Abstract
The mechanisms which determine the nuclear accumulation and inactivation of the extracellular signal-regulated kinase 1 (ERK1) or ERK2 mitogen-activated protein (MAP) kinases are poorly understood. Here we demonstrate that DUSP5, an inducible nuclear phosphatase, interacts specifically with ERK2 via a kinase interaction motif (KIM) within its amino-terminal noncatalytic domain. This binding determines the substrate specificity of DUSP5 in vivo, as it inactivates ERK2 but not Jun N-terminal protein kinase or p38 MAP kinase. Using green fluorescent protein fusions, we identify within this same domain of DUSP5 a functional nuclear localization signal (NLS) which functions independently of the KIM. Moreover, we demonstrate that the expression of DUSP5 causes both nuclear translocation and sequestration of inactive ERK2. Nuclear anchoring is ERK2 specific and requires both interactions between the DUSP5 KIM and the common docking site of ERK2 and a functional NLS within DUSP5. Finally, the expression of a catalytically inactive mutant of DUSP5 also tethers ERK2 within the nucleus. Furthermore, this nuclear ERK2 is phosphorylated by MAP kinase kinase in response to growth factors and also activates transcription factor Elk-1. We conclude that DUSP5 is an inducible nuclear ERK-specific MAP kinase phosphatase that functions as both an inactivator of and a nuclear anchor for ERK2 in mammalian cells. In addition, our data indicate that the cytoplasm may not be an exclusive site of MAP kinase activation.
The Ras/extracellular signal-regulated kinase (ERK) mitogen-activated protein (MAP) kinase signaling pathway mediates a diverse array of distinct cellular responses to extracellular stimuli (7, 34, 38). These include cell proliferation, differentiation, transformation, and survival. These responses are mediated via the ERK-dependent phosphorylation of both cytoplasmic proteins, such as p90rsk, and a number of nuclear transcription factors, including the Ets family member Elk-1 (11, 48, 49). Because of the differential localization of ERK substrates, the correct spatial-temporal regulation of MAP kinase signaling is a crucial determinant of biological outcome. Inactive MAP kinase is localized predominantly in the cytoplasm (1, 18). However, on activation, MAP kinase translocates to the nucleus, and this event is essential for growth factor-induced gene expression and cell cycle entry (3, 9, 19, 31).
One mechanism for the nuclear translocation of MAP kinase involves its phosphorylation and subsequent dimerization, but the precise nature of the transport process and its molecular mechanism are not yet clear and may be complex (1, 26). Despite evidence for the growth factor-regulated nuclear-cytoplasmic shuttling of MAP kinase, the protein lacks any obvious sequence motifs corresponding to either nuclear localization signals (NLSs) or nuclear export signals (NESs). These data suggest that transit across the nuclear membrane may be achieved through interactions with either NLS- or NES-containing partner proteins, and increasing attention has been focused on the possible roles of MAP kinase activators, regulators, and substrates in mediating these transport events.
The first evidence for such a role came with the discovery of a leucine-rich NES within the amino terminus of MAP kinase kinase (MEK) (17). Subsequent work showed that this export signal, in conjunction with an ERK2 binding site, served to anchor inactive ERK2 in the cytoplasm and that the MEK-ERK2 complex was dissociated on activation of the MAP kinase pathway (1, 18). More recently, we demonstrated that MAP kinase phosphatase 3 (MKP-3/DUSP6, an ERK-specific M)/kAP, also contains a leucine-rich NES and can play a role in the cytoplasmic retention of inactive ERK2 (24). Finally, two other proteins, PEA-15 and SEF, have been implicated as cytoplasmic anchors for ERK2 (16, 45). However, in contrast to MEK and MKP-3/DUSP6, both PEA-15 and SEF are able to retain ERK2 in the cytoplasm in the activated state, indicating that these proteins may act to restrict the activity of ERK2 to cytoplasmic targets, such as p90rsk.
In contrast to the understanding of the proteins involved in mediating the cytoplasmic localization of ERK2, relatively little is known about the factors influencing the nuclear residence of ERK2. Some insight into the mechanism by which MAP kinase accumulates in the cell nucleus came with the observation that, in addition to the requirement for phosphorylation and activation, this process required the de novo synthesis of short-lived proteins which were postulated to act as nuclear anchors (1, 30). More recently, it was shown that MAP kinase which accumulated in the nucleus on prolonged exposure to growth factors was in the inactive form (47). Furthermore, the phosphatase(s) required for the nuclear inactivation of ERK2 was vanadate sensitive, was newly synthesized in response to the activation of the Ras/MAP kinase pathway, and interacted with ERK2 through the common docking (CD) site. Based on these properties, it was proposed that the nuclear inactivation and anchoring of MAP kinase were likely to be regulated by the inducible nuclear phosphatases MKP-1/DUSP1 and MKP-2/DUSP4 and that the nucleus represents a site for ERK action, sequestration, and inactivation (36, 47).
Here we studied DUSP5 (hVH-3/B23), a nuclear MKP which is inducible by both heat shock and growth factors in mammalian cells (22, 28). In contrast to MKP-1/DUSP1, MKP-2/DUSP4, and PAC-1/DUSP2, which interact with and inactivate both mitogen- and stress-activated MAP kinase isoforms (10), we found that DUSP5 binds specifically and directly to the ERK1 and ERK2 MAP kinases via a conserved kinase interaction motif (KIM) within its amino-terminal noncatalytic domain. This binding reflects the substrate specificity of DUSP5 in vivo, as it inactivates ERK2 but not Jun N-terminal protein kinase (JNK) or p38 MAP kinase in mammalian cells. Using green fluorescent protein (GFP) fusions to study the localization of DUSP5, we identified within the amino-terminal domain a lysine- and arginine-rich motif which functions as an NLS and show that this NLS functions independently of the KIM, which is also located in this domain of the protein. Moreover, we found that the expression of DUSP5 in mammalian cells causes both nuclear translocation and sequestration of inactive ERK2. This nuclear anchoring is ERK2 specific and requires both interactions between the DUSP5 KIM and the CD motif of ERK2 and a functional NLS within DUSP5. Finally, we found that the expression of a catalytically inactive form of DUSP5 also tethers ERK2 within the nucleus. However, this nuclear ERK2 can still be activated by MEK in response to epidermal growth factor and can also phosphorylate and activate transcription factor Elk-1. We conclude that DUSP5 represents a distinct inducible nuclear ERK-specific MKP and that it functions as both a nuclear inactivator and a bona fide nuclear anchor for MAP kinase in mammalian cells and tissues. In addition, our data indicate that the cytoplasm is not an exclusive site for the activation of MAP kinase by MEK.
MATERIALS AND METHODS
Reagents.
Monoclonal antibodies to Myc (9E10) and hemagglutinin (HA) (12CA5) were obtained from Cancer Research UK. Antibodies to ERK, phospho-ERK, JNK, and p38 were purchased from Cell Signaling. The polyclonal antibody to DUSP5 was raised in sheep by using recombinant DUSP5 as an antigen. The antibody to MEK was purchased from Santa Cruz. The antibody to upstream binding factor (UBF) was kindly provided by Brian McStay (Biomedical Research Centre, University of Dundee). For immunocytochemical analysis, the antibody to MAP kinase 1 or MAP kinase 2 was obtained from Upstate, and the antibody to active MAP kinase was obtained from Promega. All chemicals were purchased from Sigma, unless indicated otherwise. All tissue culture reagents were obtained from Gibco BRL.
DNA constructs.
The human DUSP5 cDNA was amplified by PCR with Image clone 5808876 (29) as a template. The PCR product was subcloned as an NdeI-XhoI fragment into bacterial expression vector pET15b (Novagen) and Matchmaker System 3 DNA binding domain and activation domain fusion vectors pGBKT7 and pGADT7 (Clontech). The DUSP5 cDNA was ligated into mammalian expression vector pSG5 (Stratagene) in frame with the Myc tag sequence by using EcoRI and XhoI restriction sites. GFP expression constructs were generated by cloning the DUSP5 cDNA as an XhoI-KpnI fragment into vector pECFP-N1 (Clontech).
The various point mutations of DUSP5 were produced by overlap extension PCR (21) with the following complementary primers (only the forward primer is shown) (mutations are in bold type): C263S, 5′-AAGGTCCTGGTCCACAGTGAGGCTGGGATCTCCCGT-3′; D232N, 5′-CACTACAAATGGATCCCTGTGGAAAACAGCCACACG-3′; R53,54A, 5′-GTCAACCTCAACTCGGTGGTGCTGGCGGCGGCCCGGGGCGGCGCGGTGTCG-3′; R53A, 5′-GTCAACCTCAACTCGGTGGTGCTGGCGCGGGCCCGGGGCGGCGCGGTGTCG-3′; and R54A, 5′-GTCAACCTCAACTCGGTGGTGCTGCGGGCGGCCCGGGGCGGCGCGGTGTCG-3′; appropriate external flanking primers also were used. Following digestion, the resulting PCR products were cloned into pET15b and pGBKT7 as NdeI-XhoI fragments. The resulting plasmids were used as templates to amplify mutant DUSP5 cDNAs containing the appropriate flanking restriction sites for cloning into pSG5 and pECFP-N1.
C-terminal deletion mutants of DUSP5 were made by PCR with a forward primer having the sequence 5′-CCGCTCGAGATGAAGGTCACGTCGCTC-3′ (restriction site is underlined) and the following reverse primers (restriction sites are underlined): 1-61, 5′-CGGGGTACCGTCGACACCGCGCCGCCCCGGGCCCGCCGCAG-3′; 1-127, 5′-CGGGGTACCGTCCCTTTGAGGAAGTAGACCCG-3′; 1-178, 5′-CGGGGTACCGTGCCACCCTGGTCATAAGC-3′; and 1-193, 5′-CGGGGTACCGTATGGTAGGCACTTCCAAGGTA-3′. To generate the N-terminal deletion mutant of DUSP5 (residues 194 to 384), primers with the sequences 5′-GAAGATCTGCATCCAAGTGCGAGTTCCTCGCC-3′ and 5′-CCGCTCGAGTTAGCAGGATGTGGCCGTTGCCACAGGGCT-3′ (restriction sites are underlined) were used. The C-terminal deletion mutants were cloned as XhoI-KpnI fragments into pECFP-N1, and the N-terminal deletion mutant was cloned as a BglII-XhoI fragment into BglII-SalI-digested vector pECFP-C1 (Clontech).
