Abstract
To explore the role of mPer2 in the circadian oscillation in the mammalian cellular clock, we established fibroblast cell lines in which expression of mPer2 is controlled through a tetracycline-regulatable promoter. We revealed that constitutive expression and overexpression of mPer2 mRNA severely impair serum shock-induced cyclic circadian clock gene expression. Moreover, under conditions of lower mPer2 mRNA expression, mPER2 protein accumulation in these cells showed clear circadian oscillation even in constitutive mPer2 mRNA expression, suggesting that the protein cycling of mPER2 was required for oscillation of the circadian feedback loop. Since the rhythms of gene expression driven by the intrinsic clock oscillation system dampen rapidly in the absence of cyclic expression of mPer2, the transcriptional rhythm helps to sustain the clock oscillation.
Circadian rhythmicity is observed in many aspects of basic cellular functions such as membrane excitation and energy metabolism as well as behavioral events such as sleep-waking cycles. In mammals, all these circadian rhythms are generated at the cellular level by the circadian core oscillator composed of an autoregulatory transcription-(post)translation-based feedback loop involving a set of clock genes. In the past several years, molecular understanding of the mammalian circadian system has progressed considerably. Indeed, genetic dissection of mammalian genes has revealed that mPer2 (4, 33), mCry (24), Clock (25), and Bmal1 (7) genes were indispensable components of the clock. The transcription-(post)translation feedback loop, which is supposed to exist in mammals as in other organisms, is understood mainly from the scheme of Drosophila melanogaster (3, 10, 20, 31), since many of the components are common in mammals and in Drosophila (10, 32).
However, there is a considerable difference in the roles of molecules in Drosophila and in mammals; e.g., Drosophila cry and mammalian mCry1/mCry2 (32). Particularly, the central role of per in the negative feedback loop is established in Drosophila (13, 32), but the role of its mammalian homologue mPer genes was not determined yet. The negative effects of mPer gene products were detected by luciferase reporter gene assay in mammalian cell lines, but the suppression ability of mPER1, mPER2, and mPER3 on BMAL1/CLOCK was far weaker than those of mCRY1 and mCRY2 (15). This was completely different from the suppression ability of Drosophila per, which shows a very strong suppressive effect at the transcription level (31). No further data suggesting that the mPer genes were negative components in mammals were reported, although considerable evidence was accumulated for Drosophila (32).
As in Drosophila, mPer genes are speculated to be crucial for the generation of rhythms. Among mPer genes, mPer2 is considered to be the most important gene since gene targeting studies have demonstrated that the deletion of mPer2 induced arrhythmicity at both the behavioral and molecular levels, although the deletion of mPer1 only shortened the period length, and the deletion of mPer3 yielded almost normal locomotor activity (4, 33). Furthermore, the introduction of the mPer2 gene as well as the mPer1 gene into the arrhythmic per01 mutant of Drosophila, which are otherwise arrhythmic due to a lack of endogenous PER protein, restored rhythm (22). Thus, mPer2 is thought to be a counterpart of Drosophila per among mammalian mPer genes. Unfortunately, despite a number of studies, no data showing that mPer2 is the crucial molecule for determining the state of circadian rhythms are available.
To prove that mPer2 is the central molecule which determines the state of the rhythms, the handling of the level of expression of this gene is crucial. As reported previously, serum shock can induce circadian gene expression of a variety of genes in cultured fibroblasts (1, 6, 18, 27). Since it has been demonstrated that the oscillation in this in vitro system occurs from the basically common core circadian feedback loop as in vivo (27), this culture system can be used as a model to address mPer2 function at a cellular level.
For this purpose, we established subsets of fibroblast cell lines in which expression of mPer2 is controlled through a tetracycline-regulatable (Tet-Off) transcription factor with the application of a high concentration of horse serum to generate clock gene oscillation (1, 6, 18, 27). In these cell lines, exogenously expressed mPer2 is driven by a tetracycline response element (TRE)-containing constitutive promoter which is not under the control of circadian molecular feedback loops. To establish this Tet-Off system as a mammalian model system of circadian rhythm in the present study, we evaluated the effect of induced mPer2 expression in NIH 3T3 fibroblast cells with the identical genetic background before and after exposure to doxycycline, a derivative of tetracycline.
