Abstract
Komagataeibacter nataicola is an acetic acid bacterium (AAB) that can produce abundant bacterial cellulose and tolerate high concentrations of acetic acid. To globally understand its fermentation characteristics, we present a high-quality complete genome sequence of K. nataicola RZS01. The genome consists of a 3,485,191-bp chromosome and 6 plasmids, which encode 3,514 proteins and bear three cellulose synthase operons. Phylogenetic analysis at the genome level provides convincing evidence of the evolutionary position of K. nataicola with respect to related taxa. Genomic comparisons with other AAB revealed that RZS01 shares 36.1%~75.1% of sequence similarity with other AAB. The sequence data was also used for metabolic analysis of biotechnological substrates. Analysis of the resistance to acetic acid at the genomic level indicated a synergistic mechanism responsible for acetic acid tolerance. The genomic data provide a viable platform that can be used to understand and manipulate the phenotype of K. nataicola RZS01 to further improve bacterial cellulose production.
Introduction
Acetic acid bacteria (AAB) are a group of microorganisms that belong to the family Acetobacteraceae of the class Alphaproteobacteria. These bacteria are widely found on fruits, flowers, and rotten food1. Their metabolic uniqueness has been utilized in the industrial production of sorbose, vitamin C, dihydroxyacetone, d-gluconic acid, and bacterial cellulose (BC)2–5. Furthermore, AAB can also act as fermentative organisms, therefore, they are applied in beer, wine, and vinegar production6, 7. A remarkable feature of these strains is their ability to survive under extreme environments, such as high sugar concentrations and low pH values, which makes AAB suitable for various industrial applications8.
Currently, AAB include 14 genera, namely Acetobacter, Gluconobacter, Gluconacetobacter, Komagataeibacter, Granulibacter, Asaia, Acidomonas, Kozakia, Swaminathania, Saccharibacter, Neoasaia, Tanticharoenia, Ameyamaea, and Neokomagataea 1. Classification within the Acetobacteraceae depends on the ubiquinone type, including the Q-9 and Q-10 types. Acetobacter species use the Q-9–type ubiquinone, whereas the AAB genera of Gluconacetobacter and Gluconobacter contain mainly the Q-10–type ubiquinone9. The genus Komagataeibacter, a gram-negative, obligately aerobic and rod-shaped acidophilic organism, was initially proposed by Yamada in 2012, with 12 other strains on the basis of their taxonomic characteristics10. Komagataeibacter gen. nov. originated from the Gluconacetobacter xylinus (previously named Acetobacter xylinus) group, which is first separated from the Gluconacetobacter liquefaciens group at the genus level. These 2 groups are significantly different in phylogenetic and phenotypic characteristics11. Thus far, 66 complete genomes of Acetobacteraceae have been published in NCBI databases (http://www.ncbi.nlm.nih.gov/), most of which just present draft genome sequences. The genomes of 3 species of Komagataeibacter, K. xylinus E25, K. hansenii ATCC 23769, and K. medellinensis NBRC3288, have been sequenced completely. However, draft genomes are available for K. europaeus LMG18494, K. intermedius AF2, K. rhaeticus AF1, K. kakiaceti JCM 25156, and K. oboediens 174Bp212, 13.
Generally, Komagataeibacter species possess the ability to secrete exopolysaccharides (EPS), especially BC14. Notably, K. medellinensis NBRC3288 is the only cellulose-nonproducing member of Komagataeibacter species, isolated from vinegar15. BC has been the focus of research owing to its excellent properties, such as high purity, predominant three-dimensional structure, high degree of crystallinity, and superior biocompatibility16–18. In view of these qualities, it is utilized commercially in the development and production of health food, flexible electrodes, acoustic speakers, and tissue scaffolds16, 19–21.
K. nataicola RZS01 originates from rotten apples and is regarded as the model organism for BC production, owing to its high resistance to ethanol and acetic acid, which makes it an ideal strain for industrial applications22, 23. In this study, we present the complete genome sequence and sequence analysis of K. nataicola RZS01 to obtain genetic information for further insights into its biochemical features. Comparison of K. nataicola RZS01 with other AAB strains will address the unique functional characteristics of this strain, as well as the common properties within the Acetobacteraceae. Furthermore, the genome data will also provide a rich resource for guiding future research in industrial applications.
Results
General features of the K. nataicola RZS01 genome
The complete genome of RZS01 is composed of 1 circular chromosome of 3,485,191 bp with a G + C content of 61.49%, and 6 plasmids ranging in size from 25,766 bp to 102,282 bp (Table 1; Fig. 1). Gene prediction and annotation of the RZS01 genome resulted in 3,609 open reading frames. The number of genes in the plasmids occupies 6.5% of the total genes in the genome, and genes with predicted function are assigned to 68.86% of the genome (2,485 genes). Furthermore, 5 ribosomal RNA operons (16S-23S-5S) were detected and 62 tRNA genes were predicted. From the genome data, we also identified 66 transposase genes, which might be responsible for the genetic instability leading to deficiencies in various physiological properties as reported in other AAB24.