The DUSP5 NLS mutant (12-RKMLRK-17 to 12-GAMLGA-17; mutations are in bold type) was generated by PCR with a forward primer having the sequence5′-CCGCTCGAGATGAAGGTCACGTCGCTCGACGGGCGCCAGCTGGCTAGCATGCTCGCTAGCGAGGCGGCGGCGCGCTGCG-3′ and a reverse primer having the sequence 5′-CGGGGTACCGTGCAGGATGTGGCCGTTGC-3′ (mutations are in bold type and restriction sites are underlined). The PCR product was ligated into pECFP-N1 by using XhoI and KpnI restriction sites. The resulting plasmid was used as a template in PCRs with the appropriate flanking primers to subclone the DUSP5 NLS into pET15b, pGBKT7, and pSG5.
Mouse p38α was amplified with primers having the sequences 5′-CCGCTCGAGATGTCGCAGGAGAGGCCCACGTTC-3′ and 5′-GCGCGGATCCCGGGACTCCATTTCTTCTTGGTCAAGG-3′ (restriction sites are underlined) and then subcloned as an XhoI-BamHI fragment into pDsRed2-N1 (Clontech). JNK1 was amplified with primers having the sequences 5′-CCGCTCGAGCTATGAGCAGAAGCAAGCGTG-3′ and 5′-CGCGGATCCTCACTGCTGCACCTGTGC-3′ (restriction sites are underlined). Following digestion, the JNK1 cDNA was cloned as an XhoI-BamHI fragment into pDsRed-C2 (Clontech).
All constructs were verified by DNA sequencing, and expression was verified by Western blotting (data not shown).
Yeast expression vectors encoding GAL4 DNA binding domain and activation domain fusions of MKP-1/DUSP1, ERK1, ERK2, ERK3, ERK5, JNK1, p38α, p38δ, and p38γ and mammalian expression constructs encoding HA-tagged ERK2, p38α, and JNK1 and Myc-tagged MKP-1/DUSP1 were described previously (39). Vector pRedF2C1-ERK2 is described elsewhere (24). Plasmid MLVGALELK, which encodes the C-terminal activation domain of Elk-1 fused to the GAL4 DNA binding domain, plasmid G5E4Luc, a GAL4-dependent luciferase reporter plasmid, and pAG(1-147), which encodes the GAL4 DNA binding domain alone, were described previously (39). The vector encoding GFP-PEA-15 was kindly provided by H. Chneiweiss (INSERM U114, College de France, Paris, France).
Yeast two-hybrid analysis.
GAL4 DNA binding domain and activation domain fusion plasmids were transformed into yeast strains PJ69-4A and PJ69-4α, respectively, according to the manufacturer's instructions (Clontech). Semiquantitative analysis of two-hybrid interactions was performed by using a β-galactosidase assay according to the manufacturer's instructions (Clontech).
Kinase assays.
The activity of DUSP5 toward HA-tagged ERK2, JNK1, and p38α MAP kinases was determined by cotransfection of Cos-1 cells with 0.5 μg of expression vector encoding HA-tagged MAP kinase isoforms in combination with increasing amounts of expression vector encoding either wild-type or mutant forms of DUSP5. Following transfection, cells were exposed to an appropriate activating stimulus (serum for ERK2 or anisomycin for JNK1 and p38α), followed by lysis, immunoprecipitation, and assays of kinase activities exactly as described previously (14).
Protein expression and purification.
pET15b-derived expression vectors encoding His-tagged wild-type and mutant forms of DUSP5 were transformed into Escherichia coli strain BLR(DE3) by using standard techniques. Following expression, proteins were purified by using nickel-agarose resin (Ni2+-nitrilotriacetate; Qiagen) according to the manufacturer's instructions. Protein-containing fractions were pooled and dialyzed overnight at 4°C against a buffer containing 50 mM HEPES (pH 7.5), 200 mM KCl, 200 μM dithiothreitol, and 20% glycerol.
Protein phosphatase assays.
Phosphatase activities and catalytic activation of wild-type and mutant forms of DUSP5 were assayed by using para-nitrophenyl phosphate (p-NPP) hydrolysis exactly as described previously (12).
GST pull-down assays and coimmunoprecipitation.
Glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) were blocked by incubation for 3 h in buffer A (20 mM HEPES [pH 7.6], 1 mM EDTA, 1 mM dithiothreitol, 15% [vol/vol] glycerol, 100 mM potassium acetate, 0.1% [vol/vol] Triton X-100, 0.01% [vol/vol] NP-40) containing 2 mg of bovine serum albumin (BSA)/ml. Recombinant GST or GST-ERK2 (0.5 μg) was immobilized on 25 μl of settled beads by incubation with gentle agitation at 4°C for 2 h. Beads were washed three times with buffer A containing 0.75 mg of BSA/ml and incubated with either wild-type or mutant DUSP5 (0.2 μg) with gentle agitation at 4°C for 1 h. Following incubation, beads were washed five times with buffer A containing 0.75 mg of BSA/ml, and 25 μl of sodium dodecyl sulfate (SDS) sample buffer was added. Samples were heated to 95°C for 5 min, and 20 μl of supernatant was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting with an anti-DUSP5 antibody. Coimmunoprecipitation of Myc-tagged DUSP5 and MKP-1/DUSP6 with endogenous MAP kinase was performed exactly as described previously (20).
Fluorescence microscopy.
The subcellular localizations of GFP and red fluorescent protein (RFP) fusions in Cos-1 and NIH 3T3 cells were determined exactly as described previously (24). For immunofluorescence, Cos-1 cells were fixed for 10 min at −20°C with methanol, followed by acetone permeabilization for 3 min at −20°C. Coverslips were blocked by incubation with 10% horse serum and 1% BSA in Tris-buffered saline for 1 h. Cells were incubated with primary antibodies (anti-Myc, 1/1,000; anti-ERK, 1/1,000; or anti-phospho-ERK, 1/500) overnight at 4°C. Secondary antibodies—goat anti-mouse fluorescein isothiocyanate conjugated (1/1,000; Sigma) or goat anti-rabbit Texas Red conjugated (1/1,000; Vector Laboratories, Inc.)—were left on cells for 1 h. Fluorescence was observed by using an Olympus BX-60 microscope with the appropriate excitation and emission filters. At least 200 cells from each transfection were examined and scored for the subcellular localizations of the relevant proteins. Staining patterns were entirely consistent with the representative cells photographed.
Cell fractionation.
NIH 3T3 cells in a 10-cm-diameter dish were serum starved overnight and then stimulated with 15% serum for 2 h to induce DUSP5 expression. Cytoplasmic and nuclear fractions were prepared by using NE-PER nuclear and cytoplasmic extraction reagents (Pierce). A total of 10 μg of fractionated protein lysate was separated by PAGE and analyzed by Western blotting.
Cell culture and transfection and reporter assays.
Cos-1 and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and with newborn calf serum (both from Gibco BRL), respectively, penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were transfected by using FuGENE-6 (Roche Diagnostics) according to the manufacturer's instructions. Reporter gene assays for the detection of luciferase activities were performed by using a dual-luciferase reporter assay system (Promega).
RESULTS
DUSP5 interacts specifically and directly with ERK1 and ERK2.
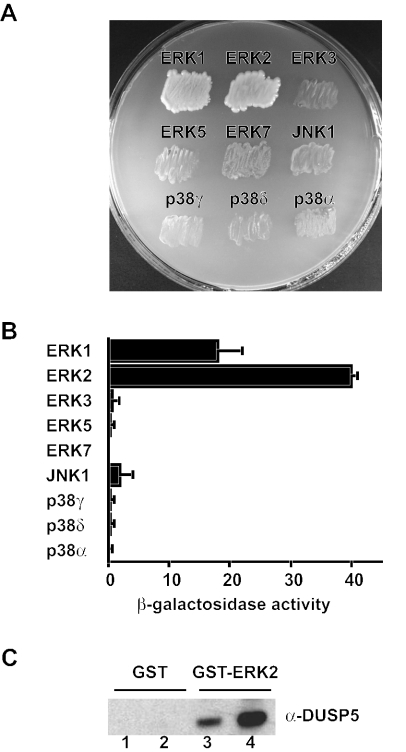
A yeast two-hybrid assay was used to determine the ability of DUSP5 to interact with a panel of nine distinct mitogen- and stress-activated MAP kinase isoforms. Binding was determined by the activation of GAL4-dependent ADE2-HIS3-lacZ reporters. Activation of HIS3 and ADE2 was assessed by growth on synthetic dropout medium deficient for histidine and adenine, respectively, while the strength of interactions was assessed by β-galactosidase assays. Full-length wild-type DUSP5 interacted specifically with the classical ERK1 and ERK2 MAP kinases (Fig. 1A and B). In contrast, only a very weak interaction was detected with JNK1, while no significant interaction was seen with ERK3, ERK5, ERK7, p38α, p38γ, or p38δ (Fig. 1A and B). Western blot analysis with GAL4-specific antibodies verified the expression of DUSP5 and each of the MAP kinase fusion proteins used in the screen (data not shown). The interaction between DUSP5 and ERK2 was direct, as we could readily detect the binding of either purified wild-type DUSP5 or a catalytically inactive mutant of DUSP5 to ERK2 in a GST pull-down assay (Fig. 1C). We conclude that unlike the closely related nuclear phosphatase MKP-1/DUSP1, which interacts with both mitogen- and stress-activated MAP kinase isoforms (39), DUSP5 is highly selective in binding to the ERK1 and ERK2 MAP kinases. This binding specificity is reminiscent of that of the cytosolic ERK-specific phosphatase MKP-3/DUSP6 (20, 32).
FIG. 1.
