Abstract
Homologous recombinational repair (HRR) of DNA damage is critical for maintaining genome stability and tumor suppression. RAD51 and BRCA2 colocalization in nuclear foci is a hallmark of HRR. BRCA2 has important roles in RAD51 focus formation and HRR of DNA double-strand breaks (DSBs). We previously reported that BCCIPα interacts with BRCA2. We show that a second isoform, BCCIPβ, also interacts with BRCA2 and that this interaction occurs in a region shared by BCCIPα and BCCIPβ. We further show that chromatin-bound BRCA2 colocalizes with BCCIP nuclear foci and that most radiation-induced RAD51 foci colocalize with BCCIP. Reducing BCCIPα by 90% or BCCIPβ by 50% by RNA interference markedly reduces RAD51 and BRCA2 foci and reduces HRR of DSBs by 20- to 100-fold. Similarly, reducing BRCA2 by 50% reduces RAD51 and BCCIP foci. These data indicate that BCCIP is critical for BRCA2- and RAD51-dependent responses to DNA damage and HRR.
DNA double-strand breaks (DSBs) are induced by exogenous agents, such as ionizing radiation (IR), and arise spontaneously during normal DNA metabolism, such as at blocked or collapsed replication forks (9, 10, 39, 45). Defects in DSB repair confer genome instability associated with tumorigenesis. In mammalian cells, DSBs are repaired by nonhomologous end-joining and by homologous recombinational repair (HRR) (60, 62, 65). RAD51 binds single-stranded DNA (ssDNA) to form nucleoprotein filaments that are essential for strand transfer during HRR (23, 44, 61, 66). RAD51 is normally dispersed in the nucleus, but upon DNA damage induction, it redistributes to nuclear foci that are presumed sites of HRR (6, 7, 14, 20, 31, 46). RAD51 foci have been shown to be associated with ssDNA regions after DNA damage (46). Several HRR proteins, including XRCC2, XRCC3, RAD51B, RAD51C, RAD51D, and BRCA2, are important for RAD51 focus formation (1, 5, 7, 43, 55, 56).
BRCA2 has nine RAD51 binding regions, including eight BRC repeats encoded by exon 11 and a distinct RAD51 binding region encoded by exon 27 (8, 33, 69). Expression of individual BRC repeats interferes with RAD51 focus formation and HRR (5, 53, 70), indicating that RAD51-BRCA2 interactions are important for both processes. The C-terminal half of BRCA2 has three regions that are structurally related to the ssDNA binding region of RPA and bind ssDNA in vitro, suggesting that ssDNA binding is also important for BRCA2 function in HRR (71). These ssDNA binding regions occur in a region called conserved domain IV (30, 48, 73) or the BRCA2 C-terminal domain (71), which is the longest and most evolutionarily conserved BRCA2 domain (32, 57). This domain also has binding sites for several proteins including DSS1, BUBR1, ABP-280/filamin-A, and BCCIPα (16, 30, 34, 73).
BCCIPα is a BRCA2 and CDKN1A (p21, Cip1, and Waf1) interaction protein (30); it has also been called Tok-1α (42). A second isoform, BCCIPβ, shares an N-terminal acidic domain and a central conserved domain but has a distinct C-terminal domain (Fig. 1A). In this report, BCCIP indicates both proteins. The BCCIP proteins share no significant homology to other mammalian proteins. Although the interaction between BCCIPα and BRCA2 implicates BCCIP in RAD51-dependent processes, there is no prior direct support for this idea. In this report, we show that BCCIPβ interacts with BRCA2 in a region shared with the BCCIPα isoform. We further show that BCCIP colocalizes with BRCA2 in the nucleus and is associated with RAD51. To further analyze BCCIP function, we reduced BCCIP protein levels by RNA interference (RNAi) and found that this reduces RAD51 and BRCA2 focus formation in response to IR and sharply reduces HRR of DSBs. These results suggest a critical role for BCCIP in HRR regulation.
FIG. 1.
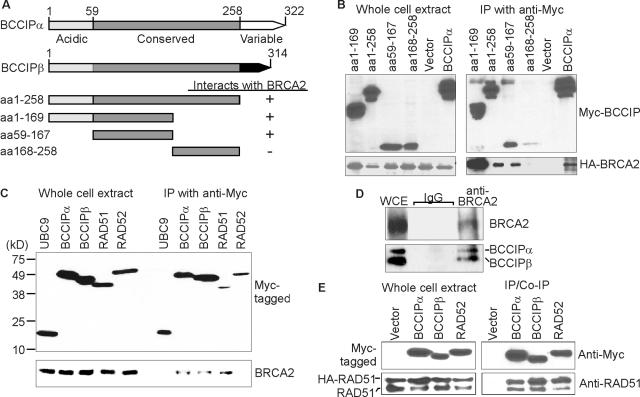
BCCIP interactions with BRCA2 and RAD51. (A) BCCIPα and BCCIPβ share a 258-N-terminal-amino-acid region including an acidic domain and a central conserved domain. Alternative mRNA splicing produces distinct C termini. BCCIP fragments used in immunoprecipitation assays (B) are shown below along with a summary of those results. (B) Mapping the BCCIP domain that interacts with BRCA2. Myc-tagged BCCIP fragments were coexpressed with a HA-tagged fragment of BRCA2 (aa 2883 to 3194) shown previously to interact with full-length BCCIPα in HeLa cells (30). Myc-tagged proteins were precipitated with mouse anti-Myc antibody and detected by rabbit anti-Myc antibodies in a Western blot (top panels). Coprecipitated HA-tagged BRCA2 fragment was detected by rabbit anti-HA antibody (bottom panels). (C) BCCIPβ interacts with BRCA2. Myc-tagged UBC9, BCCIPα, BCCIPβ, RAD51, and RAD52 were expressed in HeLa cells. Myc-tagged proteins were detected by Western blotting in whole-cell extracts or after precipitation with anti-Myc antibodies (top panel). Coprecipitated endogenous BRCA2 protein (bottom panel) was detected with anti-BRCA2 antibodies. Note that BCCIPα and BCCIPβ coprecipitate BRCA2 with approximately the same affinity. (D) Endogenous BCCIPα and BCCIPβ coprecipitate with endogenous BRCA2. Agarose-conjugated anti-BRCA2 beads were used to precipitate endogenous BRCA2 (top panel), and coprecipitated BCCIPα and BCCIPβ were detected by anti-BCCIP blotting (bottom panel). Nonspecific immunoglobulin G (IgG)-agarose beads were used as negative controls. WCE, whole-cell extract; IP, immunoprecipitation. (E) BCCIPα and BCCIPβ coprecipitate RAD51. Myc-tagged BCCIPα, BCCIPβ, and RAD52 were coexpressed with HA-tagged RAD51 in HeLa cells and precipitated with anti-Myc antibodies (top panels). Coprecipitated HA-RAD51 and endogenous RAD51 were detected by anti-RAD51 blotting (bottom panels), indicating that RAD51 is in complex with BCICPα and BCCIPβ.
MATERIALS AND METHODS
Plasmids, cell culture, and plasmid transfection.
