Abstract
The differentiation potential of stem cells is determined by the ability of these cells to establish and maintain developmentally regulated gene expression programs that are specific to different lineages. Although transcriptionally potentiated epigenetic states of genes have been described for haematopoietic progenitors, the developmental stage at which the formation of lineage-specific gene expression domains is initiated remains unclear. In this study, we show that an intergenic cis-acting element in the mouse λ5-VpreB1 locus is marked by histone H3 acetylation and histone H3 lysine 4 methylation at a discrete site in embryonic stem (ES) cells. The epigenetic modifications spread from this site toward the VpreB1 and λ5 genes at later stages of B-cell development, and a large, active chromatin domain is established in pre-B cells when the genes are fully expressed. In early B-cell progenitors, the binding of haematopoietic factor PU.1 coincides with the expansion of the marked region, and the region becomes a center for the recruitment of general transcription factors and RNA polymerase II. In pre-B cells, E2A also binds to the locus, and general transcription factors are distributed across the active domain, including the gene promoters and the intergenic region. These results suggest that localized epigenetic marking is important for establishing the transcriptional competence of the λ5 and VpreB1 genes as early as the pluripotent ES cell stage.
Mammalian development is a spatially and temporally regulated process that depends on the differentiation potential of individual cells. This potential is determined by the ability of the cells to establish gene expression programs that are specific to different cell lineages. The patterns of expression of individual genes are regulated by the interaction of transcription factors with a set of cis-acting sequences that combine to form a genetic functional unit (reviewed in reference 12). Modification of the amino-terminal regions of the core histones has also been proposed to play an essential role in gene regulation by establishing domains of active or silent chromatin that would mediate the cellular memory of gene expression states (4, 24, 44, 70). The histone modifications identified to date include acetylation, methylation, ubiquitination, and ADP ribosylation (reviewed in references 28 and 80). Of these, histone H3 and H4 acetylation and histone H3 lysine 4 (K4) methylation have been shown to be associated with the transcriptional activation of genes (69). Although patterns of histone modifications have been analyzed for several gene loci, the relationship between the organization of genetic functional units and domains of histone modifications is still not well understood.
Stem cells have the ability to differentiate into multiple cell types as well as to self-renew and maintain their multipotential capacity. In mammals, a specific type of stem cell, the embryonic stem (ES) cell, has been shown to be pluripotent (i.e., able to generate all somatic lineages) (8, 64). ES cells are found in the inner cell mass at the blastocyst stage of development and retain their differentiation potential when grown in culture under appropriate conditions. The pluripotency of ES cells is of immense biological and clinical interest, but little is known about the basis for this phenomenon. Multipotent stem cells are partially committed cells with a relatively broad differentiation capacity, but they can only give rise to a subset of differentiated cell types. An example is the haematopoietic stem cell, which can differentiate into all of the cells of the myeloid and lymphoid lineages but does not normally develop into other cell types (for a review, see reference 16).
Low-level transcription of a number of lineage-specific genes has been observed in haematopoietic progenitors, leading to the suggestion that genes that are destined for activation are already in a transcriptionally permissive configuration (26, 50, 85). Prior to high-level transcription of β-globin genes in erythroid cells, histone H3 acetylation and histone H3 K4 methylation are present at the gene promoters in multipotent haematopoietic progenitors (6). It has also been shown that transcription factor complexes are already partially assembled on regulatory elements of macrophage- and neuron-specific genes in progenitor cells (39, 42, 62, 76, 77). Transcription from cis-acting regulatory elements (intergenic transcription) has also been proposed to be important for the generation of potentiated epigenetic states in developmentally regulated chromatin domains (19, 48, 63, 65). The presence of epigenetic modifications in multipotent progenitor cells that specify the activation of genes at later stages suggests that the existence of these marks could be fundamental to the ability of stem cells to activate multiple lineage-specific gene expression programs. If this suggestion is correct, then it would also raise the possibility that gene-specific epigenetic marks play a similar role in pluripotent ES cells.
As a model system for investigating the epigenetic regulation of gene expression during B-cell commitment, we are studying the mouse λ5-VpreB1 locus. Transcription of the VpreB1 and λ5 genes is activated during the pro-B-cell stage prior to heavy-chain rearrangement (43, 49). Their protein products associate to form the surrogate light chain which chaperones newly synthesized μ chains to the cell surface to form the pre-B-cell receptor. This receptor is thought to mediate signaling which leads to the proliferation of pre-B cells that have a productive heavy-chain rearrangement. The expression of the VpreB1 and λ5 genes is downregulated at the time of immunoglobulin (Ig) light-chain rearrangement, and the genes are silent in mature B cells. The functional unit of the VpreB1 and λ5 genes has been localized within a 19-kb fragment containing both genes and 12 DNase I-hypersensitive sites (HS) (47, 49a, 66). The HS are spread over the 19-kb region, where they form a multicomponent locus control region. The expression of the VpreB1 and λ5 genes is regulated by interactions between trans-acting factors and the locus control region, but the initial events that allow the activators to gain access to regulatory elements at early stages of differentiation have not been defined.
Here we show that the λ5-VpreB1 domain is already marked by histone H3 acetylation and histone H3 K4 methylation at a discrete site in ES cells and that these modifications are not present in the rest of the locus. The marked region expands in early B-cell progenitors and becomes a localized center for transcription factor and RNA polymerase II (Pol II) recruitment. An extended, transcriptionally active chromatin domain is established and maintained in pre-B cells when the λ5 and VpreB1 genes are fully active. Our results provide evidence that epigenetic marking of cis-acting elements of tissue-specific genes that are expressed at later stages can already be set up as early as the undifferentiated ES cell stage.
MATERIALS AND METHODS
Sequence of the λ5-VpreB1 locus.
The sequence of the entire 19-kb λ5-VpreB1 locus has been determined elsewhere (accession number AJ852426 [49a] ). The positions of restriction sites were calculated relative to that of an EcoRI site (see Fig. 4D) 2.8 kb upstream from the initiation site for VpreB1.
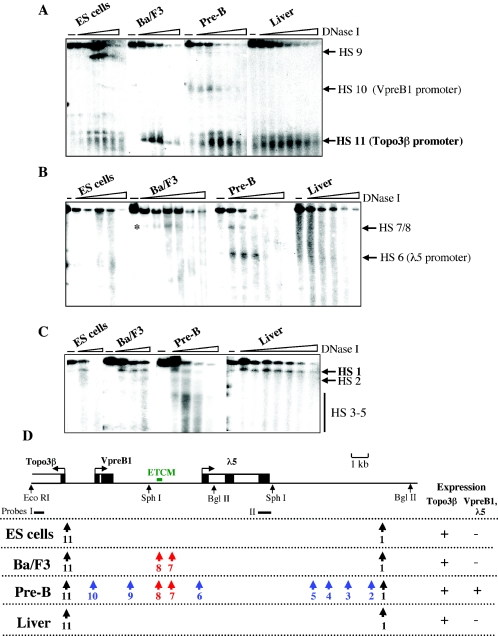
FIG.4.
Changes in patterns of DNase I HS during differentiation. The 12 HS in pre-B cells were mapped elsewhere ( 49a). (A) Pattern of HS at the 5′ region of the λ5-VpreB1 domain. DNA was digested with EcoRI and SphI, Southern blotted, and hybridized with probe I (see the map in panel D). (B) Pattern of HS in the central part of the λ5-VpreB1 domain. DNA was digested with SphI, Southern blotted, andhybridized with probe II (see the map in panel D). The appearance of HS7 and HS8 in Ba/F3 early pro-B cells is indicated by an asterisk. (C) Mapping of HS downstream of the λ5 gene. DNA was digested with BglII, Southern blotted, and hybridized with probe II. (D) Summary of HS patterns at successive differentiation stages. Restriction sites and locations of probes used for mapping are shown below the locus map. Color key: black, constitutive HS; blue, pre-B-cell-specific HS; red, HS found in early pro-B and pre-B cells.
Cells and tissues.
E14 and CJ7 ES cells were cultured in Dulbecco minimal essential medium (Sigma) supplemented with 15% fetal calf serum (FCS), 2 mM l-glutamine, 1% sodium pyruvate, 1% nonessential amino acid solution, 50 μM 2-mercaptoethanol, and 2,400 U of leukemia inhibitory factor (LIF)/ml in 0.1% gelatin-coated tissue culture flasks. ES cell cultures were always started on mitomycin C-treated SNL feeder layers until the first passage; no more feeder cells were added afterward when the cultures were expanded. To verify the undifferentiated state, the expression of Oct-4 was monitored by fluorescence-activated cell sorting analysis. Ba/F3 cells were grown in RPMI medium (Sigma) supplemented with 15% fetal FCS, 2 mM l-glutamine, 1% nonessential amino acid solution, 50 μM 2-mercaptoethanol, and 10% conditioned medium from WEHI 3 cells as a source of interleukin 3 (IL-3). Primary cultures of pre-B cells from 16.5-day-old fetal livers were established as described previously (66). To generate primary cultures of mature B cells, spleens were disaggregated, and the single-cell suspension was subjected to centrifugation on Ficoll to remove erythrocytes. Cells from the interface were collected and activated in RPMI medium supplemented with 15% FCS, 50 μM 2-mercaptoethanol, 2 mM l-glutamine, 50 μg of gentamicin/ml, and 2.5 μg of lipopolysaccharide/ml for 3 days. Primary mouse embyonic fibroblasts were obtained from whole 16.5-day-old embryos by trypsin treatment and subsequent culturing in Dulbecco minimal essential medium supplemented with 15% FCS, 50 μM 2-mercaptoethanol, 2 mM l-glutamine, 1% sodium pyruvate, and 50 μg of gentamicin/ml.
In vitro differentiation of ES cells.