Table 1.
General features of the Komagataeibacter nataicola RZS01 genome.
| Features | |
|---|---|
| Size of the chromosome (bp) | 3,485,191 |
| Size of the plasimds (bp) | 102,282 (pKNA01) |
| 39,914 (pKNA02) | |
| 38,682 (pKNA03) | |
| 37,912 (pKNA04) | |
| 30,554 (pKNA05) | |
| 25,766 (pKNA06) | |
| DNA scaffolds | 7 |
| G + C content (%) | 61.49 |
| Total genes | 3,609 |
| Protein coding genes | 3,514 |
| RNA genes | 95 |
| Genes with function prediction | 2,485 |
Figure 1.
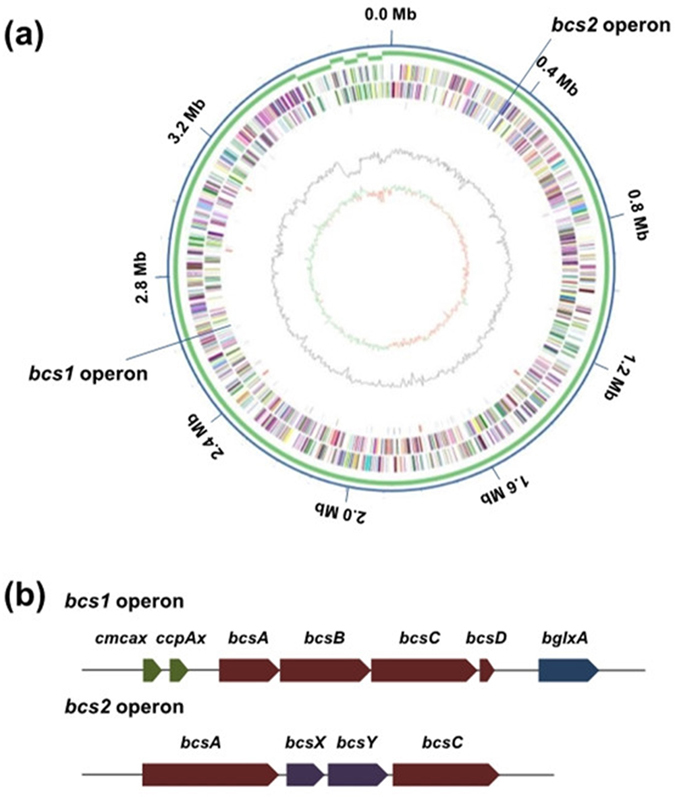
Overview of K. nataicola RZS01 genome. (a) The circles represent (from the outside to the inside): circle 1, DNA base position (bp); circle 2, contig components; circle 3, protein-coding regions transcribed clockwise; circle 4, protein-coding regions transcribed anti-clockwise; circle 5, distribution of genes encoding ncRNA (black), tRNA (blue) and rRNA (red); circle 6, G + C content; circle 7, GC skew. (b) The compositions of 2 bcs operons, which differ from each other in gene content.
The complete genome has a total of 3,514 putative coding sequences, among which 2,485 are assigned a putative function, and 1,029 encode hypothetical proteins. The 6 plasmids have 228 coding DNA sequences (CDSs), of which approximately 47.8% encode hypothetical proteins. The remaining 42 CDSs encode mobile element proteins. Figure 2 summarizes the distribution of RZS01 proteins among 21 functional groups. Statistically, nearly 38% CDSs are completely uncharacterized, which is similar to the proportion of unassigned CDSs in some other sequenced bacterial genomes, such as in Escherichia coli (40%), Haemophilus influenzae (43%), and Mycoplasma genitalium (32%)25. The 2 largest functional groups contain 277 and 231 proteins involved in carbohydrate and amino acid metabolism, respectively, which correspond to the physiological functions in RZS01.
Figure 2.
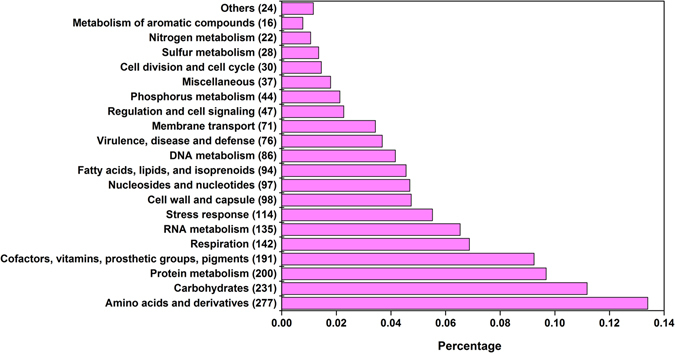
Subsystem category distributions of RZS01 proteins.
