Abstract
Eukaryotic Y-box proteins are nucleic acid-binding proteins implicated in a wide range of gene regulatory mechanisms. They contain the cold shock domain, which is a nucleic acid-binding structure also found in bacterial cold shock proteins. The Y-box protein YB-1 is known to be a core component of messenger ribonucleoprotein particles (mRNPs) in the cytoplasm. Here we disrupted the YB-1 gene in chicken DT40 cells. Through the immunoprecipitation of an epitope-tagged YB-1 protein, which complemented the slow-growth phenotype of YB-1-depleted cells, we isolated YB-1-associated complexes that likely represented general mRNPs in somatic cells. RNase treatment prior to immunoprecipitation led to the identification of a Y-box protein-associated acidic protein (YBAP1). The specific association of YB-1 with YBAP1 resulted in the release of YB-1 from reconstituted YB-1-mRNA complexes, thereby reducing the translational repression caused by YB-1 in the in vitro system. Our data suggest that YBAP1 induces the remodeling of YB-1-mRNA complexes.
Posttranscriptional regulation constitutes one of the principal regulatory steps in eukaryotic gene expression. A number of multifunctional RNA-binding proteins that play roles in both the nucleus and the cytoplasm are involved in this process (14). Some of these RNA-binding proteins have regulatory functions through association with specific sequences or regions of mRNA, while others may have structural roles as constituents of messenger ribonucleoprotein particles (mRNPs).
Cytoplasmic mRNPs are substrates of the translational apparatus in eukaryotic cells. At present, the full complement of mRNPs is not known for any specific mRNA. Two proteins, of 70 and 50 kDa, are common major components of cytoplasmic mRNPs in somatic cells of different organisms. Whereas the 70-kDa protein has long been known as a poly(A)-binding protein (PABP) (6), the 50-kDa protein was only cloned and identified as a Y-box protein, YB-1, in the 1990s (19). The assembly of mRNA into mRNP by these proteins is expected to have an influence on the translational activity of the mRNA. PABP selectively associates with poly(A) tails of mRNAs to organize poly(A) RNPs in repeating structures of 27 nucleotides (4), whereas Y-box proteins are presumed to bind to the mRNA body. PABP is highly conserved in most eukaryotic organisms. The yeast homologue of PABP, Pab1p, is essential for cell survival (59). Genetic studies carried out with yeasts and supported by biochemical experiments have provided strong evidence that PABP/Pab1p is a key regulator in the translation, stability, and 3′-end processing of mRNA (39). In contrast to PABP, it is known that there are no yeast homologues of Y-box proteins.
Eukaryotic Y-box proteins were originally identified as transcription factors which bind to Y-box (inverted CCAAT-box) sequences in promoters of a variety of genes (75). They are able to bind to single-stranded and double-stranded DNAs and RNAs. Y-box proteins have been proven to function in various aspects of gene regulation in both the nucleus and the cytoplasm; these include transcriptional activation or repression, regulation of alternative splicing, mRNA stability, and translational activation or repression (18, 32, 45, 63). Y-box proteins FRGY2 and mRNP3 are major RNA-binding components of storage mRNPs or informosomes in Xenopus oocytes, in which the majority of maternal mRNAs are sequestered from the translational apparatus (13, 49, 66, 70). In the cytoplasm of somatic cells, YB-1/p50 is known to be a core component of both translationally active and inactive mRNPs (19, 47). It has been proposed that Y-box proteins are likely responsible for the packaging of mRNAs into mRNPs (18, 63). Consistent with this proposition, the visualization of FRGY2-mRNA complexes by electron microscopy revealed that FRGY2 proteins package mRNAs into compact RNPs (43). Interestingly, in rabbit reticulocyte lysates, a low level of YB-1/p50 (less than 10 molecules of p50 per mRNA molecule) is found in polysomal globin mRNPs, while mRNPs are inactive at a high ratio of YB-1/p50 to mRNA (∼20 molecules of p50 per mRNA molecule) (17, 19, 54). The overexpression of Y-box proteins in somatic cells as well as in oocytes results in a substantial repression of translation (8, 12, 56). Thus, in addition to functioning as a structural component of cytoplasmic mRNPs, Y-box proteins are likely to have an impact on the fate of mRNAs in the cytoplasm.
Y-box proteins consist of three domains: the N-terminal domain, the cold shock domain (CSD), and the C-terminal tail domain. The CSD is a nucleic acid-binding domain exceptionally highly conserved among eukaryotic Y-box proteins and is so named because of its homology to bacterial major cold shock proteins in both primary sequence and three-dimensional structure (24). The CSD consists of a five-stranded β barrel containing the well-characterized RNA-binding motifs RNP-1 and RNP-2. In Xenopus FRGY2, the CSD has sequence-specific RNA-binding activity (7). The CSD of vertebrate Y-box proteins is >40% identical to Escherichia coli cold shock protein A (CspA). In E. coli, CspA was the first known cold shock protein, and there are nine CspA homologues, CspA to CspI, four of which are induced by cold shock (22, 74, 77). Members of this family of proteins bind to RNA and single-stranded DNA to regulate transcription and translation. CspA is proposed to function as an RNA chaperone that destabilizes secondary structures of RNA to facilitate translation (27). In E. coli, deletion of the CspA gene did not result in any growth defect, even at low culture temperatures (3). This finding could be explained by the presence of CspA homologues in E. coli. In a CspA deletion strain, the production of cold-inducible CspA homologues CspB and CspG increased with a downshift in temperature, suggesting that these homologues compensate for the loss of CspA (3). In contrast to the functions of bacterial cold shock proteins containing the CSD, the cellular functions of eukaryotic Y-box proteins in the cold have not been explored.
The C-terminal tail domain of Y-box proteins also has RNA-binding activity, although its sequence selectivity has not been clarified (7, 36, 48). The tail domain in vertebrate Y-box proteins consists of islands of acidic and basic amino acid clusters, and its amino acid sequence is less well conserved among organisms. The tail domain is also required for homomultimerization and association with various proteins (for a recent review, see reference 32). Its presence is sufficient for YB-1 to be incorporated into mRNPs and repress translation (16, 41). On the other hand, the short, N-terminal domain has been poorly studied, although the N-terminal domain of rabbit YB-1 is known to be capable of associating with actin (58).
The domain structure of vertebrate Y-box proteins is conserved among organisms. Based on the amino sequence of the C-terminal tail domain, vertebrate Y-box proteins can be further classified into two groups, germ cell type and somatic cell type. For instance, in Xenopus, the tail domains of germ cell-specific FRGY2 and mRNP3 are highly homologous to each other but show only about 30% homology to that of FRGY1, a somatic cell-type Y-box protein. In the present study, we explored the cellular functions of Y-box proteins in somatic cells by gene disruption in chicken DT40 cells. We found that a downshift in the cell culture temperature led to the growth arrest of YB-1-depleted cells. Through the expression of an epitope-tagged YB-1 protein in the gene disruptants, cell growth in the cold was restored. We isolated RNPs containing the YB-1 protein through immunoprecipitation of the epitope-tagged YB-1 protein. RNase treatment prior to immunoprecipitation led to the identification of an acidic YB-1-associated protein. We provide evidence that the acidic protein directly bound to YB-1 may cause mRNP remodeling.
MATERIALS AND METHODS
Construction of targeting vectors.
The genomic chicken YB-1 locus was cloned from DT40 genomic DNA by long-range PCR, and the positions of the exons and introns were determined by sequencing. The histidinol or blasticidin S resistance selection marker genes under the control of the chicken β-actin promoter were inserted between the left and the right arms derived by PCR amplification of the constructs.
Construction of the YB-1 expression vector.
A partial chicken YB-1 cDNA was obtained from DT40 total RNA by reverse transcription (RT)-PCR with primers having the sequences 5′-ATGAATTCGCAAACAGCCAGTCACCACCA-3′ and 5′-ATCCGCGGATCCCTCAGCCCCGCCCTGCTCGGCCTCGGGAGCGGACGTG-3′ (23). The PCR product was digested with EcoRI and BamHI, and the resulting DNA fragment was inserted into vector p3×FLAG-CMV-14 (Sigma) to yield a FLAG-tagged chicken YB-1 (chkYB-1) expression plasmid under the control of the human cytomegalovirus immediate-early promoter.
Cell culture and DNA transfection.
DT40 cells and all subclones were cultured in growth medium (RPMI 1640 medium supplemented with 10% fetal calf serum, 1% chicken serum, and 0.1 mM 2-mercaptoethanol) at 39.5°C in a 5% CO2 incubator. To count cell numbers, 5 × 104 cells which had been cultured at 39.5°C were inoculated with 1 ml of growth medium and incubated at 39.5 or 33°C. At various time points, cell numbers were determined. Viable and dead cells were assessed by trypan blue exclusion.
DNA transfection was performed essentially as described previously (11). Briefly, 107 cells were electroporated with 25 μg of linearized plasmids by using a Gene Pulser apparatus (Bio-Rad). Drug-resistant colonies were selected in 96-well plates with medium containing 25 μg of blasticidin S/ml, 1 mg of histidinol/ml, or 1.5 mg of G418/ml at 39.5°C. Gene disruption was confirmed by Southern blotting.
Southern blotting.
Genomic DNA was isolated from drug-resistant clones. DNA samples (10 μg each) were digested with EcoRI, and the resulting fragments were electrophoresed in 1.1% agarose gels and transferred to nylon membranes. Blots were hybridized with a DNA probe synthesized by PCR. The sequences of the primers used were as follows: forward primer, 5′-CCCTCAGTGTGTTAATTTGAAGA-3′; and reverse primer, 5′-GAGCTCACGCCGCATTTCTCATTAG-3′ (see Fig. 1A for the position of the probe).