DUSP5 interacts specifically and directly with the ERK1 and ERK2 MAP kinases. (A) Yeast two-hybrid assays. pGBKT7.DUSP5 was transformed into PJ69-4A and mated with PJ69-4α expressing the GAL4 activation domain (AD) fusions pGADT7.ERK1, pGADT7.ERK2, pGADT7.ERK3, pGADT7.ERK5, pGADT7.ERK7, pGADT7.JNK1, pGADT7.p38γ, and pGADT7.p38δ. p38α was expressed in PJ69-2A as a GAL4 binding domain (BD) fusion (pGBKT7.p38α) and mated with pGADT7.DUSP5 that had been transformed into PJ69-4α. Yeast diploids expressing both BD and AD fusions were selected on synthetic dropout (SD) medium deficient for leucine (Leu) and tryptophan (Trp) (SD−Leu/−Trp). Leu- and Trp-positive colonies were restreaked onto SD−Leu/−Trp/−His/−Ade, and protein-protein interactions were assessed by growth on this selective medium. (B) Semiquantitative analysis of the two-hybrid interactions based on the level of induction of the β-galactosidase gene. Assays were performed in triplicate, and means are presented with associated errors. (C) To show that interactions between DUSP5 and ERK2 are direct, either GST alone (lanes 1 and 2) or GST-ERK2 (lanes 3 and 4) was mixed with purified recombinant wild-type (lanes 1 and 3) or mutant (D232N) DUSP5 (lanes 2 and 4), and protein complexes were pulled down with glutathione-agarose. Protein complexes then were analyzed by SDS-PAGE and Western blotting with anti-DUSP5 antiserum.
DUSP5 is an ERK-specific phosphatase in vivo.
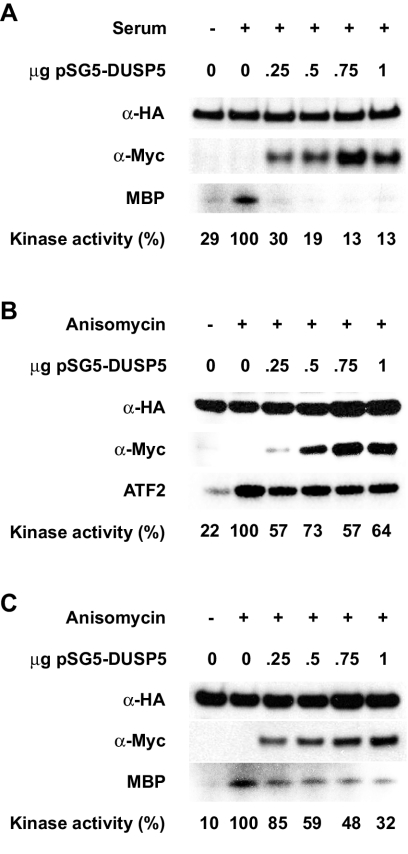
Since DUSP5 exhibits restricted abilities to interact with various MAP kinase isoforms, we set out to determine whether this property reflects the substrate selectivity of this phosphatase in vivo. Cos-1 cells were cotransfected with plasmids encoding Myc epitope-tagged wild-type DUSP5 and HA epitope-tagged ERK2, JNK1, or p38α. Transfected cells then were exposed to an appropriate stimulus to induce MAP kinase activation, either serum (ERK2) or anisomycin (JNK1 and p38α). Finally, MAP kinases were immunoprecipitated with anti-HA monoclonal antibody 12CA5 and assayed directly for kinase activity toward an appropriate substrate, either myelin basic protein (ERK2 and p38α) or ATF2 (JNK1). The expression of DUSP5 led to the complete inactivation of ERK2, even at the lowest levels (0.25 to 0.5 μg of input DNA) of DUSP5 expressed (Fig. 2A). In contrast, at equivalent expression levels, DUSP5 reduced the activities of JNK1 by approximately 30 to 40% and p38α by only 15 to 40%. Furthermore, significant levels of both JNK1 and p38α activities were detected even at the highest levels of DUSP5 expressed (Fig. 2B and C). We conclude that in contrast to other inducible nuclear MKPs, such as MKP-1/DUSP1, MKP-2/DUSP4, and PAC1/DUSP2, DUSP5 is highly selective in its ability to bind to and inactivate the ERK2 MAP kinase in vivo.
FIG. 2.
DUSP5 specifically inactivates ERK2 in vivo. (A) Cos-1 cells were transfected with HA-tagged ERK2 (0.5 μg) together with the indicated amount of plasmid encoding Myc-tagged DUSP5. Cells then were stimulated with serum before lysis, immunoprecipitation, and a kinase assay with myelin basic protein (MBP) as a substrate (bottom panel). Kinase activities were quantitated by using a PhosphorImager and are expressed here as percentages of the activity in stimulated cells in the absence of transfected DUSP5. Immunoblotting was performed with anti-HA (top panel) and anti-Myc (middle panel) antibodies to verify the expression of ERK2 and DUSP5, respectively. (B) Cos-1 cells were transfected with HA-tagged JNK1 together with the indicated amount of plasmid encoding Myc-tagged DUSP5. Cells then were stimulated with anisomycin before lysis, immunoprecipitation, and a kinase assay with ATF2 as a substrate (bottom panel). Kinase activities were quantitated by using a PhosphorImager and are expressed here as percentages of the activity in stimulated cells in the absence of transfected DUSP5. Immunoblotting was performed with anti-HA (top panel) and anti-Myc (middle panel) antibodies to verify the expression of JNK1 and DUSP5, respectively. (C) Cos-1 cells were transfected with HA-tagged p38α together with the indicated amount of plasmid encoding Myc-tagged DUSP5. Cells then were stimulated with anisomycin before lysis, immunoprecipitation, and a kinase assay with MBP as a substrate (bottom panel). Kinase activities were quantitated by using a PhosphorImager and are expressed here as percentages of the activity in stimulated cells in the absence of transfected DUSP5. Immunoblotting was performed with anti-HA (top panel) and anti-Myc (middle panel) antibodies to verify the expression of p38α and DUSP5, respectively.
Identification of a functional KIM within the amino-terminal noncatalytic domain of DUSP5.
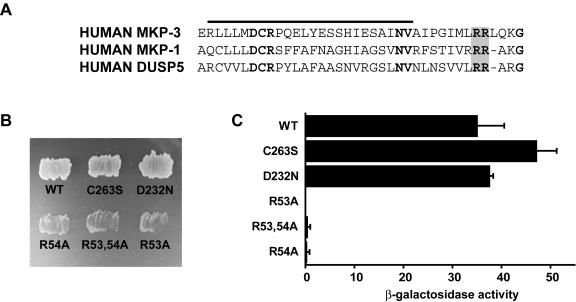
A CD site within MAP kinases is responsible for mediating interactions with activators, substrates, and regulators of these enzymes (41). The cognate MAP kinase docking site within the dual-specificity MKPs MKP-1/DUSP1, MKP-2/DUSP4, and MKP-3/DUSP6 is a cluster of positively charged amino acids located within the amino-terminal noncatalytic domain of these proteins and lying immediately adjacent to a region of sequence homology with the cell cycle regulatory phosphatase Cdc25 (8, 33, 39). Based on sequence comparisons with MKP-1/DUSP1 and MKP-3/DUSP6, we predict that arginine residues 53 and 54 may be involved in MAP kinase recognition by DUSP5 (Fig. 3A). Arginine residues 53 and 54 were replaced with alanine, either singly or in combination, and the abilities of these mutants to bind to ERK2 were compared with those of both wild-type and catalytically inactive forms of DUSP5 in yeast two-hybrid assays (Fig. 3B and C). Mutation of either or both arginine residues abolished the ability of DUSP5 to interact with ERK2. In contrast, mutation of key catalytic residues (C263S or D232N), which led to the loss of enzyme activity, had no effect on ERK2 binding.
FIG. 3.
A conserved KIM within the amino-terminal noncatalytic domain of DUSP5 mediates specific binding to ERK2. (A) Alignment of sequences within the amino-terminal noncatalytic domains of MKP-3/DUSP6, MKP-1/DUSP1, and DUSP5. Conserved residues within the Cdc25 homology domain (overlined) are shown in bold type, and conserved arginine residues within the KIM are shaded. (B) Yeast two-hybrid assays. Either wild-type (WT) pGBKT7.DUSP5 or the indicated mutants were transformed into PJ69-4A and mated with PJ69-4α expressing pGADT7.ERK2. Yeast diploids expressing both binding domain and activation domain fusions were selected on SD−Leu/−Trp (see the legend to Fig. 1 for an explanation of the medium). Leu- and Trp-positive colonies were restreaked onto SD−Leu/−Trp/−His/−Ade, and protein-protein interactions were assessed by growth on this selective medium. (C) Semiquantitative analysis of the two-hybrid interactions based on the level of induction of the β-galactosidase gene. Assays were performed in triplicate, and means are presented with associated errors.
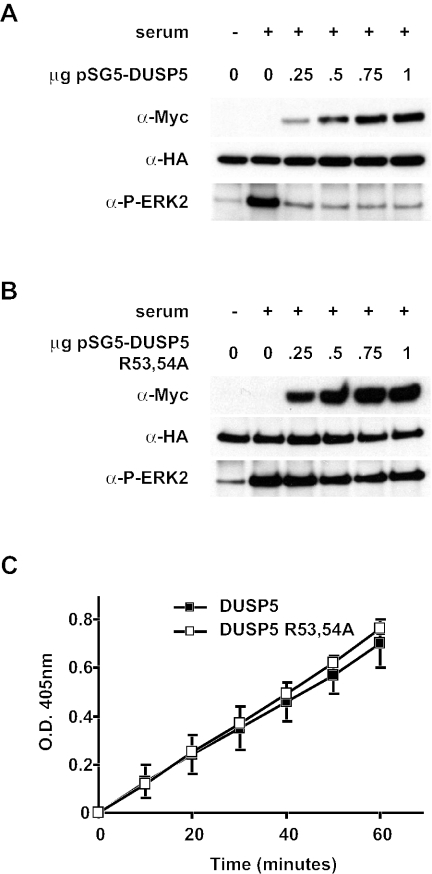
We next performed cotransfection experiments in which Myc-tagged wild-type or mutant (R53,54A) DUSP5 was expressed together with HA-tagged ERK2. Following activation of the MAP kinase by the addition of serum, the phosphorylation state of ERK2 was monitored by immunoprecipitation with anti-HA monoclonal antibody 12CA5 and Western blotting with an antibody that specifically recognizes phosphorylated ERK1 or ERK2. Wild-type DUSP5 efficiently dephosphorylated ERK2. In contrast, the mutant protein was unable to dephosphorylate the MAP kinase even when expressed at the highest levels (Fig. 4A and B). Finally, it is formally possible that the loss of function observed in both of our yeast two-hybrid assays and the cotransfection experiments was due to gross disruption of the tertiary structure of DUSP5. To investigate this possibility, we expressed and purified both wild-type and mutant (R53,54A) forms of DUSP5 in E. coli and assayed the intrinsic phosphatase activities of these proteins by hydrolysis of p-NPP in vitro. The activities of these proteins were identical, indicating that replacement of arginine residues 53 and 54 with alanine had no effect on the folding of DUSP5 (Fig. 4C). We conclude that the specific recognition and inactivation of ERK2 by DUSP5 are mediated by a conserved functional KIM located within the amino-terminal noncatalytic domain of the protein.