E14 ES cells were plated at high densities in uncoated tissue culture flasks by using the medium described above, except that it contained 10% FCS and no LIF. After 3 days of culturing, floating ES cell aggregates were transferred to fresh medium. After another 5 days of culturing, embryoid bodies were disaggregated by trypsin treatment, plated at low densities, and allowed to grow for a further 5 days. At this point, the cell cultures were confluent, with differentiated, flattened fibroblast-like cells, and only a small population of ES cell colonies (<15%) remained.
Reverse transcription (RT).
Total RNA was extracted from various cells and tissues by using an RNeasy minikit (Qiagen). Total RNA (1 μg) was treated with 10 U of DNase I (Roche) in a 20-μl volume containing 10 mM Tris-HCl (pH 8.0), 0.5 mM MgCl2, and 1 mM dithiothreitol for 1 h at 37°C. DNase I-treated RNA (100 ng) was reverse transcribed in a 20-μl volume with 20 U of Moloney murine leukemia virus reverse transcriptase (Gibco) and 100 ng of random primers according to the manufacturer's instructions.
DNase I hypersensitivity assay.
Whole livers were disaggregated in ice-cold phosphate-buffered saline by 30 strokes of a Dounce homogenizer with a “loose” pestle. The cell suspension was washed with phosphate-buffered saline, homogenized, and washed two more times. Preparation of nuclei from all cell types and mapping of DNase I HS were performed as described previously (66) (see Fig. 4D for the HS mapping strategy). Southern blot probe I was generated by PCR with primers PI5′ and PI3′ (Table 1). Probe II was a SacI-SphI restriction fragment (66).
TABLE 1.
PCR primers and conditions used for ChIP in this study
| Primer | Primer sequence (5′-3′) | Position (kb)a | Concn (nM)b | Annealing temp (°C) | Primer | Primer sequence (5′-3′) | Position (kb)a | Concn (nM)b | Annealing temp (°C) | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | TTC TTT GCC ACA CCT CAC TAA T | 0.204 | 900 | 52 | 30 | GGG GGA TAT TAC CTG CTC TTT | 7.394 | 300 | 52 | |
| 2 | CCT TTT GGG GCT CCT CA | 0.31 | 900 | 52 | 31 | TGG ATA TCA GTC AGG CAG AG | 7.869 | 900 | 60 | |
| 3 | TGC CGG TCA CAG GTC AG | 0.522 | 900 | 57 | 32 | CCC GGT TGT GGT TGG | 7.968 | 900 | 60 | |
| 4 | GAT GGC TCA GCG CTC TTA CAA | 0.623 | 900 | 57 | 33 | CTA AGA GGA GGG GGC TGT GC | 8.487 | 900 | 70 | |
| 5 | TAC CCA AGT TTT CTC CTA ATC TGC | 0.856 | 900 | 57 | 34 | GGA GGT GGG CTA TGG ATG TGG | 8.586 | 900 | 70 | |
| 6 | GGC CTG CGG TTT GCG TTC T | 0.974 | 900 | 57 | 35 | TCA GCA ACC ATT CAT CTA CTA TCA | 9.001 | 900 | 65 | |
| 83 | AGT CCG AGA ACA GCC TGG GT | 0.888 | 300 | 68 | 36 | CCT GCC TCT TCC TCC CAA CT | 9.119 | 900 | 65 | |
| 84 | AGT TGT GCT GCC CAC AGA GG | 0.998 | 300 | 68 | 37 | AGG CCC TAA CAG CTT CAT CTA CTC | 9.777 | 300 | 55 | |
| 708 | TTT ATT TTT ATT TGA TAT GCA TTG GT | 1.587 | 900 | 57 | 38 | GCA TCT GGG CCT CGG TTT A | 9.894 | 900 | 55 | |
| 808 | ATA TGG CAG CTC ACA ACT ATC CC | 1.665 | 400 | 57 | 39 | GGC TGC ACC TGG AAA ACC TTA | 11.701 | 900 | 55 | |
| 11 | TAG AAG GTT GAG ACA GCG AGT TAG | 1.831 | 900 | 58 | 40 | ATC AAA ATC TTC CCC TCA ATC TGT | 11.801 | 300 | 55 | |
| 12 | AGG TGC TTT GTT TTG TTT TGT CTT | 1.934 | 900 | 58 | 43 | AGG AGA CCA GAA GGG GCA GTT | 13.382 | 900 | 53 | |
| 13 | CAA ACC CAG AGC CAC AAA G | 2.759 | 900 | 62 | 44 | GGT TCA ATG TTT AAG AGC AAG TTT | 13.491 | 900 | 53 | |
| 14 | GGG GAG AGG GTC ACT GC | 2.875 | 900 | 62 | 45 | ACC TTT GCT TTC TTG TTG TT | 14.357 | 900 | 50 | |
| 15 | GGG CTT CTT TGC TCC TCC TAT GG | 3.027 | 900 | 60 | 46 | TTG AAA TTT TTA GGG ACT GTG AGA | 14.456 | 900 | 50 | |
| 16 | GAA GAT GCT AAT GGT GGC TGA TGC | 3.137 | 900 | 60 | 47 | CAC AGA AGA GCA GAG AAC AGA | 15.322 | 900 | 60 | |
| 17 | CAA GTG ACA GGT GTG GAG CAA GTT | 3.939 | 900 | 60 | 48 | CAC CCC ATG AGA CAA CCA G | 15.432 | 900 | 60 | |
| 18 | GGA GAG CAC ACC CCA GTA GGA TTA | 4.046 | 900 | 60 | 49 | ACC CAG TAA GCA AGT TTT CA | 16.262 | 900 | 55 | |
| 19 | GTG ACA GAC CCG TTA CCA A | 4.445 | 900 | 52 | 50 | ATA AGC TCT CCT CCC TCA AG | 16.372 | 300 | 55 | |
| 20 | AGA AAG AGA AGG GGA AAA AGA G | 4.589 | 300 | 52 | 51 | TGG AGT ATG AGG CAG TGA TTG TTA | 16.731 | 300 | 53 | |
| 25 | TCC CCA TTG CCA GAT AGA GAC ACA | 5.434 | 900 | 61 | 52 | AGA GTT TTG ACG GCT TCC AGA | 16.861 | 900 | 53 | |
| 26 | TGG GCC CAA CAG ATT AAC ACA GAG | 5.538 | 900 | 61 | 53 | TGT AAG CTC CTG TAT TTG GTT TTT | 16.977 | 900 | 54 | |
| 27 | TGA CTT GCT TGT GCT TGC CTG GAC | 5.732 | 300 | 58 | 54 | GCC CTA CAG AGC TTT TGG AGA | 17.103 | 900 | 54 | |
| 28 | ATA ATA ACA AAG TAC TGA GAA AAC | 5.841 | 900 | 58 | 55 | ACC CCT CCC TCC CCG TAT G | 17.927 | 900 | 62 | |
| 57 | TGG ATA ACT GCA GGA AGC TGT | 6.608 | 600 | 68 | 56 | TCC TTC CCT GAC CAC TCC | 18.031 | 900 | 62 | |
| 58 | GCA GTG CCA GAT CTC CAG AC | 6.728 | 600 | 68 | P15′ | TTC TGC CAA GGC TCT GAC ACTA | 0.098 | 400 | 57 | |
| 29 | GGA TAG ACT TTG CAT GTT TTT GAT | 7.285 | 300 | 52 | P13′ | GCA ATG TTG GTA GAC AGA GCT G | 0.424 | 400 | 57 |
Positions correspond to the 5′ end of each primer in the 19-kb locus sequence.
Final concentration of each primer in the PCR.
Immunoprecipitation of unfixed chromatin.
Preparation of unfixed chromatin and chromatin immunoprecipitation (ChIP) were carried out as described previously (56). At least two independent immunoprecipitations were performed for each cell type and antibody, and 150 μg of unfixed chromatin (measured as DNA at an optical density at 260 nm) was used in each immunoprecipitation. The volume of the samples was adjusted to 1 ml with ChIP buffer 1 (50 mM NaCl, 10 mM Tris-HCl [pH 7.4], 5 mM sodium butyrate, 1 mM EDTA, EDTA-free protease inhibitor cocktail [Roche]), and the samples were precleared for 1 h with 100 μl of protein A-agarose (Sigma) and then centrifuged at 1,400 rpm (Eppendorf Centrifuge 5417R) for 5 min. The supernatants were collected, and 50 to 100 μl was kept as input material. Immunoprecipitation was carried out with rotation at 4°C overnight and the following antibodies: 50 μl of antiserum to tetraacetylated histone H4 (Upstate), 20 μl of antibody to diacetylated histone H3 (Upstate), 50 μl of antiserum to dimethylated histone H3 K4 (Upstate), and 15 μg of nonspecific anti-rabbit immunoglobulin G (IgG) (Santa Cruz Biotechnology). Immunocomplexes were collected 3 h after the addition of 100 μl of protein A-agarose at 4°C and then centrifuged for 5 min at 1,400 rpm. Beads were washed once with ChIP buffer 1, once with ChIP buffer 2 (same as ChIP buffer 1 except for 100 mM NaCl), and then twice with ChIP buffer 3 (same as ChIP buffer 1 except for 150 mM NaCl). The bound fraction was eluted twice with 250 μl of ChIP buffer 1 containing 1% sodium dodecyl sulfate. Immunoprecipitated and input samples were extracted twice with phenol-chloroform, ethanol precipitated, and resuspended in 100 μl of Tris-EDTA buffer.
Immunoprecipitation of fixed chromatin.