Phylogenetic analysis and comparative genomics
Analysis of the most conservative sequence in the process of evolution, the 16S rRNA gene, is widely used in taxonomic resolution in bacteria. To ascertain the phylogenetic positions of RZS01 in Acetobacteraceae, a phylogenetic tree was constructed using 16S rRNA gene sequences (Fig. 3). The phylogenetic tree summarizes 13 other Acetobacteraceae, the genomes of which have been completely sequenced. The tree suggests that RZS01 is most closely related to Komagataeibacter xylinus E25, Komagataeibacter medellinensis LMG 1693, and Gluconacetobacter hansenii ATCC 23769. Furthermore, comparative analysis of RZS01 in relation to other members of the Acetobacteraceae family reveals that this strain has 36.1% shared genes with the human pathogen Granulibacter bethesdensis CGDNIH1 (Table 2). Approximately 40% of the genes are in common with members of the genus Acidiphilium. Thus, RZS01 has more genes in common with Komagataeibacter species than with Gluconacetobacter, which corresponds with the phylogenetic relationship.
Figure 3.
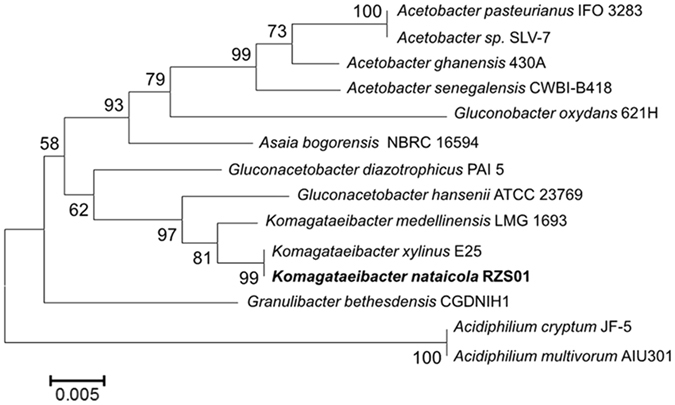
A phylogenetic tree based on 16S rRNA sequences constructed by the neighbour-joining method of MEGA5.1.
Table 2.
Comparative analysis of Komagataeibacter nataicola RZS01. with members of the Acetobacteracea family.
| Acetic acid bacterium strain | Number of shared genes | Percentage of shared genes |
|---|---|---|
| Gluconacetobacter diazotrophicus Pal5 | 5031 | 72.0 |
| Komagataeibacter medellinensis NBRC 3288 | 4742 | 75.0 |
| Acetobacter senegalensis 108B | 4339 | 61.7 |
| Komagataeibacter xylinus E25 | 4335 | 65.0 |
| Komagataeibacter hansenii ATCC 23769 | 3592 | 52.4 |
| Acetobacter pasteurianus IFO 3283 | 3216 | 51.1 |
| Acidiphilium multivorum AIU301 | 3098 | 44.5 |
| Gluconobacter oxydans 621H | 2993 | 50.3 |
| Acetobacter sp. SLV-7 | 2893 | 48.1 |
| Acetobacter ghanensis 430A | 2774 | 46.0 |
| Asaia bogorensis NBRC 16594 | 2742 | 43.7 |
| Acidiphilium cryptum JF-5 | 2656 | 39.4 |
| Granulibacter bethesdensis CGDNIH1 | 2146 | 36.1 |
Cellulose production
Acetobacteraceae are characterized by their ability to synthesize high yields of cellulose. Maria José Valera et al.26 investigated the cellulose-producing ability of 77 strains of 35 species of AAB, of which 19 demonstrated high yields of cellulose after cultivation. Species of the genus Komagataeibacter are considered to be the main producers of BC, particularly strains of the species K. xylinus, K. hansenii, K. swingsii, K. rhaeticus, and K. nataicola 27, 28. In fact, K. xylinus is considered a model microorganism for BC production29, 30.
On the RZS01 chromosome, we found the full set of cellulose synthase genes in the bcs1 operon, which comprised the cellulose synthase genes bcsA1 (B0W47_12635), bscB1 (B0W47_12640), bcsC1 (B0W47_12645), and bcsD1 (B0W47_12650), as well as cmcax (B0W47_12625), ccpAx (B0W47_12630) upstream, and bglxA (B0W47_12655) downstream. cmcax encodes endo-β-1,4-glucanase, and bglxA encodes β-glucosidase, both of which have been suggested to assist cellulose biosynthes is by hydrolysing tangled glucan chains when there is a failure in chain arrangement, and all these genes are necessary for cellulose production31. In addition to the bcs1 operon, we found an additional bcs operon (bcs2) at the genomic position 363284–373685, and it differs in structure from the bcs1 operon. The bcs2 operon is composed of bcsA and bcsC, with 2 additional genes homologous to bcsX and bcsY in the middle of bcsA and bcsC. The protein encoded by bcsA contains the catalytically active subunit with a PilZ domain, which is responsive to c-di-GMP32. An additional bcsA (bcsA3) is also located on the reverse strand. Unlike the bcsA1 gene, the bcsA2 and bcsA3 genes do not contain a PilZ domain.