Plasmid vectors expressing Myc-tagged BCCIPα, BCCIPβ, RAD52, UBC9, and RAD51 and the hemagglutinin (HA)-tagged fragment of BRCA2 (amino acids [aa] 2883 to 3194) were described previously (27, 29, 30, 36, 50, 73). Plasmid pMSGneo2S12His (54) carrying a neo direct repeat homologous recombination (HR) substrate was transfected into HT1080 cells, mycophenolic acid-resistant derivatives were isolated, and Southern hybridization and PCR assays were used to identify transfectants with a single, intact copy of the HR substrate as described previously (54). The I-SceI nuclease expression vector, pCMV(HA-3xNLS)I-SceI, was described previously (54). Plasmid pPUR/U6 carries a U6 promoter for short RNA transcription and a puromycin resistance cassette (38). HT1080 cells (American Type Culture Collection) were cultured in αMEM (Gibco BRL, Grand Island, N.Y.) with 10% fetal bovine serum (Biowhitaker, Walkersville, Md.), 20 mM glutamine, and 1% penicillin-streptomycin (Gibco BRL). Plasmids were transfected into cells by using the Geneporter transfection kit (Gene Therapy Systems, Inc., San Diego, Calif.).
Antibodies, Western blotting, and immunoprecipitation.
Rabbit anti-BCCIPα/β antibodies were reported previously (30). Other antibodies were purchased: anti-BRCA2 (Ab2) and anti-RAD51 from Oncogene Science (Cambridge, Mass.) and anti-Myc/anti-HA from Clontech Laboratories (Palo Alto, Calif.). Protein extracts were prepared from cells lysed with 50 mM HEPES (pH 7.6), 250 mM NaCl, 5 mM EDTA, and 0.1% Nonidet P-40. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blotting was performed as described previously (36, 38). For immunoprecipitation, cells were washed with phosphate-buffered saline (PBS), harvested by centrifugation, treated with lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Tween 20, 0.25% NP-40, 1 mM Na3VO4, 100 μg of phenylmethylsulfonyl fluoride/ml, 20 μg of aprotinin/ml, 10 μg of leupeptin/ml), sonicated, and centrifuged for 10 min at 20,000 × g. The supernatant (whole-cell extract) was treated with Benzonase nuclease (Novagene, Madison, Wis.) to digest DNA and RNA and prevent their interfering with protein complex formation. To precipitate the Myc-tagged proteins, agarose-conjugated anti-Myc antibodies (Clontech Laboratories) were incubated with whole-cell extracts for 4 h at 4°C on a rotating platform. The Ab2 anti-BRCA2 antibodies (Oncogene Science) were conjugated with agarose beads by using the Seize X mammalian immunoprecipitation kit (Pierce, Rockford, Ill.) and used to precipitate endogenous BRCA2 protein. The agarose beads were washed 10 times with radioimmunoprecipitation assay buffer (1× PBS, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM Na3VO4, 100 μg of phenylmethylsulfonyl fluoride/ml, 20 μg of aprotinin/ml, 10 μg of leupeptin/ml), and precipitated proteins were detected with appropriate antibodies as noted in the figure legends.
Stable short hairpin RNA (shRNA) interference of BCCIP.
Specific sequences targeting BCCIPα and/or BCCIPβ were inserted into pPUR/U6: 5′-GGG AAC CTT CAT GAC TGT TGG targets BCCIPα, 5′-GGG AAG CAA ATG GTC TTT TGA targets BCCIPβ, and 5′-GGG AAG TGC TAC TTT TAC CTT-3′ targets both proteins. The resulting vectors (pPUR/U6/RNAi-BCCIPα, pPUR/U6/RNAi-BCCIPβ, and pPUR/U6/RNAi-BCCIPαβ) were transfected into cells, stable transfectants were selected in growth medium with 1.0 μg of puromycin/ml, and BCCIP expression was measured by Western blotting.
Adenovirus-mediated shRNA interference of BCCIP and BRCA2.
The BCCIP shRNA expression cassettes in pPUR/U6/RNAi-BCCIPα, pPUR/U6/RNAi-BCCIPβ, and pPUR/U6/RNAi-BCCIPαβ were amplified by PCR with primers 5′-ATA AGA ATG CGG CCG CAT AGG CGT ATC ACG AGG CCC and 5′-CCC AAG CTT GAC GCC GCC ATC TCT AGG CCC-3′ and inserted into the pAdTrack-CMV shuttle vector (21) in the opposite direction to the cytomegalovirus (CMV) promoter-driven green fluorescent protein (GFP) gene. Adenovirus stocks were prepared and used to infect cells as described previously (21). A shRNA sequence targeted to BRCA2 (5′-GGG AAA CAC TCA GAT TAA AGA) was inserted into pPUR/U6, transferred to pAdTrack-CMV, and used to downregulate BRCA2 as described for BCCIP above.
Immunocytochemistry.
After infection with adenovirus shRNA vectors and/or treatment with gamma rays, cells were fixed by one of two procedures. The first procedure was used to fix total cellular protein: cells on coverslips were washed with PBS, fixed with PBS-buffered 4% paraformaldehyde in a 2% sucrose solution for 10 min at room temperature, and permeabilized for 10 min in 0.2% Triton X-100 in TBS-T buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Tween 20) at room temperature. This procedure was used to detect GFP, BCCIP, and RAD51, as it retains total cellular proteins and preserves signals from GFP and RAD51 nuclear foci. The second procedure removes soluble proteins and fixes chromatin-bound proteins: cells on coverslips were washed with PBS, treated for 5 min with Triton buffer (0.5% Triton X-100 in 20 mM HEPES [pH 7.4], 50 mM NaCl, 3 mM MgCl2, and 300 mM sucrose) on ice to remove soluble proteins. Cells with the remaining chromatin-bound proteins were fixed with PBS-buffered 4% paraformaldehyde in 2% sucrose solution for 10 min at room temperature. This procedure was used to detect BRCA2 and BCCIP foci.
Fixed cells were washed with TBS-T three times and blocked for 30 min with 3% bovine serum albumin in TBS-T. Cells were incubated in anti-BCCIP (1:500), anti-BRCA2 (1:100 or 1:200), or anti-RAD51 (1:1,000) primary antibody diluted in 3% bovine serum albumin blocking buffer at 4°C overnight. After washing three times with TBS-T, cells on coverslips were incubated with appropriate secondary antibodies conjugated with fluorescein (FITC) or Texas Red (Jackson ImmunoResearch, West Grove, Pa.) for 1 h at room temperature in the dark and then washed and mounted by using mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI). The images were recorded with a LSM510 Zeiss laser scanning microscope and a Zeiss fluorescence microscope with an Axioskop digital camera. BCCIP, BRCA2, and RAD51 foci were scored in 100 to 300 cells on at least three independent slides. Our criteria for colocalization required a minimum of three coincident green and red signals per cell.
DSB-induced HRR assay.
HRR was assayed essentially as described previously (54). Briefly, HT256 cells or its derivatives expressing BCCIP shRNAs were transfected with 2.5 μg of pCMV(HA-3xNLS)I-SceI that expresses a HA epitope- and nuclear localization signal-tagged I-SceI nuclease. Forty-eight hours later, ∼100,000 viable cells were transferred to 10-cm-diameter dishes, and HRR products were selected in growth medium with 325 μg of G418/ml. G418-resistant colonies were scored 12 to 14 days later. The plating efficiency was determined by seeding the appropriate numbers of cells to 10-cm-diameter dishes with growth medium lacking G418. HRR frequencies were calculated as the number of G418-resistant colonies per viable cell plated in G418 medium. Background HR levels were estimated in parallel experiments, except that cells were transfected with an empty pCMV vector. Statistical analyses were performed by t tests. To distinguish gene conversion and deletion products, the TripleMaster PCR system (Eppendorf, Hamburg, Germany) was used to amplify the entire 7.5-kbp HR substrate with the following primers: 5′-CGC CAT CCC GTC TCC GCT CGT CAC and 5′-CTC ATG AGC GGA TAC ATA TTT GAA TGT ATT TAG. PCR products were digested with I-SceI (New England BioLabs, Beverly, Mass.) to confirm the loss of the I-SceI site. Gene conversions result in 7.5-kbp PCR products, and deletions result in 2.5-kbp PCR products.