FIG. 4.
The KIM is essential for the dephosphorylation of ERK2 by DUSP5 in vivo. (A) Cos-1 cells were transfected with HA-tagged ERK2 (0.5 μg) together with the indicated amount of plasmid encoding Myc-tagged DUSP5. Cells then were stimulated with serum before lysis, immunoprecipitation, and detection of phosphorylated ERK2 (P-ERK2) by Western blotting (bottom panel). Immunoblotting was performed with anti-HA (middle panel) and anti-Myc (top panel) antibodies to verify the expression of ERK2 and DUSP5, respectively. (B) Cos-1 cells were transfected with HA-tagged ERK2 (0.5 μg) together with the indicated amount of plasmid encoding Myc-tagged DUSP5 R53,54A. Cells then were stimulated with serum before lysis, immunoprecipitation, and detection of phosphorylated ERK2 by Western blotting (bottom panel). Immunoblotting was performed with anti-HA (middle panel) and anti-Myc (top panel) antibodies to verify the expression of ERK2 and DUSP5, respectively. (C) Mutation of the KIM does not affect the intrinsic phosphatase activity of DUSP5. Time-dependent hydrolysis of p-NPP by either wild-type (▪) or mutant (□) DUSP5 was monitored by measuring the change in the optical density (O.D.) at 405 nm. Mean values with associated errors are presented.
Binding to ERK2 is not accompanied by the catalytic activation of DUSP5.
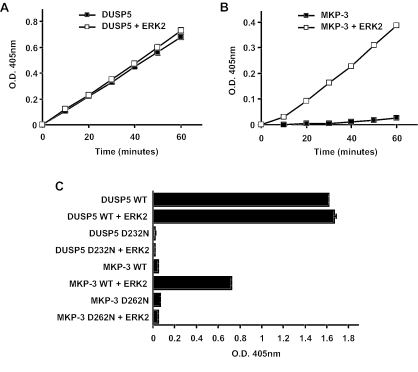
The cytosolic ERK-specific phosphatase MKP-3/DUSP6 exhibited catalytic activation on binding to ERK2, as measured by a greatly increased ability to hydrolyze p-NPP in vitro (6). Subsequent work showed that other dual-specificity phosphatases were also activated on binding to MAP kinases, including MKP-1/DUSP1, MKP-2/DUSP4, MKP-X/DUSP7, and MKP-4/DUSP9 (8, 12, 14, 39). To determine whether DUSP5 is also activated, we measured the hydrolysis of p-NPP by recombinant DUSP5 both in the absence and in the presence of recombinant ERK2 (Fig. 5A). It was clear that the activity of DUSP5 was unaffected by the addition of ERK2 to the reaction mixture. In contrast, the activity of recombinant MKP-3/DUSP6 was greatly increased in the presence of recombinant ERK2 when incubated under identical conditions (Fig. 5B). From these experiments, it was also clear that the basal activity of DUSP5 in the absence of ERK2 was much greater than that of MKP-3/DUSP6 and actually exceeded the activity of the latter enzyme when fully activated.
FIG. 5.
DUSP5 does not undergo catalytic activation on incubation with recombinant ERK2, and the conserved general acid residue is fully engaged in catalysis in the absence of substrate. (A) Time-dependent hydrolysis of p-NPP by 2.5 μg of DUSP5 either in the absence (▪) or in the presence (□) of 10 μg of ERK2 was monitored by measuring the change in the optical density (O.D.) at 405 nm. (B) Time-dependent hydrolysis of p-NPP by 2.5 μg of MKP-3 either in the absence (▪) or in the presence (□) of 10 μg of ERK2 was monitored by measuring the change in the optical density at 405 nm. (C) Mutation of the conserved general acid (Asp232) abolishes the basal phosphatase activity of DUSP5, while mutation of the corresponding residue in MKP-3 abolishes only its ERK2-dependent phosphatase activity. Recombinant DUSP5, DUSP5 D232N, MKP-3, or MKP-3 D262N (5 μg) was incubated for 1 h either in the absence or in the presence of 10 μg of ERK2. WT, wild type. Hydrolysis of p-NPP was monitored by measuring the change in the optical density at 405 nm. Assays were performed in triplicate, and means are presented with associated errors.
Structural and biochemical studies revealed that the catalytic activation of MKP-3/DUSP6 is mediated by an ERK-induced conformational change which results in the movement of a conserved general acid residue (aspartic acid residue 262) into a more favorable conformation to participate in catalysis (37, 40, 50). To determine the role of this general acid in DUSP5, the corresponding residue (aspartic acid residue 232) was mutated to asparagine, and the activity of the mutant was compared with those of both wild-type DUSP5 and the corresponding forms of MKP-3/DUSP6. Mutation of this residue abolished the basal activity of DUSP5, reducing it to a level comparable to that of MKP-3/DUSP6 in the absence of ERK2, i.e., in a conformation in which aspartic acid plays no role in catalysis (Fig. 5C). However, it did not affect the ability of DUSP5 to interact with ERK2 either in vitro or in vivo (Fig. 1C and 3C and D). In contrast, the role of the general acid in MKP-3 was manifested only in the presence of ERK2 (Fig. 5C). We conclude that the general acid residue in DUSP5 is already in an optimal conformation for catalysis in the basal (unbound) state. Therefore, in contrast to that of MKP-3/DUSP6, the substrate selectivity of DUSP5 is determined solely by specific recognition of and interaction with ERK2.
DUSP5 contains a functional NLS.
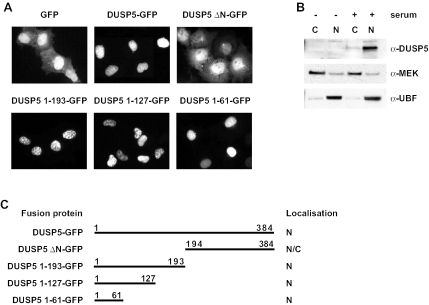
DUSP5, like MKP-1/DUSP1, MKP-2/DUSP4, and PAC-1/DUSP2, is a nuclear protein when expressed in mammalian cells (28). However, nothing is known about the mechanisms governing the subcellular distribution of nuclear MKPs or its physiological significance. To study this distribution further, we constructed an expression vector in which DUSP5 was expressed as an amino-terminal fusion with GFP. Like MKP-3/DUSP6 fused to GFP (15), DUSP5-GFP is active and readily dephosphorylates ERK2 when expressed in mammalian cells (data not shown). We then compared the localization of the fusion protein with that of GFP alone when expressed in Cos-1 cells. Unlike GFP, which was found in both the nucleus and the cytoplasm of expressing cells, DUSP5-GFP was localized exclusively in the nucleus (Fig. 6A and C). Identical results were obtained when these proteins were expressed in HeLa and NIH 3T3 cells (data not shown). To be certain that the subcellular distribution of the DUSP5-GFP fusion accurately reflected that of the endogenous protein, we performed cell fractionation studies. Endogenous DUSP5 could be readily detected in serum-stimulated NIH 3T3 cells and, like the DUSP5-GFP fusion, was found exclusively in the nuclear fraction (Fig. 6B). We conclude that the localization of the DUSP5-GFP fusion mirrors that of endogenous DUSP5, indicating that this system constitutes an appropriate model system for studying the mechanisms determining the subcellular localization of this protein.
FIG. 6.
The amino-terminal noncatalytic domain of DUSP5 is both necessary and sufficient for the nuclear localization of DUSP5. (A) Cos-1 cells were transfected with an expression vector encoding either GFP alone or the indicated DUSP5-GFP fusion, and the subcellular distribution of GFP was determined by fluorescence microscopy. Experiments were performed at least three times, and representative images are shown. (B) NIH 3T3 cells either were left untreated or were serum stimulated and then subjected to subcellular fractionation. Fractions (C, cytoplasmic fraction; N, nuclear fraction) were analyzed by Western blotting with a DUSP5-specific antibody (top panel). Antibodies to MEK (middle panel) and UBF (bottom panel) were used to verify the efficient fractionation of cytoplasmic and nuclear proteins, respectively. (C) Schematic summary of the results from panel A, indicating the localization of the GFP fusions analyzed (N, nuclear; N/C, nuclear and cytoplasmic).
To identify which domain of DUSP5 might be responsible for nuclear localization, we expressed either the amino-terminal noncatalytic domain (residues 1 to 193) or the carboxyl-terminal catalytic domain (residues 194 to 384; DUSP5 ΔN-GFP) fused to GFP. While the carboxyl-terminal domain of DUSP5 was present in both the nucleus and the cytoplasm of expressing cells, the amino-terminal domain of DUSP5 was sufficient to drive GFP into the cell nucleus (Fig. 6A and C). Furthermore, additional truncations of the amino terminus clearly showed that the first 61 amino acids of DUSP5 are sufficient for the nuclear targeting of GFP (Fig. 6A and C).