Generally, AAB possess the ability to secrete other polysaccharides, including capsular polysaccharides, lipopolysaccharides, and water-soluble EPS. One of the most studied polysaccharides is a xanthan gum-like EPS, named ‘acetan’, the structure of which is shown in Supplementary Information. The chemical repeat unit consists of a cellobiose unit solubilized by attachment of a charged pentasaccharide sidechain to one of the glucose residues33, 34. Genetic analysis of RZS01 shows that the biosynthesis of acetan occurs via a pathway similar to that for BC biosynthesis. Because it is the composite of different monosaccharides, 2 CDSs are responsible for encoding UDP-glycosyl transferase (B0W47_01855, B0W47_03955, and B0W47_12635), GDP-mannosyl transferase (B0W47_09075, and B0W47_11925), and phosphomannose isomerase (B0W47_06245). The enzymes UDP-glycosyl transferase and GDP-mannosyl transferase transfer a glucose and mannose residues from UDP-Glc and GDP-Man, respectively, to one of the intermediates of acetan35. However, not all genes from the gum operon are present in RZS01. Only 2 CDSs (B0W47_14415 and B0W47_14460) represent the genes of gumK and gumH, respectively. The genes for gumB, C, D, E, G, I, J, and M are absent in the RZS01 genome.
Regulation of cellulose biosynthesis
BC biosynthesis is regulated by the expression of bcs genes, which appear to be expressed in response to the second messenger cyclic-di-GMP (c-di-GMP)36. An earlier study revealed that cellulose biosynthesis could be stimulated almost 100-fold by c-di-GMP37. c-di-GMP is produced from 2 molecules of GTP by diguanylate cyclases (DGCs) and is broken down into 5′-phosphoguanylyl-(3′-5′)-guanosine by specific phosphodiesterases (PDEs). DGC activity is associated with the GGDEF domain, whereas PDE activity is associated with EAL or HD-GYP domains; all these domains are essential for enzymatic activity38. We found 3 cdg operons (cdg1, cdg2, and cdg3) containing a c-di-GMP PDE gene followed by a DGC gene. Besides, 4 standalone c-di-GMP PDEs (pde1–4) and 1 standalone DGC are present in the genome, which share 32–40% amino acid sequence identity with each other. These multiple genes together control the intracellular c-di-GMP levels.
Genes for substrate metabolism
On the basis of the analysis of gene annotation for the complete genome, K. nataicola RZS01 possesses various transporters for the uptake of substrates and ions. We identified 49 ABC transporters, 10 symporters, and 47 permeases, which are responsible for the transportation of sugars (e.g. glucose and ribose), polyols (e.g. mannitol and sorbitol), sugar acids (e.g. gluconate and acetate), amino acids, purines, pyrimidines, phosphate, sulphate, NH4 +, and metal ions. In addition, some components of the phosphotransferase system are also found. As the most widely used substrate for industrial production, glucose could be taken up by a sugar symporter (B0W47_14370). Owing to the lack of the gene encoding phosphofructokinase (EC 2.7.1.11), RZS01 has an incomplete Embden-Meyerhof-Parnas pathway, as has been reported in Acetobacter pasteurianus 386B6, Gluconacetobacter diazotrophicus Pal539, and Gluconobacter oxydans 621H2. However, the genes encoding the enzymes of the pentose-phosphate (PPP) pathway are all found, suggesting that glucose might be degraded via the PPP pathway. Carbohydrates are generally metabolized by dehydrogenases and isomerases after phosphorylation by specific kinases. In addition, RZS01 possesses an orthologous cluster for gluconate utilization similar to the GntU, GntK, and GntR cluster found in E.coli 40. However, RZS01 does not contain a GntK gene, which encodes gluconate kinase. As for disaccharides, such as sucrose and lactose, substrates can be utilized after being decomposed to monosaccharides. RZS01 contains levansucrase (B0W47_08430), which converts sucrose into glucose and fructose for further oxidation, suggesting that this strain can take up sucrose from the environment to use it as a carbon source. Sucrose is able to adjust the osmotic pressure of the medium, which has been verified by Velasco-Bedrán41.
Genome analysis illustrated that RZS01 possesses all the genes catalysing metabolic pathways involved in the de novo synthesis of all nucleotides, amino acids, phospholipids and many vitamins. Ammonia, which participates in the activity of glutamate synthase (B0W47_13175 and B0W47_13180) and glutamine synthase (B0W47_07980), can be taken up by a specific ammonia transporter (B0W47_07675).
Membrane-associated primary dehydrogenases and the respiratory chain
K. nataicola RZS01 contains several genes that encode membrane-associated primary quinone reductases coupling substrate oxidation with quinone reduction, which can be classified into 2 groups (Table 3)2. The first group of dehydrogenases depends on the cofactor pyrroloquinoline quinine (PQQ). The most representative dehydrogenases are PQQ-dependent alcohol dehydrogenase (PQQ-ADH) and aldehyde dehydrogenase (PQQ-ALDH), which convert alcohol into acetaldehyde and acetaldehyde into acetate, respectively. The genes encoding PQQ-ADHs are present as 8 copies distributed throughout the chromosomes, and just is localized in the membrane. Moreover, RZS01 possesses 1 membrane-bound PQQ-ALDH, whereas 5 other copies are dispersed in the cytoplasm. Furthermore, the genome of RZS01 contains glucose dehydrogenase and glycerol dehydrogenase.
