noy, J. Kwon, R. L. Levine, T. C. Stadtman, and S. G. Rhee. 2000. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc. Natl. Acad. Sci. USA 97:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lincoln, D. T., E. M. Ali Emadi, K. F. Tonissen, and F. M. Clarke. 2003. The thioredoxin-thioredoxin reductase system: over-expression in human cancer. Anticancer Res. 23:2425-2433. [PubMed] [Google Scholar]
- 24.Matsui, M., M. Oshima, H. Oshima, K. Takaku, T. Maruyama, J. Yodoi, and M. M. Taketo. 1996. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev. Biol. 178:179-185. [DOI] [PubMed] [Google Scholar]
- 25.Muller, E. G. 1991. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J. Biol. Chem. 266:9194-9202. [PubMed] [Google Scholar]
- 26.Mustacich, D., and G. Powis. 2000. Thioredoxin reductase. Biochem. J. 346:1-8. [PMC free article] [PubMed] [Google Scholar]
- 27.Nalvarte, I., A. E. Damdimopoulos, C. Nystrom, T. Nordman, A. Miranda-Vizuete, J. M. Olsson, L. Eriksson, M. Bjornstedt, E. S. Arner, and G. Spyrou. 2004. Overexpression of enzymatically active human cytosolic and mitochondrial thioredoxin reductase in HEK-293 cells: effect on cell growth and differentiation. J. Biol. Chem. 279:54510-54517. [DOI] [PubMed] [Google Scholar]
- 28.Nalvarte, I., A. E. Damdimopoulos, and G. Spyrou. 2004. Human mitochondrial thioredoxin reductase reduces cytochrome c and confers resistance to complex III inhibition. Free Radic. Biol. Med. 36:1270-1278. [DOI] [PubMed] [Google Scholar]
- 29.Nishinaka, Y., H. Nakamura, and J. Yodoi. 2002. Thioredoxin cytokine action. Methods Enzymol. 347:332-338. [DOI] [PubMed] [Google Scholar]
- 30.Nonn, L., R. R. Williams, R. P. Erickson, and G. Powis. 2003. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol. 23:916-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordberg, J., and E. S. Arner. 2001. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 31:1287-1312. [DOI] [PubMed] [Google Scholar]
- 32.Pekkari, K., and A. Holmgren. 2004. Truncated thioredoxin: physiological functions and mechanism. Antioxid. Redox Signal. 6:53-61. [DOI] [PubMed] [Google Scholar]
- 33.Powis, G., D. Mustacich, and A. Coon. 2000. The role of the redox protein thioredoxin in cell growth and cancer. Free Radic. Biol. Med. 29:312-322. [DOI] [PubMed] [Google Scholar]
- 34.Qin, J., G. M. Clore, W. M. Kennedy, J. R. Huth, and A. M. Gronenborn. 1995. Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B. Structure 3:289-297. [DOI] [PubMed] [Google Scholar]
- 35.Rhee, S. G., S. W. Kang, T. S. Chang, W. Jeong, and K. Kim. 2001. Peroxiredoxin, a novel family of peroxidases. IUBMB Life 52:35-41. [DOI] [PubMed] [Google Scholar]
- 36.Rundlof, A. K., and E. S. Arner. 2004. Regulation of the mammalian selenoprotein thioredoxin reductase 1 in relation to cellular phenotype, growth, and signaling events. Antioxid. Redox Signal. 6:41-52. [DOI] [PubMed] [Google Scholar]
- 37.Rundlof, A. K., M. Janard, A. Miranda-Vizuete, and E. S. Arner. 2004. Evidence for intriguingly complex transcription of human thioredoxin reductase 1. Free Radic. Biol. Med. 36:641-656. [DOI] [PubMed] [Google Scholar]
- 38.RZPD. http://www.rzpd.de.
- 39.Schuhmacher, M., F. Kohlhuber, M. Holzel, C. Kaiser, H. Burtscher, M. Jarsch, G. W. Bornkamm, G. Laux, A. Polack, U. H. Weidle, and D. Eick. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sporle, R., and K. Schughart. 1998. Paradox segmentation along inter- and intrasomitic borderlines is followed by dysmorphology of the axial skeleton in the open brain (opb) mouse mutant. Dev. Genet. 22:359-373. [DOI] [PubMed] [Google Scholar]
- 41.Sun, Q. A., L. Kirnarsky, S. Sherman, and V. N. Gladyshev. 2001. Selenoprotein oxidoreductase with specificity for thioredoxin and glutathione systems. Proc. Natl. Acad. Sci. USA 98:3673-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, Q. A., F. Zappacosta, V. M. Factor, P. J. Wirth, D. L. Hatfield, and V. N. Gladyshev. 2001. Heterogeneity within animal thioredoxin reductases. Evidence for alternative first exon splicing. J. Biol. Chem. 276:3106-3114. [DOI] [PubMed] [Google Scholar]
- 43.Wei, Y., C. A. Mizzen, R. G. Cook, M. A. Gorovsky, and C. D. Allis. 1998. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc. Natl. Acad. Sci. USA 95:7480-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wettschureck, N., H. Rutten, A. Zywietz, D. Gehring, T. M. Wilkie, J. Chen, K. R. Chien, and S. Offermanns. 2001. Absence of pressure overload induced myocardial hypertrophy after conditional inactivation of Galphaq/Galpha11 in cardiomyocytes. Nat. Med. 7:1236-1240. [DOI] [PubMed] [Google Scholar]
- 45.Xia, L., T. Nordman, J. M. Olsson, A. Damdimopoulos, L. Bjorkhem-Bergman, I. Nalvarte, L. C. Eriksson, E. S. Arner, G. Spyrou, and M. Bjornstedt. 2003. The mammalian cytosolic selenoenzyme thioredoxin reductase reduces ubiquinone. A novel mechanism for defense against oxidative stress. J. Biol. Chem. 278:2141-2146. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto, M., G. Yang, C. Hong, J. Liu, E. Holle, X. Yu, T. Wagner, S. F. Vatner, and J. Sadoshima. 2003. Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J. Clin. Investig. 112:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamawaki, H., J. Haendeler, and B. C. Berk. 2003. Thioredoxin: a key regulator of cardiovascular homeostasis. Circ. Res. 93:1029-1033. [DOI] [PubMed] [Google Scholar]
- 48.Zhong, L., and A. Holmgren. 2000. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J. Biol. Chem. 275:18121-18128. [DOI] [PubMed] [Google Scholar]