RESULTS
Mapping the BRCA2-interacting domain of BCCIPα and BCCIPβ.
We showed previously that BCCIPα interacts with amino acids 2883 to 3053 of BRCA2, centering around amino acids 2973 to 3001 (30). However, the corresponding interaction region in BCCIPα was not determined. To map this region, we constructed a series of plasmids that express Myc-tagged fragments of BCCIPα (Fig. 1A). The BCCIPα fragments were coexpressed with a HA-tagged BRCA2 fragment (aa 2883 to 3194) that contains the BCCIP interaction region. We found that BCCIPα fragments corresponding to amino acids 1 to 258, 1 to 167, and 59 to 167 coprecipitate BRCA2 (Fig. 1B). Thus, the minimal region of BCCIPα required to interact with BRCA2 resides in amino acids 59 to 167 in the N-terminal half of the internal conserved domain. The C-terminal half of this BCCIP domain (amino acids 161 to 259) interacts with p21 (42). Thus, BRCA2 and p21 interact with distinct regions of BCCIPα.
Because the BRCA2 interacting domain of BCCIPα was mapped to a region shared by BCCIPα and BCCIPβ (Fig. 1A), we tested whether BCCIPβ also interacts with BRCA2. This was done by immunoprecipitating full-length BCCIP and then determining whether endogenous BRCA2 was coprecipitated (Fig. 1C). Human RAD51 was used as a positive control for coprecipitating BRCA2; RAD52 and UBC9 were used as negative controls. To ensure equal affinity for each protein during precipitation, proteins were Myc tagged and expressed in HeLa cells. In addition, DNA and RNA were removed by nuclease digestion to eliminate potential interference with protein complex formation. As expected, Myc-RAD51 coprecipitated endogenous BRCA2. Although UBC9 and RAD52 interact with RAD51 (26, 50, 51), they did not coprecipitate BRCA2. Both Myc-BCCIPα and Myc-BCCIPβ coprecipitated endogenous BRCA2, and each isoform has approximately the same affinity toward BRCA2. Furthermore, endogenous BCCIPα and BCCIPβ coprecipitate with endogenous BRCA2 (Fig. 1D). Thus, BCCIPβ interacts with BRCA2; the similar affinities of the two isoforms with BRCA2 are consistent with the finding that BRCA2 interacts with a region shared by BCCIPα and BCCIPβ.
BCCIP is in complex with RAD51.
Because RAD51 and BCCIPα/β each interact with BRCA2, we tested whether RAD51 also associates with BCCIP. HA-tagged RAD51 was coexpressed with Myc-tagged BCCIPα, BCCIPβ, or RAD52 in HeLa cells. RAD52 is a positive control, since it binds to RAD51 (50). When these Myc-tagged proteins were precipitated, both endogenous and HA-tagged RAD51 coprecipitated with BCCIPα and BCCIPβ (Fig. 1E). These data indicate that BCCIP forms protein complexes with RAD51. BRCA2 and RAD51 are known to be involved in HRR, thus the interactions between BCCIP and these proteins implicate BCCIP in HRR.
BCCIP colocalizes with BRCA2 and RAD51 nuclear foci.
RAD51 and BRCA2 colocalize in nuclear foci after ionizing radiation (6, 8, 69, 72). Because BCCIP interacts with BRCA2 and (perhaps indirectly) with RAD51, we tested whether BCCIP forms nuclear foci and, if so, whether these colocalize with BRCA2/RAD51 foci. We previously showed that most BCCIP resides in the nucleus (30). When total cellular protein was examined by immunofluorescence, BCCIP and BRCA2 nuclear foci were blurry (data not shown) and difficult to count accurately. To overcome this problem, cells were treated with Triton X-100 before paraformaldehyde fixation, which removes soluble nuclear proteins and retains chromatin-bound proteins (see Materials and Methods). Immunofluorescence revealed both BCCIP and BRCA2 nuclear foci in unirradiated cells (Fig. 2A). Interestingly, ∼80% of cells showed colocalized BCCIP and BRCA2 foci, but this was not altered by irradiation (Fig. 2C). Irradiation did not increase the number of BCCIP foci (see below) nor the percentage of cells with colocalized BCCIP and BRCA2 foci, although BCCIP foci became more sharply defined. Among cells treated with 0, 2, or 8 Gy of gamma rays that showed BCCIP foci, 100% had colocalized BRCA2 foci (Fig. 2D) and vice versa (Fig. 2E). These results suggest that BRCA2 and BCCIP are associated in the absence of exogenous DNA damage and remain so after DNA damage.
FIG. 2.
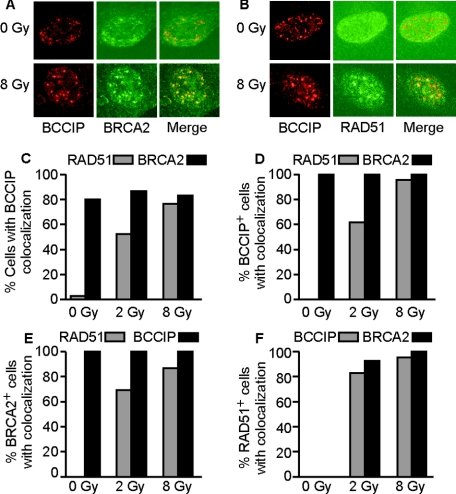
BCCIP colocalizes with BRCA2 and RAD51. (A) Colocalization of BCCIP and BRCA2 in HT1080 cells. Chromatin-bound endogenous BCCIP was stained with anti-BCCIP (Texas Red, red) and BRCA2 was stained with anti-BRCA2 (FITC, green) in untreated cells or 8 h after an 8-Gy dose of gamma rays. Yellow foci in the merged images indicate colocalization. (B) Colocalization of BCCIP and RAD51. Cells were treated, and BCCIP was stained as described for panel A. RAD51 was stained with FITC. (C) Percentage of HT1080 cells with BCCIP foci colocalized with RAD51 or BRCA2 foci in cells treated with 0, 2, or 8 Gy of gamma rays and scored 8 h later. Cells with at least three colocalized BCCIP/BRCA2 or BCCIP/RAD51 foci were counted. (D) Percentage of BCCIP focus-positive (BCCIP+) cells with at least three colocalized RAD51 or BRCA2 foci in cells treated with 0, 2, or 8 Gy of gamma rays and scored 8 h later. (E, F) Percentages of BRCA2 focus-positive (BRCA2+) or RAD51 focus-positive (RAD51+) cells with colocalized foci. Cells were treated as described for panel D.
As shown in Fig. 2B, significant numbers of RAD51 foci are apparent only after irradiation, and the majority colocalize with BCCIP. Among cells with BCCIP foci after a 2- or 8-Gy dose of gamma rays, 60 to 95% show colocalized RAD51 foci (Fig. 2D). Similar percentages of irradiated cells with BRCA2 foci show colocalized RAD51 foci (Fig. 2E). These results indicate an increased association of BCCIP with RAD51 in response to DNA damage. BRCA2 foci have not previously been detected in the absence of exogenous DNA damage. Note that we detected BRCA2 foci without exogenous damage only when soluble protein was removed, suggesting a certain level of constitutive association of BRCA2 with chromatin, perhaps to resolve endogenous DNA damage.