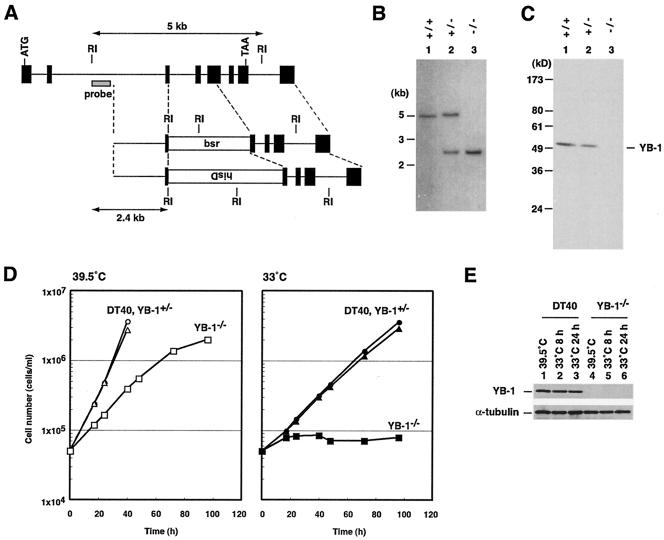
FIG. 1.
Generation of a YB-1−/− clone. (A) Schematic representation of part of the YB-1 locus, the gene disruption constructs, and the configuration of the targeted loci. Black boxes indicate the positions of the exons. RI, EcoRI recognition sites. The sizes of the hybridized fragments of the wild-type and targeted loci in Southern blotting (B) are shown. (B) Southern blot analysis of EcoRI-digested genomic DNA from the indicated genotypes with the probe shown in panel A. (C) Immunoblot analysis of total cell lysates with anti-YB-1 antibodies. The positions of molecular mass markers are shown on the left. (D) Growth curves for DT40 cells and YB-1 gene disruptants. DT40 cells (circles), YB-1+/− cells (triangles), and YB-1−/− cells (squares), which had been maintained at 39.5°C, were inoculated at 5 × 104 cells/ml and incubated at 39.5°C (left panel) or 33°C (right panel). The cell numbers were determined at the time points indicated. (E) Protein expression after the temperature shift. DT40 and YB-1−/− cells were cultured at 39.5 or 33°C for the times indicated and harvested, and 5 μg of total protein from the cell lysates was analyzed by immunoblotting with anti-YB-1 and anti-α-tubulin antibodies.
Immunoblotting.
Cells were lysed by sonication of approximately 2 × 107 cells/ml of hypotonic buffer (5 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 0.25 mM phenylmethylsulfonyl fluoride). A total of 5 or 10 μg of total protein from the cell lysates was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Immunoblotting was performed essentially as described previously (42). A rabbit immunoglobulin G fraction containing anti-FRGY1 antibodies was used as anti-YB-1 antibodies in this study (69). Rabbit anti-chicken 38,000-molecular weight (38K) protein antibodies (a gift from T. Okagaki) (52), mouse anti-HuR antibodies (3A2; Santa Cruz), anti-FLAG antibodies (M2; Sigma), and anti-α-tubulin antibodies (ICN) were used as primary antibodies.
Sucrose gradient centrifugation.
Sucrose gradient fractionation of cell lysates was performed essentially as described previously (28, 51). Prior to harvest, cycloheximide was added to the medium at a concentration of 100 μg/ml for 3 min at 37°C. Cells were harvested, washed with phosphate-buffered saline containing 100 μg of cycloheximide/ml, and lysed by the addition of 1.0 × 108 to 1.5 × 108 cells/ml of buffer A (15 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 60 mM NaCl, 0.5% Triton X-100, 1 mM DTT, 200 U of RNase inhibitor/ml, 100 μg of cycloheximide/ml). The lysates were passaged three times through a 27-gauge needle. Nuclei were removed by centrifugation at 12,000 × g for 5 min at 4°C. The supernatant was centrifuged as described above one more time, and the resulting supernatant was layered on a 15 to 40% sucrose gradient (5 ml) in buffer A lacking Triton X-100. For analysis in the presence of EDTA, cells were washed with phosphate-buffered saline, and the cell lysis buffer and the sucrose gradient buffer included 15 mM EDTA instead of cycloheximide. The gradient was centrifuged at 280,000 × g for 50 min (cycloheximide) or 80 min (EDTA) in a Hitachi P50S2 rotor. Fractions (300 μl) were collected from the top of the gradient. Sucrose gradient fractionation of the reconstituted YB-1-mRNPs is described in the figure legends.
Immunoprecipitation.
Cells at a concentration of 1.0 × 108 cells/ml were lysed by the addition of buffer B (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Nonidet P-40) containing 5 mM MgCl2 and Complete EDTA-free protease inhibitor cocktail (Roche). The lysates were passaged three times through a 27-gauge needle. Nuclei were removed by centrifugation at 12,000 × g for 5 min at 4°C. The supernatant was centrifuged one more time to ensure the removal of any nuclei. The resulting cell lysates (200 μl) were mixed with 800 μl of buffer B containing 50 mM EDTA, yielding a final EDTA concentration of 40 mM. For treatment with RNase, RNase A was added to the reaction at a final concentration of 50 μg/ml, and the mixture was incubated for 10 min at 37°C. Cell lysates were incubated with 20 μl of anti-FLAG M2 affinity gel (Sigma) for 2 h at 4°C. The affinity gel was extensively washed with buffer B containing 40 mM EDTA, and complexes containing FLAG-tagged protein were eluted with 150 μg of 3× FLAG peptide (Sigma)/ml in 50 mM Tris-HCl (pH 7.5)-150 mM NaCl for 5 min at room temperature. Tandem mass spectrometric analyses of the tryptic peptides were carried out at the Research Resources Center at our institute.
To prepare RNA, eluates were treated with 200 μg of proteinase K/ml for 1 h at 37°C and extracted with phenol-chloroform. RNA was precipitated with ethanol and used for RT-PCR with primers specific for chicken β-actin mRNA. The sequences of the primers used were as follows: for the β-actin coding sequence, 5′-CAGCCATGTATGTAGCCATCCAG-3′ and 5′-TAGGGATCCGGTTTAGAAGCATTTGCGGTGGACAATG-3′; and for the β-actin 3′ untranslated region, 5′-CGCGAGCTCACCGGACTGTTACCAACACCCACACCCCTGTG-3′ and 5′-CGCGAGCTCATACTGGCACTTTACTCCTAGACTG-3′.
Recombinant proteins.
Plasmid pET21d containing cDNA encoding the chicken 38K protein (a gift from T. Okagaki) was transformed into E. coli strain BL21(DE3) (52). Recombinant 38K protein (Y-box protein-associated acidic protein [YBAP1]) overexpressed in bacterial cells was purified by ammonium sulfate fractionation, followed by DEAE chromatography as described previously (52). To generate expression plasmids for glutathione S-transferase (GST)-chkYB-1 and GST-YBAP1 fusions, full-length YB-1 and YBAP1 coding sequences, respectively, were inserted into pGEX-4T-3 (Amersham Biosciences). Recombinant GST fusion proteins were overexpressed in E. coli and purified as described previously (42).
To generate expression plasmids for His6-tagged chkYB-1 and YBAP1, full-length YB-1 and YBAP1 coding sequences, respectively, were inserted into pET24b (Novagen). His6-chkYB-1 protein was prepared as described previously for FRGY2 protein (40). The expression plasmid for His6-YBAP1 was transformed into E. coli strain BL21(DE3). Recombinant His6-YBAP1 was overexpressed by the addition of isopropyl-β-d-thiogalactopyranoside to the bacterial culture. Bacterial cells were sonicated in 20 ml of sonication buffer, consisting of 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Cell lysates were centrifuged at 12,000 × g for 10 min, and the supernatant was applied to a TALON metal affinity column (Clontech). His6-YBAP1 was eluted with sonication buffer containing 100 mM imidazole and dialyzed against 20 mM Tris-HCl (pH 7.5)-100 mM NaCl-1 mM DTT-10% glycerol-0.5 mM phenylmethylsulfonyl fluoride.
GST pull-down assays.
For GST pull-down assays with purified proteins, 2 μg of recombinant YBAP1 was incubated with 2 μg of GST or GST-chkYB-1 bound to glutathione-Sepharose in buffer B containing 20 mM EDTA for 1 h at 4°C. After extensive washes with the same buffer, the bound materials were analyzed by immunoblotting with anti-38K protein antibodies (52).
To determine the domain of YB-1 interacting with YBAP1, full-length and deletion constructs of YB-1 with a C-terminal FLAG tag were generated by PCR amplification of the respective cDNA sequences and subcloned into vector pTNT (Promega) (2). YB-1 proteins were expressed in a TNT T7-coupled wheat germ extract system (Promega) by using each construct. Aliquots of the reaction mixtures were incubated with 4 μg of GST or GST-YBAP1 bound to glutathione-Sepharose, and assays were performed as described above. The bound materials were analyzed by immunoblotting with anti-FLAG M2 antibodies.
Gel retardation assays.
Chicken β-actin cDNA was obtained from DT40 total RNA by RT-PCR with primers having the sequences 5′-AATAAGCTTAAAGATGGATGATGATATTGCTGCGCTCGTTGTTGAC-3′ and 5′-TAGGGATCCGGTTTAGAAGCATTTGCGGTGGACAATG-3′. The PCR product was digested with HindIII and BamHI and cloned into pSP64 poly(A) (Promega) to obtain plasmid pSPcACT1. Plasmid pSPCAT was described previously (8). Linearized plasmids pSPcACT1 and pSPCAT were used for in vitro transcription with an SP6 mMessage mMachine kit (Ambion) in the presence of [α-32P]UTP. The formation of YB-1-mRNA complexes and gel retardation assays were carried out as described previously with modifications (40, 43). His6-chkYB-1 and His6-YBAP1 were incubated in 23.5 μl of reaction mixture (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 4 mM MgCl2, 1 mM DTT, 0.8 U of RNase inhibitor/μl) for 15 min at 30°C. mRNA (1 pmol; 1.5 μl) was added, and the mixture was further incubated for 20 min at 30°C. A 10-μl sample of each reaction mixture was electrophoresed in a 1% agarose gel in 0.5× Tris-borate-EDTA buffer.