Table 3.
Primary membrane-bound dehydrogenases in K. nataicola RZS01.
| Family | Cofactor | Genes |
|---|---|---|
| Glucose dehydrogenase | PQQ | B0W47_01230, B0W47_02520, B0W47_10950 |
| Glycerol dehydrogenase | PQQ | B0W47_01005, B0W47_01010 |
| Alcohol dehydrogenase | PQQ | B0W47_13410 |
| Aldehyde dehydrogenase | PQQ | B0W47_16410 |
| 2-Keto-d-gluconate dehydrogenase | FAD | B0W47_11030, B0W47_11035, B0W47_11040 |
| Gluconate 2-dehydrogenase | FAD | B0W47_05395, B0W47_05400, B0W47_05405, B0W47_13405 |
Adaptation to extreme conditions
K. nataicola RZS01 possesses the ability to tolerate low pH and high concentrations of organic acid in the environment42. One strategy for this tolerance is that RZS01 has a cytosolic acetate-assimilating detoxification pathway, whereby acetate is first converted into acetyl-CoA, which is catalysed by acetyl-CoA synthetase (acn; B0W47_06505). The product is further oxidized by the citric acid cycle to water and carbon dioxide. Overoxidation occurs in Acetobacter and Gluconacetobacter but not in Gluconobacter, which exhibits relatively weak acetic acid resistance8, 43. The second mechanism for acid tolerance may involve an ABC transporter gene, aatA (B0W47_06270), which functions as an efflux pump of acetic acid44. In another similar efflux mechanism, a proton motive force-dependent efflux system occurs in the cytoplasmic membrane, which is capable of exporting acetic acid and is dependent on the proton motive force but not on ATP45. Besides, there is also an operon involved in high resistance to acetic acid, a trait that can be a suitable target for enhancement by breeding. This operon is composed of the genes groE (B0W47_13360 and B0W47_13365), groS (B0W47_00055), and groL (B0W47_02405, B0W47_03785, and B0W47_09650), and the overexpression of the groESL operon increases acetic acid resistance in Acetobacter 46. A previous thorough genome analysis revealed the presence of a gene cluster encoding a urease (ureDABCEFG; B0W47_16420, B0W47_16425, B0W47_16430, B0W47_16435, B0W47_16440, B0W47_16445, and B0W47_16450), a urea transporter (urtABCDE; B0W47_16475, B0W47_16480, B0W47_16485, B0W47_16490, and B0W47_16495), and an allophanate hydrolase (B0W47_14240, B0W47_14245, B0W47_16295, and B0W47_16300). These proteins transport urea and catalyse it to ammonia, which enables the survival of RZS01 in an acidic environment, such as in vinegar fermentation. This mechanism is also present in A. pasteurianus 386B and G. bethesdensis CGDNIH1 but is not widespread among AAB strains6, 47.
Discussion
K. nataicola is widely distributed in nature and displays strong abilities of producing BC and tolerating acetic acid. In this study, we sequenced, annotated, and analysed the complete genome sequence of K. nataicola RZS01 and found that RZS01 possesses a 3.5-Mb chromosome and 6 plasmids. The global overview of all genes provided comprehensive insights into the metabolic features, including the uptake of different substrates and intolerance of acidic stress during BC production.
Comparative genome analysis of 13 AAB revealed that the genome of K. nataicola RZS01 is most closely related to that of K. medellinensis NBRC 3288, with 75.0% shared genes, which is congruent with the phylogenetic relationship. On the basis of the genomic sequence of RZS01, we can now more extensively describe the process of cellulose biosynthesis and the physiological basis of the underlying pathways in this organism. For other AAB species whose genomes have been sequenced, 2 bcs operons are present in RZS01, which may explain the high cellulose productivity observed. However, the regulation of AcsAB activity by c-di-GMP may also play important roles. We identified 3 cdg operons containing a DGC and a c-di-GMP PDE. Four standalone c-di-GMP PDEs and 1 standalone DGC are also present in the genome. This regulatory mechanism has also been found in other bacteria38.
K. nataicola RZS01 contains many membrane-bound dehydrogenases, which are responsible for the assimilation of substrates and contribution to acetic acid resistance, resisting the presence of high concentrations of acetic acid. The metabolism of several amino acids, such as threonine, glycine, and ornithine, produces a large amount of ammonia, which decreases the intracellular pH value. Certain other mechanisms also participate in acetic acid tolerance, including alcohol overoxidation, an acetate assimilation-related mechanism, an ABC transporter, and a proton motive force-dependent efflux system.