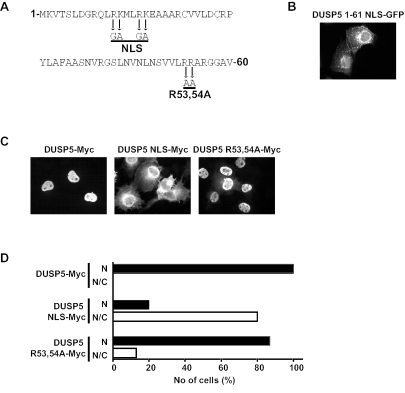
Examination of the sequence of DUSP5 indicates that the first 61 residues of the protein contain both a lysine- and arginine-rich sequence (residues 9 to 22) and the two arginine residues (residues 53 and 54) previously identified as the functional KIM (Fig. 7A). We first introduced mutations into the amino terminus of DUSP5 by replacing arginine residues 12 and 16 and lysine residues 13 and 17 with glycine and alanine, respectively (DUSP5 1-61 NLS-GFP). This mutant was distributed in both the nucleus and the cytoplasm, indicating that the lysine- and arginine-rich motif constitutes a functional NLS (Fig. 7B). Because GFP alone was found in both the nuclear and the cytoplasmic compartments in Cos-1 cells (Fig. 6A), we wanted to be certain that the localization pattern for DUSP5 was not influenced by its fusion partner. To this end, we expressed full-length DUSP5 tagged at the carboxyl terminus with a single copy of the Myc epitope tag. This protein, just like the DUSP5-GFP fusion, was found in the nucleus in 100% of expressing cells (Fig. 7C and D). Furthermore, mutation of the putative NLS within this protein (DUSP5 NLS-Myc) caused a profound change in the localization of DUSP5, with 80% of expressing cells showing staining in both the cytoplasm and the nucleus (Fig. 7C and D). Finally, mutation of the KIM (DUSP5 R53,54A-Myc) did not have a profound influence on the localization of DUSP5, with 85% of expressing cells still showing exclusively nuclear staining (Fig. 7C and D).
FIG. 7.
Identification of a functional NLS within the amino terminus of DUSP5. (A) Amino acid sequence of the first 61 residues of DUSP5. Mutations introduced into the putative arginine- and lysine-rich NLS and the conserved arginine residues of the KIM are indicated by arrows. (B) Mutation of the putative NLS within the DUSP5 1-61 NLS-GFP fusion prevents nuclear localization. (C) Myc-tagged wild-type DUSP5 is a nuclear protein, and mutation of the NLS but not the KIM prevents the nuclear accumulation of DUSP5. (D) The distribution of Myc-tagged proteins in at least 200 expressing cells for each transfection in panel C was recorded and is presented graphically as the percentage of cells in which fluorescence either was predominantly nuclear (N) or was found in both the nucleus and the cytoplasm (N/C). Experiments were performed at least three times, and representative images and cell counting results are shown.
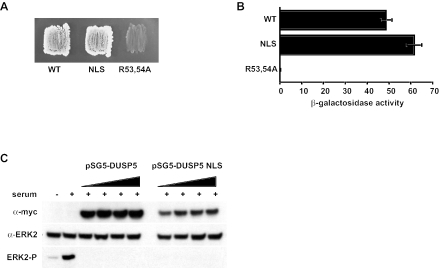
It is possible that the introduction of mutations into the putative NLS caused the misfolding of DUSP5 and that this, rather than interference with a bona fide NLS function, prevented the nuclear entry of DUSP5. To examine this possibility, we used a yeast two-hybrid assay to determine whether these mutations interfere with the ability of DUSP5 to interact with ERK2. We found that the NLS mutant bound its substrate kinase as strongly as the wild-type protein (Fig. 8A and B). In addition, the intrinsic phosphatase activity of the NLS mutant was comparable to that of the wild-type protein in vitro (data not shown). Finally, cotransfection experiments demonstrated that the NLS mutant of DUSP5 could dephosphorylate an RFP-ERK2 fusion as efficiently as the wild-type protein (Fig. 8C). We conclude that the arginine- and lysine-rich motif (residues 9 to 22 of DUSP5) constitutes a bona fide NLS and that this sequence functions independently of the KIM to drive the nuclear localization of DUSP5.
FIG. 8.
Mutation of the NLS within DUSP5 does not affect the ability of DUSP5 to either interact with or dephosphorylate ERK2. (A) Yeast two-hybrid assays. Either wild-type (WT) pGBKT7.DUSP5 or the indicated mutants were transformed into PJ69-4A and mated with PJ69-4α expressing pGADT7.ERK2. Yeast diploids expressing both binding domain and activation domain fusions were selected on SD−Leu/−Trp (see the legend to Fig. 1 for an explanation of the medium). Leu- and Trp-positive colonies were restreaked onto SD−Leu/−Trp/−His/−Ade, and protein-protein interactions were assessed by growth on this selective medium. (B) Semiquantitative analysis of the two-hybrid interactions based on the level of induction of the β-galactosidase gene. Assays were performed in triplicate, and means are presented with associated errors. (C) Cos-1 cells were transfected with ERK2 fused to RFP together with increasing amounts of a plasmid encoding either Myc-tagged wild-type DUSP5 or the NLS mutant. Cells then were stimulated with serum before lysis and detection of phosphorylated ERK2 (ERK2-P) by Western blotting (bottom panel). Immunoblotting was performed with anti-Myc (top panel) and anti-ERK2 (middle panel) antibodies to verify the expression of DUSP5 and RFP-ERK2, respectively.
DUSP5 acts as a specific nuclear anchor for ERK2.
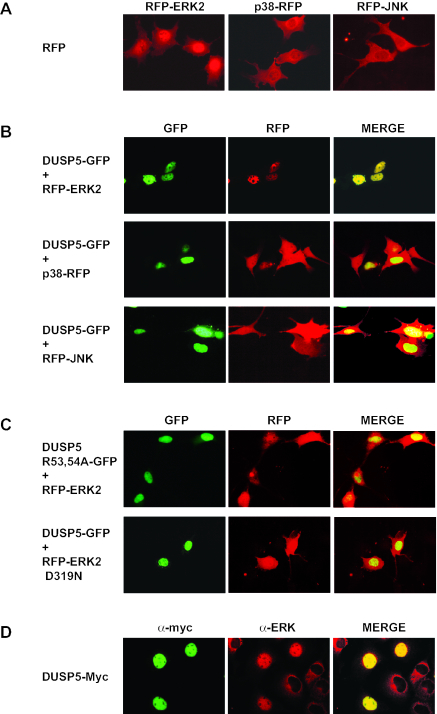
The activation of ERK2 in response to a variety of stimuli is accompanied by the nuclear translocation of the kinase; this translocation is due, at least in part, to phosphorylation of the regulatory T-X-Y motif (26, 31). Thus, the expression of ERK-specific phosphatase MKP-3/DUSP6 prevents both the phosphorylation and the nuclear translocation of ERK2 (3, 20). To study the effects of DUSP5 expression on the localization of MAP kinases, we constructed expression vectors encoding RFP fused to ERK2, p38, or JNK1. When expressed in NIH 3T3 cells, all three MAP kinases were found in both the nucleus and the cytoplasm (Fig. 9A).
FIG. 9.
Expression of DUSP5 causes the specific nuclear translocation of both RFP-ERK2 and endogenous ERK2. (A) NIH 3T3 cells were transfected with an expression plasmid encoding RFP-ERK2, p38-RFP, or RFP-JNK, and the subcellular distribution of RFP was determined by fluorescence microscopy. (B) NIH 3T3 cells were cotransfected with wild-type DUSP5-GFP and RFP-ERK2, p38-RFP, or RFP-JNK, and the subcellular distributions of both GFP and RFP were determined by fluorescence microscopy. (C) NIH 3T3 cells were cotransfected with either DUSP5 R53,54A-GFP and RFP-ERK2 or wild-type DUSP5-GFP and RFP-ERK2 D319N (sevenmaker mutant), and the subcellular distributions of both GFP and RFP were determined by fluorescence microscopy. (D) Cos-1 cells were transfected with Myc-tagged DUSP5, and the subcellular distributions of DUSP5 and endogenous ERK were monitored by immunofluorescence with anti-Myc and anti-ERK antibodies, respectively. Experiments were performed at least three times, and representative images are shown.
We next examined the effect of coexpressing DUSP5-GFP with each of these MAP kinases. The coexpression of DUSP5 had no effect on the localization of either p38 or JNK. However, to our surprise, rather than preventing the nuclear translocation of ERK2, the expression of DUSP5 caused inactive (dephosphorylated) ERK2 to be relocalized to the nucleus in every cell in which the two proteins were coexpressed (Fig. 9B). Furthermore, ERK2 translocation did not occur either when the KIM within DUSP5 was mutated or when a sevenmaker mutant of ERK2 with an amino acid substitution within the CD motif (D319N) was coexpressed with wild-type DUSP5 (Fig. 9C).
Finally, to ensure that the nuclear accumulation of RFP-ERK2 was not an artifact associated with the use of the RFP fusion partner, we expressed Myc-tagged DUSP5 in Cos-1 cells and examined the localization of endogenous ERK2. ERK2 was predominantly cytoplasmic in cells that did not express DUSP5; in contrast, endogenous ERK2 was found exclusively in the nucleus in cells expressing this phosphatase (Fig. 9D).
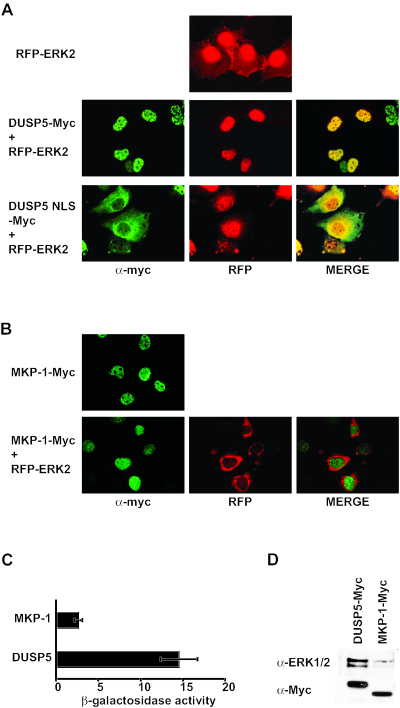
To examine the role of the NLS in the ability of DUSP5 to cause the nuclear translocation of ERK2, Myc-tagged DUSP5 (wild type or NLS mutant) was cotransfected with RFP-ERK2 into Cos-1 cells. Just as in NIH 3T3 cells, the expression of wild-type DUSP5 caused the nuclear translocation of RFP-ERK2 (Fig. 10A). However, the NLS mutant of DUSP5 failed to cause the nuclear translocation of ERK2. We conclude that a specific interaction between ERK2 and DUSP5 is required for the abilities of DUSP5 to inactivate and anchor ERK2 within the nucleus and that the nuclear translocation of DUSP5 in complex with ERK2 is driven by the functional NLS within DUSP5.
FIG. 10.