In vitro translation.
His6-chkYB-1 and His6-YBAP1 were incubated in 20 μl of reaction mixture containing 12.5 μl of wheat germ extracts (Promega) for 15 min at 25°C. Template-amino acid mixture (5 μl; 1 pmol of 32P-labeled mRNA, 2 μl of 1 mM amino acid mixture minus methionine, 6 μCi of [35S]methionine [>1,000 Ci/mmol], 20 U of RNase inhibitor) was added, and the mixture was further incubated for 120 min at 25°C. For analysis of protein products, a 2.5-μl aliquot of the mixture was treated with RNase A and analyzed by SDS-PAGE. The radioactivities of translational products were quantified by using a BAS2500 image analyzer (Fuji Film). For analysis of the integrity of mRNA after the translation reaction, a 2-μl aliquot of the mixture was treated with proteinase K, and the purified RNA was analyzed in an agarose gel containing formaldehyde (71).
RESULTS
Growth defects of YB-1−/− cells.
To investigate the role of YB-1 in DT40 cells, the YB-1 locus was disrupted by homologous recombination. A genomic DNA fragment containing the YB-1 locus was amplified by long-range PCR with primers synthesized on the basis of the chicken YB-1 cDNA sequences. To generate YB-1 targeting constructs, either histidinol or blasticidin S resistance genes were inserted into the genomic sequences (Fig. 1A). Two YB-1 targeting constructs were sequentially transfected into DT40 cells. YB-1−/− cells were viable. Disruption of the YB-1 gene was verified by genomic Southern blotting (Fig. 1B). Immunoblotting indicated the depletion of YB-1 protein in YB-1−/− cells (Fig. 1C).
When isolating YB-1−/− clones after transfection of the second targeting construct into YB-1+/− cells, we noted that some colonies arose more slowly than others. The slowly growing colonies were proved to be YB-1−/− cells by genomic Southern blotting (data not shown). While the growth curve for YB-1+/− cells was indistinguishable from that for wild-type DT40 cells, YB-1−/− cells proliferated considerably more slowly than wild-type and YB-1+/− cells at 39.5°C (Fig. 1D). The doubling times were 7.5, 7.5, and 14 h for wild-type, YB-1+/−, and YB-1−/− cells, respectively.
Although Y-box proteins contain the CSD that is also present within bacterial cold shock proteins, which are overexpressed upon a downshift of temperature, the roles of Y-box proteins in the presence of cold stress have not been explored. We therefore examined the consequences of the depletion of YB-1 upon a temperature downshift. Wild-type DT40 and YB-1+/− cells exhibited slower growth at 33°C than at 39.5°C, with a doubling time at 33°C of 15 to 16 h. Notably, YB-1−/− cells showed no increase in viable cell numbers after 4 days at 33°C (Fig. 1D). Next, we examined the expression of YB-1 upon a temperature downshift (Fig. 1E). Immunoblotting revealed that the YB-1 protein levels in DT40 cells cultured at 33°C were indistinguishable from those in cells cultured at 39.5°C. Reprobing with anti-α-tubulin antibodies showed no change in the expression of α-tubulin. Our results suggest that a lack of YB-1 protein led to the growth defect in the presence of cold stress, although YB-1 was not overexpressed upon a temperature downshift in wild-type cells.
Complementation of YB-1−/− cells by exogenous YB-1 expression.
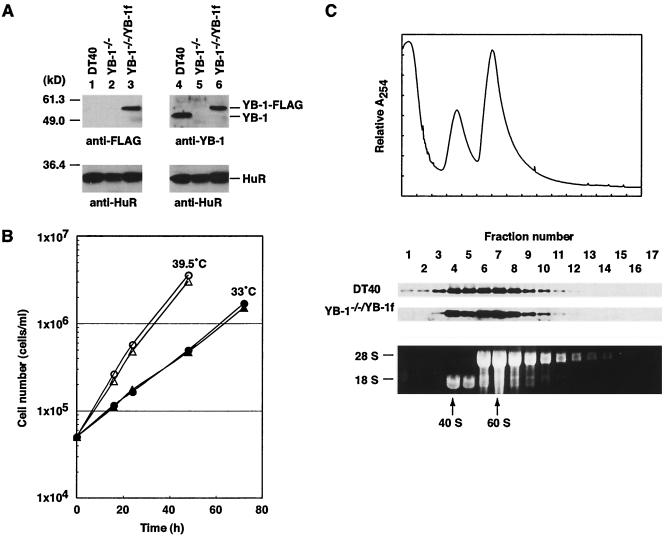
In order to determine whether the growth defects of YB-1−/− cells could be attributable to the loss of YB-1 protein and to facilitate the isolation of complexes containing YB-1, an expression vector encoding FLAG epitope-tagged YB-1 was introduced into YB-1−/− cells. We obtained a stable clone, which we designated YB-1−/−/YB-1f; this clone expressed the FLAG-tagged YB-1 protein at a level similar to that of the endogenous YB-1 protein in DT40 cells, as shown by immunoblotting with anti-FLAG and anti-YB-1 antibodies (Fig. 2A). Reprobing with anti-HuR antibodies served as the loading control. The rate of growth of YB-1−/−/YB-1f cells at either 39.5 or 33°C was indistinguishable from that of DT40 cells (Fig. 2B), indicating that the slow growth and the growth defect at low temperatures were complemented by exogenous YB-1 expression. Analysis with a sucrose gradient showed that the distribution of the FLAG-tagged YB-1 protein in the cytoplasm of YB-1−/−/YB-1f cells was similar to that of the endogenous YB-1 protein in wild-type cells (Fig. 2C). We conclude that the FLAG-tagged YB-1 protein can functionally replace the wild-type protein.
FIG. 2.
Complementation of the growth defects of the YB-1 gene disruptants by FLAG-tagged YB-1 expression. (A) Lysates of DT40, YB-1−/−, and YB-1−/−/YB-1f cells were examined for the expression of YB-1 by immunoblotting with anti-FLAG (top left panel) or anti-YB-1 (top right panel) antibodies. The same membranes were reprobed with anti-HuR antibodies (bottom panels). (B) Growth curves. DT40 cells (circles) and YB-1−/−/YB-1f cells (triangles), which had been maintained at 39.5°C, were inoculated at 5 × 104 cells/ml and incubated at 39.5°C (open symbols) or 33°C (closed symbols). The cell numbers were determined at the time points indicated. (C) DT40 and YB-1−/−/YB-1f cell lysates were fractionated with sucrose gradients in the presence of EDTA. Fractions were collected from the top of the gradients. Proteins from each fraction were analyzed by immunoblotting with anti-YB-1 (DT40) and anti-FLAG (YB-1−/−/YB-1f) antibodies. (Top panel) Absorbance profile at 254 nm for the gradient of DT40 cell lysates. (Bottom panel) Gel electrophoresis pattern of total RNA from each fraction. The positions of 28S and 18S rRNAs are indicated; 40S and 60S indicate the positions of ribosomal subunits.
Isolation of YB-1-containing RNPs.
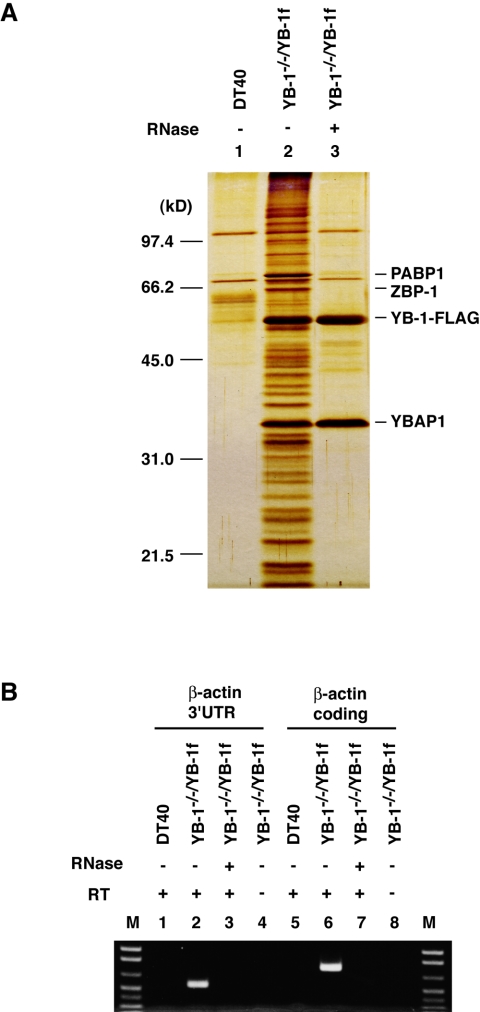
Having established a cell line expressing FLAG-tagged YB-1 in place of endogenous YB-1, we searched for proteins and RNAs that interact with the YB-1 protein in cells. YB-1 was immunoprecipitated from YB-1−/−/YB-1f cell lysates with anti-FLAG antibodies (Fig. 3A). We estimated that the efficiency of the immunoprecipitation was 30 to 50% (data not shown). A number of proteins that were not present in immunoprecipitates isolated from DT40 cell lysates were specifically copurified with YB-1 from YB-1−/−/YB-1f cell lysates when immunoprecipitation was performed under conditions that preserved RNA (Fig. 3A, lanes 1 and 2). Among them, proteins with molecular masses of 70, 66, and 35 kDa were abundant. To test whether RNA was required for the copurification of these proteins with YB-1, cell lysates were treated with RNase A prior to immunoprecipitation. RNase digestion disrupted the complex(es), indicating that the complex(es) isolated by immunoprecipitation contains RNA (Fig. 3A, lane 3). Moreover, the associations between most of the proteins and YB-1 are likely to be mediated primarily by their binding to the same RNA molecules. It is notable that among the abundant YB-1-associated proteins, only the 35-kDa protein was still copurified with YB-1 and the amount of this protein appeared to be increased after RNase digestion.