ChIP of formaldehyde-cross-linked chromatin was carried out essentially as described previously (72). Fixed chromatin from 3 × 108 to 5 × 108 cells was sonicated by using Soniprep 150 equipment to obtain DNA fragments of between 200 and 500 bp. Chromatin (500 μg) (measured as described above) was immunoprecipitated with mouse monoclonal antibodies overnight in sonication buffer (72) containing 0.1% bovine serum albumin. The antibodies used were as follows: 50 μg of purified anti-PU.1 antibody (BD Pharmingen), 50 μg of purified E2A (E12/E47) antibody (BD Pharmingen), 50 μl of anti-TATA-binding protein (TBP)-associated factor 5 (TAF5) antibody solution, 50 μg of purified anti-TAF10 antibody, 50 μg of purified anti-TBP antibody (84), and 50 μl each of anti-Brg1 antibody solution (54), anti-TRRAP antibody solution (22), and anti-Pol II antibody solution (5). To demonstrate the specific binding of transcription factors above the background, precipitations were also carried out with 50 μl each of anti-Gal4 antibody solution and anti-VP16 antibody solution (83) and without antibody. Eluates were resuspended in 20 to 50 μl of Tris-EDTA buffer.
Real-time PCR and data analysis.
The total amount of DNA in each sample was determined by Pico Green fluorimetry (Molecular Probes). cDNA and ChIP material were analyzed by using a DNA Engine Opticon system (MJ Research Inc.) and SYBR green master mix (Applied Biosystems). Reactions were carried out in duplicate in 20- to 25-μl volumes. Primers were designed by using Primer Select or Primer Express software (see Fig. 2A for positions of primers that were used in ChIP); sequences, concentrations, and annealing temperatures are shown in Table 1. RT-PCR primer sequences are available upon request. The thermal cycling conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, annealing at a primer-dependent temperature for 15 s, and extension at 72°C for 15s. For each experiment, the threshold was set to cross a point at which real-time PCR amplification was linear (0.02 to 0.05 for the majority of the experiments). To test the efficiency of different primer pairs in real-time amplification, we carried out reactions with various known concentrations of a plasmid containing the appropriate region. Calibration curves were generated by plotting the cycle number at which the PCR signal rose above the background (Ct value) against the logarithm of the number of template molecules. The slope of the standard curve was used to determine the efficiency of the PCR (75), which was above 70% with all primer pairs.
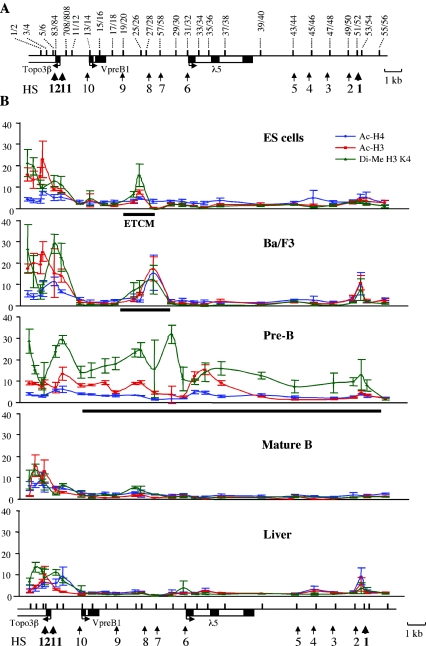
FIG. 2.
Patterns of histone modifications in stem cells and differentiated cells. (A) Positions of primers that were used in this study. Thick vertical lines in the locus map indicate PCR amplicon positions; numbers above the locus map correspond to primer pairs. Primer sequences and PCR conditions are shown in Table 1. (B) Histone modification state of the λ5-VpreB1 locus at different stages of B-cell development. ChIP analysis was performed with unfixed chromatin and antibodies against acetylated (Ac) histones H4 and H3 and dimethylated (Di-Me) histone H3 K4 as described in Materials and Methods. A similar analysis with nonspecific IgG antibody or no antibody confirmed that histone modification signals were detected above the background (data not shown). Fold enrichment of target sequences in the immunoprecipitated material relative to the input material is shown on the y axis of each plot. x axes represent positions across the λ5-VpreB1 domain. Error bars indicate standard deviations. A locus map is shown below the plots; amplicon positions are indicated by black vertical lines. A thick black horizontal line below a plot indicates the region in the λ5-VpreB1 domain that is marked by acetylation and K4 dimethylation of histone H3. The tightly localized mark in ES cells was termed the ETCM.
To quantify mRNA, a 2.5-μl sample from the RT reaction was added to each PCR. Immunoprecipitated material from ChIP assays was quantified by using a previously described method (45). From the samples (input or immunoprecipitated), equal amounts (0.5 to 2 ng) of DNA were analyzed in every PCR. The enrichment of a given target sequence precipitated by an antibody was determined as the fold difference between the amount of target sequence in the immunoprecipitated fraction and the amount of target sequence in the input DNA (fold enrichment). Average fold enrichment was calculated from at least two independent immunoprecipitations and PCR duplicates for each immunoprecipitation. Normalized values reflecting the binding of transcription factors were expressed as a percentage of input material (fold enrichment multiplied by 100). Error bars in the plots represent the standard deviations of the whole set of values obtained for each PCR amplicon position. The background immunoprecipitation of unfixed chromatin with nonspecific rabbit Ig or no antibody was determined for each cell type (data not shown). Quantification of transcription factor binding above the background was achieved by subtracting an average of normalized values obtained from three negative controls (treatment of fixed chromatin with anti-Gal4 and anti-VP16 antibodies and without antibody). Thus, enrichment (percentage of input material) reflects only the specific binding of transcription factors and not the affinity of antibodies for different sites.
RACE.
Rapid amplification of cDNA ends (RACE) was carried out by using a SMART RACE cDNA amplification kit (Clontech) according to the manufacturer's instructions (see Fig. 7A for positions of primers that were used in RT and PCR). Primer sequences are available upon request. For mapping of initiation sites for intergenic transcripts, nested PCR products were analyzed by blotting and hybridization with a restriction fragment (SphI-BamHI; 5.4 to 6.8 kb) prior to cloning. Nested PCR products were directly cloned into the pGEM-T Easy vector system (Promega) and bacterial strain DH5α. Colonies containing a fragment from the intergenic regions were identified by colony hybridization with the SphI-BamHI probe. At least 10 independent clones were sequenced with each strategy.
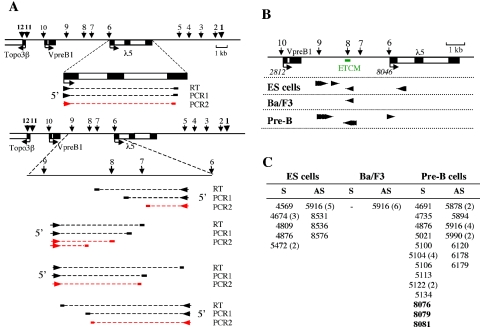
FIG. 7.
Mapping of transcription initiation sites by RACE. (A) Mapping of λ5 transcription start sites (upper panel) and intergenic initiation sites (lower panel). RT and PCR approaches are shown for each mapping strategy. 3′ primers (RT, PCR1, and PCR2) that were used for each region are indicated as short thick horizontal lines extended by dotted lines that represent the part of the transcript that was converted into cDNA. RT was performed with locus-specific 3′ primers to detect RNAs transcribed from either the leading or the lagging DNA strand. The orientation of transcription corresponding to each strategy is indicated by arrowheads. Nested PCR products (PCR2; red lines) were cloned and sequenced. (B) Pattern of intergenic transcription initiation sites. Numbers in italics below the locus map indicate positions of transcription initiation sites for the three genes relative to the EcoRI site 2.8 kb upstream from VpreB1. Arrowheads indicate the transcription initiation sites that were mapped in various cell types. The direction of the transcripts is also represented by these arrowheads. (C) Positions of transcription initiation sites. Numbers correspond to positions in the 19-kb locus sequence (as in panel B). Correct initiation sites for the λ5 gene are shown in bold type. Numbers in parentheses after position numbers indicate how many sequenced clones contained that particular initiation site when there was more than one. S, sense; AS, antisense (relative to the directions of VpreB1 and λ5 transcription).
RESULTS
Expression status of the λ5-VpreB1 locus at successive stages of B-cell differentiation.
The different stages of B-cell development have been extensively characterized on the basis of the expression of cell surface markers and the rearrangement status of the Ig loci. We investigated gene expression and epigenetic characteristics of the entire λ5-VpreB1 locus from undifferentiated ES cells through successive stages of B-cell differentiation. Differentiation stages were represented by primary cells from fetal and adult mice and by nontransformed cell lines that provided enough material for the analysis of chromatin structure. The system is shown in Fig. 1A. When cultured under the appropriate conditions (see Materials and Methods), ES cells remain undifferentiated and retain their capacity to give rise to every cell type in the animal (64). Ba/F3 cells are nontransformed, IL-3-dependent bone marrow-derived early B-cell precursors (14, 23, 57) that have been used as models to represent the early pro-B-cell stage of B-cell development (38, 68, 82). They have been found to be positive for very early B-cell markers, such as Mb-1, B29, and E47 monomers and germ line μ transcripts, but do not express markers of later stages of B-cell development, such as CD19, B220, Pax5, EBF, TdT, Rag1, IL-7R, VpreB, and λ5 (71). To study chromatin structure in pre-B cells, primary cultures were established from 16.5-day-old fetal livers and grown on ST-2 feeder cell layers in the presence of IL-7. Primary mature B cells were obtained by lipopolysaccharide activation of cells from adult spleens.
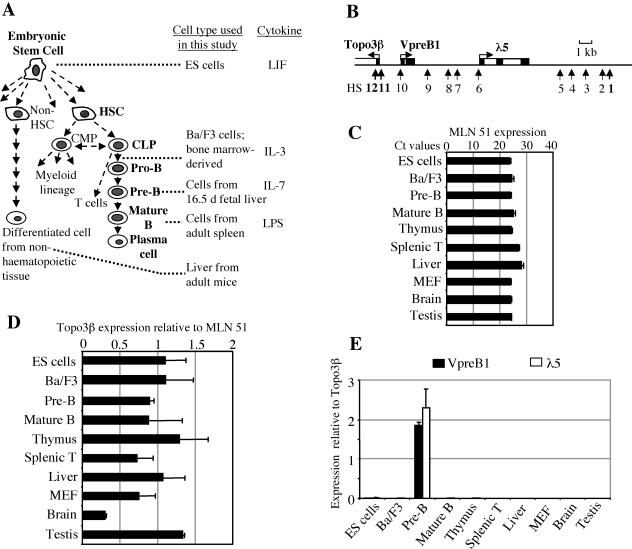
FIG. 1.