To gain further insight into the association of RAD51, BRCA2, and BCCIP, we scored the number of cells with at least three foci in HT1080 cells treated with 2 or 8 Gy of gamma rays. Eight hours after irradiation, we found that ∼80% of cells with RAD51 foci also have BCCIP or BRCA2 foci (Fig. 2F). Thus, in ∼20% of cells with at least three RAD51 foci, these foci apparently form independently of BRCA2 or BCCIP. This is consistent with a recent report showing some RAD51 foci forming independently of BRCA2 (58). In contrast, in both untreated and irradiated cells, 100% of cells with BCCIP foci have BRCA2 foci and vice versa (Fig. 2D and E). We obtained similar results 2 h after irradiation (data not shown). Together, these results suggest that chromatin-bound BRCA2 and BCCIP are constitutively associated, that RAD51 is recruited to these complexes after DNA damage, and that RAD51 self-associates at some sites independently of BCCIP and BRCA2.
RNAi silencing of BCCIP inhibits the formation of BRCA2 and RAD51 nuclear foci.
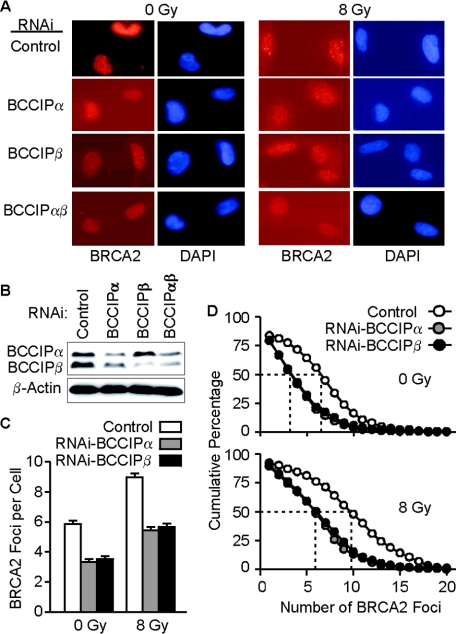
Based on the results above, we hypothesized that BCCIP is required for most RAD51 and BRCA2 focus formation. To test this, we downregulated BCCIPα and/or BCCIPβ by expressing shRNAs targeted to shared or unique sequences in these isoforms by using adenovirus vectors that coexpress GFP. When soluble protein is removed by Triton X-100 treatment, the GFP signal indicative of adenovirus infection is destroyed. Therefore, we used sufficient adenovirus to infect an entire HT1080 cell population, irradiated the cells with 8 Gy of gamma rays or mock treated them 72 h later, and visualized chromatin-bound BRCA2 2 or 8 h after irradiation by immunostaining, as described above. After irradiation, cells infected with the control virus showed well-defined BRCA2 foci (Fig. 3A, top row). Reduced BCCIP expression was confirmed by Western blot analysis (Fig. 3B). We scored BRCA2 foci in >300 cells per treatment group and found that the average number of BRCA2 foci per cell was reduced by twofold upon BCCIPα or BCCIPβ downregulation, and these reductions were apparent in both irradiated and untreated cells (Fig. 3A, bottom 3 rows, and C). In addition, BRCA2 foci were less intense in cells with reduced BCCIP (Fig. 3A, bottom 3 rows). The number of BRCA2 foci per cell varied widely within each treatment group; this is apparent in plots of the cumulative percentages of cells with more than a specific number of BRCA2 foci (Fig. 3D). For example, 50% of unirradiated cells had ∼6 or more BRCA2 foci, and the number of foci per cell ranged from 0 to 15. Following irradiation, a wider distribution was seen: 50% of cells had 10 or more foci and ∼10% had more than 15 foci. Downregulation of BCCIPα or BCCIPβ narrowed the distributions of foci in both unirradiated and irradiated cells to similar degrees. For example, 50% of irradiated cells with downregulated BCCIPα or BCCIPβ had 6 or more BRCA2 foci, compared to 10 or more foci in control cells (Fig. 3D, bottom panel). The similar effects of BCCIPα and BCCIPβ on BRCA2 foci distributions is consistent with our finding that BRCA2 interacts with a region common to the BCCIP isoforms (Fig. 1A and B).
FIG. 3.
BCCIP is important for BRCA2 focus formation. (A) Entire populations of HT1080 cells were infected with adenoviruses expressing shRNAs targeted to BCCIPα, BCCIPβ, of both isoforms or with an empty vector as a control and stained with anti-BRCA2 antibodies (red) or DAPI to identify nuclei. Images were recorded 8 h after irradiation. (B) BCCIP expression in adenovirus-infected cells. Seventy-two hours after infection with adenoviruses expressing BCCIP-shRNAs, BCCIP expression was detected by anti-BCCIP blotting (top panel); anti-β-actin was used as loading control (bottom panel). (C) Average number of BRCA2 foci (± standard error) in cells with normal or reduced levels of BCCIPα or BCCIPβ. For each data point, foci were scored in ∼300 cells. BCCIP downregulation reduced BRCA2 foci significantly with or without irradiation, and irradiation increased BRCA2 foci significantly with or without BCCIP downregulation (all P < 0.0001). (D) Cumulative percentage of cells with at least a specified number of BRCA2 foci (x axis) in control and BCCIP-downregulated cells. BRCA2 foci were scored in unirradiated cells or 8 h after irradiation.
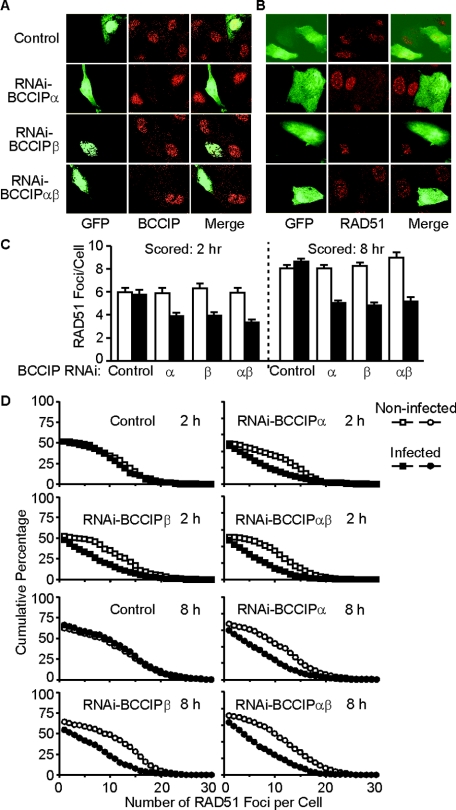
We next investigated whether BCCIP silencing affected RAD51 focus formation in irradiated cells. Unlike BRCA2 and BCCIP, RAD51 foci can be visualized without removing soluble nuclear protein. Therefore, the GFP signals in adenovirus-infected cells can be preserved. This allowed us to score RAD51 foci in BCCIP silenced cells (GFP+) and in cells with normal BCCIP levels (GFP−) simultaneously on the same slide. We treated HT1080 cells with an appropriate virus titer to infect ∼50% of the cells and then fixed cells with paraformaldehyde before treating them with Triton X-100, which preserves the GFP signal. In cell populations infected with a control vector, endogenous BCCIP protein was readily detected whether cells were infected (GFP+) or not (GFP−) (Fig. 4A, top row). Infection with viruses expressing BCCIP shRNAs substantially reduced BCCIP protein levels in GFP+ cells but not in GFP− cells (Fig. 4A, bottom three rows), confirming the efficacy of these silencing vectors. As expected, RAD51 focus formation was unaffected by infection with a control vector (compare RAD51 signals in GFP+ and GFP− cells in Fig. 4B, top row). However, downregulation of BCCIPα and/or BCCIPβ (GFP+ cells) resulted in substantially fewer RAD51 foci than in neighboring, uninfected (GFP−) cells (Fig. 4B, bottom 3 rows). We scored RAD51 foci after 8-Gy gamma ray exposures and found that BCCIP downregulation reduced the average number of RAD51 foci by ∼2-fold when scored 2 or 8 h after irradiation (Fig. 4C). As seen with BRCA2 foci above, downregulation of BCCIPα and/or BCCIPβ had similar effects on the distribution of RAD51 foci (Fig. 4D).