FIG. 3.
Isolation of YB-1-associated complexes. (A) DT40 cell lysates (lane 1) and YB-1−/−/YB-1f cell lysates before (lane 2) or after (lane 3) RNase A treatment were incubated with anti-FLAG antibody beads, and bound proteins were eluted as described in Materials and Methods. Eluted proteins were resolved by SDS-PAGE and detected by silver staining. (B) RNA in immunoprecipitates obtained with anti-FLAG antibodies was used as the template for RT-PCR to detect chicken β-actin mRNA. In lanes 4 and 8, reverse transcriptase was omitted during RT-PCR. 3′UTR, 3′ untranslated region. Lane M, markers.
We identified some of the YB-1-associated proteins by mass spectrometry. The relevant bands were excised from a Coomassie brilliant blue-stained gel and analyzed by tandem mass spectrometry. The abundant 70-kDa protein was PABP 1 (PABP1). PABP1 binds to the poly(A) tail at the 3′ termini of mRNAs to package the poly(A) tail by cooperative binding (see above). As PABP1 is known to be a major component of mRNPs from various sources, its abundance in our preparations indicates that the YB-1-containing RNPs contain poly(A)+ RNPs and also validates the purification approach taken. The 66-kDa protein was chicken “zipcode”-binding protein 1 (ZBP-1); ZBP-1 was identified as an RNA-binding protein that binds to the zipcode segment in the 3′ untranslated region of β-actin mRNA, which directs the localization of mRNA (57). This finding raised the possibility that although YB-1 has been known to be a general mRNA-binding protein (18), one of the mRNAs interacting with YB-1 in DT40 cells is that encoding β-actin. Accordingly, RT-PCR clearly showed that RNAs prepared from immunoprecipitates included β-actin mRNA (Fig. 3B). RNase A treatment of cell lysates prior to immunoprecipitation resulted in the disappearance of RT-PCR products (Fig. 3B, lanes 3 and 7), demonstrating the efficacy of RNase digestion. The 35-kDa protein was identical to chicken 38K protein, which assembles skeletal muscle myosin (52). We hereafter refer to this protein as YBAP1, for Y-box protein-associated acidic protein (see below). The human homologue of YBAP1 was originally isolated by copurification with splicing factor ASF/SF2 (34, 35).
We further tried to identify protein components that constitute YB-1-containing complexes. We cut the Coomassie brilliant blue-stained gel into slices 2 mm in height and subjected them to tryptic digestion. The resultant tryptic peptides were analyzed by tandem mass spectrometry (Table 1). Among the YB-1-associated proteins were a number of hnRNP proteins and ribosomal proteins. To test whether ribosomes were copurified with YB-1, we analyzed RNAs prepared from immunoprecipitates. Both 28S and 18S rRNAs were specifically detected in immunoprecipitates from YB-1−/−/YB-1f cells (data not shown). Since immunoprecipitation with anti-FLAG antibodies was performed in the presence of 40 mM EDTA, the association of YB-1 with ribosomes was likely to be direct rather than mediated by mRNA.
TABLE 1.
Proteins associating with YB-1, as identified by mass spectrometric analysis
| Fraction | Apparent mass (kDa) | Protein(s) |
|---|---|---|
| 1 | 120-100 | hsp105,a Upf1, hnRNP U, KIAA0183 |
| 2 | 100-86 | Nucleolin |
| 3 | 86-76 | hnRNP R |
| 4 | 76-66 | hsc70,a PABP1 |
| 5 | 66-60 | ZBP-1, hnRNP Q1/Q2 |
| 6 | 60-56 | YB-1b |
| 7 | 56-52 | YB-1b |
| 8 | 52-48 | YB-1, L4 |
| 9 | 48-44 | hnRNP A/B, L3 |
| 10 | 44-41 | hnRNP A3, hnRNP UP2 |
| 11 | 41-38 | hnRNP A2/B1, L6 |
| 12 | 38-35 | P0, L5 |
| 13 | 35-33 | 38K protein,b S3A, L5 |
| 14 | 33-30 | S6, L8, S2 |
| 15 | 30-28 | S4 |
| 16 | 28-26 | L19, S8 |
| 17 | 26-24 | L10 |
| 18 | 24-23 | S9, L17, L18 |
| 19 | 23-21 | L17, L23A, L24 |
| 20 | 21-19 | L26, L23A |
| 21 | 19 | S13, S17, S24 |
Also detected in DT40 and RNase-treated samples.
Also detected in RNase-treated samples.
Interaction between YB-1 and YBAP1.
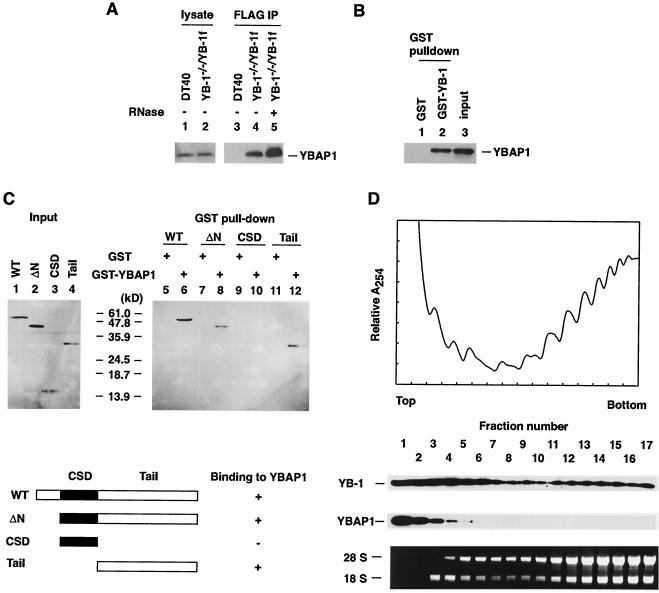
YBAP1 is distinct from other YB-1-associated proteins identified here in that it was copurified with YB-1 either before or after RNase A digestion of cell lysates (Fig. 4A). This result suggests a direct interaction between YBAP1 and YB-1. To test this suggestion, we performed a GST pull-down assay with recombinant proteins produced in E. coli. YBAP1 interacted with GST-YB-1 but not with GST, demonstrating a direct protein-protein interaction between YBAP1 and YB-1 (Fig. 4B).
FIG. 4.
Interaction between YB-1 and YBAP1. (A) Coimmunoprecipitation of chicken YBAP1 with YB-1. Lysates of DT40 (lane 1) and YB-1−/−/YB-1f (lane 2) cells and immunoprecipitates (IP) obtained with anti-FLAG antibodies from DT40 and YB-1−/−/YB-1f cell lysates before or after RNase A treatment (lanes 3 to 5) (Fig. 3A shows silver staining of total protein) were examined by immunoblotting with anti-38K protein/YBAP1 antibodies. (B) GST pull-down assay. Recombinant YBAP1 was incubated with GST (lane 1) or GST-YB-1 (lane 2) bound to glutathione-Sepharose. The bound materials were analyzed by immunoblotting with anti-38K protein/YBAP1 antibodies. An aliquot of recombinant YBAP1 was analyzed in parallel (lane 3). (C) Domain of YB-1 required for interaction with YBAP1. Full-length YB-1 (wild type [WT]) and deletion mutants of YB-1 with a C-terminal FLAG tag were generated in an in vitro transcription-translation system. Each reaction mixture was incubated with GST or GST-YBAP1 bound to glutathione-Sepharose. The bound materials were analyzed by immunoblotting with anti-FLAG antibodies. Schematic diagrams of the constructs are shown. (D) Sucrose gradient fractionation of DT40 cell lysates in the presence of cycloheximide. Proteins from each fraction were analyzed by immunoblotting with anti-38K protein/YBAP1 and anti-YB-1 antibodies. (Top panel) Absorbance profile at 254 nm. (Bottom panel) Gel electrophoresis pattern of total RNA from each fraction. The positions of 28S and 18S rRNAs are indicated.
YBAP1 is an acidic protein with a pI of 4.4, while YB-1 has clusters of basic amino acid residues in its C-terminal tail domain. To define the region(s) of YB-1 required for the association with YBAP1, fragments of YB-1 were prepared in vitro and tested for their ability to bind to GST-YBAP1 that had been produced in E. coli (Fig. 4C). Full-length YB-1 and the N-terminally truncated form were able to bind to GST-YBAP1 but not to GST. The CSD itself did not bind to GST-YBAP1, while the C-terminal tail domain efficiently bound to GST-YBAP1. These results suggest that the association of YB-1 with YBAP1 is mediated through the basic C-terminal tail domain.
To examine whether YBAP1 is present in a large complex containing RNAs in cell lysates, the distribution of YBAP1 was examined by using sucrose gradient fractionation, which distributed YB-1 ubiquitously (Fig. 4D). YBAP1 was detected only in the top fractions of the gradient. We therefore argue that YBAP1 is not likely to be a bona fide mRNP component and that it associates with YB-1 that is not bound to RNA.
Effect of YBAP1 on translational repression mediated by YB-1.