MATERIALS AND METHODS
Cell culture and generation of cell lines.
To obtain mPer2-expressing cell lines regulated by tetracycline, we used the Tet-Off system (BD Biosciences Clontech, Palo Alto, Calif.). Before transfection, 4 × 105 NIH 3T3 cells were plated onto a 6-cm-diameter dish. NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS), and pTet-Off vector was transfected. After 2 days, we started selection for 2 weeks with 1 mg of G418 (GIBCO)/ml. We picked up colonies and transfected pTRE2hyg-Luc into cloned cells transiently. A colony that has low background with doxycycline (+Dox) and that has high luciferase activity without doxycycline (−Dox) was selected. Secondly, mPer2 pTRE2 and pTK-Hyg were cotransfected into cloned NIH 3T3 cells that contained pTet-Off, and cells were selected for 2 weeks with 400 μg of hygromycin B (Invivogen)/ml and 200 μg of G418/ml. Colonies were picked up and cultured for ∼4 to 5 days with 2 μg of doxycycline/ml or without doxycycline. Induction of mPER2 protein was detected by Western blotting assay, and some cell lines were generated. The serum shock was done as follows. A total of 5 × 105 NIH 3T3 cells were plated onto a 6-cm-diameter dish 3 days before the experiment. Cells were cultured in DMEM-10% FBS supplemented with 100 μg of hygromycin B/ml and 200 μg of G418/ml and with 2 μg of doxycycline/ml or without doxycycline, respectively. Twelve hours before serum shock, the medium was exchanged with DMEM-5% FBS with or without doxycycline. At time zero, the medium was exchanged with DMEM and 50% horse serum (GIBCO), and after 1 h, this medium was replaced with serum-free DMEM with or without doxycycline. At time zero, cells were harvested before serum shock, and at the indicated times, the whole-cell RNA or protein was collected from cultured cells.
Northern blot analysis.
Cultured cells were washed three times with ice-cold phosphate-buffered saline (PBS) and harvested in 1 ml of TRIzol reagent (Invitrogen). These samples were frozen and stored at −70°C until whole-cell RNA was extracted. Ten micrograms of total RNA was electrophoresed in a 1.2% agarose gel containing 2% formaldehyde. RNAs were transferred to Byodyne membrane (PALL BioSupport, New York, N.Y.) and hybridized with probes. To compare between Dox+ and Dox− conditions, RNAs obtained under each culture condition were applied onto the same gel and transferred as one membrane. For mPer2, we used 1 to 878 bp (GenBank accession number NM_011066) of mPer2 cDNA for the 5′ probe. To detect endogenous mPer2, 3,939 to 5,816 bp (GenBank accession number NM_011066) of the 3′ noncoding region of mPer2 cDNA were used for probes. For dbp, the total coding region of mouse dbp (GenBank accession number U29762) was cloned by reverse transcription-PCR and ligated into the pCR2.1 TOPO vector (Invitrogen). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Clontech) was used as a control. Probes were labeled with [32P]dCTP using a TaKaRa (Tokyo, Japan) random primer labeling kit. Hybridization was performed at 42°C for 16 h, and membranes were washed twice in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 60°C for 30 min. Membranes were exposed to an imaging plate and analyzed by BAS 5000 (Fuji Film, Tokyo, Japan). For rehybridization purposes, probes were stripped in water at 95°C for 3 min.
Western blot analysis.
At the indicated times, cells were washed three times with ice-cold PBS and harvested in 50 μl of SDS sample buffer (125 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 0.05% bromophenol blue, 1 μM phenylmethylsulfonyl fluoride, 50 mM NaF, 100 μM NaVO3, 40 mM dithiothreitol). After these samples were boiled for 3 min, 20 μl of these samples was separated by 6.5% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride membrane (Immunoblot-P membrane; Atto, Tokyo, Japan). As primary antibodies, rabbit anti-mPER2 (affinity purified, 1:500; Alpha Diagnostic International, San Antonio, Tex.) and anti-ACTINE (1:1,000; Santa Cruz) antibodies were used. Cy3 anti-rabbit immunoglobulin G antibody (1:2,000; Jackson Immuno Research Laboratories) was used as a secondary antibody. Chemiluminescence was performed by using Western Blotting Luminol reagent (Santa Cruz) and analyzed by LAS 1000 (Fuji Film).