FIG. 4.
BCCIP is important for RAD51 focus formation. (A) About50% of HT1080 cell populations were infected with adenoviruses expressing GFP and shRNAs targeted to BCCIPα, BCCIPβ, or both isoforms or GFP alone as a control. Cells were irradiated with 8 Gy of gamma rays and prepared for immunofluorescence 8 h later. Cells were stained with anti-BCCIP antibodies (red); infected cells show green fluorescence. Within each field are infected and noninfected cells. Cells infected with BCCIP shRNA vectors (bottom three rows) show reduced BCCIP levels. (B) Cells from panel A were stained with anti-RAD51 antibodies (red). Infected (green fluorescence) and noninfected cells are shown in each panel. Cells infected with viruses expressing BCCIP shRNAs show reduced RAD51 foci. (C) Average number of RAD51 foci (± standard error) in cells with normal or reduced levels of BCCIPα or BCCIPβ (uninfected or adenovirus infected, respectively), 2 and 8 h after treatment with 8 Gy of gamma rays. For each data point, foci were scored in at least 300 cells. RAD51 foci are reduced only in cells infected with adenoviruses expressing BCCIP shRNAs. (D) Cumulative percentage of cells with at least a specified number of RAD51 foci (x axis) in control and BCCIP-downregulated cells. RAD51 foci were scored in uninfected or adenovirus-infected cells 2 and 8 h after treatment with 8 Gy of gamma rays.
Role of BRCA2 in BCCIP and RAD51 focus formation.
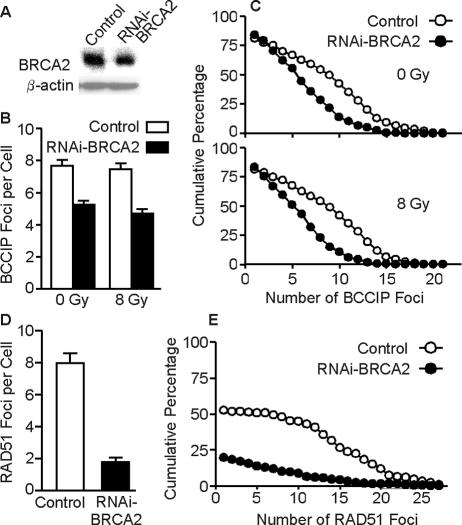
The data above indicate that BCCIP plays a role in the formation of BRCA2 and RAD51 foci. It was previously established that BRCA2 is required for RAD51 focus formation (58, 72). To determine whether BCCIP focus formation also depends on BRCA2, we downregulated BRCA2 by RNAi and examined BCCIP foci. Consistent with BRCA2 being essential for cell viability (49), we were able to reduce BRCA2 to only ∼50% of wild-type levels (Fig. 5A), yet this was sufficient to reduce the average number of BCCIP foci per cell by ∼40 to 50% in unirradiated cells and 8 h after an 8-Gy gamma ray exposure (Fig. 5B and C); similar results were obtained when foci were scored 2 h after irradiation (data not shown). Thus, BRCA2 downregulation reduced but did not eliminate BCCIP focus formation. However, unlike BRCA2 foci, which increased after irradiation when BCCIP was downregulated (Fig. 3C and D), BCCIP foci did not increase after irradiation in BRCA2 downregulated cells (Fig. 5B and C). Consistent with prior reports (58, 72), BRCA2 downregulation sharply reduced RAD51 focus formation, evident by the fourfold reduction in the average number of RAD51 foci per cell (Fig. 5D) and by the dramatic change in the distribution of RAD51 foci (Fig. 5E). These results suggest a degree of mutual dependence among BRCA2, BCCIP, and RAD51 for focus formation. However, because the amount of RNAi reduction varied among the BCCIP isoforms and BRCA2, it is difficult to quantify the magnitude of the various dependencies.
FIG. 5.
BRCA2 is important for formation of BCCIP and RAD51 foci. (A) HT1080 cells were infected with adenovirus expressing shRNA targeted to BRCA2, and 72 h later, BRCA2 was detected by Western blotting. Empty virus was used as control; blots were also hybridized with anti-β-actin antibody as a loading control. (B) Average number of BCCIP foci (± standard error) in cells expressing normal or low levels of BRCA2, either unirradiated or irradiated with 8 Gy of gamma rays; foci were scored 8 h after irradiation. (C) Distribution of BCCIP foci in unirradiated or irradiated cells expressing normal or low levels of BRCA2. These data are plotted as described for Fig. 3D. (D, E) Average number and distribution of RAD51 foci in irradiated cells expressing normal or low levels of BRCA2. Data are presented as described for panels B and C, respectively.
Role for BCCIP in HRR of DNA DSBs.
Based on the results above, we hypothesized that BCCIP is involved in HRR. We tested this by using a derivative of HT1080 called HT256, which carries a single integrated copy of a neo direct repeat HR substrate. One of the neo repeats is driven by the mouse mammary tumor virus promoter but is inactive due to the insertion of an I-SceI recognition site, and the second allele (neo12) has 12 single-base polymorphic markers but codes for wild-type protein and lacks a promoter (54) (Fig. 6A). Transfection of HT256 cells with an I-SceI expression vector creates DSBs at the I-SceI site. Repair of these DSBs by HRR can result in gene conversion without a crossover, which preserves the gross structure of the HR substrate, or deletions by crossing over or single-strand annealing. These outcomes can produce a functional neo that confers G418 resistance and are distinguishable by a PCR assay (see Materials and Methods). As in CHO cells carrying this same HR substrate (54), spontaneous HR in HT256 cells is below the limit of detection (<10−7) and expression of I-SceI increases HR by several orders of magnitude (data not shown).
FIG. 6.
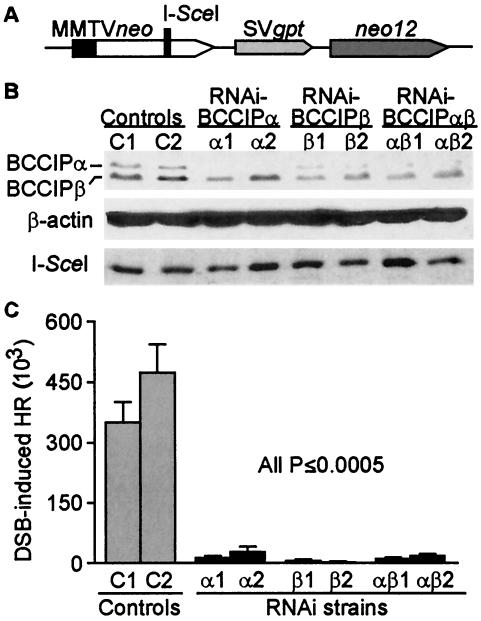
BCCIP is critical for HRR. (A) Structure of HR substrate in HT256 cells. (B) Downregulation of BCCIP in HT256 cells. Cells were stably transfected with vectors expressing shRNAs targeted to BCCIPα, BCCIPβ, or both isoforms or with an empty vector as a control. Expression of both isoforms was monitored in two independent transfectants by Western blotting with anti-BCCIP antibodies (top panel). β-Actin serves as a loading control (middle panel), and expression of HA-tagged I-SceI nuclease was detected by anti-HA blotting (bottom panel). (C) HRR frequencies calculated as the number of G418-resistant colonies per viable cell plated in selective medium. HT256 cells expressed normal (control) or reduced levels of BCCIPα, BCCIPβ, or both isoforms. Values are averages ± standard errors for 4 determinations per cell line.