Based on the results described above, we reasoned that the interaction with YBAP1 may affect the RNA-binding activity of YB-1 and thereby regulate the translational activity of the mRNA with which YB-1 associates. To examine the consequence of the YB-1-YBAP1 interaction, we performed a gel retardation assay to show the RNA-binding activity of YB-1 (Fig. 5). We used β-actin mRNA as the template for RNA binding, because we found that this mRNA was present in the YB-1-containing RNPs isolated from YB-1−/−/YB-1f cells. His6-tagged YB-1 and YBAP1 were overexpressed in E. coli and purified (Fig. 5A). Titration of YB-1 revealed the formation of complexes between mRNA and YB-1, which we refer to as YB-1-mRNPs (Fig. 5B). Increasing the amount of YB-1 in the reaction caused a decrease in the mobility of the complexes, suggesting that multiple YB-1 proteins assembled with a single RNA molecule (43). To examine the effect of the formation of YB-1-mRNPs on translation, we used an in vitro translation system with wheat germ extracts. Consistent with the results of previous studies (31, 41, 71, 78), the addition of YB-1 to the translation system with wheat germ extracts inhibited β-actin protein synthesis in a dose-dependent manner (Fig. 6A).
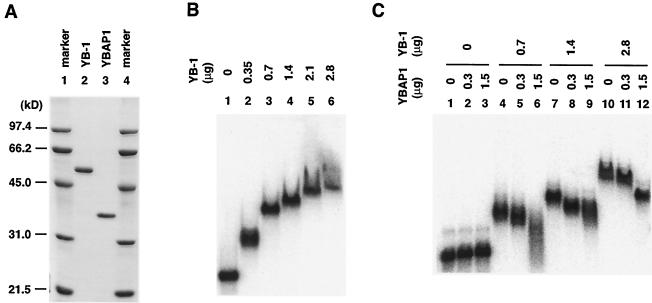
FIG. 5.
YBAP1 remodels YB-1-mRNPs. (A) One microgram each of purified His6-YB-1 and His6-YBAP1 was analyzed by SDS-PAGE and stained with Coomassie brilliant blue. The sizes of the molecular mass markers (Bio-Rad) are shown on the left. (B) Gel retardation assay of reconstituted YB-1-mRNPs. One picomole of 32P-labeled β-actin mRNA and 0 μg (lane 1), 0.35 μg (10 pmol; lane 2), 0.7 μg (20 pmol; lane 3), 1.4 μg (40 pmol; lane 4), 2.1 μg (60 pmol; lane 5), and 2.8 μg (80 pmol; lane 6) of recombinant His6-YB-1 were incubated and electrophoresed in an agarose gel. (C) His6-YB-1 and His6-YBAP1 were incubated, and then β-actin mRNA was added to the reaction mixtures as described in Materials and Methods. The mixtures were electrophoresed in an agarose gel.
FIG. 6.
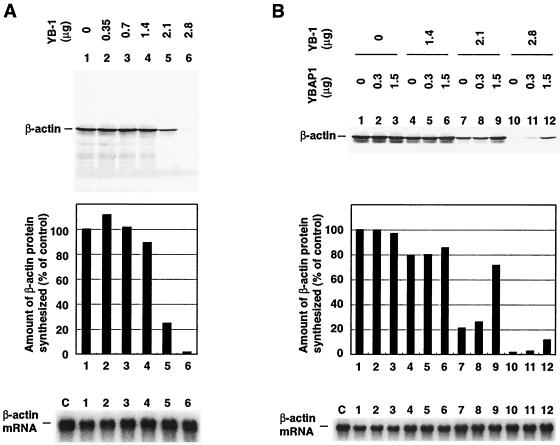
YBAP1 relieves translational repression mediated by YB-1. (A) Translational repression by YB-1. 32P-labeled β-actin mRNA and the indicated amounts of His6-YB-1 were incubated with wheat germ extracts in the presence of [35S]methionine. Translational products were analyzed by SDS-PAGE and quantified. The graph shows relative [35S]methionine incorporation into synthesized β-actin. The values were normalized to the value in lane 1, which was 100%. After the translation reaction, β-actin mRNA was recovered and analyzed in an agarose gel containing formaldehyde. Lane C shows an aliquot of the template RNA. (B) β-actin mRNA was incubated with YB-1-YBAP1 complexes and wheat germ extracts. Translational products and RNA were analyzed as described for panel A.
Having established that YB-1 binds to and inhibits the translation of β-actin mRNA, we examined the effect of the addition of YBAP1 on RNA binding and translational repression by YB-1. First, YBAP1 itself showed neither RNA-binding activity nor any effect on the translational activity of β-actin mRNA (Fig. 5C and 6B, lanes 1 to 3). With the addition of YBAP1 to the RNA-binding reaction, YB-1-mRNPs migrated faster in the gel retardation assay, suggesting either a decrease in the amount of YB-1 in the complexes or a structural change in the complexes (Fig. 5C, lanes 4 to 12). This change in the mobility of YB-1-mRNPs correlated well with the effect of YBAP1 on translational repression by YB-1 (Fig. 6B, lanes 4 to 12). YBAP1 restored protein synthesis when YB-1 inhibited translation by 80% (Fig. 6B, lanes 7 to 9).
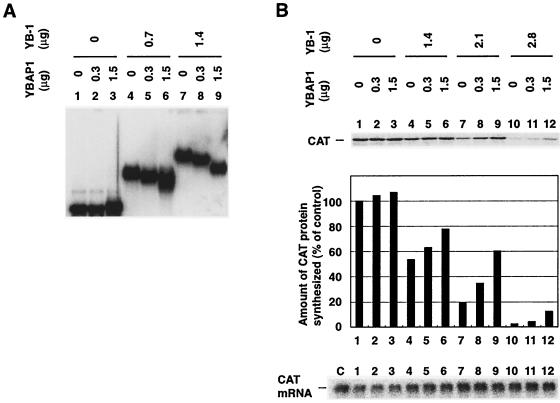
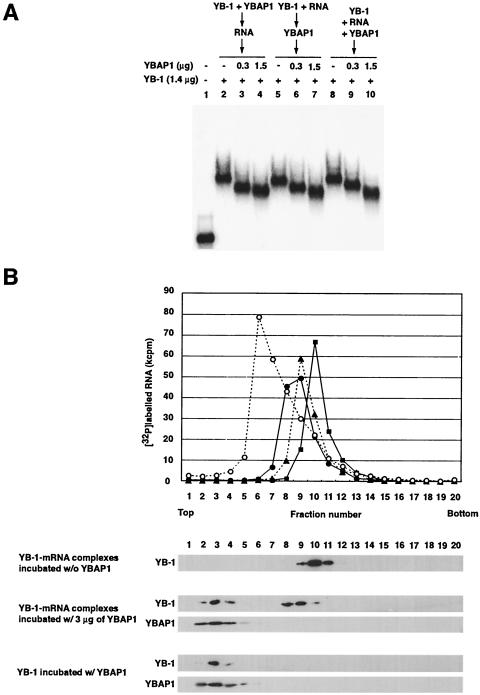
We examined whether YBAP1 would affect the translation of another mRNA. When YB-1 was incubated with bacterial chloramphenicol acetyltransferase (CAT) mRNA, the formation of complexes were observed (Fig. 7A). As shown with β-actin mRNA, the addition of YBAP1 resulted in the formation of complexes that migrated faster than those formed in the absence of YBAP1. YBAP1 also relieved the translational repression of CAT mRNA mediated by YB-1 (Fig. 7B), suggesting that the effect of YBAP1 on RNA binding and translational repression by YB-1 is not dependent on specific mRNAs that are associated with YB-1 in cells. Furthermore, YBAP1 affected the RNA-binding activity of YB-1 irrespective of the timing of its addition; a similar change in the mobility of YB-1-mRNPs was observed when YBAP1 was added before or after the formation of YB-1-mRNPs (Fig. 8A).
FIG. 7.
YBAP1 relieves translational repression of CAT mRNA mediated by YB-1. (A) His6-YB-1 and His6-YBAP1 were incubated, and then 1 pmol of 32P-labeled CAT mRNA was added to the reaction mixtures as described in Materials and Methods. The mixtures were electrophoresed in an agarose gel. (B) CAT mRNA was incubated with YB-1-YBAP1 complexes and wheat germ extracts. Translational products and RNA were analyzed as described in the legend to Fig. 6A.
FIG. 8.
YBAP1 displaces YB-1 molecules from YB-1-mRNPs. (A) YBAP1 remodels preformed YB-1-mRNPs. Lane 1, naked 32P-labeled β-actin mRNA. Lanes 2 to 4, His6-YB-1 (1.4 μg) was preincubated for 15 min at 30°C with the indicated amounts of His6-YBAP1. β-actin mRNA then was added, and the mixtures were incubated for an additional 20 min at 30°C. Lanes 5 to 7, His6-YB-1 was preincubated for 15 min at 30°C with β-actin mRNA. His6-YBAP1 then was added, and the mixtures were incubated for an additional 20 min at 30°C. Lanes 8 to 10, His6-YB-1, His6-YBAP1, and β-actin mRNA were mixed and incubated for 20 min at 30°C. Aliquots of the mixtures were analyzed in an agarose gel. (B) YB-1-mRNPs were prepared by incubating 20 pmol of 32P-labeled β-actin mRNA and 42 μg of His6-YB-1 for 20 min at 30°C and purified through a sucrose gradient (data not shown). Purified YB-1-mRNPs containing 3 pmol of mRNA were incubated for 20 min at 30°C without His6-YBAP1 (closed squares) or with 0.9 μg (closed triangles) or 3 μg (closed circles) of His6-YBAP1. The mixtures were centrifuged in 15 to 40% sucrose gradients at 50,000 rpm for 4 h in an SW55 Ti rotor (Beckman). Two control reactions, (i) His6-YB-1 and His6-YBAP1 incubated without RNA and (ii) naked β-actin mRNA (open circles), were analyzed in parallel. (Top panel) The gradient was collected into 20 fractions from the top, and the radioactivity in each fraction was quantified. (Bottom panels) YB-1 and YBAP1 in each fraction were analyzed by immunoblotting.