Experimental system and gene expression analysis. (A) Scheme showing haematopoietic cell differentiation and cell types that were used to represent differentiation stages. ES cells give rise to more specialized, multipotent stem cells, such as haematopoietic stem cells (HSC). HSC develop into common myeloid progenitors (CMP), which generate the myeloid lineage. Common lymphoid progenitors (CLP) give rise to the lymphoid lineage. The relationship between CMP and CLP is controversial (reviewed in reference 32). The B-cell lineage differentiates through progenitor B cells (pro-B cells) and precursor B cells (pre-B cells) and finally generates antibody-producing plasma cells. Ba/F3 cells are IL-3-dependent early pro-B cells. d, day; LPS, lipopolysaccharide. (B) Organization of the λ5-VpreB1 locus. VpreB1 and λ5 are transcribed on the same DNA strand (sense direction), whereas Topo3β is transcribed on the opposite strand (antisense direction). Black and white boxes symbolize exons and introns, respectively. Vertical arrows show the positions of the 12 previously mapped DNase I HS (49a, 66). Constitutive HS1, HS11, and HS12 are shown in bold type; the other HS (HS2 to HS10) are pre-B-cell specific. (C) Pattern of expression of the MLN51 gene. Ct values represent the cycle numbers in real-time PCR at which signals start to be detectable above the threshold. Accordingly, the higher the Ct value, the lower the expression level. MEF, mouse embryonic fibroblasts. (D) Expression of the Topo3β gene. Real-time RT-PCR signals for Topo3β were normalized to those for MLN51. (E) Expression of VpreB and λ5. Signals were normalized to those for Topo3β. Error bars in panels D and E indicate standard deviations.
VpreB1 and λ5 have been shown to be expressed at the pro- and pre-B-cell stages of B-cell development (49) (Fig. 1E). The two genes are located within a 10-kb genomic fragment on mouse chromosome 16 (Fig. 1B). Upstream from the transcription start site for VpreB1, sequence analysis identified another gene, Topoisomerase 3β (Topo3β), which is transcribed on the strand opposite that of VpreB1 and λ5. The promoter of the Topo3β gene resembles that of a housekeeping gene and overlaps with a CpG island. To determine the expression pattern for Topo3β, we performed quantitative real-time RT-PCR. PCR signals were normalized to those obtained for the metastatic lymph node 51 (MLN51) gene (20), which was chosen as a reference because its expression is constant in every cell type examined (Fig. 1C). The results of RT-PCR showed that Topo3β was expressed at every differentiation stage and in every tissue that we analyzed (Fig. 1D). Expression varied by less than twofold, with the exception of expression in the brain, which was present at a lower level. These results show that VpreB1 and Topo3β have entirely different expression patterns (Fig. 1E), despite the fact that their transcription start sites are separated by only 1.5 kb.
Histone modifications mark a discrete intergenic site in the λ5-VpreB1 locus in ES cells.
Current evidence indicates that histone modifications mark differentially expressed chromatin domains and also shows that these epigenetic modifications are present in multipotent haematopoietic stem cells before transcription is fully activated (3, 6). We set out to investigate at which differentiation stage these epigenetic marks are established. The small size of the λ5-VpreB1 domain allowed us to carry out a high-resolution search for histone modifications across the entire locus by ChIP analysis.
The pattern of histone modifications across the λ5-VpreB1 locus was determined at five differentiation stages (Fig. 2). Unfixed chromatin was isolated from cells and tissues and digested with micrococcal nuclease to produce fragments containing one to three nucleosomes. Digested chromatin was subjected to immunoprecipitation with antibodies recognizing diacetylated histone H3, tetraacetylated histone H4, and dimethylated histone H3 K4. PCR primers were designed to cover the entire 19-kb region at intervals of 0.2 to 1 kb (Fig. 2A and Table 1). The specificity of ChIP analysis was monitored by using either nonspecific rabbit IgG antibody or no antibody. The results of ChIP analysis are shown in Fig. 2B.
Previous studies with immunostaining suggested that the ES cell genome is subject to global histone acetylation. Our ChIP analysis revealed no evidence of generalized acetylation or histone H3 K4 methylation across the λ5-VpreB1 locus. Instead, we detected a tightly localized peak of histone H3 acetylation and histone H3 K4 dimethylation between the VpreB1 and λ5 genes in ES cells (Fig. 2B). In Ba/F3 early pro-B cells, the histone modification mark expands in both directions from this discrete site toward the VpreB1 and λ5 genes. In pre-B cells, in which the VpreB1 and λ5 genes are fully active, H3 K4 dimethylation extends across the rest of the 19-kb region. Acetylation of H3 in pre-B cells shows a more localized pattern of peaks and troughs that extend only as far as the 3′ end of the λ5 gene. A low and uniform level of histone H4 acetylation is observed across the locus, in keeping with previously published results describing relatively constant levels of H4 acetylation across the human growth hormone and human β-globin loci (36, 70) but in contrast to the nonuniform state of H4 acetylation across the murine β-globin locus (17, 34). Our results suggest that H4 acetylation is not involved in the establishment of the active λ5-VpreB1 domain during B-cell development. In mature B cells and adult liver, histone modifications are largely absent from the locus. Peaks of histone acetylation and H3 K4 dimethylation are present around the initiation site for the Topo3β gene in all cell types examined. This finding is consistent with the observation that the Topo3β gene is ubiquitously expressed. We observed a sharp transition between the histone modification states of the Topo3β promoter and the silenced VpreB1 promoter in cells that do not express the VpreB1 and λ5 genes. The transition occurs in the region between the transcription initiation site for VpreB1 and a point approximately 600 bp upstream of the VpreB1 start site.
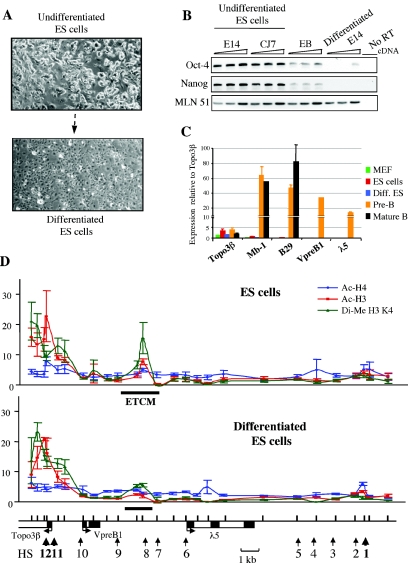
To further investigate the relationship between the localized intergenic histone modification mark and B-cell lineage-specific expression of the λ5-VpreB1 locus, ES cells were allowed to differentiate in vitro into nonlymphoid cells (see Materials and Methods) (Fig. 3). After 2 weeks of differentiation, the cell cultures were confluent, with flattened fibroblast-like cells, and only a small population of ES cell colonies remained (Fig. 3A). Differentiation was confirmed by showing that the expression of the known ES cell-specific factors Oct-4 (55) and Nanog (9) was silenced in in vitro differentiated ES cells (Fig. 3B). The facts that these cells were adherent in culture and that B-cell-specific transcripts of the Mb-1 (67), B29 (27), and VpreB1 and λ5 genes were absent (Fig. 3C) showed that the ES cells did not differentiate into the B-cell lineage. ChIP analysis of the λ5-VpreB1 locus indicated that the intergenic histone H3 acetylation and histone H3 K4 dimethylation marks largely disappeared in differentiated ES cells (Fig. 3D). The low residual level of modification that was still detectable was likely due to a small number of undifferentiated ES cells remaining in the cultures.
FIG. 3.
Loss of ETCM during in vitro differentiation of ES cells. (A) Morphology of ES cells after in vitro differentiation. (B) Expression of ES cell factors during in vitro differentiation of ES cells. The cells were tested for Oct-4 and Nanog expression by RT-PCR. The measurements for each stage were obtained from three sequential fivefold dilutions. Two independent ES cell lines, E14 and CJ7, expressed these ES cell factors, and their expression was downregulated in embryoid bodies (EB) and lost in differentiated E14 ES cells. MLN51 served as a loading control. (C) In vitro differentiated (Diff.) ES cells do not express B-cell-specific genes. Real-time RT-PCR signals were normalized to those for MLN51. MEF, mouse embryonic fibroblasts. Error bars indicate standard deviations. (D) ChIP analysis of the λ5-VpreB1 domain in in vitro differentiated ES cells. Explanations of the plots are given in the legend to Fig. 2B. Note the reduction in histone modifications at the ETCM in differentiated ES cells. The low residual signal could be attributed to a small number of undifferentiated ES cells that remained in the differentiated culture.
These results show that a localized epigenetic mark is present in the λ5-VpreB1 domain in pluripotent ES cells. The mark expands during commitment to the B-cell lineage and disappears during differentiation into nonlymphoid cells. Functional analysis has shown that the region containing the mark is able to enhance the transcription of a transgene driven by the VpreB1 promoter in pre-B cells (49a). Colocalization of the mark with a transcriptional enhancer suggests that this region is involved in initiating the formation of a transcriptionally active chromatin structure in the λ5-VpreB1 domain. Therefore, we propose the name early transcription competence mark (ETCM) to describe this type of modification.
The ETCM becomes hypersensitive to DNase I digestion prior to the activation of VpreB1 and λ5 transcription.