Immunoprecipitation.
Immunoprecipitation was performed 3 days after the cell culture without doxycycline by use of whole-cell lysates harvested with 0.2 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 50 mM NaF, 100 μM NaVO3, and Complete Mini Protease Inhibitor [Roche, Mannheim, Germany]). Total cell lysate was centrifuged at top speed for 10 min at 4°C. The supernatant was transferred to a fresh microtube, and anti-FLAG M2 antibodies (Sigma) were added and incubated for 2 h at 4°C with mild agitation. After adding 30 μl of protein G-agarose equilibrated with lysis buffer, beads were collected and washed twice with wash buffer (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.1% NP-40, 0.05% sodium deoxycholate). After removing the supernatant completely, 8 μl of 3× SDS sample buffer was added, and samples were boiled for 5 min.
Pulse-chase.
P2-#19 cells were cultured in doxycycline-free DMEM-10% FBS for 3 days. Cells were then cultured in methionine- and cysteine-free medium with [35S]methionine and cysteine (250 mCi/ml) for 1 h and then chased for the indicated times. Cells were lysed and immunoprecipitated with anti-mPER2 antibody, and the mPER2 protein was resolved by 6.5% SDS-PAGE. Radiolabeled signals were analyzed by BAS5000.
Comparative immunofluorescence.
A total of 106 Per2-#19 cells were plated onto a 10-cm-diameter dish and cultured in DMEM-10% FBS with 2 μg of doxycycline/ml or without doxycycline. After 12 h, 400 μl of bead solution (Polybeads, 0.7-μm latex microsphere; Polyscience, Inc., Warrington, Pa.) was added to medium and cultured for 2 days. Cells were washed with PBS, trypsinized, and centrifuged at 1,200 × g for 3 min. The Per2-#19 cells labeled with beads and NIH 3T3 cells without labeling were mixed, and 2.5 × 105 cells were plated onto coverslips and cultured overnight in serum-free DMEM with or without 2 μg of doxycycline/ml. Serum shock was performed, and cells were fixed with 4% paraformaldehyde at the indicated times. mPER2 protein was detected by using an anti-mPER2 antibody (affinity purified, 1:500) followed by a Cy3 anti-rabbit immunoglobulin G antibody (1:2,000). We used DAPI (4′,6′-diamidino-phenylindole) for staining of the nuclei.
Transcriptional assay.
Luciferase reporter gene assays were performed with NIH 3T3 cells. Cells (2 × 105) were seeded onto a 6-well plate and transfected the following day. In this study, we used the Dual Luciferase Reporter assay system (Promega). Each transfection reaction mixture contained a pGL3 promoter vector subcloned into the DNA fragment of E-box enhancer elements. Human Clock and human Bmal1 were used at 750 ng per transfection. One hundred nanograms of pTet-Off vector and 400 ng of pTRE2-mPer2 were used per transfection. The total amount of DNA per dish was adjusted to 2 μg by adding pcDNA3 vector as carrier. Forty-eight hours after transfection, cells were harvested to determine luciferase activity by illuminometer.
RESULTS
Establishment of doxycycline-regulatable mPer2 stable cell lines.
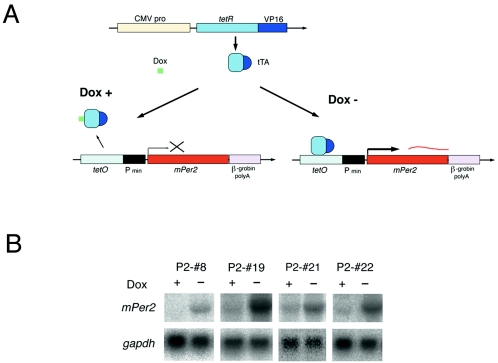
We first established subsets of NIH 3T3 lines in which expression of mPer2 is controlled through the Tet-Off promoter (Fig. 1). We chose Tet-Off-regulated mPer2 double stable cell lines showing various types of induction levels. The P2-#8 line exhibited almost no basal expression of exogenous mPer2 mRNA in the presence of doxycycline (Dox+), but after removal of doxycycline (Dox−), a moderate level of mPer2 mRNA was induced (Fig. 1B). On the other hand, other cell lines (P2-#19, P2-#21, and P2-#22) exhibited weak basal expression of mPer2 mRNA under Dox+ conditions, and the removal of doxycycline induced a high level of mPer2 (Fig. 1B). Among these lines, P2-#19 lines showed the highest level of mPer2 mRNA under Dox− conditions. By using these various cell lines, we address the dose effect of constitutively expressed mPer2 in the circadian feedback loop in the same genetic backgrounds.