YBAP1 displaces YB-1 from reconstituted mRNPs.
We next wanted to examine whether the increase in the mobility of YB-1-mRNPs in the gel retardation assay was due to the partial release of YB-1 from the complexes upon incubation with YBAP1. To do so, we first purified reconstituted YB-1-mRNPs and incubated them with or without YBAP1. The mixtures then were fractionated through sucrose gradients (Fig. 8B). YB-1-mRNPs incubated without YBAP1 sedimented as a single peak at fraction 10. The distribution of YB-1-mRNPs incubated with YBAP1 shifted to fraction 8 or 9, which is between the naked RNA (fraction 6) and YB-1-mRNPs incubated without YBAP1. Consistent with these findings, upon incubation with YBAP1, about half of the YB-1 molecules were detected in fractions 2 to 4 together with YBAP1. This distribution indicates the release of YB-1 from mRNA, as we found that YB-1 and YBAP1 were distributed in fractions 2 to 4 when incubated in the absence of mRNA (Fig. 8B, bottom). We noted that a tiny amount of YBAP1 was incorporated into YB-1-mRNPs in this purified system, possibly due to the high affinity for YB-1. Taken together, our results indicate that YBAP1 displaces YB-1 from YB-1-mRNPs and consequently relieves the translational repression caused by YB-1.
DISCUSSION
YB-1 is required for cell proliferation at low temperatures.
We revealed that the depletion of YB-1 in DT40 cells results in a slow-growth phenotype and that YB-1 is essential for cell proliferation at low culture temperatures (Fig. 1). However, we did not detect any overexpression of YB-1 upon cold shock in DT40 cells. This finding is in sharp contrast to the results obtained with bacterial cold shock proteins containing the CSD (22). In plants, glycine-rich proteins with the CSD are also induced by cold stress (29). Fluorescence-activated cell sorting analysis revealed that in a given population of YB-1−/− cells, about 60% of the cells underwent apoptosis after 5 days of culturing at 33°C, while the rest survived (data not shown). Thus, the apparent inhibition of cell proliferation can be recognizable as a combined phenotype of apoptotic cell death and slow growth.
After we completed generating YB-1−/− cells, disruption of the YB-1b gene in DT40 cells was reported (68). The heterozygous disruption of one YB-1b allele leads to a slow-growth phenotype, increased cell size, and increased DNA content, suggesting major defects in the G2/M phase of the cell cycle. In contrast, when we performed the first round of transfection of the YB-1 targeting construct, all of the clones with the heterozygous disruption of the YB-1 gene formed colonies comparable in size to those of wild-type DT40 cells, indicating no growth defect in the heterozygous disruptants (Fig. 1D). The reason for this discrepancy is unknown. A recent report showed that heterozygous disruption of mouse YB-1 results in no growth defects, consistent with our data (60).
A downshift in the cell culture temperature generally induces the cold shock response (20, 73). Therefore, YB-1 may regulate an essential component(s) of the cold shock response pathway. The cold shock response includes slow growth and responses to alterations in membrane fluidity by changes in the degree of saturation of the hydrocarbon chains of membrane phospholipids. Another major response to low temperatures is a change in translational activity (20, 73). At low temperatures, the mRNA secondary structure is stabilized and will significantly impair protein synthesis in cells.
A plausible explanation for how YB-1 regulates cell proliferation at low temperatures, which we favor and which already has been proposed for E. coli CspA (27), is that YB-1 functions as an RNA chaperone. RNA chaperones melt the secondary structure of mRNA and thereby make the mRNA accessible to the translational apparatus (38). Rabbit YB-1/p50 has been reported to melt the secondary structure of globin mRNA (19). However, it also stimulates the annealing of complementary single-stranded RNAs or DNAs (62). These results seem to reveal two opposite effects on the mRNA structure. Interestingly, which of these two activities YB-1 displays depends on the YB-1/mRNA ratio: at a low YB-1/mRNA ratio, YB-1 stimulates annealing, whereas at a high YB-1/mRNA ratio, it causes RNA to melt (62). Therefore, YB-1 may contribute to the appropriate packaging of mRNA to facilitate translation at low temperatures.
YB-1-containing RNPs.
In order to obtain RNPs formed under physiological conditions, it is important to allow tagged proteins to assemble into RNPs without disruption of endogenous complexes if the endogenous counterpart is present in parental cells. FLAG-tagged YB-1 in YB-1−/−/YB-1f cells was present in amounts similar to those of YB-1 in wild-type cells and sedimented in sucrose gradients in a manner similar to that of YB-1 in parental wild-type cells. YB-1−/−/YB-1f cells grew at a rate identical to that of wild-type DT40 cells, indicating that the FLAG tag did not measurably affect the function of YB-1. With YB-1−/−/YB-1f cells, we were able to examine the proteins or RNPs associated with YB-1 (Fig. 3). Many RNA-binding proteins were identified in our preparation of YB-1-associated complexes; these included PABP1, ZBP-1, hnRNP proteins, and ribosomal proteins.
Given that pre-mRNA is never naked and must be packaged into an RNP throughout its life (14, 76), it is of great importance to understand when and how a set of proteins become associated with or released from a given mRNA. Although until recently hnRNPs were considered to be reorganized into cytoplasmic mRNPs upon nuclear export, it is known that many hnRNPs accompany mRNAs from the nucleus into the cytoplasm. Regardless of their active roles in mRNA export, evidence is accumulating that many hnRNP proteins shuttle between the nucleus and the cytoplasm and that some of these proteins indeed have cytoplasmic functions (14, 61). In accordance with this notion, this study revealed that a number of hnRNP proteins were coimmunoprecipitated with YB-1 under conditions that preserved RNA (Table 1). It has been recognized that the consequences of the organization of mRNPs, which reflect the nuclear history of mRNAs, have a major influence on the translational activity of mRNAs in the cytoplasm (9, 44, 51). Our preparation of YB-1-associated RNPs likely represents general and abundant components of cytoplasmic mRNPs. However, our preparation also contains proteins that are not components of mRNPs but rather directly associate with YB-1 (see below). Further purification of YB-1-associated RNPs is in progress.
Interaction of YB-1 with YBAP1.
We identified YBAP1 as a protein interacting with YB-1 in an RNA-independent manner. Under the conditions used here to immunoprecipitate FLAG-tagged YB-1, only YBAP1 was reproducibly and stoichiometrically coimmunoprecipitated from RNase-treated lysates of YB-1−/−/YB-1f cells (Fig. 3A). This observation demonstrates that the interaction between YB-1 and YBAP1 is highly specific. YBAP1 seems to be a multifunctional protein. Okagaki et al. detected the colocalization of 38K protein/YBAP1 with myosin and α-actin in mouse smooth muscle cells (52). The human homologue of chicken YBAP1, human p32, was originally copurified with splicing factor ASF/SF2 (35). Human p32 was independently identified as a cell surface glycoprotein that binds to the plasma complement component C1q (21). p32 was found to localize in the nucleus, cell surface, cytoplasm, and mitochondria and to shuttle between the nucleus and the cytoplasm or mitochondria in different cell types and conditions (10, 21, 35, 50, 52). p32 was reported to modulate RNA-binding or RNA-processing activities in several situations. p32 regulates RNA splicing by inhibiting RNA-binding activity and phosphorylation of ASF/SF2 (53). p32 also interacts with the Rev protein of human immunodeficiency virus type 1 and modulates the Rev-dependent splicing inhibition of Rev-responsive element-containing pre-mRNAs (72, 79). Moreover, in Trypanosoma brucei, the homologue of chicken YBAP1 interacts with the Y-box protein RBP16 and stimulates the binding activity of RBP16 to guide RNA for RNA editing (25).
In this study, using sucrose gradient fractionation under conditions which separate polysomes and mRNPs, we found that YBAP1 was present only in the top fractions, indicating that YBAP1 is not an integral component of mRNPs (Fig. 4D). Although the physiological roles of the association of YB-1 with YBAP1 in cells are totally unknown, our analysis revealed that YBAP1 negatively regulates the RNA-binding activity of YB-1 and relieves the translational repression of mRNA by associating with YB-1 (Fig. 5 to 8). We found that YBAP1 displaces YB-1 from YB-1-mRNPs (Fig. 8). Thus, given that Y-box proteins are the core components of cytoplasmic mRNPs in a variety of cells, YBAP1 may play a role in the remodeling of mRNPs. The displacement of YB-1 from mRNAs would bring about structural changes in mRNPs, which in turn would have an impact on the stability and translational activity of mRNAs.
In addition, further insight into the role of YBAP1 in modulating the activity of YB-1 during various steps in mRNA metabolism may come from consideration of the acidic nature of YBAP1 and our identification of the basic tail domain of YB-1 associating with YBAP1 (Fig. 4C). First, YBAP1 may serve as a repository for YB-1 proteins that are not bound to mRNAs. It is presumed that Y-box proteins are not associated with RNAs when they are synthesized de novo or when they are released from stored mRNPs upon translational activation or perhaps from mRNPs during mRNA degradation. We detected stoichiometric amounts of YBAP1 in immunoprecipitates from RNase-treated cell lysates. The association with YBAP1 would therefore prevent the undesirable aggregation of YB-1 proteins until the next usage.