(A) The nuclear accumulation of ERK2 is driven by the functional NLS within DUSP5. Cos-1 cells were cotransfected with either Myc-tagged wild-type DUSP5 and RFP-ERK2 or the NLS mutant of DUSP5 and RFP-ERK2, and the subcellular distributions of DUSP5 and RFP were determined by immunofluorescence with anti-Myc antibodies and by fluorescence microscopy, respectively. (B) The expression of the related nuclear phosphatase MKP-1 does not cause the nuclear accumulation of ERK2. Cos-1 cells were either transfected with Myc-tagged MKP-1 or cotransfected with Myc-tagged MKP-1 and RFP-ERK2, and the subcellular distributions of MKP-1 and RFP were determined by immunofluorescence with anti-Myc antibodies and by fluorescence microscopy, respectively. (C) Yeast two-hybrid assays. Either pGBKT7.DUSP5 or pGBKT7.MKP-1 was transformed into PJ69-4A and mated with PJ69-4α expressing pGADT7.ERK2. Semiquantitative analysis of these interactions was based on the level of induction of the β-galactosidase gene. Assays were performed in triplicate, and means are presented with associated errors. (D) DUSP5 interacts with endogenous MAP kinase in Cos-1 cells. Cells were transfected with an expression vector encoding either Myc-tagged DUSP5 or MKP-1. At 24 h after transfection, cells were lysed, and phosphatases were immunoprecipitated with anti-Myc monoclonal antibody 9E10. MAP kinase was detected in these immunoprecipitates by Western blotting with an anti-MAP kinase antibody (top panel). Cell lysates also were analyzed by Western blotting with anti-Myc monoclonal antibody 9E10 to verify the expression of both DUSP5 and MKP-1 (bottom panel).
The nuclear phosphatase MKP-1/DUSP1 is unable to cause the nuclear accumulation of ERK2.
Based on the observation that new protein synthesis is required for the nuclear sequestration of inactive ERK2 and the demonstration that peptides which can compete with endogenous proteins for binding to the CD site of ERK2 prevented the nuclear anchoring of the kinase, the inducible phosphatases MKP-1/DUSP1 and MKP-2/DUSP4 were identified as likely nuclear anchors for ERK2 (30, 47). We previously demonstrated that MKP-1/DUSP1 binds to and inactivates ERK2 but, like MKP-2/DUSP4, is also capable of interacting with and inactivating both JNK and p38 MAP kinases (39). To determine whether the ability to anchor ERK2 in the nucleus is a general property of the inducible nuclear MKPs, we coexpressed Myc epitope-tagged wild-type MKP-1/DUSP1 and RFP-ERK2 in Cos-1 cells. As previously reported, MKP-1/DUSP1 was localized exclusively in the nucleus in Cos-1 cells. However, unlike that of DUSP5, the expression of MKP-1/DUSP1 had no effect on the localization of ERK2 in these cells (Fig. 10B). One possible explanation for this result is that despite the ability of MKP-1/DUSp1 to recognize and dephosphorylate ERK2, the strength of the interaction between the two proteins might be insufficient for nuclear anchoring. To examine this possibility, we used yeast two-hybrid assays to determine the relative strengths of binding of MKP-1/DUSP1 and DUSP5 to ERK2 (Fig. 10C). This analysis clearly showed that the interaction between DUSP5 and ERK2 was significantly stronger (≈5- to 6-fold) than that between MKP-1/DUSP1 and ERK2. Finally, we expressed Myc-tagged DUSP5 or MKP-1/DUSP1 in Cos-1 cells and, following immunoprecipitation, analyzed these phosphatases by Western blotting with an anti-ERK1 or -ERK2 antibody. In agreement with the data from the yeast two-hybrid assays, we readily detected endogenous MAP kinases in DUSP5 immunoprecipitates, while these proteins were present at much lower levels in immunoprecipitates from cells transfected with Myc-tagged MKP-1/DUSP1 (Fig. 10D). We conclude that DUSP5 is highly specific in its ability to sequester ERK2 but not JNK1 or p38α in the cell nucleus and that it may be unique among the inducible dual-specificity MKPs in performing this function.
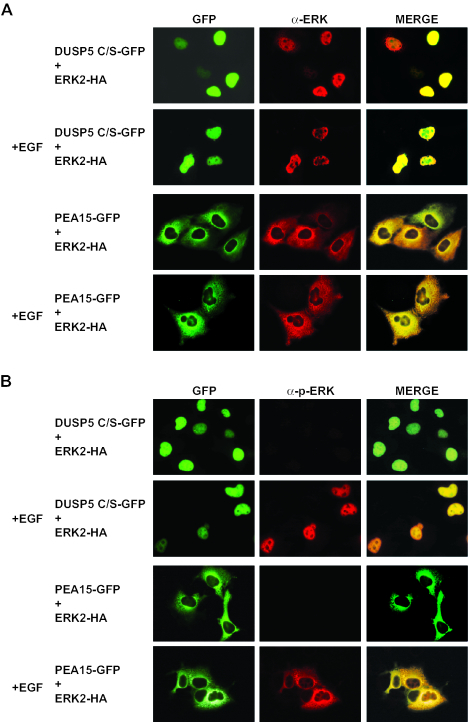
An inactive mutant of DUSP5 can sequester activated, functional ERK2 in the nucleus.
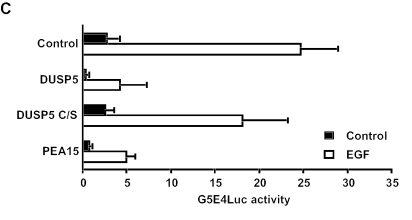
A catalytically inactive mutant of the dual-specificity phosphatase MKP-3/DUSP6 can anchor ERK2 in the cytoplasm (3, 24). To examine the effects of expressing a catatytically inactive mutant of DUSP5 on ERK2 localization and activity, we coexpressed RFP-ERK2 with a fusion of GFP to DUSP5 in which the essential catalytic cysteine residue was mutated to a serine residue (C263S). Like the wild-type protein, the mutant protein also caused the complete relocalization of ERK2 to the nucleus (Fig. 11A). However, in contrast to what was seen in cells expressing the wild-type form of DUSP5, this nuclear ERK2 was readily phosphorylated in response to epidermal growth factor (Fig. 11B). As a control, we expressed the PEA-15 protein, which acts as a cytoplasmic anchor for ERK2 (16). In contrast to DUSP5 C263S, PEA-15 restricted both ERK2 and phospho-ERK2 to the cytoplasm (Fig. 11A and B). To determine whether ERK2 tethered in the nucleus by DUSP5 C263S was functional, we used a nuclear GAL4-Elk-1 fusion protein, the ERK2-dependent phosphorylation and activation of which can be assayed by using a GAL4-dependent luciferase reporter. As expected, wild-type DUSP5, which potently inactivates ERK2, readily blocked GAL4-Elk-1 transcriptional activation in response to the treatment of cells with epidermal growth factor. In addition, PEA-15, which restricts the nuclear entry of activated ERK2, also blocked this response. In contrast, DUSP5 C263S did not prevent epidermal growth factor-stimulated Elk-1-dependent transcription. We conclude that ERK2 sequestered in the cell nucleus by this inactive form of DUSP5 has ready access to both its upstream activator MEK and downstream nuclear substrates exemplified by transcription factor Elk-1.
FIG. 11.
(A and B) A catalytically inactive (active-site Cys-to-Ser) mutant of DUSP5 causes the nuclear translocation and accumulation of HA-tagged ERK2 in Cos-1 cells, and this nuclear MAP kinase is phosphorylated in response to epidermal growth factor (EGF). Cos-1 cells were cotransfected with a catalytically inactive mutant of DUSP5 (DUSP5 C/S-GFP) and HA-tagged ERK2. At 24 h after transfection, cells either were left untreated or were stimulated with EGF (0.1 μg/ml) for 15 min. As a control, the ERK2 cytoplasmic anchoring protein GFP-PEA-15 was cotransfected with HA-tagged ERK2. The subcellular distributions of unphosphorylated ERK2 and GFP (A) and of phosphorylated ERK2 and GFP (B) were determined by immunofluorescence with anti-ERK2 and anti-phosphorylated ERK2 antibodies and by fluorescence microscopy, respectively. (C) Endogenous ERK2, retained in the nucleus by DUSP5, can phosphorylate Elk-1 and activate an Elk-1-dependent transcriptional reporter in response to EGF. Cos-1 cells were cotransfected with empty vector (control), wild-type DUSP5, catalytically inactive DUSP5 C/S, or PEA-15 together with the GAL4-dependent reporter plasmid G5E4Luc and plasmid MLVGALELK, which encodes the GAL4 DNA binding domain fused to Elk-1. pRL-TK, which encodes Renilla luciferase, was included in each transfection as a control. At 24 h after transfection, cells either were left untreated or were stimulated for 4 h with EGF (0.1 μg/ml). GAL4-dependent luciferase activity was monitored by using a Luminometer, and Elk-1-mediated expression levels were determined by correcting for transfection efficiency with the Renilla control. Experiments were performed at least three times, and the results of a representative experiment are shown. Luciferase assays were performed in triplicate, and means are presented with associated errors.
DISCUSSION
The inducible nuclear MKPs comprise a subfamily of four related enzymes, MKP-1/DUSP1, MKP-2/DUSP4, PAC-1/DUSP2, and DUSP5. All are encoded by immediate-early genes, which are also inducible by various forms of cellular stress (5, 25, 43). Of these enzymes, MKP-1/DUSP1, MKP-2/DUSP4, and PAC-1/DUSP2 are characterized by the abilities to interact with and inactivate all three major classes of MAP kinases in mammalian cells, showing activities toward ERK1, ERK2, JNK, and p38 (8, 10, 39). This apparent promiscuity has led to the suggestion that their specificity in vivo is dictated mainly by access to substrate (43). In contrast, relatively little is known about DUSP5. This MKP was originally characterized as a serum- and heat shock-inducible phosphatase which could inactivate ERK2 in vitro (22, 28). More recently, DUSP5 was found to be inducible by interleukin 2 in T cells and to be a direct transcriptional target of the p53 tumor suppressor (27, 46). However, the substrate selectivity, mechanism of catalysis, and significance of the nuclear localization of DUSP5 remain unexplored.