FIG. 1.
Conditional expression of mPer2 in NIH 3T3 fibroblasts using the Tet-Off system. (A) Schematic drawing of the tetracycline-regulatable mPer2 expression system. Dox represents doxycycline, which is a derivative of tetracycline. In this Tet-Off system (BD Biosciences Clontech), the tetracycline-controlled transactivator (tTA), which is composed of tetracycline repressor protein (tetR) and the VP16 activation domain of herpes simplex virus, is expressed from the constitutive cytomegalovirus promoter (CMV pro). When Dox is absent (Dox−), tTa binds to TetO (tetracycline operator) and Pmin (CMV minimum promoter) activates mPer2 expression. In the presence of Dox (Dox+), mPer2 expression is not activated (B) Expression of exogenous mPer2 mRNA in pTet-Off and pTRE2-mPer2 double stable NIH 3T3 cell lines. The P2-#8 cell line shows almost no expression of mPer2 under Dox+ conditions, whereas apparent induction of mPer2 is seen under Dox− conditions. P2-#19, P2-#21, and P2-#22 cell lines represent weak basal expression under Dox+ conditions and strong induction of mPer2 under Dox− conditions.
pTet-Off-induced mPER2 suppresses CLOCK/BMAL1-mediated transcription.
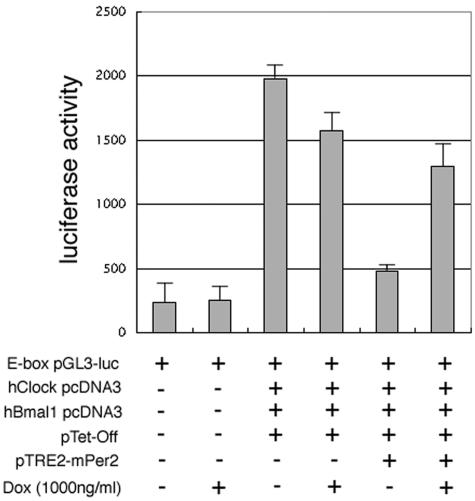
Before analyzing the effect of pTet-Off on circadian rhythms, we first confirmed whether our mPER2 produced by the pTet-Off vector is a functionally active negative regulator to inhibit the BMAL1/CLOCK-mediated transcriptional activity in the luciferase reporter gene assay (Fig. 2). We cotransfected pTet-Off- and mPer2-subcloned pTRE2 vectors in this assay because we used these constructs to generate Tet-Off/mPer2 NIH 3T3 stable cell lines. To eliminate the possibility of some unexpected effects by the pTet-Off vector, we checked the effect of expression of the Tet-Off vector-derived product with and without doxycycline. Since CLOCK/BMAL1 dramatically increased the luciferase activity in pTet-Off-transfected cells as described in previous reports (15), the coexistence of the pTet-Off vector does not interfere with the increase of transcription by CLOCK/BMAL1 (Fig. 2). During cotransfection of both pTet-Off and mPer2-pTRE2, luciferase activity was suppressed under Dox− conditions, whereas much less inhibitory effect was observed under Dox+ conditions (Fig. 2). Thus, pTet-Off-induced mPER2 is functionally active for suppressing E-box-mediated CLOCK/BMAL1 transcriptional activation.
FIG. 2.