Second, YBAP1 may be involved in the nuclear transport of YB-1. Although several studies have demonstrated that YB-1 is predominantly localized in the cytoplasm, nuclear YB-1 has been detected under specific conditions (5, 33, 67). Antibodies that bind to Y-box proteins relieved the inhibition of translation when introduced into the nucleus of a Xenopus oocyte (9). Immunostaining with the same antibodies detected FRGY2 or mRNP3 in lampbrush chromosomes of oocytes (64). Moreover, immunoelectron microscopy revealed that, in the salivary gland cells of the dipteran Chironomus tentans, Y-box proteins became associated with nascent transcripts and also were detected in nuclear pre-mRNPs (65). These studies suggested that Y-box proteins associate with pre-mRNA cotranscriptionally and accompany mRNA into the cytoplasm. The nuclear accumulation of the otherwise cytoplasmic human p32 is observed when cells are treated with leptomycin B, which inhibits CRM1-dependent nuclear export, suggesting that p32 is a nuclear-cytoplasmic shuttling protein (10). The formation of a complex with YBAP1 may therefore facilitate the nuclear translocation of YB-1. Another splicing factor, SRp30c, has been implicated in a similar process (55).
These considerations lead to the idea that YBAP1 is functionally similar to histone chaperones, many of which are acidic proteins involved in the storage of histones and the formation of nucleosomes (1). Several histone chaperones are also implicated in chromatin remodeling as well as the nuclear import of histones. Nucleoplasmin is the prototype of such histone chaperones (37). Importantly, nucleoplasmin was shown to be involved in the unmasking of stored mRNAs in Xenopus early embryos and to partially relieve the translational repression caused by the FRGY2 tail domain in the in vitro system (46). Nucleoplasmin forms a pentamer, which is required for interactions with histones (15). In this regard, it is worth noting that the trimer of human p32 forms a doughnut-shaped quaternary structure, in which negatively charged residues are distributed highly asymmetrically (26). This arrangement may contribute to the highly specific interaction between YBAP1 and YB-1. Specific acidic proteins may function in modulating the RNA-binding activity of Y-box proteins to alter structures of mRNPs and thereby make them accessible to the translational machinery. Conversely, the assembly of mRNPs may be facilitated by the functions of such acidic proteins.
Given that PABP is the other major protein in mRNPs, it should be noted that a PABP-interacting protein, Paip2, is another acidic protein (30). Paip2 inhibits the interaction of PABP with poly(A) and preferentially represses poly(A)-dependent translation, further exemplifying the role of acidic proteins in the reorganization of mRNPs.
In conclusion, our identification of YBAP1 as a protein that interacts with and relieves the translational repression caused by YB-1 suggests a novel step by which the activity of YB-1 is regulated. It remains to be examined whether the amount of YBAP1 influences the availability of Y-box proteins in cells, which in turn would have an influence on mRNA metabolism and which might be involved in the control of cell proliferation mediated by YB-1. Possible roles played by acidic proteins in the remodeling of mRNPs require further research to improve the understanding of how general translational efficiency is regulated.
Acknowledgments
We thank Alan P. Wolffe, Shunichi Takeda, Masayuki Seki, Nobuhiro Morishima, Kazuhiro Kohama, and Tsuyoshi Okagaki for reagents. We are grateful to Kyosuke Nagata and Akira Hattori for stimulating discussions. We also thank Yasue Ichikawa and Rie Nakazawa for DNA sequencing and M. Usui and K. Otsuki for mass spectrometric analysis.
This work was supported in part by a grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a grant for the Bioarchitect Research Program from RIKEN.
Footnotes
K.M. dedicates this work to the memory of Alan P. Wolffe.
REFERENCES
- 1.Akey, C. W., and K. Luger. 2003. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 13:6-14. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, K., K. Matsumoto, and M. Tsujimoto. 2003. Xenopus cold-inducible RNA-binding protein 2 interacts with ElrA, the Xenopus homolog of HuR, and inhibits deadenylation of specific mRNAs. J. Biol. Chem. 278:48491-48497. [DOI] [PubMed] [Google Scholar]
- 3.Bae, W., P. G. Jones, and M. Inouye. 1997. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J. Bacteriol. 179:7081-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer, B. W., and R. D. Kornberg. 1983. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J. Cell Biol. 96:717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bargou, R. C., K. Jurchott, C. Wagener, S. Bergmann, S. Metzner, K. Bommert, M. Y. Mapara, K. J. Winzer, M. Dietel, B. Dorken, and H. D. Royer. 1997. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat. Med. 3:447-450. [DOI] [PubMed] [Google Scholar]
- 6.Blobel, G. 1973. A protein of molecular weight 78,000 bound to the polyadenylate region of eukaryotic messenger RNAs. Proc. Natl. Acad. Sci. USA 70:924-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouvet, P., K. Matsumoto, and A. P. Wolffe. 1995. Sequence-specific RNA recognition by the Xenopus Y-box proteins. An essential role for the cold shock domain. J. Biol. Chem. 270:28297-28303. [DOI] [PubMed] [Google Scholar]
- 8.Bouvet, P., and A. P. Wolffe. 1994. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 77:931-941. [DOI] [PubMed] [Google Scholar]
- 9.Braddock, M., M. Muckenthaler, M. R. White, A. M. Thorburn, J. Sommerville, A. J. Kingsman, and S. M. Kingsman. 1994. Intron-less RNA injected into the nucleus of Xenopus oocytes accesses a regulated translation control pathway. Nucleic Acids Res. 22:5255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brokstad, K. A., K. H. Kalland, W. C. Russell, and D. A. Matthews. 2001. Mitochondrial protein p32 can accumulate in the nucleus. Biochem. Biophys. Res. Commun. 281:1161-1169. [DOI] [PubMed] [Google Scholar]
- 11.Buerstedde, J. M., and S. Takeda. 1991. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67:179-188. [DOI] [PubMed] [Google Scholar]
- 12.Davydova, E. K., V. M. Evdokimova, L. P. Ovchinnikov, and J. W. Hershey. 1997. Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res. 25:2911-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deschamps, S., A. Viel, M. Garrigos, H. Denis, and M. le Maire. 1992. mRNP4, a major mRNA-binding protein from Xenopus oocytes is identical to transcription factor FRG Y2. J. Biol. Chem. 267:13799-13802. [PubMed] [Google Scholar]
- 14.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 15.Dutta, S., I. V. Akey, C. Dingwall, K. L. Hartman, T. Laue, R. T. Nolte, J. F. Head, and C. W. Akey. 2001. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8:841-853. [DOI] [PubMed] [Google Scholar]
- 16.Evdokimova, V., P. Ruzanov, H. Imataka, B. Raught, Y. Svitkin, L. P. Ovchinnikov, and N. Sonenberg. 2001. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 20:5491-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evdokimova, V. M., E. A. Kovrigina, D. V. Nashchekin, E. K. Davydova, J. W. Hershey, and L. P. Ovchinnikov. 1998. The major core protein of messenger ribonucleoprotein particles (p50) promotes initiation of protein biosynthesis in vitro. J. Biol. Chem. 273:3574-3581. [DOI] [PubMed] [Google Scholar]
- 18.Evdokimova, V. M., and L. P. Ovchinnikov. 1999. Translational regulation by Y-box transcription factor: involvement of the major mRNA-associated protein, p50. Int. J. Biochem. Cell. Biol. 31:139-149. [DOI] [PubMed] [Google Scholar]
- 19.Evdokimova, V. M., C. L. Wei, A. S. Sitikov, P. N. Simonenko, O. A. Lazarev, K. S. Vasilenko, V. A. Ustinov, J. W. Hershey, and L. P. Ovchinnikov. 1995. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J. Biol. Chem. 270:3186-3192. [DOI] [PubMed] [Google Scholar]
- 20.Fujita, J. 1999. Cold shock response in mammalian cells. J. Mol. Microbiol. Biotechnol. 1:243-255. [PubMed] [Google Scholar]
- 21.Ghebrehiwet, B., B. L. Lim, E. I. Peerschke, A. C. Willis, and K. B. Reid. 1994. Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular “heads” of C1q. J. Exp. Med. 179:1809-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein, J., N. S. Pollitt, and M. Inouye. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant, C. E., and R. G. Deeley. 1993. Cloning and characterization of chicken YB-1: regulation of expression in the liver. Mol. Cell. Biol. 13:4186-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graumann, P. L., and M. A. Marahiel. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23:286-290. [DOI] [PubMed] [Google Scholar]
- 25.Hayman, M. L., M. M. Miller, D. M. Chandler, C. C. Goulah, and L. K. Read. 2001. The trypanosome homolog of human p32 interacts with RBP16 and stimulates its gRNA binding activity. Nucleic Acids Res. 29:5216-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang, J., Y. Zhang, A. R. Krainer, and R. M. Xu. 1999. Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc. Natl. Acad. Sci. USA 96:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 28.Johannes, G., and P. Sarnow. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4:1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlson, D., K. Nakaminami, T. Toyomasu, and R. Imai. 2002. A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J. Biol. Chem. 277:35248-35256. [DOI] [PubMed] [Google Scholar]
- 30.Khaleghpour, K., Y. V. Svitkin, A. W. Craig, C. T. DeMaria, R. C. Deo, S. K. Burley, and N. Sonenberg. 2001. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell 7:205-216. [DOI] [PubMed] [Google Scholar]