In summary, we uncovered global insights into BC production and acidic resistance mechanisms by genome analysis. These results provide useful information for further studies on evolution and genetic variation in Komagataeibacter and provide a valuable resource for biological research. Furthermore, comparative genomics analysis and functional genomics analysis can also be performed to trace the origin and evolution of this organism.
Methods
Bacterial strain and isolation of chromosomal DNA
K. nataicola RZS01 (CGMCC number 10961) used in this study was isolated from rotten apples48. For genomic DNA extraction, RZS01 was grown from a single colony in HZ medium, containing 20 g·L−1 glucose, 6 g·L−1 ammonium sulphate, 1 g·L−1 monopotassium phosphate, 0.4 g·L−1 magnesium sulphate, 3 g·L−1 peptone, 2.25 g·L−1 yeast extract, and 0.4 g·L−1 carboxymethylcellulose sodium at 30 °C for 24 h. The produced cellulose was digested by adding 0.2% cellulase and maintained at 30 °C for 2 h. After the cells were harvested, chromosomal DNA was isolated using the QiaAmp DNA Mini kit (Qiagen, Valencia, Germany) following the manufacturer’s instructions. The quality of the DNA was evaluated using a Nanodrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) after isolation.
Genome sequencing and assembly
The wholegenome of K. nataicola RZS01 was sequenced at HangZhou GeneRui Biotechnology Co., Ltd. (Hangzhou, China) using a PacBio RS DNA sequencer. The average insert size was 550 bp. Data from 515-Mb paired-end reads were delivered by Baseclear as 2 fastq files. The FastQC procedure was used to verify the raw read data, and QC metrics including insert-sizes, mapped reads, unmapped reads and reads that align with a deviated pattern were examined49. The sequencing errors were discarded using the error-correction module of Allpaths-LG. For single-molecule real-time sequencing (SMRT), a SMRT bell DNA template library with an insert size of 2~40 kb was prepared. Subsequently, the fragmented DNA was end-repaired and ligated into hairpin adapters. Sequence reads have been deposited in the ENA Sequence Read Archive (EMBL: ERS550016). Sequencing reads were corrected using the HGAP pipeline50. Genomic sequences were assembled de novo using PacBio analysis RS_HGAP_Assembly.3 (Pacific Biosciences, Menlo Park, CA, USA).
Bioinformatics analysis
Automatic gene prediction and annotation were performed using Glimmer3.0. The genome was visualized using Circos (http://circos.ca/tutorials/lessons/). 16S rRNA phylogeny was constructed by the neighbour-joining method of MEGA5.1 at default settings. Metabolic enzymes were identified from UniProt (http://www.uniprot.org/) and BLASTp analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify orthologues in the genomes.
Nucleotide sequence accession numbers
The accession numbers of the complete sequences of K. nataicola RZS01 determined in this study and of the 6 plasmids from NCBI can be found in GenBank (http://www.ncbi.nlm.nih.gov) under the accession no. CP019875 to CP019881.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21206076), Natural Science Foundation of Jiangsu Province (No. BK2012401), the Fundamental Research Funds for the Central Universities (No. 30920130121001), the Science and Technology Innovation Fund of QiXia District (201528) and a project funded by the Surface Program of Natural Science Research of Jiangsu Higher Education Institutions (No. 16KJB180034).
Author Contributions
X.X. and B.S. contributed to the annotation of the genome; X.C. and F.Y. helped to analyse the genomic information; H.Z. and Y.X. wrote the first draft of the manuscript; and J.Y. and D.S. contributed substantially to revisions.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04589-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiazhi Yang, Email: jiazhiyang@sina.com.
Dongping Sun, Email: sundpe301@163.com.
References
- 1.Saichana N, Matsushita K, Adachi O, Frébort I, Frebortova J. Acetic acid bacteria: A group of bacteria with versatile biotechnological applications. Biotechnol. Adv. 2014;33:1260–1271. doi: 10.1016/j.biotechadv.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Prust C, et al. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 2005;23:195–200. doi: 10.1038/nbt1062. [DOI] [PubMed] [Google Scholar]
- 3.Adachi O, et al. New developments in oxidative fermentation. Appl. Microbiol. Biotechnol. 2003;60:643–653. doi: 10.1007/s00253-002-1155-9. [DOI] [PubMed] [Google Scholar]
- 4.Lazarini SC, et al. Characterization of bilayer bacterial cellulose membranes with different fiber densities: a promising system for controlled release of the antibiotic ceftriaxone. Cellulose. 2016;23:737–748. doi: 10.1007/s10570-015-0843-4. [DOI] [Google Scholar]
- 5.Liu M, et al. Metabolomic profiling coupled with metabolic network reveals differences in Gluconacetobacter xylinus from static and agitated cultures. Biochem. Eng. J. 2015;101:85–98. doi: 10.1016/j.bej.2015.05.002. [DOI] [Google Scholar]
- 6.Illeghems K, Vuyst LD, Weckx S. Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genomics. 2013;14:42–45. doi: 10.1186/1471-2164-14-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battey AS, Schaffner DW. Modelling bacterial spoilage in cold-filled ready to drink beverages by Acinetobacter calcoaceticus and Gluconobacter oxydans. J. Appl. Microbiol. 2001;91:237–247. doi: 10.1046/j.1365-2672.2001.01381.x. [DOI] [PubMed] [Google Scholar]
- 8.Kersters, K., Lisdiyanti, P., Komagata, K. & Swings, J. The Family Acetobacteraceae: The genera Acetobacter, Acidomonas, Asaia, Gluconacetobacter, Gluconobacter, and Kozakia. (Springer New York, 2006).