Doxycycline-dependent suppression of CLOCK/BMAL1-mediated transcription by Tet system-driven mPER2. NIH 3T3 cells were transfected with human Clock (hClock), hBmal1, pTet-Off, and pTRE2-mPer2 expression vectors. E-box-mediated transcriptional activity was measured in the absence (−) or presence (+) of doxycycline (Dox) treatment using E-box luciferase reporter plasmid (E-box pGL3-luc). In the absence of doxycycline, relative luciferase activity was suppressed under both pTRE2-mPer2- and pTet-Off-cotransfected conditions. CLOCK/BMAL1-mediated transcriptional activity was not suppressed only under pTet-Off-transfected conditions. In the presence of 1,000 ng of doxycycline/ml, the transcriptional activity rose again. Each value is the mean ± standard error of the mean of data from three replicate experiments.
Impairment of cyclic expression of endogenous mPer2 by Tet-Off-induced overexpressed mPer2.
We then analyzed the effect of constitutively expressed mPer2 on the core oscillation of the circadian feedback loop itself and its outputs. After the serum shock, we examined the expression profile of the 3′ noncoding region of mPer2 as a state marker of core clock oscillator and dbp as a representative of the clock-controlled gene directly regulated by the circadian feedback loops (21, 29).
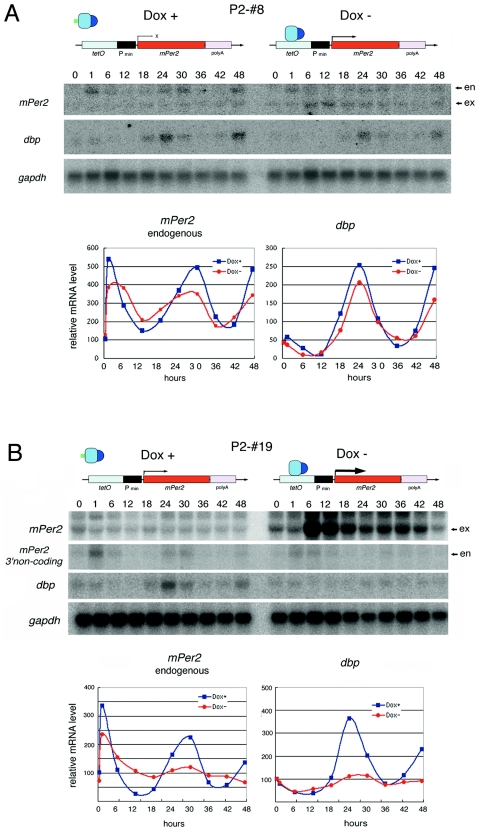
In the P2-#8 line under DOX+ conditions, dbp and endogenous mPer2 mRNA (Fig. 3A) showed robust circadian rhythm as expected (Fig. 2A). Under Dox− conditions, moderate levels of Tet-Off system-driven mPer2 were expressed without showing circadian change throughout the experiments (Fig. 3A). Even under this constitutive (at least noncircadian in Northern blot analysis) mPer2 expression condition, dbp mRNA and endogenous mPer2 mRNA (Fig. 3A) showed clear circadian rhythms (Fig. 3A, right diagram). We then analyzed the P2-#19 line, which exhibited constitutive but weaker basal exogenous mPer2 expression even under Dox+ conditions and much stronger mPer2 induction under Dox− conditions (Fig. 3B). Under Dox+ conditions, molecular rhythms of dbp and endogenous mPer2 mRNA are still obvious until the second cycle. On the other hand, Dox− conditions induced constitutive high levels of mPer2 expression which severely impaired the circadian rhythm of both dbp and endogenous mPer2 mRNA (Fig. 3B). Under these conditions, peaks of dbp and endogenous mPer2 expression are suppressed, suggesting that overexpressed products of the mPer2 gene inhibited the rise of their expression. Suppression of endogenous mPer2 and dbp peaks at high levels of constitutive mPer2 expression under Dox− conditions was also observed in P2-#21 and P2-#22 Tet-Off/mPer2 cell lines (data not shown). Thus, the inhibition of cyclicity of internal circadian clock oscillation by the mPer2 overexpression induced under Dox− conditions is the first demonstration which suggests that mammalian circadian oscillation is generated through the cyclically regulated mPer2 gene products.
FIG. 3.