It was of particular interest that the marked site is a cis-acting regulatory element that is likely to be involved in the assembly of transcription factor complexes during B-cell differentiation. It has been reported that DNase I HS formation precedes the activation of the β-globin gene during erythroid cell commitment in multilineage progenitors (29). Two DNase I HS (HS7 and HS8) have been mapped to the ETCM region in pre-B cells (49a, 66). In ES cells, the mark coincides with the region in which HS8 is located; it then spreads to encompass HS7 and HS8 in early pro-B cells. To investigate the timing of HS formation during differentiation, the DNase I hypersensitivity of the entire locus was analyzed in ES cells, Ba/F3 early pro-B cells, and pre-B cells (Fig. 4). In Ba/F3 early pro-B cells, in which VpreB1 and λ5 are not yet expressed, an HS appeared between the genes near the locations of HS7 and HS8 in pre-B cells (Fig. 4B and D). The mapping strategy used in this study did not discriminate between HS7 and HS8 due to their close proximity; they are separated by less than 100 bp. In pre-B cells, multiple HS, including HS7 and HS8 and the promoters of VpreB1 (HS10) and λ5 (HS6), were observed. A previously characterized 3′ HS (HS1) (66) and an HS at the ubiquitously expressed Topo3β promoter (HS11) were detected in all cell types examined. The appearance of the otherwise pre-B-cell-specific HS7 and HS8 in early pro-B cells shows that marking of the intergenic cis-acting regulatory element by histone modifications is followed by further structural changes that precede transcriptional activation.
The ETCM is a center for general transcription factor recruitment during B-cell development.
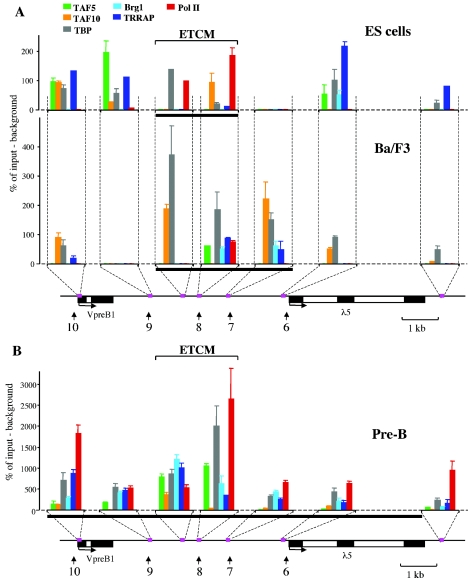
There is evidence that general transcription factors and RNA Pol II can bind to cis-acting regulatory elements, such as enhancers (30, 35, 48, 62, 73). Therefore, we investigated the recruitment of the general transcription machinery (including factors that are involved in transcriptional initiation and others that are parts of chromatin-modifying complexes) to the ETCM by ChIP analysis. TBP, TAF5, and TAF10 are components of the transcription factor IID (TFIID) complex, which plays a role in the formation of the preinitiation complex (52). TAF5 and TAF10 are subunits of the TBP-free TAF-containing (TFTC) complex, which contains GCN5 histone acetyltransferase and plays a role similar to that of the yeast SAGA complex in gene regulation (7, 84). Brg1 is a subunit of the SWI/SNF ATP-dependent chromatin-remodeling complex (51). Transformation-transactivation domain-associated protein (TRRAP) is a coactivator in histone acetyltransferase complexes, such as STAGA, PCAF, TFTC, and TIP60 (7, 18, 46).
The results of ChIP analysis are shown in Fig. 5. They reveal a complex relationship between binding of these factors and changes in epigenetic marking of the locus during B-cell differentiation. In undifferentiated ES cells, we detected TAF10, TBP, and Pol II but not Brg1 in the ETCM region (Fig. 5A). Surprisingly, at this stage, Pol II binding was observed only in the ETCM region and not at the promoters of the VpreB1 and λ5 genes. The binding of TAFs, TBP, and TRRAP was also observed at the promoters of the genes and in a region immediately downstream from the VpreB1 gene. It is notable that the neighboring HS7 region, which is not part of the ETCM, showed no evidence of factor binding in ES cells.
FIG. 5.
The ETCM is a center for general transcription factor recruitment during B-cell development. (A) Binding profiles for general transcription factors at early stages of B-cell differentiation. ChIP analysis was performed with fixed chromatin as described in Materials and Methods. The percentage of target sequences in the immunoprecipitated material relative to the input material is shown on the y axis of each plot. Background immunoprecipitation (an average normalized value obtained by treatment of chromatin with two nonspecific antibodies and with no antibody) was subtracted from normalized specific ChIP signals (obtained by immunoprecipitation with antibodies to general transcription factors) at each position. x axes represent positions across the λ5-VpreB1 domain. A locus map is shown below the plots; amplicon positions are indicated by pink boxes. A thick black horizontal line below each plot indicates the region in the λ5-VpreB1 domain that is marked by acetylation and K4 dimethylation of histone H3. (B) Binding profiles for general transcription factors at the pre-B-cell stage. Quantification, symbols, and color codes are explained in panel A. Because of the higher level of transcription factor binding in pre-B cells, the scale on the y axis is different from that in panel A. Error bars in panels A and B indicate standard deviations.
In Ba/F3 early pro-B cells, in which λ5 and VpreB1 are not yet expressed, the presence of the transcription factors investigated was less detectable at many sites (including the gene promoters) than it was in ES cells (Fig. 5A). In contrast, in the ETCM region and at HS7, increased levels of TBP and TAFs were observed. Interestingly, the recruitment of transcription factors to HS7 coincided with the expansion of the region of histone modifications (Fig. 5A). At this stage, the chromatin-modifying factors Brg1 and TRRAP became localized to the ETCM region, and DNase I HS7 and HS8 were detected for the first time. Another important observation is that Pol II still bound only to the ETCM region and was absent from the rest of the locus. These results suggest that the HS7-HS8 region is a center for transcription factor recruitment and that the increasing complexity of factor binding correlates with localized changes in the chromatin structure of the region immediately prior to the activation of the locus in early pro-B cells.
In pre-B cells, in which the locus is fully active, the binding of general transcription factors and Pol II was detected across the λ5-VpreB1 domain (Fig. 5B). A dramatic increase was observed in the abundance of factors such as TBP, Brg1 and, most interestingly, Pol II, in the ETCM region and at the active gene promoters (compare scales in Fig. 5A and B). In contrast to the findings at the earlier differentiation stages, in pre-B cells, TAF5 and TAF10 were preferentially detected at HS8. The finding that this substantial increase in factor binding to the locus was observed at HS8 and in the ETCM region at the pre-B-cell stage indicates that the region continues to be a major center for the recruitment of general transcription factors when the λ5-VpreB1 domain is active.
Expansion of the region of histone modifications correlates with the binding of transcription factors PU.1 and E2A.
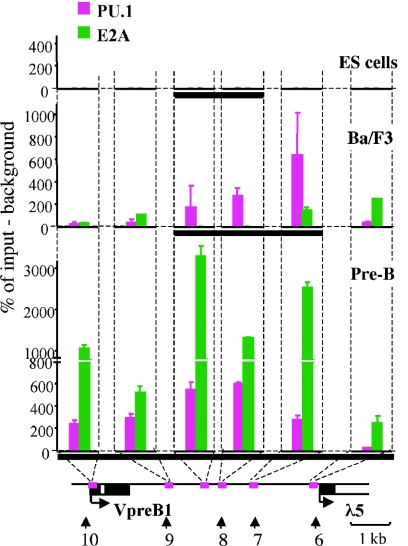
The expansion of the marked region and the formation of the general transcription factor recruitment center in early pro-B cells suggest that chromatin-modifying complexes and basal factors are recruited to the λ5-VpreB1 locus by lineage-specific factors that can recognize a specific sequence in the intergenic region. E2A is a ubiquitously expressed factor which is upregulated in pro- and pre-B cells (33) and has been shown to be directly involved in activating λ5 and VpreB1 expression (71). PU.1 is expressed in haematopoietic stem cells and in lymphoid and myeloid progenitors, in which it plays an important role in specifying the choice between lymphoid and myeloid lineages (10). Its expression is maintained in all differentiating haematopoietic cells except for T cells (1, 2, 25). Multiple consensus recognition sequences for E2A and PU.1 are found across the locus, and the HS7-HS8 region also contains conserved binding sites for E2A and PU.1, with a cluster of PU.1 binding sites at HS7 (49a).
ChIP analysis was used to assess the binding of these factors to the intergenic region and the gene promoters at different stages of development (Fig. 6). Neither factor bound to the locus in ES cells, despite the fact that E2A is expressed at this stage. In Ba/F3 early pro-B cells, the binding of PU.1 coincided with the expansion of the region of histone modifications. Interestingly, the highest level of PU.1 binding at HS7 correlated with the expansion of the histone modification mark and the recruitment of general transcription factors at the early pro-B-cell stage. In pre-B cells, in which the λ5 and VpreB1 genes are active, continued binding of PU.1 was accompanied by a striking increase in the binding of E2A in the intergenic region. These results suggest that lineage-specific factors, such as PU.1 and E2A, contribute to the establishment of the active λ5-VpreB1 domain by directing the recruitment of chromatin-modifying and general transcription factor complexes during B-cell differentiation.
FIG. 6.
ChIP analysis of binding of lineage-specific transcription factors E2A and PU.1. ChIP was performed with fixed chromatin as described in Material and Methods. Explanations are given in the legend to Fig. 5. A thick black horizontal line below each plot indicates the region in the λ5-VpreB1 domain that is marked by acetylation and K4 dimethylation of histone H3.
The intergenic region contains active promoters.
The observation that general transcription factors and Pol II bind to the ETCM region raised the possibility that factor recruitment is due to the presence of intergenic promoters. Sense and antisense intergenic transcription was described previously for several tissue-specific domains in mammals (19, 48, 60, 63, 65, 78, 79) and Drosophila (13, 61). Quantitative RT-PCR analysis detected significant levels of intergenic transcripts in the λ5-VpreB1 locus in pre-B cells (data not shown). To test whether sites of recruitment of general transcription factors coincided with intergenic promoters, transcription initiation sites were mapped by RACE (Fig. 7A). RACE-PCR products were cloned, and bacterial colonies were screened for the presence of locus sequences by colony hybridization. Positive clones were sequenced to determine initiation sites. Significant changes were observed in the patterns of transcription initiation sites during differentiation from ES cells to pre-B cells (Fig. 7B and C).