- 31.Kick, D., P. Barrett, A. Cummings, and J. Sommerville. 1987. Phosphorylation of a 60 kDa polypeptide from Xenopus oocytes blocks messenger RNA translation. Nucleic Acids Res. 15:4099-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohno, K., H. Izumi, T. Uchiumi, M. Ashizuka, and M. Kuwano. 2003. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays 25:691-698. [DOI] [PubMed] [Google Scholar]
- 33.Koike, K., T. Uchiumi, T. Ohga, S. Toh, M. Wada, K. Kohno, and M. Kuwano. 1997. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 417:390-394. [DOI] [PubMed] [Google Scholar]
- 34.Krainer, A. R., G. C. Conway, and D. Kozak. 1990. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 4:1158-1171. [DOI] [PubMed] [Google Scholar]
- 35.Krainer, A. R., A. Mayeda, D. Kozak, and G. Binns. 1991. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66:383-394. [DOI] [PubMed] [Google Scholar]
- 36.Ladomery, M., and J. Sommerville. 1994. Binding of Y-box proteins to RNA: involvement of different protein domains. Nucleic Acids Res. 22:5582-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laskey, R. A., B. M. Honda, A. D. Mills, and J. T. Finch. 1978. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275:416-420. [DOI] [PubMed] [Google Scholar]
- 38.Lorsch, J. R. 2002. RNA chaperones exist and DEAD box proteins get a life. Cell 109:797-800. [DOI] [PubMed] [Google Scholar]
- 39.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto, K., K. Aoki, N. Dohmae, K. Takio, and M. Tsujimoto. 2000. CIRP2, a major cytoplasmic RNA-binding protein in Xenopus oocytes. Nucleic Acids Res. 28:4689-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto, K., F. Meric, and A. P. Wolffe. 1996. Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA. Role of the cold shock domain, tail domain, and selective RNA sequence recognition. J. Biol. Chem. 271:22706-22712. [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto, K., K. Nagata, M. Miyaji-Yamaguchi, A. Kikuchi, and M. Tsujimoto. 1999. Sperm chromatin decondensation by template activating factor I through direct interaction with basic proteins. Mol. Cell. Biol. 19:6940-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto, K., K. J. Tanaka, K. Aoki, M. Sameshima, and M. Tsujimoto. 2003. Visualization of the reconstituted FRGY2-mRNA complexes by electron microscopy. Biochem. Biophys. Res. Commun. 306:53-58. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto, K., K. M. Wassarman, and A. P. Wolffe. 1998. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 17:2107-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto, K., and A. P. Wolffe. 1998. Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 8:318-323. [DOI] [PubMed] [Google Scholar]
- 46.Meric, F., K. Matsumoto, and A. P. Wolffe. 1997. Regulated unmasking of in vivo synthesized maternal mRNA at oocyte maturation. A role for the chaperone nucleoplasmin. J. Biol. Chem. 272:12840-12846. [DOI] [PubMed] [Google Scholar]
- 47.Minich, W. B., N. L. Korneyeva, and L. P. Ovchinnikov. 1989. Translational active mRNPs from rabbit reticulocytes are qualitatively different from free mRNA in their translatability in cell-free system. FEBS Lett. 257:257-259. [DOI] [PubMed] [Google Scholar]
- 48.Murray, M. T. 1994. Nucleic acid-binding properties of the Xenopus oocyte Y box protein mRNP3+4. Biochemistry 33:13910-13917. [DOI] [PubMed] [Google Scholar]
- 49.Murray, M. T., D. L. Schiller, and W. W. Franke. 1992. Sequence analysis of cytoplasmic mRNA-binding proteins of Xenopus oocytes identifies a family of RNA-binding proteins. Proc. Natl. Acad. Sci. USA 89:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muta, T., D. Kang, S. Kitajima, T. Fujiwara, and N. Hamasaki. 1997. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 272:24363-24370. [DOI] [PubMed] [Google Scholar]
- 51.Nott, A., H. Le Hir, and M. J. Moore. 2004. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 18:210-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okagaki, T., A. Nakamura, T. Suzuki, K. Ohmi, and K. Kohama. 2000. Assembly of smooth muscle myosin by the 38k protein, a homologue of a subunit of pre-mRNA splicing factor-2. J. Cell Biol. 148:653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petersen-Mahrt, S. K., C. Estmer, C. Ohrmalm, D. A. Matthews, W. C. Russell, and G. Akusjarvi. 1999. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 18:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pisarev, A. V., M. A. Skabkin, A. A. Thomas, W. C. Merrick, L. P. Ovchinnikov, and I. N. Shatsky. 2002. Positive and negative effects of the major mammalian messenger ribonucleoprotein p50 on binding of 40 S ribosomal subunits to the initiation codon of beta-globin mRNA. J. Biol. Chem. 277:15445-15451. [DOI] [PubMed] [Google Scholar]
- 55.Raffetseder, U., B. Frye, T. Rauen, K. Jurchott, H. D. Royer, P. L. Jansen, and P. R. Mertens. 2003. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J. Biol. Chem. 278:18241-18248. [DOI] [PubMed] [Google Scholar]
- 56.Ranjan, M., S. R. Tafuri, and A. P. Wolffe. 1993. Masking mRNA from translation in somatic cells. Genes Dev. 7:1725-1736. [DOI] [PubMed] [Google Scholar]
- 57.Ross, A. F., Y. Oleynikov, E. H. Kislauskis, K. L. Taneja, and R. H. Singer. 1997. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17:2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruzanov, P. V., V. M. Evdokimova, N. L. Korneeva, J. W. Hershey, and L. P. Ovchinnikov. 1999. Interaction of the universal mRNA-binding protein, p50, with actin: a possible link between mRNA and microfilaments. J. Cell Sci. 112:3487-3496. [DOI] [PubMed] [Google Scholar]
- 59.Sachs, A. B., R. W. Davis, and R. D. Kornberg. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shibahara, K., T. Uchiumi, T. Fukuda, S. Kura, Y. Tominaga, Y. Maehara, K. Kohno, Y. Nakabeppu, T. Tsuzuki, and M. Kuwano. 2004. Targeted disruption of one allele of the Y-box binding protein-1 (YB-1) gene in mouse embryonic stem cells and increased sensitivity to cisplatin and mitomycin C. Cancer Sci. 95:348-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shyu, A. B., and M. F. Wilkinson. 2000. The double lives of shuttling mRNA binding proteins. Cell 102:135-138. [DOI] [PubMed] [Google Scholar]
- 62.Skabkin, M. A., V. Evdokimova, A. A. Thomas, and L. P. Ovchinnikov. 2001. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J. Biol. Chem. 276:44841-44847. [DOI] [PubMed] [Google Scholar]
- 63.Sommerville, J. 1999. Activities of cold-shock domain proteins in translation control. Bioessays 21:319-325. [DOI] [PubMed] [Google Scholar]
- 64.Sommerville, J., and M. Ladomery. 1996. Transcription and masking of mRNA in germ cells: involvement of Y-box proteins. Chromosoma 104:469-478. [DOI] [PubMed] [Google Scholar]
- 65.Soop, T., D. Nashchekin, J. Zhao, X. Sun, A. T. Alzhanova-Ericsson, B. Bjorkroth, L. Ovchinnikov, and B. Daneholt. 2003. A p50-like Y-box protein with a putative translational role becomes associated with pre-mRNA concomitant with transcription. J. Cell Sci. 116:1493-1503. [DOI] [PubMed] [Google Scholar]
- 66.Spirin, A. S. 1969. The second Sir Hans Krebs Lecture. Informosomes. Eur. J. Biochem. 10:20-35. [DOI] [PubMed] [Google Scholar]
- 67.Stein, U., K. Jurchott, W. Walther, S. Bergmann, P. M. Schlag, and H. D. Royer. 2001. Hyperthermia-induced nuclear translocation of transcription factor YB-1 leads to enhanced expression of multidrug resistance-related ABC transporters. J. Biol. Chem. 276:28562-28569. [DOI] [PubMed] [Google Scholar]
- 68.Swamynathan, S. K., B. R. Varma, K. T. Weber, and R. V. Guntaka. 2002. Targeted disruption of one allele of the Y-box protein gene, Chk-YB-1b, in DT40 cells results in major defects in cell cycle. Biochem. Biophys. Res. Commun. 296:451-457. [DOI] [PubMed] [Google Scholar]
- 69.Tafuri, S. R., M. Familari, and A. P. Wolffe. 1993. A mouse Y box protein, MSY1, is associated with paternal mRNA in spermatocytes. J. Biol. Chem. 268:12213-12220. [PubMed] [Google Scholar]
- 70.Tafuri, S. R., and A. P. Wolffe. 1990. Xenopus Y-box transcription factors: molecular cloning, functional analysis and developmental regulation. Proc. Natl. Acad. Sci. USA 87:9028-9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka, K. J., K. Matsumoto, M. Tsujimoto, and T. Nishikata. 2004. CiYB1 is a major component of storage mRNPs in ascidian oocytes: implications in translational regulation of localized mRNAs. Dev. Biol. 272:217-230. [DOI] [PubMed] [Google Scholar]
- 72.Tange, T. O., T. H. Jensen, and J. Kjems. 1996. In vitro interaction between human immunodeficiency virus type 1 Rev protein and splicing factor ASF/SF2-associated protein, p32. J. Biol. Chem. 271:10066-10072. [DOI] [PubMed] [Google Scholar]
- 73.Thieringer, H. A., P. G. Jones, and M. Inouye. 1998. Cold shock and adaptation. Bioessays 20:49-57. [DOI] [PubMed] [Google Scholar]
- 74.Wang, N., K. Yamanaka, and M. Inouye. 1999. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 181:1603-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolffe, A. P. 1994. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays 16:245-251. [DOI] [PubMed] [Google Scholar]
- 76.Wolffe, A. P., and F. Meric. 1996. Coupling transcription to translation: a novel site for the regulation of eukaryotic gene expression. Int. J. Biochem. Cell. Biol. 28:247-257. [DOI] [PubMed] [Google Scholar]
- 77.Yamanaka, K., L. Fang, and M. Inouye. 1998. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol. 27:247-255. [DOI] [PubMed] [Google Scholar]
- 78.Yu, J., N. B. Hecht, and R. M. Schultz. 2002. RNA-binding properties and translation repression in vitro by germ cell-specific MSY2 protein. Biol. Reprod. 67:1093-1098. [DOI] [PubMed] [Google Scholar]
- 79.Zheng, Y. H., H. F. Yu, and B. M. Peterlin. 2003. Human p32 protein relieves a post-transcriptional block to HIV replication in murine cells. Nat. Cell Biol. 5:611-618. [DOI] [PubMed] [Google Scholar]