Constitutive expression and overexpression of mPer2 mRNA severely impair serum shock-induced circadian gene expression in NIH 3T3 fibroblast cells. (A) Serum shock-induced circadian gene expression in the P2-#8 cell line. Clear circadian gene expression of endogenous mPer2 and dbp mRNAs are observed under both Dox+ and Dox− conditions. Quantitative results are shown on the right graphs. en indicatesendogenous mPer2 mRNA; ex indicates moderate levels of Tet-Off system-driven mPer2 expressed without showing circadian change throughout the experiments. (B) Serum shock-induced circadian gene expressions in the P2-#19 cell line. Apparent circadian cycles of endogenous mPer2 and dbp mRNA expression were seen under Dox+ conditions. However, circadian gene expression is severely impaired under Dox− conditions. Quantitative results are shown in the right graph.
Accumulation profile of mPER2 protein in Tet-Off/mPer2 cells.
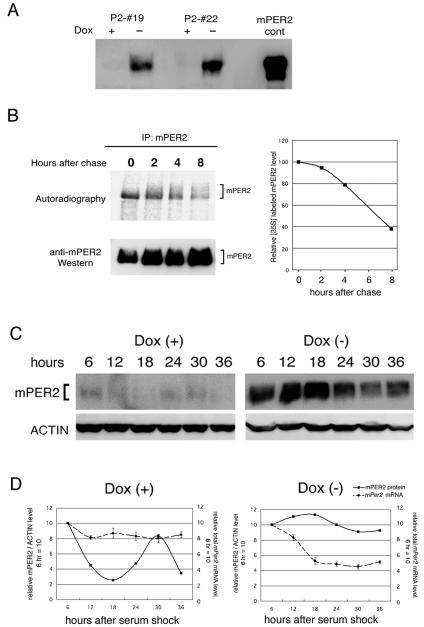
Next, we analyzed the fate of the forced expression of mPER2 protein in Tet-Off/mPer2 cells. We first characterized mPER2 protein induced under Dox− conditions in various Tet-Off/mPer2 cell lines (Fig. 4A). The P2-#19 cell line strongly induced mPER2 protein under Dox− conditions, whereas only faint signals of mPER2 were detected under Dox+ conditions. It has been known that the progressive phosphorylation following degradation of mPER2 protein is an important process within the circadian feedback loop (17, 23). Thus, we characterized the induced mPER2 protein by the pulse-chase method, where cells cultured in Dox− medium were pulse-labeled for 1 h with [35S]methionine and then chased. The 35S-labeled mPER2 band was sharp, and no shifted band was seen at the time point just after the chase (Fig. 4B, left panel). The shifted band appeared after 2 h and developed over time (Fig. 4B, left panel). The shifted band is thought to be phosphorylated mPER2 because it disappeared after incubation with calf intestinal phosphatase (data not shown) (8). Total 35S-labeled mPER2 signals weakened over time as quantified in right panel of Fig. 4B, which indicated that expressed mPER2 not only phosphorylated but also degraded. Thus, the present Tet-Off cell lines undergo progressive phosphorylation and degradation of mPER2, similar to what is seen in vivo (16, 19).
FIG. 4.
Rhythmicity of gene expression depends on cyclic mPER2 accumulation in Tet-Off/mPer2 cells that constitutively express mPer2 mRNA. (A) P2-#19 and P2-#22 Tet-Off/mPer2 cell lines exhibit doxycycline-dependent mPER2 protein expression. mPER2 cont represent control sample extracted from mPer2-pcDNA3-transfected COS7 cells. (B) Pulse-chase labeling of mPER2. P2-#19 cells without doxycycline were pulse-labeled with [35S]methionine and cysteine (250 mCi/ml) for 1 h and then chased for the times (hours) indicated at the top of each lane. Cells were lysed and immunoprecipitated (IP) with anti-mPER2 antibody and resolved on 5.5% SDS-PAGE. The right graph represents quantitative results of immunoprecipitated 35S-labeled mPER2 levels. (C) Western blot analysis of temporal mPER2 accumulation patterns in whole-cell lysates of the P2-#19 cell line with (+) or without (−) doxycycline (Dox) after the serum shock. Whole-cell lysates were obtained from P2-#19 cells cultured with or without doxycycline at the indicated times after serum shock. Western blot with anti-actin antibody was done as a control. Under Dox+ conditions, robust circadian mPER2 accumulation rhythm was observed, whereas mPER2 accumulation under Dox− conditions was constitutively high. (D) Quantitative results of mPER2 protein accumulation and total mPer2 mRNA accumulation patterns. Total mPER2 accumulation represents the sum of endogenous and exogenous mPer2 mRNA signal intensity obtained from Northern blot analysis (n = 3). Relative densitometric units of mPER2/actin at 6 h after the serum shock under Dox+ conditions were set to 10.