- 9.Yamada Y, Hoshino K, Ishikawa T. The phylogeny of acetic acid bacteria based on the partial sequences of 16S ribosomal RNA: The elevation of the subgenus to the generic level. Biosci. Biotechnol. Biochem. 1997;61:1244–1251. doi: 10.1271/bbb.61.1244. [DOI] [PubMed] [Google Scholar]
- 10.Yamada Y, et al. Subdivision of the genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: the proposal of Komagatabacter gen. nov., for strains accommodated to the Gluconacetobacter xylinus group in the α-Proteobacteria. Ann. Microbiol. 2012;62:849–859. doi: 10.1007/s13213-011-0288-4. [DOI] [Google Scholar]
- 11.Yamada Y, et al. Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). J. Gen. Appl. Microbiol. 2012;58:397–404. doi: 10.2323/jgam.58.397. [DOI] [PubMed] [Google Scholar]
- 12.Santos RACD, et al. Draft genome sequence of Komagataeibacter intermedius strain AF2, a producer of cellulose, isolated from kombucha tea. Genome Announcements. 2015;3:1–2. doi: 10.1128/genomeA.01404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos RACD, et al. Draft genome sequence of Komagataeibacter rhaeticus strain AF1, a high producer of cellulose, isolated from kombucha tea. Genome Announcements. 2014;2:1–2. doi: 10.1128/genomeA.00731-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mamlouk D, Gullo M. Acetic acid bacteria: Physiology and carbon sources oxidation. Indian J. Microbiol. 2013;53:377–384. doi: 10.1007/s12088-013-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogino H, et al. Complete genome sequence of NBRC 3288, a unique cellulose-nonproducing strain of Gluconacetobacter xylinus isolated from vinegar. J. Bacteriol. 2011;193:6997–6998. doi: 10.1128/JB.06158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KY, Buldum G, Mantalaris A, Bismarck A. More Than Meets the Eye in Bacterial Cellulose: Biosynthesis, Bioprocessing, and Applications in Advanced Fiber Composites †. Macromol. Biosci. 2014;14:10–32. doi: 10.1002/mabi.201300298. [DOI] [PubMed] [Google Scholar]
- 17.Sun S, et al. Comparison of the mechanical properties of cellulose and starch films. Biomacromolecules. 2009;11:126–132. doi: 10.1021/bm900981t. [DOI] [PubMed] [Google Scholar]
- 18.Na Y, Stilwell MD, Santos TMA, Wang H, Weibel DB. Agarose particle-templated porous bacterial cellulose and its application in cartilage growth in vitro. Acta Biomater. 2014;12:129–138. doi: 10.1016/j.actbio.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemm D, Schumann D, Udhardt U, Marsch S. Bacterial synthesized cellulose—artificial blood vessels for microsurgery. Prog Polym Sci. Prog. Polym. Sci. 2001;26:1561–1603. doi: 10.1016/S0079-6700(01)00021-1. [DOI] [Google Scholar]
- 20.Yadav V, et al. Novel in vivo-degradable cellulose-chitin copolymer from metabolically engineered Gluconacetobacter xylinus. Appl. Environ. Microbiol. 2010;76:6257–6265. doi: 10.1128/AEM.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ummartyotin S, Juntaro J, Sain M, Manuspiya H. Development of transparent bacterial cellulose nanocomposite film as substrate for flexible organic light emitting diode (OLED) display. Ind. Crops Prod. 2012;35:92–97. doi: 10.1016/j.indcrop.2011.06.025. [DOI] [Google Scholar]
- 22.Mehta K, Pfeffer S, Malcolm Brown R., Jr. Characterization of an acsD disruption mutant provides additional evidence for the hierarchical cell-directed self-assembly of cellulose in Gluconacetobacter xylinus. Cellulose. 2015;22:119–137. doi: 10.1007/s10570-014-0521-y. [DOI] [Google Scholar]
- 23.Zhong C, et al. Revealing differences in metabolic flux distributions between a mutant strain and its parent strain Gluconacetobacter xylinus CGMCC 2955. PLoS One. 2014;9:e98772. doi: 10.1371/journal.pone.0098772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo K, Horinouchi S. Characterization of an insertion sequence, IS12528, from Gluconobacter suboxydans. Appl. Environ. Microbiol. 1997;63:1139–1142. doi: 10.1128/aem.63.3.1139-1142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Sci. 2010;5331:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 26.Valera MJ, Torija MJ, Mas A, Mateo E. Cellulose production and cellulose synthase gene detection in acetic acid bacteria. Appl. Microbiol. Biotechnol. 2015;99:1–13. doi: 10.1007/s00253-014-6198-1. [DOI] [PubMed] [Google Scholar]