Here we demonstrate that, in contrast to the other members of this MKP subfamily, DUSP5 is an ERK-specific phosphatase in vivo. DUSP5 can thus be regarded as a nuclear equivalent of the cytoplasmic ERK-specific phosphatase MKP-3/DUSP6. Like MKP-3/DUSP6, DUSP5 specifically recognizes and binds to ERK2 via a conserved KIM located within the amino-terminal noncatalytic domain of the protein. However, unlike the situation for MKP-3/DUSP6, binding to ERK2 does not increase the catalytic activity of DUSP5 in vitro. Furthermore, mutation of the conserved general acid residue in DUSP5 (D232N) demonstrates that, even in the absence of bound substrate, this residue is already in an optimal conformation for catalysis. Previous work had indicated that specific binding to MAP kinase coupled with catalytic activation was a general mechanism underlying the substrate selectivity of mammalian MKPs (6, 14, 39). However, possible indications of discordance between in vivo activity and catalytic activation were reported previously. For instance, MKP-5/DUSP10, which selectively inactivates stress-activated MAP kinases, does not appear to be activated on incubation with either p38 or JNK in vitro (42), and the nuclear phosphatase MKP-2/DUSP4 interacts strongly with p38 MAP kinase, but its activity is not increased on incubation with this MAP kinase (8). Our results clearly indicate that for DUSP5, the recognition and inactivation of ERK2 require only a specific protein-protein interaction with its MAP kinase substrate and that this binding occurs without any concomitant change in the conformation of catalytic residues. Why certain phosphatases, such as MKP-3/DUSP6, undergo catalytic activation, whereas others do not, is unclear, but our experiments provide a mechanistic explanation for the lack of any such requirement for DUSP5.
The molecular mechanism underlying the nuclear localization of MKPs and its physiological significance have not yet been addressed. We have demonstrated that an arginine- and lysine-rich sequence located within the amino-terminal noncatalytic domain of DUSP5 constitutes a bona fide NLS. In addition, we have shown that this NLS functions independently of the conserved KIM, which is also located in this domain of DUSP5. We recently reported the identification of a functional leucine-rich NES within the corresponding domain of the cytoplasmic ERK-specific phosphatase MKP-3/DUSP6, which mediates the CRM1-dependent nuclear export of this protein (24). Our results serve to emphasize the importance of the amino-terminal noncatalytic domain of MKPs as a regulatory region which is responsible for determining both the substrate selectivity and the subcellular localization of these key regulators of MAP kinase signaling.
The existence of nuclear anchors for MAP kinase was inferred from the observations that inhibitors of protein synthesis can both prevent the nuclear accumulation of ERK2 and accelerate the nuclear efflux of MAP kinase. Furthermore, the inhibition of targeted proteolysis by the proteasome both potentiated the nuclear retention of MAP kinase in response to nonmitogenic agonists and markedly prolonged the nuclear localization of MAP kinase after mitogenic stimulation. Taken together, these observations suggest that the de novo synthesis of short-lived proteins plays a key role in the nuclear anchoring and retention of MAP kinase (30). Subsequent work further defined these putative anchors by showing that the MAP kinase which accumulated in the nucleus was in the inactive form and that inactivation was mediated by a vanadate-sensitive (tyrosine-specific or dual-specificity) protein phosphatase. Furthermore, this enzyme(s) was newly synthesized in response to MAP kinase activation and interacted with MAP kinase via the CD site (47). Based on these properties, the inducible nuclear phosphatases MKP-1/DUSP1 and MKP-2/DUSP6 were suggested to both inactivate and anchor ERK2 in the nucleus (36, 47). However, although MKP-1/DUSP1 and MKP-2/DUSP4 are possible candidates, both proteins also interact with and inactivate the stress-activated MAP kinases JNK and p38 (8, 10, 39), and it is difficult to envisage how they might fulfill a specific role with respect to ERK2. Moreover, the inactivation of the MKP-1/DUSP1 gene does not modify the time course of ERK1 or ERK2 MAP kinase activation (13).
The specificity of DUSP5 for the inactivation of ERK1 or ERK2 coupled with its nuclear localization prompted us to examine the effects of DUSP5 on the activity and localization of its substrate. To our surprise, the expression of DUSP5 resulted in both the specific inactivation and the nuclear translocation of ERK2. Furthermore, the nuclear translocation of ERK2 was dependent on both the KIM within DUSP5 and the conserved CD site of ERK2, while the nuclear import of the DUSP5-ERK2 complex required the functional NLS within DUSP5. DUSP5 therefore fulfills several of the key criteria for a specific nuclear anchor for ERK2. (i) It is synthesized in response to growth factors. (ii) It is a nuclear protein. (iii) It interacts with ERK2 via a specific docking domain. (iv) It is a vanadate-sensitive phosphatase. Our observation that the expression of DUSP5 causes the specific nuclear translocation of ERK2 while the expression of MKP-1/DUSP1 does not strongly suggests that in addition to its role in the regulated dephosphorylation and inactivation of ERK2, DUSP5 acts as an anchor to retain inactive ERK2 in the cell nucleus.
The prevailing model for the spatial-temporal regulation of ERK2 MAP kinase is based on the ability of MEK to anchor ERK2 in the cytoplasm in resting cells. On activation, this complex is thought to dissociate, with ERK2 now free to enter the nucleus, where its activity can be expressed toward nuclear substrates (1). Once the phosphorylation of nuclear targets has occurred, the nucleus is proposed to be the critical site for mitogenic signal termination through a combination of dephosphorylation of ERK2 and sequestration of ERK2 away from MEK, its cytoplasmic activator (1, 35, 47). Our data strongly suggest that DUSP5 may fulfill the latter function. Although this model is supported by many experimental observations, it is still unclear whether MEK activity is absolutely restricted to the cytoplasm. The fact that MEK contains an NES suggests that it must reside within the nuclear compartment at some point. Furthermore, experiments in which the NES within MEK was disrupted by mutation revealed that MEK is translocated into the nucleus either in response to growth factor stimulation or during G2-M progression (23, 44).
To examine the ability of MEK to function within the nuclear compartment, we expressed an inactive mutant of DUSP5. Like wild-type DUSP5, this protein acts to retain inactive ERK2 in the nucleus. However, on stimulation by epidermal growth factor, this nuclear ERK2 is readily phosphorylated, as visualized by immunofluorescence with anti-phosphorylated ERK2 antibodies. In addition, nuclear ERK2 tethered by DUSP5 can phosphorylate and activate a nuclear target in the form of transcription factor Elk-1. Overall, our results demonstrate that activated MEK can phosphorylate and activate ERK2, even when it is located in the cell nucleus. In addition, despite the occupancy of the CD site of ERK2 by DUSP5, ERK2 can readily access a downstream substrate such as Elk-1. This latter finding is surprising, as it had been assumed that prior occupancy of the docking site by a protein such as DUSP5 would block the access of ERK2 to both MEK and substrates such as Elk-1 (41). This view was reinforced by biochemical experiments in which MEK, MKPs, and transcription factor Elk-1 were shown to compete for binding to the same region of ERK2 in vitro (2). However, one paradox has always been that an ERK2 CD site mutant (sevenmaker mutant; D319N) exhibits a gain of function (4) and, while this mutation severely affects interactions between ERK2 and MKPs such as DUSP5, the MAP kinase itself is still activated in response to physiological stimuli and is clearly able to express its activity toward downstream substrates. Our results strongly suggest either that additional protein-protein interactions can mediate both the docking of activated MEK and the recognition of Elk-1 by ERK2 or that a dynamic exchange of both regulatory and substrate proteins bound to ERK2 facilitates multiple interactions through the CD site.
In conclusion, we have identified DUSP5 as an inducible nuclear ERK-specific MKP in mammalian cells. Furthermore, we have determined the molecular mechanisms which underlie its substrate specificity, catalytic mechanisms, and nuclear localization. In addition, we have provided compelling evidence that DUSP5 performs a dual role in inactivating ERK2 and in acting as a nuclear anchor for this kinase. Finally, the use of a catalytically inactive mutant to anchor ERK2 in the nucleus reveals that MEK function is not restricted to the cytoplasmic compartment and that the occupancy of the ERK2 CD site by DUSP5 is no barrier to an interaction with either activated MEK or transcription factor Elk-1.
Acknowledgments
We acknowledge Iain Goldsmith for oligonucleotide synthesis and Andy Cassidy for DNA sequencing. We thank Brian McStay for providing the anti-UBF antibody, H. Chneiweiss for providing the expression vector encoding GFP-PEA-15, Mark Didmon for assistance in performing the GST pull-down assays, and Robin J. Dickinson for assistance with bioinformatics.
This work was supported by Cancer Research UK.