Multiple initiation sites were detected in ES cells, indicating that these cells are transcriptionally permissive. Correct initiation from the λ5 promoter was not observed at this stage. In Ba/F3 early pro-B cells, only a single intergenic initiation site was detected; it was located in the ETCM region and was transcribed in the antisense direction relative to the VpreB1 and λ5 genes. Two main clusters of start sites were observed in pre-B cells. One group of transcripts (antisense relative to the genes) initiated at HS8, where the ETCM is located. The other major cluster of start sites, transcribing in the same direction as the VpreB1 and λ5 genes (sense direction), was located near HS9. This analysis showed that intergenic promoters are located at HS8 and HS9. However, the fact that levels of Pol II at HS8 in ES cells were 20-fold higher than at HS9 suggests that high-level Pol II recruitment is a specific feature of the ETCM and is not due solely to the presence of an intergenic promoter. Taken together, our results suggest that the ETCM is a localized center for the recruitment of transcription factors, including RNA Pol II, from undifferentiated ES cells to (and through) the pre-B-cell stage.
DISCUSSION
Localized epigenetic marking of the λ5-VpreB1 domain in ES cells.
A defining characteristic of stem cells is their ability to activate multiple gene expression programs that give rise to different specialized cell types. Stem cells also have the important property of being able to divide and self-renew while maintaining their multipotential capacity. Differentiation potential can be described in terms of the number of genes that can be activated as the cell (and its daughter cells) progress through a particular lineage. A fundamental question in stem cell biology concerns the nature of the epigenetic mechanisms that regulate this potential. Analysis of global histone modification patterns in ES cells has suggested that the ES cell genome is subject to generalized histone acetylation and histone H3 K4 methylation (37, 41). Our high-resolution analysis of the histone modification pattern across a specific gene expression domain raises doubts about this conclusion, as most of the λ5-VpreB1 domain shows no evidence of general histone H3 and H4 acetylation and histone H3 K4 methylation at the ES cell stage. A similar absence of general acetylation and H3 K4 methylation was observed at the α-globin locus in ES cells (3).
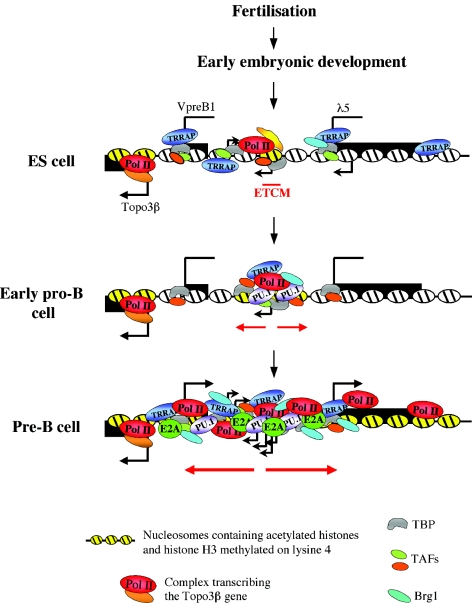
Instead of global histone modifications, our results demonstrate the existence of a tightly localized mark of H3 acetylation and histone H3 K4 methylation (the ETCM) which is already established in the λ5-VpreB1 locus in ES cells. Several lines of evidence suggest that the ETCM is likely to contribute to the establishment of the active epigenetic state of the λ5-VpreB1 domain. A key observation is the fact that the region of histone modifications expands as the cells progress toward the B-cell lineage and disappears when they differentiate into nonlymphoid cells. In addition, the ETCM region is a center for the recruitment of transcription factors and RNA Pol II at all stages from ES cells to pre-B cells. The progressive recruitment of lineage-specific and general transcription factors during B-cell differentiation correlates with the formation of the first B-cell lineage-specific DNase I HS in the locus. In pre-B cells, in which the VpreB1 and λ5 genes are expressed, the HS7-HS8 region shows the highest level of transcription factor and Pol II binding and also has enhancer activity. A schematic representation of the successive changes in the epigenetic state of the λ5-VpreB1 locus during B-cell differentiation is shown in Fig. 8.
FIG. 8.
Schematic illustration of the formation of the active λ5-VpreB1 domain. The Topo3β gene is expressed throughout the entire differentiation program. VpreB1 and λ5 are in a transcriptionally potentiated but inactive state in ES cells and in early pro-B cells. Black arrows represent intergenic transcription initiation sites. The ETCM is a localized histone modification mark in ES cells that is likely to be established by sequence-specific transcription factors (yellow shapes). In addition to factor recruitment to the ETCM region, the λ5-VpreB1 domain is permissive for the binding of general transcription factors at the ES cell stage. Note that Pol II binds only to the ETCM. Expansion of the mark in early pro-B cells (red arrows) would facilitate the recruitment of more transcription factors, the binding of which is reflected in the appearance of DNase I HS in the ETCM region. In early pro-B cells, before the activation of VpreB1 and λ5 transcription, the region forms a center for the recruitment of PU.1 and general transcription factors, and the rest of the locus becomes less permissive for factor binding. At the pre-B-cell stage, the epigenetically modified region expands, and additional transcription factor complexes and Pol II are recruited to the entire λ5-VpreB1 domain. The binding of E2A is likely to be important for the generation of the active epigenetic state of the locus. This process leads to the activation of the VpreB1 and λ5 promoters.
Establishment of the active λ5-VpreB1 domain during B-cell development.
The establishment of a localized epigenetic mark at early stages of development implies that sequence-specific factors bind to the region and initiate the recruitment of histone-modifying enzymes and general transcription factors. The factors that establish the ETCM in ES cells have not been identified, but we do have evidence that the expansion of the mark in early pro-B cells involves the action of transcription factor PU.1, which is known to play a key role in determining the choice between lymphoid and myeloid lineages. High levels of PU.1 binding are observed at HS7 at the time when histone acetylation and K4 methylation and recruitment of general transcription factors expand into this region. It is interesting that Brg1 and TRRAP, factors that are involved in modifying chromatin structure, became detectable at HS7 and HS8 at this stage. PU.1 has been shown to cooperate with various transcription factors and chromatin-remodeling factors to activate or repress transcription (15, 40, 53, 59, 74).
As the cells progress to the pre-B-cell stage, expansion of the region of histone modifications across the λ5-VpreB1 domain is accompanied by high-level binding of E2A to the locus (Fig. 8). The highest level of E2A binding is observed at HS7 and HS8, and PU.1 also continues to bind to the same region. Interestingly, E2A has been shown to act in synergy with PU.1 to activate transcription in B lymphocytes (81). These data, together with the fact that the ETCM region is a center for general transcription factor binding, suggest that the factor recruitment center at HS8 is likely to be formed by cooperation between lineage-specific activators, chromatin modifications, and the general transcription machinery. The presence of a factor recruitment center at HS7-HS8 could facilitate the binding of transcription factors to other regions of the locus both by creating a locally accessible chromatin structure and by direct contacts with these regions through loop formation (21, 31, 58).
The fact that active intergenic promoters have been detected in the locus raises the question as to whether intergenic transcription has a role in generating the active, open λ5-VpreB1 domain during B-cell development (19, 48, 60, 65). Theoretically, the factor recruitment that we observed at HS7-HS8 might be due partly to the presence of intergenic promoters. However, two lines of evidence suggest that intergenic transcription on its own may not be sufficient for the establishment of the active epigenetic structure of the λ5-VpreB1 domain. First, the fact that histone modifications and high-level Pol II binding were detected only at the ETCM region and not at the other intergenic initiation sites in ES cells suggests that the presence of an intergenic promoter is not in itself sufficient to generate the mark and establish a factor recruitment center during B-cell development. Second, expansion of the region of histone modifications does not coincide with spreading of the transcribed region in early pro-B cells. Nevertheless, the possibility remains that intergenic transcription facilitates the distribution of transcriptionally active complexes across the active λ5-VpreB1 locus and plays a role in the maintenance of the active state of the domain in pre-B cells.
Assembly of general transcription factor complexes on an epigenetically marked intergenic regulatory element.
The binding of TFIID subunits, such as TBP and TAFs, to the intergenic region was surprising, as the recruitment of these factors was believed to be specific to gene promoters. As discussed above, our results suggest that high-level general transcription factor binding is a specific characteristic of the ETCM region and is not due solely to the presence of an active intergenic promoter. There are several possible explanations for the quantitative and qualitative differences in general transcription factor binding at different sites in the locus and at different stages of development (Fig. 5 and 8). The process of assembly and disassembly of general transcription factor complexes is very dynamic, and the rates of turnover of different complexes can vary between different sites (e.g., at HS7-HS8 versus gene promoters). Moreover, TBP occupancy and TRRAP occupancy at the same site can indicate the presence of both TFIID and TFTC-like complexes, as TRRAP has been shown to be a subunit of both TFIID and TFTC but TBP is found only in TFIID (see Results for a more detailed description of these factors and complexes). The fact that we detected components of different complexes at the same site could be due to the sequential recruitment of complexes during the transcriptional initiation and elongation cycle (reviewed in reference 11) or to cells within the population being at different stages of the cell cycle.
In summary, our data provide evidence for the formation of a dynamically regulated transcription factor recruitment center in the ETCM region from the early stages of B-cell differentiation. These data are consistent with a model proposing that an important function of the epigenetically marked intergenic enhancer is to act as a nucleation center for the assembly of preinitiation complexes prior to their recruitment (or transfer) to the promoters of the genes.
Differential regulation of the closely linked λ5-VpreB1 and Topo3β domains.