We transfected HT256 cells with plasmid vectors that express a puromycin-resistance gene and shRNAs to downregulate BCCIPα, BCCIPβ, or both isoforms or with an empty vector as a control. Stably transfected derivatives were selected with puromycin, and BCCIP expression was measured by Western blotting with anti-BCCIP antibodies. As shown in Fig. 6B, BCCIP downregulation varied among these derivatives, with BCCIPα reduced by ∼90% and BCCIPβ reduced by ∼50%. These lower BCCIP levels were stable for 15 to 20 population doublings (three to four passages), and all HRR assays were performed within the first three passages. RNAi knockdown of BCCIP did not affect the expression of HA-tagged I-SceI (Fig. 6B, bottom panel). It is worth noting that several alternative strategies to further reduce BCCIP expression were unsuccessful, probably because one or both BCCIP isoforms are essential for viability. As shown in Fig. 6C, downregulation of BCCIPα reduced HRR by ∼20-fold, and downregulation of BCCIPβ reduced HRR by ∼100-fold. We analyzed genomic DNA isolated from 44 G418-resistant products from the control strains with PCR (see Materials and Methods for details) and found that 42 arose by gene conversion without crossover, 1 arose by deletion, and 1 had a complex pattern. Similarly, among 17 G418-resistant products from cells with reduced BCCIP expression, 15 arose by gene conversion, 1 arose by deletion, and 1 had a complex pattern. The marked reduction in HRR efficiency with no change in HRR product spectrum suggests that BCCIP plays a critical role early in HRR.
DISCUSSION
The present study revealed several critical links between BCCIP and the HRR proteins BRCA2 and RAD51. We found that a region shared by BCCIPα and BCCIPβ interacts with BRCA2, that BCCIP is in complex with RAD51, that chromatin-bound BRCA2 consistently colocalizes with BCCIP, and that the majority of radiation-induced RAD51 nuclear foci colocalize with BCCIP. Furthermore, downregulating BCCIP reduces both BRCA2 and RAD51 focus formation and markedly reduces HRR of DNA DSBs. Although the molecular mechanism by which BCCIP regulates HRR remains to be elucidated, our results establish BCCIP as a critical component of the DNA damage response network and HRR pathway.
Two key findings are that BCCIP is present in chromatin-bound nuclear foci even in the absence of induced DNA damage and that all BRCA2 foci colocalize with BCCIP (Fig. 2E). Although RAD51 foci also colocalize with BRCA2 (5, 7, 69) and BCCIP foci (Fig. 2B), RAD51 foci appear only after induced DNA damage. These results suggest the possibility that BCCIP and BRCA2 are normally associated in chromatin and that RAD51 is recruited to these repair-complex centers when DNA is damaged.
Studies of mouse embryonic stem cells expressing BRCA2 lacking exon 27 revealed twofold (64) and five- to sixfold (40) reductions in HRR. In human Capan-1 cells, which express truncated BRCA2 lacking BRC repeats 7 and 8, conserved region IV, and exon 27, HRR is reduced by 2- to 12-fold (70). In the present study, moderate decreases in BCCIP levels reduce HRR by up to 100-fold (Fig. 6). This surprising result may reflect a threshold effect, i.e., BCCIP-BRCA2-RAD51 function in HRR requires proper BCCIP stoichiometry. We note also that reducing BCCIP reduces both the number and intensity of BRCA2 foci (Fig. 3A), raising the possibility that the foci that do form are not fully functional. In any case, our results indicate a critical role for BCCIP in HRR. BCCIP most likely functions in HRR through its direct interaction with BRCA2 in or near the BRCA2-ssDNA interaction domains and through its indirect interactions with RAD51 (discussed further below). However, we cannot exclude the possibility that BCCIP regulates HRR indirectly because BCCIP is also involved in cell cycle control (36, 37, 42).
The BCCIPα and BCCIPβ isoforms are 80% identical, and our biochemical results indicate that these isoforms interact with BRCA2 through a shared domain of BCCIPα and BCCIPβ (Fig. 1A and B). The very similar effects of BCCIPα and/or BCCIPβ downregulation on BRCA2 and RAD51 focus formation (Fig. 3 and 4) suggest that both isoforms are capable of mediating focus formation, probably through this specific BRCA2 interaction. Note also that there was an apparent decrease of BCCIPα protein when BCCIPβ was silenced and vice versa (Fig. 6B). Thus, silencing of one isoform may affect the stability of the other, and this may account for the similar effects when either isoform is reduced.
There is only one BCCIP homolog in Saccharomyces cerevisiae, Arabidopsis thaliana, Caenorhabditis elegans, and mice. The C terminus of human BCCIPα (aa 259 to 322) has no homology with BCCIP homologs in other organisms, but the C terminus of human BCCIPβ (aa 258 to 314) shows various degrees of homology with all known BCCIP homologs (data not shown). Thus, BCCIPβ appears to be the conserved isoform. The S. cerevisiae BCCIP homolog, Bcp1 (open reading frame number YDR361C), is essential for viability (17) (Saccharomyces Genome Database at http://www.yeastgenome.org). Silencing of the C. elegans 2H343 gene (the ortholog of human BCCIPβ) leads to embryonic lethality (24). In addition, BCCIPα is expressed at lower levels than BCCIPβ, and we were able to silence BCCIPα much more than BCCIPβ. These results suggest that BCCIPβ is essential in human cells. RAD51 and BRCA2 are both required for cell viability (28, 49, 52), and RAD51 defects in particular result in proliferation-dependent defects and altered DNA replication (22, 52). Given that HR proteins function in restarting blocked replication forks (11, 12, 47) and the importance of BRCA2, RAD51, and BCCIP in HRR, it is possible that these proteins function together in recombinational restart of blocked replication forks and that this function is essential for cell viability.
Biochemical and structural studies revealed several features of BRCA2 critical for HRR. The interactions between RAD51 and the BRCA2 BRC repeats and/or exon 27 may promote RAD51 loading onto ssDNA (68). BRCA2 appears to interact with ssDNA via three OB domains that mimic the structure of the ssDNA binding domain of RPA. Protruding from the second ssDNA binding region, OB2 is a 3-helix bundle domain that has structural similarity to proteins that bind double-stranded DNA (71). Interestingly, BCCIP interacts with aa 2973 to 3001 in the OB2 region, but this binding site lies opposite the ssDNA binding surface (30). Thus, BCCIP may play a critical role in regulating ssDNA binding by BRCA2, perhaps by stabilizing the OB2-ssDNA interaction. BCCIP may also influence RAD51 function indirectly through its interactions with BRCA2. We recently found that expression of an OB2 peptide has strong dominant-negative effects on HRR (M. A. Brenneman, X. Guo, Z. Shen, D. J. Chen, and J. A. Nickoloff, submitted for publication), indicating that OB2 is critical for HRR. This result, together with the present results indicating a critical role for BCCIP in HRR, further supports the notion that BCCIP functions in HRR through its interaction with OB2. Note, however, that BCCIP downregulation strongly inhibits HRR but has only a moderate effect on RAD51 focus formation. Thus, it appears that BRCA2 can promote RAD51 focus formation in the absence of BCCIP, but such foci may be very inefficient repair centers.
Genome instability, manifested as aneuploidy, broken chromosomes, translocations, and chromosome fusions, is common in cells with defects in HRR proteins, including RAD51, RAD51 paralogs, BRCA1, and BRCA2 (2, 3, 13, 56, 59). Defects in checkpoint proteins also cause genome instability (25, 41). We have found that only 5 to 10% is chromatin-bound (unpublished data), suggesting that BCCIPβ has chromatin-independent functions. We recently showed that BCCIP directly regulates p21 activity and contributes to G1-S progression (36, 37). Thus, BCCIP may influence genome stability by promoting HRR of DSBs, restarting blocked replication forks, and regulating checkpoint activity. BCCIP is therefore likely to be a tumor suppressor. This idea is supported by recent evidence showing that BCCIP expression is often reduced in kidney tumors (35) and with the observed loss of single BCCIP alleles in some brain tumors (unpublished data).