REFERENCES
- 1.Adachi, M., M. Fukuda, and E. Nishida. 1999. Two co-existing mechanisms for nuclear import of MAP kinase: passive diffusion of a monomer and active transport of a dimer. EMBO J. 18:5347-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, A. J., M. Abdollahi, and L. Bardwell. 2003. Docking sites on mitogen-activated protein kinase (MAPK) kinases, MAPK phosphatases and the Elk-1 transcription factor compete for MAPK binding and are crucial for enzymic activity. Biochem. J. 370:1077-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunet, A., D. Roux, P. Lenormand, S. Dowd, S. Keyse, and J. Pouyssegur. 1999. Nuclear translocation of p42/p44 mitogen-activated protein kinase is required for growth factor-induced gene expression and cell cycle entry. EMBO J. 18:664-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunner, D., N. Oellers, J. Szabad, W. H. Biggs III, S. L. Zipursky, and E. Hafen. 1994. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell 76:875-888. [DOI] [PubMed] [Google Scholar]
- 5.Camps, M., A. Nichols, and S. Arkinstall. 2000. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 14:6-16. [PubMed] [Google Scholar]
- 6.Camps, M., A. Nichols, C. Gillieron, B. Antonsson, M. Muda, C. Chabert, U. Boschert, and S. Arkinstall. 1998. Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science 280:1262-1265. [DOI] [PubMed] [Google Scholar]
- 7.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P., D. Hutter, X. Yang, M. Gorospe, R. J. Davis, and Y. Liu. 2001. Discordance between the binding affinity of mitogen-activated protein kinase subfamily members for MAP kinase phosphatase-2 and their ability to activate the phosphatase catalytically. J. Biol. Chem. 276:29440-29449. [DOI] [PubMed] [Google Scholar]
- 9.Chen, R. H., C. Sarnecki, and J. Blenis. 1992. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol. Cell. Biol. 12:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu, Y., P. A. Solski, R. Khosravi-Far, C. J. Der, and K. Kelly. 1996. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J. Biol. Chem. 271:6497-6501. [DOI] [PubMed] [Google Scholar]
- 11.Davis, R. J. 1993. The mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 268:14553-14556. [PubMed] [Google Scholar]
- 12.Dickinson, R. J., D. J. Williams, D. N. Slack, J. Williamson, O. M. Seternes, and S. M. Keyse. 2002. Characterization of a murine gene encoding a developmentally regulated cytoplasmic dual-specificity mitogen-activated protein kinase phosphatase. Biochem. J. 364:145-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorfman, K., D. Carrasco, M. Gruda, C. Ryan, S. A. Lira, and R. Bravo. 1996. Disruption of the erp/mkp-1 gene does not affect mouse development: normal MAP kinase activity in ERP/MKP-1-deficient fibroblasts. Oncogene 13:925-931. [PubMed] [Google Scholar]
- 14.Dowd, S., A. A. Sneddon, and S. M. Keyse. 1998. Isolation of the human genes encoding the Pyst1 and Pyst2 phosphatases: characterisation of Pyst2 as a cytosolic dual-specificity MAP kinase phosphatase and its catalytic activation by both MAP and SAP kinases. J. Cell Sci. 111:3389-3399. [DOI] [PubMed] [Google Scholar]
- 15.Eblaghie, M. C., J. S. Lunn, R. J. Dickinson, A. E. Munsterberg, J. J. Sanz-Ezquerro, E. R. Farrell, J. Mathers, S. M. Keyse, K. Storey, and C. Tickle. 2003. Negative feedback regulation of FGF signaling levels by Pyst1/MKP3 in chick embryos. Curr. Biol. 13:1009-1018. [DOI] [PubMed] [Google Scholar]
- 16.Formstecher, E., J. W. Ramos, M. Fauquet, D. A. Calderwood, J. C. Hsieh, B. Canton, X. T. Nguyen, J. V. Barnier, J. Camonis, M. H. Ginsberg, and H. Chneiweiss. 2001. PEA-15 mediates cytoplasmic sequestration of ERK MAP kinase. Dev. Cell 1:239-250. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda, M., I. Gotoh, Y. Gotoh, and E. Nishida. 1996. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem. 271:20024-20028. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda, M., Y. Gotoh, and E. Nishida. 1997. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 16:1901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, F. A., A. Seth, D. L. Raden, D. S. Bowman, F. S. Fay, and R. J. Davis. 1993. Serum-induced translocation of mitogen-activated protein kinase to the cell surface ruffling membrane and the nucleus. J. Cell Biol. 122:1089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groom, L. A., A. A. Sneddon, D. R. Alessi, S. Dowd, and S. M. Keyse. 1996. Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 15:3621-3632. [PMC free article] [PubMed] [Google Scholar]
- 21.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 22.Ishibashi, T., D. P. Bottaro, P. Michieli, C. A. Kelley, and S. A. Aaronson. 1994. A novel dual specificity phosphatase induced by serum stimulation and heat shock. J. Biol. Chem. 269:29897-29902. [PubMed] [Google Scholar]
- 23.Jaaro, H., H. Rubinfeld, T. Hanoch, and R. Seger. 1997. Nuclear translocation of mitogen-activated protein kinase kinase (MEK1) in response to mitogenic stimulation. Proc. Natl. Acad. Sci. USA 94:3742-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson, M., J. Mathers, R. J. Dickinson, M. Mandl, and S. M. Keyse. 2004. Both nuclear-cytoplasmic shuttling of the dual specificity phosphatase MKP-3 and its ability to anchor MAP kinase in the cytoplasm are mediated by a conserved nuclear export signal. J. Biol. Chem. 279:41882-41891. [DOI] [PubMed] [Google Scholar]
- 25.Keyse, S. M. 2000. Protein phosphatases and the regulation of mitogen-activated protein kinase signalling. Curr. Opin. Cell Biol. 12:186-192. [DOI] [PubMed] [Google Scholar]
- 26.Khokhlatchev, A. V., B. Canagarajah, J. Wilsbacher, M. Robinson, M. Atkinson, E. Goldsmith, and M. H. Cobb. 1998. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 93:605-615. [DOI] [PubMed] [Google Scholar]
- 27.Kovanen, P. E., A. Rosenwald, J. Fu, E. M. Hurt, L. T. Lam, J. M. Giltnane, G. Wright, L. M. Staudt, and W. J. Leonard. 2003. Analysis of gamma c-family cytokine target genes. Identification of dual-specificity phosphatase 5 (DUSP5) as a regulator of mitogen-activated protein kinase activity in interleukin-2 signaling. J. Biol. Chem. 278:5205-5213. [DOI] [PubMed] [Google Scholar]
- 28.Kwak, S. P., and J. E. Dixon. 1995. Multiple dual specificity protein tyrosine phosphatases are expressed and regulated differentially in liver cell lines. J. Biol. Chem. 270:1156-1160. [DOI] [PubMed] [Google Scholar]
- 29.Lennon, G., C. Auffray, M. Polymeropoulos, and M. B. Soares. 1996. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics 33:151-152. [DOI] [PubMed] [Google Scholar]
- 30.Lenormand, P., J. M. Brondello, A. Brunet, and J. Pouyssegur. 1998. Growth factor-induced p42/p44 MAPK nuclear translocation and retention requires both MAPK activation and neosynthesis of nuclear anchoring proteins. J. Cell Biol. 142:625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenormand, P., C. Sardet, G. Pages, G. L'Allemain, A. Brunet, and J. Pouyssegur. 1993. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J. Cell Biol. 122:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muda, M., A. Theodosiou, N. Rodrigues, U. Boschert, M. Camps, C. Gillieron, K. Davies, A. Ashworth, and S. Arkinstall. 1996. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J. Biol. Chem. 271:27205-27208. [DOI] [PubMed] [Google Scholar]
- 33.Nichols, A., M. Camps, C. Gillieron, C. Chabert, A. Brunet, J. Wilsbacher, M. Cobb, J. Pouyssegur, J. P. Shaw, and S. Arkinstall. 2000. Substrate recognition domains within extracellular signal-regulated kinase mediate binding and catalytic activation of mitogen-activated protein kinase phosphatase-3. J. Biol. Chem. 275:24613-24621. [DOI] [PubMed] [Google Scholar]
- 34.Nishida, E., and Y. Gotoh. 1993. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem. Sci. 18:128-131. [DOI] [PubMed] [Google Scholar]
- 35.Pouyssegur, J., and P. Lenormand. 2003. Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Eur. J. Biochem. 270:3291-3299. [DOI] [PubMed] [Google Scholar]
- 36.Pouyssegur, J., V. Volmat, and P. Lenormand. 2002. Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Biochem. Pharmacol. 64:755-763. [DOI] [PubMed] [Google Scholar]
- 37.Rigas, J. D., R. H. Hoff, A. E. Rice, A. C. Hengge, and J. M. Denu. 2001. Transition state analysis and requirement of Asp-262 general acid/base catalyst for full activation of dual-specificity phosphatase MKP3 by extracellular regulated kinase. Biochemistry 40:4398-4406. [DOI] [PubMed] [Google Scholar]
- 38.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 39.Slack, D. N., O. M. Seternes, M. Gabrielsen, and S. M. Keyse. 2001. Distinct binding determinants for ERK2/p38alpha and JNK map kinases mediate catalytic activation and substrate selectivity of map kinase phosphatase-1. J. Biol. Chem. 276:16491-16500. [DOI] [PubMed] [Google Scholar]
- 40.Stewart, A. E., S. Dowd, S. M. Keyse, and N. Q. McDonald. 1999. Crystal structure of the MAPK phosphatase Pyst1 catalytic domain and implications for regulated activation. Nat. Struct. Biol. 6:174-181. [DOI] [PubMed] [Google Scholar]
- 41.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2:110-116. [DOI] [PubMed] [Google Scholar]
- 42.Tanoue, T., T. Moriguchi, and E. Nishida. 1999. Molecular cloning and characterization of a novel dual specificity phosphatase, MKP-5. J. Biol. Chem. 274:19949-19956. [DOI] [PubMed] [Google Scholar]
- 43.Theodosiou, A., and A. Ashworth. 26June2002. posting date. MAP kinase phosphatases. Genome Biol. 3:reviews1-3009.10. [Online.] http://genomebiology.com/2002/3/7/reviews/3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolwinski, N. S., P. S. Shapiro, S. Goueli, and N. G. Ahn. 1999. Nuclear localization of mitogen-activated protein kinase kinase 1 (MKK1) is promoted by serum stimulation and G2-M progression. Requirement for phosphorylation at the activation lip and signaling downstream of MKK. J. Biol. Chem. 274:6168-6174. [DOI] [PubMed] [Google Scholar]
- 45.Torii, S., M. Kusakabe, T. Yamamoto, M. Maekawa, and E. Nishida. 2004. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev. Cell 7:33-44. [DOI] [PubMed] [Google Scholar]
- 46.Ueda, K., H. Arakawa, and Y. Nakamura. 2003. Dual-specificity phosphatase 5 (DUSP5) as a direct transcriptional target of tumor suppressor p53. Oncogene 22:5586-5591. [DOI] [PubMed] [Google Scholar]
- 47.Volmat, V., M. Camps, S. Arkinstall, J. Pouyssegur, and P. Lenormand. 2001. The nucleus, a site for signal termination by sequestration and inactivation of p42/p44 MAP kinases. J. Cell Sci. 114:3433-3443. [DOI] [PubMed] [Google Scholar]
- 48.Wasylyk, B., J. Hagman, and A. Gutierrez-Hartmann. 1998. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sci. 23:213-216. [DOI] [PubMed] [Google Scholar]
- 49.Yang, S. H., A. D. Sharrocks, and A. J. Whitmarsh. 2003. Transcriptional regulation by the MAP kinase signaling cascades. Gene 320:3-21. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, B., and Z. Y. Zhang. 1999. Mechanism of mitogen-activated protein kinase phosphatase-3 activation by ERK2. J. Biol. Chem. 274:35526-35534. [DOI] [PubMed] [Google Scholar]