In pre-B cells, the λ5-VpreB1 locus is part of a continuous domain of histone H3 K4 methylation that extends across the neighboring Topo3β promoter. A particularly interesting feature of the λ5-VpreB1 locus is the very short distance between the promoters of the ubiquitously expressed Topo3β gene and the developmentally regulated pre-B-cell-specific VpreB1 gene. In mature B cells and liver, the VpreB1 promoter remains silent and unaffected by the high levels of histone modifications on the nearby Topo3β promoter, despite the fact that the transcription start sites of the two genes are only 1.5 kb apart. The sharp transition of histone modifications suggests that the border of the Topo3β and VpreB1 domains falls within a very short region of approximately 600 bp and that the active Topo3β domain does not spread further from this point. These results suggest a tightly controlled epigenetic determination of differentially expressed domains over a very short genomic distance. Since this short region does not include any obvious elements, such as DNase I HS or intergenic promoters, that can have an insulator function, it is more likely that the separate histone modification profiles of the Topo3β and VpreB1 genes are generated by the differential assembly of transcription factor complexes on their cis-acting regulatory elements.
Localized epigenetic marking and regulation of differentiation potential.
It is widely accepted that the establishment of tissue-specific gene expression domains occurs through a series of sequential events that involve the potentiation of a domain before the genes located within it are fully activated. There is evidence that a specific subset of genes that are expressed late in haematopoietic differentiation are already in a permissive state in haematopoietic stem cells (26). Histone modifications and partial assembly of transcription factor complexes have also been observed on promoters in the α- and β-globin loci and at the c-fms and lysozyme genes in haematopoietic progenitors (3, 6, 39, 76, 77). These results have established a paradigm that links the epigenetic potentiation of genes to the differentiation capacity of multipotent stem cells. The basis for the pluripotency of ES cells is much less clear. Current thinking on this issue has tended to focus on the idea of nonspecific global accessibility of the ES cell genome. This accessibility would become restricted as the cells differentiate and acquire specific epigenetic modifications.
Our results suggest an alternative scenario in which there is no fundamental difference between the mechanisms that underlie the multipotency of haematopoietic stem cells and the totipotency of ES cells. According to this model, the transcriptional competence of individual gene loci would be determined by localized chromatin modifications in stem cells, and the differentiation potential of these cells would correlate with the number of marks that are present in the cells. The number and the distribution of the marks would determine the ability of the cells to differentiate along different lineages. The model predicts that more gene loci would be marked in pluripotent ES cells than in multipotent stem cells. As cells differentiated toward a specific lineage, they would lose marks on genes that belong to alternative lineages. The tightly localized nature of the ETCM suggests that such marks would be detected only by high-resolution mapping of the type described in this study. Analysis of the epigenome at this level of resolution is likely to provide further information on the distributions of the marks and their relationship to the differentiation capacities of totipotent and multipotent cells.
Acknowledgments
This work was supported by the Leukemia Research Fund and the Medical Research Council. H.S. was funded by an EU Marie Curie Intra-European Fellowship.
We thank Joel Abrahams for technical assistance, Deborah Farmer and Sophie Minaee for discussions, and Pierangela Sabbattini for comments on the manuscript.
REFERENCES
- 1.Akashi, K., D. Traver, T. Miyamoto, and I. L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404:193-197. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. K., G. Hernandez-Hoyos, R. A. Diamond, and E. V. Rothenberg. 1999. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126:3131-3148. [DOI] [PubMed] [Google Scholar]
- 3.Anguita, E., J. Hughes, C. Heyworth, G. A. Blobel, W. G. Wood, and D. R. Higgs. 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 23:2841-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anguita, E., C. A. Johnson, W. G. Wood, B. M. Turner, and D. R. Higgs. 2001. Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster. Proc. Natl. Acad. Sci. USA 98:12114-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besse, S., M. Vigneron, E. Pichard, and F. Puvion-Dutilleul. 1995. Synthesis and maturation of viral transcripts in herpes simplex virus type 1 infected HeLa cells: the role of interchromatin granules. Gene Expr. 4:143-161. [PMC free article] [PubMed] [Google Scholar]
- 6.Bottardi, S., A. Aumont, F. Grosveld, and E. Milot. 2003. Developmental stage-specific epigenetic control of human beta-globin gene expression is potentiated in hematopoietic progenitor cells prior to their transcriptional activation. Blood 102:3989-3997. [DOI] [PubMed] [Google Scholar]
- 7.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274:18285-18289. [DOI] [PubMed] [Google Scholar]
- 8.Burdon, T., A. Smith, and P. Savatier. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 12:432-438. [DOI] [PubMed] [Google Scholar]
- 9.Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643-655. [DOI] [PubMed] [Google Scholar]
- 10.DeKoter, R. P., and H. Singh. 2000. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 288:1439-1441. [DOI] [PubMed] [Google Scholar]
- 11.Dieci, G., and A. Sentenac. 2003. Detours and shortcuts to transcription reinitiation. Trends Biochem. Sci. 28:202-209. [DOI] [PubMed] [Google Scholar]
- 12.Dillon, N., and P. Sabbattini. 2000. Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. Bioessays 22:657-665. [DOI] [PubMed] [Google Scholar]
- 13.Drewell, R. A., E. Bae, J. Burr, and E. B. Lewis. 2002. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl. Acad. Sci. USA 99:16853-16858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dugray, A., J. F. Geay, A. Foudi, M. L. Bonnet, W. Vainchenker, F. Wendling, F. Louache, and A. G. Turhan. 2001. Rapid generation of a tetracycline-inducible BCR-ABL defective retrovirus using a single autoregulatory retroviral cassette. Leukemia 15:1658-1662. [DOI] [PubMed] [Google Scholar]
- 15.Eisenbeis, C. F., H. Singh, and U. Storb. 1993. PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin lambda 2-4 enhancer. Mol. Cell. Biol. 13:6452-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher, A. G. 2002. Cellular identity and lineage choice. Nat. Rev. Immunol. 2:977-982. [DOI] [PubMed] [Google Scholar]
- 17.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gribnau, J., K. Diderich, S. Pruzina, R. Calzolari, and P. Fraser. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol. Cell 5:377-386. [DOI] [PubMed] [Google Scholar]
- 20.Hamalainen, H. K., J. C. Tubman, S. Vikman, T. Kyrola, E. Ylikoski, J. A. Warrington, and R. Lahesmaa. 2001. Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal. Biochem. 299:63-70. [DOI] [PubMed] [Google Scholar]
- 21.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10:1467-1477. [DOI] [PubMed] [Google Scholar]
- 22.Helmlinger, D., S. Hardy, S. Sasorith, F. Klein, F. Robert, C. Weber, L. Miguet, N. Potier, A. Van-Dorsselaer, J. M. Wurtz, J. L. Mandel, L. Tora, and D. Devys. 2004. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 13:1257-1265. [DOI] [PubMed] [Google Scholar]
- 23.Hirota, S., T. Nishida, K. Isozaki, M. Taniguchi, K. Nishikawa, A. Ohashi, A. Takabayashi, T. Obayashi, T. Okuno, K. Kinoshita, H. Chen, Y. Shinomura, and Y. Kitamura. 2002. Familial gastrointestinal stromal tumors associated with dysphagia and novel type germline mutation of KIT gene. Gastroenterology 122:1493-1499. [DOI] [PubMed] [Google Scholar]
- 24.Ho, Y., F. Elefant, N. Cooke, and S. Liebhaber. 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9:291-302. [DOI] [PubMed] [Google Scholar]
- 25.Hromas, R., A. Orazi, R. S. Neiman, R. Maki, C. Van Beveran, J. Moore, and M. Klemsz. 1993. Hematopoietic lineage- and stage-restricted expression of the ETS oncogene family member PU.1. Blood 82:2998-3004. [PubMed] [Google Scholar]
- 26.Hu, M., D. Krause, M. Greaves, S. Sharkis, M. Dexter, C. Heyworth, and T. Enver. 1997. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11:774-785. [DOI] [PubMed] [Google Scholar]
- 27.Ishihara, K., W. J. Wood, Jr., M. Damore, G. G. Hermanson, R. Wall, and P. W. Kincade. 1992. B29 gene products complex with immunoglobulins on B lymphocytes. Proc. Natl. Acad. Sci. USA 89:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 29.Jimenez, G., S. D. Griffiths, A. M. Ford, M. F. Greaves, and T. Enver. 1992. Activation of the beta-globin locus control region precedes commitment to the erythroid lineage. Proc. Natl. Acad. Sci. USA 89:10618-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, K. D., J. A. Grass, C. Park, H. Im, K. Choi, and E. H. Bresnick. 2003. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol. Cell. Biol. 23:6484-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katsura, Y. 2002. Redefinition of lymphoid progenitors. Nat. Rev. Immunol. 2:127-132. [DOI] [PubMed] [Google Scholar]
- 33.Kee, B. L., M. W. Quong, and C. Murre. 2000. E2A proteins: essential regulators at multiple stages of B-cell development. Immunol. Rev. 175:138-149. [PubMed] [Google Scholar]