Assuming one or both BCCIP isoforms is essential for cell viability, tumors are unlikely to arise by biallelic BCCIP inactivation. Instead, downregulation by methylation or loss or inactivation of one BCCIP allele may produce a haploinsufficient phenotype that initiates or promotes tumorigenesis by destabilizing the genome. This idea gains support from our finding that a 50% reduction BCCIPβ nearly eliminates HRR (Fig. 6). Similarly, a 50% reduction of BRCA2 compromises DNA damage responses (Fig. 5), consistent with a haploinsufficient genome instability phenotype of cells heterozygous for BRCA2 mutations (4, 67). Defects in genes encoding FANCA, BLM, and other DNA damage response proteins also result in haploinsufficient genome instability (15, 18, 19, 63), suggesting that this is a general property of this class of tumor suppressors.
Acknowledgments
This research was supported by National Institute of Health NIEHS grant ES08353 and by U.S. Army Medical Research and Materiel Command grants DAMD17-02-1-0515 and DAMD17-03-1-0317 to Z.S., a U.S. Army Medical Research and Materiel Command postdoctoral fellowship (W81XWH-04-1-0566) to H.L., and grant CA77693 from the National Cancer Institute to J.A.N.
REFERENCES
- 1.Bishop, D. K., U. Ear, A. Bhattacharyya, C. Calderone, M. Beckett, R. R. Weichselbaum, and A. Shinohara. 1998. Xrcc3 is required for assembly of Rad51 complexes in vivo. J. Biol. Chem. 273:21482-21488. [DOI] [PubMed] [Google Scholar]
- 2.Brenneman, M. A., B. M. Wagener, C. A. Miller, C. Allen, and J. A. Nickoloff. 2002. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell 10:387-395. [DOI] [PubMed] [Google Scholar]
- 3.Brenneman, M. A., A. E. Weiss, J. A. Nickoloff, and D. J. Chen. 2000. XRCC3 is required for efficient repair of chromosome breaks by homologous recombination. Mutat. Res. 459:89-97. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz, T. A., X. Wu, A. Hussain, S. L. Tucker, G. B. Mills, B. Haffty, S. Bergh, M. Story, F. B. Geara, and W. A. Brock. 2002. Evidence of haplotype insufficiency in human cells containing a germline mutation in BRCA1 or BRCA2. Int. J. Cancer 97:557-561. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. F., P. L. Chen, Q. Zhong, Z. D. Sharp, and W. H. Lee. 1999. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J. Biol. Chem. 274:32931-32935. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., D. P. Silver, D. Walpita, S. B. Cantor, A. F. Gazdar, G. Tomlinson, F. J. Couch, B. L. Weber, T. Ashley, D. M. Livingston, and R. Scully. 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2:317-328. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. J., D. Silver, S. Cantor, D. M. Livingston, and R. Scully. 1999. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res. 59:1752S-1756S. [PubMed] [Google Scholar]
- 8.Chen, P. L., C. F. Chen, Y. Chen, J. Xiao, Z. D. Sharp, and W. H. Lee. 1998. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl. Acad. Sci. USA 95:5287-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, M. M. 2001. Historical overview: searching for replication help in all of the rec places. Proc. Natl. Acad. Sci. USA 98:8173-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, M. M. 2002. The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. 510:107-120. [DOI] [PubMed] [Google Scholar]
- 11.Cox, M. M. 2001. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 35:53-82. [DOI] [PubMed] [Google Scholar]
- 12.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 13.Cui, X., M. Brenneman, J. Meyne, M. Oshimura, E. H. Goodwin, and D. J. Chen. 1999. The XRCC2 and XRCC3 repair genes are required for chromosome stability in mammalian cells. Mutat. Res. 434:75-88. [DOI] [PubMed] [Google Scholar]
- 14.Essers, J., A. B. Houtsmuller, L. van Veelen, C. Paulusma, A. L. Nigg, A. Pastink, W. Vermeulen, J. H. Hoeijmakers, and R. Kanaar. 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fodde, R., and R. Smits. 2002. Cancer biology. A matter of dosage. Science 298:761-763. [DOI] [PubMed] [Google Scholar]
- 16.Futamura, M., H. Arakawa, K. Matsuda, T. Katagiri, S. Saji, Y. Miki, and Y. Nakamura. 2000. Potential role of BRCA2 in a mitotic checkpoint after phosphorylation by hBUBR1. Cancer Res. 60:1531-1535. [PubMed] [Google Scholar]
- 17.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 18.Goss, K. H., M. A. Risinger, J. J. Kordich, M. M. Sanz, J. E. Straughen, L. E. Slovek, A. J. Capobianco, J. German, G. P. Boivin, and J. Groden. 2002. Enhanced tumor formation in mice heterozygous for Blm mutation. Science 297:2051-2053. [DOI] [PubMed] [Google Scholar]
- 19.Gruber, S. B., N. A. Ellis, K. K. Scott, R. Almog, P. Kolachana, J. D. Bonner, T. Kirchhoff, L. P. Tomsho, K. Nafa, H. Pierce, M. Low, J. Satagopan, H. Rennert, H. Huang, J. K. Greenson, J. Groden, B. Rapaport, J. Shia, S. Johnson, P. K. Gregersen, C. C. Harris, J. Boyd, G. Rennert, and K. Offit. 2002. BLM heterozygosity and the risk of colorectal cancer. Science 297:2013. [DOI] [PubMed] [Google Scholar]
- 20.Haaf, T., E. I. Golub, G. Reddy, C. M. Radding, and D. C. Ward. 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 92:2298-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry-Mowatt, J., D. Jackson, J. Y. Masson, P. A. Johnson, P. M. Clements, F. E. Benson, L. H. Thompson, S. Takeda, S. C. West, and K. W. Caldecott. 2003. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol. Cell 11:1109-1117. [DOI] [PubMed] [Google Scholar]
- 23.Jasin, M. 2002. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21:8981-8993. [DOI] [PubMed] [Google Scholar]
- 24.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231-237. [DOI] [PubMed] [Google Scholar]
- 25.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 26.Kovalenko, O. V., A. W. Plug, T. Haaf, D. K. Gonda, T. Ashley, D. C. Ward, C. M. Radding, and E. I. Golub. 1996. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 93:2958-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, W., B. Hesabi, A. Babbo, C. Pacione, J. Liu, D. J. Chen, J. A. Nickoloff, and Z. Shen. 2000. Regulation of double-strand break-induced mammalian homologous recombination by UBL1, a RAD51-interacting protein. Nucleic Acids Res. 28:1145-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim, D. S., and P. Hasty. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16:7133-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, J., X. Meng, and Z. Shen. 2002. Association of human RAD52 protein with transcription factors. Biochem. Biophys. Res. Commun. 297:1191-1196. [DOI] [PubMed] [Google Scholar]
- 30.Liu, J., Y. Yuan, J. Huan, and Z. Shen. 2001. Inhibition of breast and brain cancer cell growth by BCCIPα, an evolutionarily conserved nuclear protein that interacts with BRCA2. Oncogene 20:336-345. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y., and N. Maizels. 2000. Coordinated response of mammalian Rad51 and Rad52 to DNA damage. EMBO Rep. 1:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo, T., L. Pellegrini, A. R. Venkitaraman, and T. L. Blundell. 2003. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair (Amsterdam) 2:1015-1028. [DOI] [PubMed] [Google Scholar]