- 27.Aydın YA, Aksoy ND. Isolation and characterization of an efficient bacterial cellulose producer strain in agitated culture: Gluconacetobacter hansenii P2A. Appl. Microbiol. Biotechnol. 2014;98:1065–1075. doi: 10.1007/s00253-013-5296-9. [DOI] [PubMed] [Google Scholar]
- 28.Castro C, et al. Gluconacetobacter medellinensis sp. nov., cellulose- and non-cellulose-producing acetic acid bacteria isolated from vinegar. Int. J. Syst. Evol. Microbiol. 2013;63:1119–1125. doi: 10.1099/ijs.0.043414-0. [DOI] [PubMed] [Google Scholar]
- 29.Chawla PR, Bajaj IB, Survase SA, Singhal RS. Microbial cellulose: fermentative production and applications. Food Technol. Biotechnol. 2009;47:107–124. [Google Scholar]
- 30.Römling U. Molecular biology of cellulose production in bacteria. Res. Microbiol. 2002;153:205–212. doi: 10.1016/S0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- 31.Nakai T, et al. Formation of highly twisted ribbons in a carboxymethylcellulase gene- disrupted strain of a cellulose-producing bacterium. J. Bacteriol. 2013;195:958–964. doi: 10.1128/JB.01473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nat. 2012;493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ojinnaka C, et al. Structure and conformation of acetan polysaccharide. Int. J. Biol. Macromol. 1996;19:149–156. doi: 10.1016/0141-8130(96)01120-8. [DOI] [PubMed] [Google Scholar]
- 34.Griffin AM, Morris VJ, Gasson MJ. Identification, cloning and sequencing the aceA gene involved in acetan biosynthesis in Acetobacter xylinum. FEMS Microbiol. Lett. 1996;137:115–121. doi: 10.1111/j.1574-6968.1996.tb08092.x. [DOI] [PubMed] [Google Scholar]
- 35.Colquhoun IJ, et al. Structure and conformation of a novel genetically engineered polysaccharide P2. Carbohydrate Research. 2001;330:325–333. doi: 10.1016/S0008-6215(00)00293-7. [DOI] [PubMed] [Google Scholar]
- 36.Ross P, et al. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 1990;265:18933–18943. [PubMed] [Google Scholar]
- 37.Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 39.Marcelo B, et al. Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genomics. 2009;10:43–50. doi: 10.1186/1471-2164-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong S, Porco A, Isturiz T, Conway T. Cloning and molecular genetic characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the main system for gluconate metabolism. J. Bacteriol. 1996;178:3260–3269. doi: 10.1128/jb.178.11.3260-3269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velasco-Bedrán H, López-Isunza F. The unified metabolism of Gluconacetobacter entanii in continuous and batch processes. Process Biochem. 2007;42:1180–1190. doi: 10.1016/j.procbio.2007.05.017. [DOI] [Google Scholar]
- 42.Matsushita, K. et al. Acetic acid production in acetic acid bacteria leading to their ‘death’ and survival. In: Yamada M (ed) Survival and death in bacteria 169–181 (Research Signpost, 2005).
- 43.Garrity, G. M. Bergey’s manual of systematic bacteriology (Springer, 2011).
- 44.Nakano S, Fukaya M, Horinouchi S. Putative ABC transporter responsible for acetic acid resistance in Acetobacter aceti. Appl. Environ. Microbiol. 2006;72:497–505. doi: 10.1128/AEM.72.1.497-505.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsushita K, Inoue T, Adachi O, Toyama H. Acetobacter aceti possesses a proton motive force-dependent efflux system for acetic acid. J. Bacteriol. 2005;187:4346–4352. doi: 10.1128/JB.187.13.4346-4352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakano S, Fukaya M. Analysis of proteins responsive to acetic acid in Acetobacter: molecular mechanisms conferring acetic acid resistance in acetic acid bacteria. Int. J. Food Microbiol. 2008;125:54–59. doi: 10.1016/j.ijfoodmicro.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Greenberg D, et al. Genome sequence analysis of the emerging human pathogenic acetic acid bacterium Granulibacter bethesdensis. J. Bacteriol. 2008;189:8727–8736. doi: 10.1128/JB.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou LL, Sun DP, Hu LY, Li YW, Yang JZ. Effect of addition of sodium alginate on bacterial cellulose production by Acetobacter xylinum. J. Ind. Microbiol. Biotechnol. 2007;34:483–489. doi: 10.1007/s10295-007-0218-4. [DOI] [PubMed] [Google Scholar]
- 49.Andrews, S. FQC: A quality control tool for high throughput sequence data. http://www.Bioinformaticsbabraham.ac.uk/projects/fastqc/ (2010).
- 50.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