Cyclic mPER2 protein expression is essential for cycling of circadian feedback loops.
Since the pulse-chase study has demonstrated that mPER2 proteins induced by the Tet-Off cell lines receive modifications similar to what is seen in vivo, we then analyzed the effect of overexpression of mPER2 protein on serum shock-induced circadian rhythms. Figure 4C shows the data obtained from the P2-#19 cell line (Fig. 4C). Interestingly, Western blot analysis revealed that accumulation of mPER2 protein clearly oscillated in a circadian manner under Dox+ conditions, where mPer2 mRNA is expressed constitutively, at least as determined by Northern blot analysis (Fig. 3B and 4C and D, left panels). On the other hand, under Dox− conditions, the accumulation of mPER2 was extremely strong at every time point after the serum shock and lacked circadian oscillation (Fig. 3B and 4C and D, right panels). Since dbp and endogenous mPer2 showed clear circadian rhythms, it is speculated that core clock itself oscillates and its effective outputs are conducted under these conditions. These results indicate that cyclic expression of mPER2 at the protein level but not necessarily at the transcription level is essential for cycling circadian feedback loops once or twice in cultured fibroblasts cells.
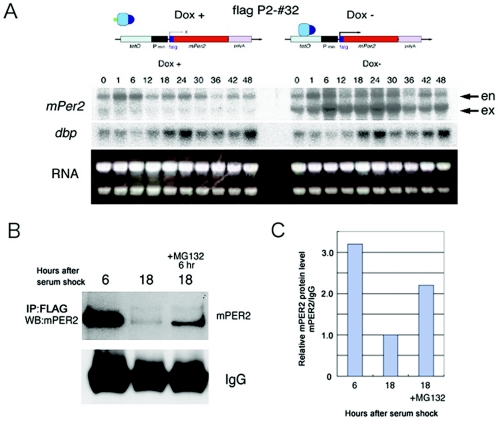
Since the ubiquitylation-proteasome system is known to be the important step for the mPER2 degradation (2, 28), we examined the involvement of this system in the mechanism generating the mPER2 protein cycle in the absence of the apparent mRNA accumulation cycle. For this purpose, we used the P2-#32 line, which carries FLAG-tagged mPer2. The P2-#32 cell line exhibited constitutive expression of exogenous FLAG-tagged mPer2 expression under Dox− conditions and faint expression of FLAG-tagged mPer2 mRNA under Dox+ conditions after the serum shock (Fig. 5A). Even though mPer2 mRNAs were constitutively expressed under Dox− conditions in this cell line, similar to what is found with the P2-#19 cell line under Dox+ conditions, dbp mRNA exhibited clear circadian oscillation. This result suggests that core clock oscillation of the P2-#32 cell line under Dox− conditions was still maintained for at least two cycles. By the immunoprecipitation assay using anti-FLAG antibodies, we detected strong accumulation of mPER2 at 6 h and very weak accumulation at 18 h after serum shock, which is compatible with results from Western blot analysis using another cell line (see Fig. 4C). Interestingly, the application of proteasome inhibitor MG132 showed the attenuation of the reduction of mPER2 at 18 h (Fig. 5B and C). This result suggests that the proteasome-mediated proteolysis plays a role in the generation of the mPER2 protein cycle in the absence of apparent mRNA accumulation.
FIG. 5.
Cyclic mPER2 protein accumulation accompanies the ubiquitin-proteasome-mediated degradation. (A) Serum shock-induced circadian gene expressions in the FLAG-tagged mPer2 P2-#32 cell line using the Tet-Off system. Under Dox+ conditions, expression of mPer2 mRNA was very faint at all points examined. Under Dox− conditions, higher levels of mPer2 mRNA were constitutively expressed without showing circadian change. Under both conditions, the expression of dbp mRNA exhibited clear circadian oscillation. en, endogenous mPer2 mRNA; ex, exogenous mPer2 mRNA. (B) FLAG-mPER2 was immunoprecipitated (IP) with anti-FLAG antibodies, and precipitates were examined by Western blot (WB) analysis using anti-mPER2 antibodies in the P2-#32 cell line without doxycycline after serum shock. The dense mPER2 accumulation found at 6 h became faint at 18 h after serum shock. Application of MG132 inhibited the reduction of mPER2 protein at 18 h. (C) Quantitative analysis of the expression of FLAG-mPER2 described above (B). IgG, immunoglobulin G.