- 33.Marmorstein, L. Y., T. Ouchi, and S. A. Aaronson. 1998. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc. Natl. Acad. Sci. USA 95:13869-13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marston, N. J., W. J. Richards, D. Hughes, D. Bertwistle, C. J. Marshall, and A. Ashworth. 1999. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol. Cell. Biol. 19:4633-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng, X., J. Liu, and Z. Shen. 2003. Genomic structure of the human BCCIP gene and its expression in cancer. Gene 302:139-146. [DOI] [PubMed] [Google Scholar]
- 36.Meng, X., J. Liu, and Z. Shen. 2004. Inhibition of G1 to S cell cycle progression by BCCIPb. Cell Cycle 3:343-357. [PubMed] [Google Scholar]
- 37.Meng, X., H. Lu, and Z. Shen. 2004. BCCIP functions through p53 to regulate p21 expression. Cell Cycle 3:1457-1462. [DOI] [PubMed] [Google Scholar]
- 38.Meng, X., Y. Yuan, A. Maestas, and Z. Shen. 2004. Recovery from DNA damage-induced G2 arrest requires actin-binding protein filamin-A/actin-binding protein 280. J. Biol. Chem. 279:6098-6105. [DOI] [PubMed] [Google Scholar]
- 39.Michel, B., M. J. Flores, E. Viguera, G. Grompone, M. Seigneur, and V. Bidnenko. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moynahan, M. E., A. J. Pierce, and M. Jasin. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7:263-272. [DOI] [PubMed] [Google Scholar]
- 41.Myung, K., and R. D. Kolodner. 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ono, T., H. Kitaura, H. Ugai, T. Murata, K. K. Yokoyama, S. M. Iguchi-Ariga, and H. Ariga. 2000. TOK-1, a novel p21Cip1-binding protein that cooperatively enhances p21-dependent inhibitory activity toward CDK2 kinase. J. Biol. Chem. 275:31145-31154. [DOI] [PubMed] [Google Scholar]
- 43.O'Regan, P., C. Wilson, S. Townsend, and J. Thacker. 2001. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J. Biol. Chem. 276:22148-22153. [DOI] [PubMed] [Google Scholar]
- 44.Orelli, B. J., and D. K. Bishop. 2001. BRCA2 and homologous recombination. Breast Cancer Res. 3:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radding, C. 2001. Colloquium introduction. Links between recombination and replication: vital roles of recombination. Proc. Natl. Acad. Sci. USA 98:8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raderschall, E., E. I. Golub, and T. Haaf. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA 96:1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 48.Sharan, S. K., and A. Bradley. 1997. Murine Brca2: sequence, map position, and expression pattern. Genomics 40:234-241. [DOI] [PubMed] [Google Scholar]
- 49.Sharan, S. K., M. Morimatsu, U. Albrecht, D. S. Lim, E. Regel, C. Dinh, A. Sands, G. Eichele, P. Hasty, and A. Bradley. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386:804-810. [DOI] [PubMed] [Google Scholar]
- 50.Shen, Z., K. G. Cloud, D. J. Chen, and M. S. Park. 1996. Specific interactions between the human RAD51 and RAD52 proteins. J. Biol. Chem. 271:148-152. [DOI] [PubMed] [Google Scholar]
- 51.Shen, Z., P. E. Pardington-Purtymun, J. C. Comeaux, R. K. Moyzis, and D. J. Chen. 1996. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics 37:183-186. [DOI] [PubMed] [Google Scholar]
- 52.Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara, H. Ogawa, M. Takata, Y. Yamaguchi-Iwai, and S. Takeda. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark, J. M., P. Hu, A. J. Pierce, M. E. Moynahan, N. Ellis, and M. Jasin. 2002. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J. Biol. Chem. 277:20185-20194. [DOI] [PubMed] [Google Scholar]
- 54.Taghian, D. G., and J. A. Nickoloff. 1997. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 17:6386-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takata, M., M. S. Sasaki, E. Sonoda, T. Fukushima, C. Morrison, J. S. Albala, S. M. Swagemakers, R. Kanaar, L. H. Thompson, and S. Takeda. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 20:6476-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 21:2858-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takata, M., S. Tachiiri, A. Fujimori, L. H. Thompson, Y. Miki, M. Hiraoka, S. Takeda, and M. Yamazoe. 2002. Conserved domains in the chicken homologue of BRCA2. Oncogene 21:1130-1134. [DOI] [PubMed] [Google Scholar]
- 58.Tarsounas, M., D. Davies, and S. C. West. 2003. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene 22:1115-1123. [DOI] [PubMed] [Google Scholar]
- 59.Tebbs, R. S., Y. Zhao, J. D. Tucker, J. B. Scheerer, M. J. Siciliano, M. Hwang, N. Liu, R. J. Legerski, and L. H. Thompson. 1995. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl. Acad. Sci. USA 92:6354-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson, L. H., and D. Schild. 1999. The contribution of homologous recombination in preserving genome integrity in mammalian cells. Biochimie 81:87-105. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, L. H., and D. Schild. 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res. 477:131-153. [DOI] [PubMed] [Google Scholar]
- 62.Thompson, L. H., and D. Schild. 2002. Recombinational DNA repair and human disease. Mutat. Res. 509:49-78. [DOI] [PubMed] [Google Scholar]
- 63.Tischkowitz, M. D., N. V. Morgan, D. Grimwade, C. Eddy, S. Ball, I. Vorechovsky, S. Langabeer, R. Stoger, S. V. Hodgson, and C. G. Mathew. 2004. Deletion and reduced expression of the Fanconi anemia FANCA gene in sporadic acute myeloid leukemia. Leukemia 18:420-425. [DOI] [PubMed] [Google Scholar]
- 64.Tutt, A., D. Bertwistle, J. Valentine, A. Gabriel, S. Swift, G. Ross, C. Griffin, J. Thacker, and A. Ashworth. 2001. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20:4704-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Gent, D. C., J. H. Hoeijmakers, and R. Kanaar. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2:196-206. [DOI] [PubMed] [Google Scholar]
- 66.Venkitaraman, A. R. 2001. Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J. Cell Sci. 114:3591-3598. [DOI] [PubMed] [Google Scholar]
- 67.Warren, M., C. J. Lord, J. Masabanda, D. Griffin, and A. Ashworth. 2003. Phenotypic effects of heterozygosity for a BRCA2 mutation. Hum. Mol. Genet. 12:2645-2656. [DOI] [PubMed] [Google Scholar]
- 68.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4:435-445. [DOI] [PubMed] [Google Scholar]
- 69.Wong, A. K., R. Pero, P. A. Ormonde, S. V. Tavtigian, and P. L. Bartel. 1997. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 272:31941-31944. [DOI] [PubMed] [Google Scholar]
- 70.Xia, F., D. G. Taghian, J. S. DeFrank, Z. C. Zeng, H. Willers, G. Iliakis, and S. N. Powell. 2001. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc. Natl. Acad. Sci. USA 98:8644-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, H., P. D. Jeffrey, J. Miller, E. Kinnucan, Y. Sun, N. H. Thoma, N. Zheng, P. L. Chen, W. H. Lee, and N. P. Pavletich. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297:1837-1848. [DOI] [PubMed] [Google Scholar]
- 72.Yuan, S. S., S. Y. Lee, G. Chen, M. Song, G. E. Tomlinson, and E. Y. Lee. 1999. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59:3547-3551. [PubMed] [Google Scholar]
- 73.Yuan, Y., and Z. Shen. 2001. Interaction with BRCA2 suggests a role for filamin-1 (hsFLNa) in DNA damage response. J. Biol. Chem. 276:48318-48324. [DOI] [PubMed] [Google Scholar]