- 34.Kiekhaefer, C. M., J. A. Grass, K. D. Johnson, M. E. Boyer, and E. H. Bresnick. 2002. Hematopoietic-specific activators establish an overlapping pattern of histone acetylation and methylation within a mammalian chromatin domain. Proc. Natl. Acad. Sci. USA 99:14309-14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, A., and A. Dean. 2004. Developmental stage differences in chromatin subdomains of the beta-globin locus. Proc. Natl. Acad. Sci. USA 101:7028-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura, A. P., S. A. Liebhaber, and N. E. Cooke. 2004. Epigenetic modifications at the human growth hormone locus predict distinct roles for histone acetylation and methylation in placental gene activation. Mol. Endocrinol. 18:1018-1032. [DOI] [PubMed] [Google Scholar]
- 37.Kimura, H., M. Tada, N. Nakatsuji, and T. Tada. 2004. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol. Cell. Biol. 24:5710-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitanaka, A., T. Suzuki, C. Ito, H. Nishigaki, E. Coustan-Smith, T. Tanaka, Y. Kubota, and D. Campana. 1999. CD38-mediated signaling events in murine pro-B cells expressing human CD38 with or without its cytoplasmic domain. J. Immunol. 162:1952-1958. [PubMed] [Google Scholar]
- 39.Kontaraki, J., H. H. Chen, A. Riggs, and C. Bonifer. 2000. Chromatin fine structure profiles for a developmentally regulated gene: reorganization of the lysozyme locus before trans-activator binding and gene expression. Genes Dev. 14:2106-2122. [PMC free article] [PubMed] [Google Scholar]
- 40.Koyama, N., D. Hoelzer, and O. G. Ottmann. 2004. Regulation of human IL-18 gene expression: interaction of PU.1 with GC-box binding protein is involved in human IL-18 expression in myeloid cells. Eur. J. Immunol. 34:817-826. [DOI] [PubMed] [Google Scholar]
- 41.Lee, J. H., S. R. Hart, and D. G. Skalnik. 2004. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis 38:32-38. [DOI] [PubMed] [Google Scholar]
- 42.Lefevre, P., S. Melnik, N. Wilson, A. D. Riggs, and C. Bonifer. 2003. Developmentally regulated recruitment of transcription factors and chromatin modification activities to chicken lysozyme cis-regulatory elements in vivo. Mol. Cell. Biol. 23:4386-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, Y. S., K. Hayakawa, and R. R. Hardy. 1993. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med. 178:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 45.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, X., J. Tesfai, Y. A. Evrard, S. Y. Dent, and E. Martinez. 2003. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J. Biol. Chem. 278:20405-20412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundgren, M., C. M. Chow, P. Sabbattini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103:733-743. [DOI] [PubMed] [Google Scholar]
- 48.Masternak, K., N. Peyraud, M. Krawczyk, E. Barras, and W. Reith. 2003. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 49.Melchers, F., H. Karasuyama, D. Haasner, S. Bauer, A. Kudo, N. Sakaguchi, B. Jameson, and A. Rolink. 1993. The surrogate light chain in B-cell development. Immunol Today. 14:60-68. [DOI] [PubMed] [Google Scholar]
- 49a. Minaee, S., D. Farmer, A. Georgiou, P. Sabbattini, Z. Webster, C.-M. Chow, and N. Dillon. Mapping and Functional analysis of regulatory sequences in the mouse λ5-VpreB1 domain. Mol. Immunol., in press. [DOI] [PubMed]
- 50.Miyamoto, T., H. Iwasaki, B. Reizis, M. Ye, T. Graf, I. L. Weissman, and K. Akashi. 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3:137-147. [DOI] [PubMed] [Google Scholar]
- 51.Muchardt, C., and M. Yaniv. 2001. When the SWI/SNF complex remodels. the cell cycle. Oncogene 20:3067-3075. [DOI] [PubMed] [Google Scholar]
- 52.Muller, F., and L. Tora. 2004. The multicoloured world of promoter recognition complexes. EMBO J. 23:2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musikacharoen, T., A. Oguma, Y. Yoshikai, N. Chiba, A. Masuda, and T. Matsuguchi. 2005.. Interleukin-15 induces IL-12 receptor {beta}1 gene expression through PU.1 and IRF 3 by targeting chromatin remodeling. Blood 105:711-720. [DOI] [PubMed]
- 54.Nielsen, A. L., C. Sanchez, H. Ichinose, M. Cervino, T. Lerouge, P. Chambon, and R. Losson. 2002. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1alpha. EMBO J. 21:5797-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 56.O'Neill, L. P., and B. M. Turner. 2003. Immunoprecipitation of native chromatin: NChIP. Methods 31:76-82. [DOI] [PubMed] [Google Scholar]
- 57.Palacios, R., and M. Steinmetz. 1985. IL-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell 41:727-734. [DOI] [PubMed] [Google Scholar]
- 58.Palstra, R. J., B. Tolhuis, E. Splinter, R. Nijmeijer, F. Grosveld, and W. de Laat. 2003. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35:190-194. [DOI] [PubMed] [Google Scholar]
- 59.Pesu, M., S. Aittomaki, T. Valineva, and O. Silvennoinen. 2003. PU.1 is required for transcriptional activation of the Stat6 response element in the Igepsilon promoter. Eur. J. Immunol. 33:1727-1735. [DOI] [PubMed] [Google Scholar]
- 60.Plant, K. E., S. J. Routledge, and N. J. Proudfoot. 2001. Intergenic transcription in the human beta-globin gene cluster. Mol. Cell. Biol. 21:6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rank, G., M. Prestel, and R. Paro. 2002. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 22:8026-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rastegar, M., L. Kobrossy, E. N. Kovacs, I. Rambaldi, and M. Featherstone. 2004. Sequential histone modifications at hoxd4 regulatory regions distinguish anterior from posterior embryonic compartments. Mol. Cell. Biol. 24:8090-8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogan, D. F., D. J. Cousins, S. Santangelo, P. A. Ioannou, M. Antoniou, T. H. Lee, and D. Z. Staynov. 2004. Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc. Natl. Acad. Sci. USA 101:2446-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rossant, J. 2001. Stem cells from the mammalian blastocyst. Stem Cells 19:477-482. [DOI] [PubMed] [Google Scholar]
- 65.Routledge, S. J., and N. J. Proudfoot. 2002. Definition of transcriptional promoters in the human beta globin locus control region. J. Mol. Biol. 323:601-611. [DOI] [PubMed] [Google Scholar]
- 66.Sabbattini, P., A. Georgiou, C. Sinclair, and N. Dillon. 1999. Analysis of mice with single and multiple copies of transgenes reveals a novel arrangement for the λ5-VpreB1 locus control region. Mol. Cell. Biol. 19:671-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakaguchi, N., S. Kashiwamura, M. Kimoto, P. Thalmann, and F. Melchers. 1988. B lymphocyte lineage-restricted expression of mb-1, a gene with CD3-like structural properties. EMBO J. 7:3457-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakamoto, N., K. Shibuya, Y. Shimizu, K. Yotsumoto, T. Miyabayashi, S. Sakano, T. Tsuji, E. Nakayama, H. Nakauchi, and A. Shibuya. 2001. A novel Fc receptor for IgA and IgM is expressed on both hematopoietic and non-hematopoietic tissues. Eur. J. Immunol. 31:1310-1316. [DOI] [PubMed] [Google Scholar]
- 69.Schneider, R., A. J. Bannister, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6:73-77. [DOI] [PubMed] [Google Scholar]
- 70.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 71.Sigvardsson, M., M. O'Riordan, and R. Grosschedl. 1997. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity 7:25-36. [DOI] [PubMed] [Google Scholar]
- 72.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 73.Spicuglia, S., S. Kumar, J. H. Yeh, E. Vachez, L. Chasson, S. Gorbatch, J. Cautres, and P. Ferrier. 2002. Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Mol. Cell 10:1479-1487. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki, M., T. Yamada, F. Kihara-Negishi, T. Sakurai, and T. Oikawa. 2003. Direct association between PU.1 and MeCP2 that recruits mSin3A-HDAC complex for PU.1-mediated transcriptional repression. Oncogene 22:8688-8698. [DOI] [PubMed] [Google Scholar]
- 75.Svanvik, N., A. Stahlberg, U. Sehlstedt, R. Sjoback, and M. Kubista. 2000. Detection of PCR products in real time using light-up probes. Anal. Biochem. 287:179-182. [DOI] [PubMed] [Google Scholar]
- 76.Tagoh, H., R. Himes, D. Clarke, P. J. Leenen, A. D. Riggs, D. Hume, and C. Bonifer. 2002. Transcription factor complex formation and chromatin fine structure alterations at the murine c-fms (CSF-1 receptor) locus during maturation of myeloid precursor cells. Genes Dev. 16:1721-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tagoh, H., S. Melnik, P. Lefevre, S. Chong, A. D. Riggs, and C. Bonifer. 2004. Dynamic reorganization of chromatin structure and selective DNA demethylation prior to stable enhancer complex formation during differentiation of primary hematopoietic cells in vitro. Blood 103:2950-2955. [DOI] [PubMed] [Google Scholar]
- 78.Tagoh, H., A. Schebesta, P. Lefevre, N. Wilson, D. Hume, M. Busslinger, and C. Bonifer. 2004. Epigenetic silencing of the c-fms locus during B-lymphopoiesis occurs in discrete steps and is reversible. EMBO J. 23:4275-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tufarelli, C., J. A. Stanley, D. Garrick, J. A. Sharpe, H. Ayyub, W. G. Wood, and D. R. Higgs. 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 34:157-165. [DOI] [PubMed] [Google Scholar]
- 80.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 81.van der Stoep, N., E. Quinten, M. M. Rezende, and P. J. van den Elsen. 2004. E47, IRF-4, and PU.1 synergize to induce B-cell-specific activation of the class II transactivator promoter III (CIITA-PIII). Blood 104:2849-2857. [DOI] [PubMed] [Google Scholar]
- 82.Volpe, C. P., A. Lundgren, A. Aints, A. J. Mohamed, P. Jaakkola, B. Christensson, G. Gahrton, M. Jalkanen, C. I. Smith, and M. S. Dilber. 2001. Proximal promoter of the murine syndecan-1 gene is not sufficient for the developmental pattern of syndecan expression in B lineage cells. Am. J. Hematol. 67:20-26. [DOI] [PubMed] [Google Scholar]
- 83.White, J., C. Brou, J. Wu, Y. Lutz, V. Moncollin, and P. Chambon. 1992. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 11:2229-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 85.Ye, M., H. Iwasaki, C. V. Laiosa, M. Stadtfeld, H. Xie, S. Heck, B. Clausen, K. Akashi, and T. Graf. 2003. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity 19:689-699. [DOI] [PubMed] [Google Scholar]