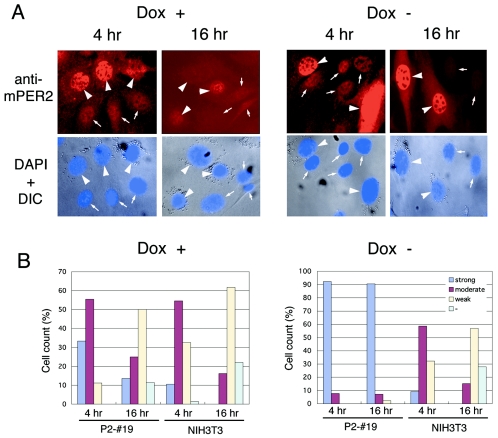
To examine the dynamics of the mPER2 protein at the cellular level, we then performed comparative immunofluorescence experiments using anti-mPER2 antibody in mixed culture of microspheric bead-incorporated P2-#19 cells and nonlabeled wild-type NIH 3T3 cells (Fig. 6). Under Dox+ conditions, expression of mPER2 in bead-labeled P2-#19 cells was slightly higher than that found with nonlabeled wild-type NIH 3T3 fibroblasts at 4 h after serum shock. However, at 16 h, cellular staining of mPER2 was weakened in both P2-#19 cells and NIH 3T3 cells (Fig. 6A and B, left panels). These results confirmed at the cellular level that the protein oscillation of mPER2 in Dox+ P2-#19 cells constitutively expressing mPer2 mRNA a at moderate level is similar to that seen in wild-type NIH 3T3 cells (Fig. 3B). On the other hand, under Dox− conditions, the expression of mPER2 protein in bead-labeled P2-#19 cells was much stronger than that found with nonlabeled wild-type NIH 3T3 cells not only at 4 h but also at 16 h after the serum shock (Fig. 6A and B, right panels). In the same mixed culture, nonlabeled wild-type NIH 3T3 cells showed an apparent decrease of mPER2 at 16 h compared to that at 4 h. Thus, mPer2-overexpressing cells lost their mPER2 protein oscillation, in contrast to the apparent cycling of mPER2 accumulation in wild-type NIH 3T3 cells, even when cocultured on the same coverslips (Fig. 6A and B right panels). These results revealed that under Dox+ conditions, P2-#19 cells behaved in a manner similar to that of wild-type NIH 3T3 cells, whereas under Dox− conditions, these Tet-Off/mPer2 cells constitutively expressed high levels of mPER2 protein in their nuclei. This result demonstrates that the level of mPer2 gene expression influences the state of oscillation of the circadian feedback loop at the cellular level.
FIG. 6.
Comparative immunofluorescence between P2-#19 cells and wild-type NIH 3T3 cells. (A) P2-#19 cells were labeled with latex microspheres and then mixed with nonlabeled wild-type NIH 3T3 cells. P2-#19 and NIH 3T3 cell mixtures were cultured on coverslips for 24 h. Fifty percent of horse serum was treated in these cells after culture for 12 additional hours in 5% fetal bovine serum-containing medium. Cells were fixed at the indicated times after serum shock, and immunofluorescence with anti-mPER2 antibody was performed. Lower panels represent merged pictures of DAPI stain and differential interference contrast (DIC). Arrowheads represent latex microspheres containing P2-#19 cells, and small arrows represent nuclei of wild-type NIH 3T3 cells. (B) Cell counts classified into four categories (strong, moderate, weak, and none [−]) from the intensity of nuclear staining of mPER2 under each experimental condition in each cell line. Data from the sum of triplicate experiments are exhibited.
Rapid dampening of the cellular rhythm in constitutively overexpressed mPer2 mRNA.
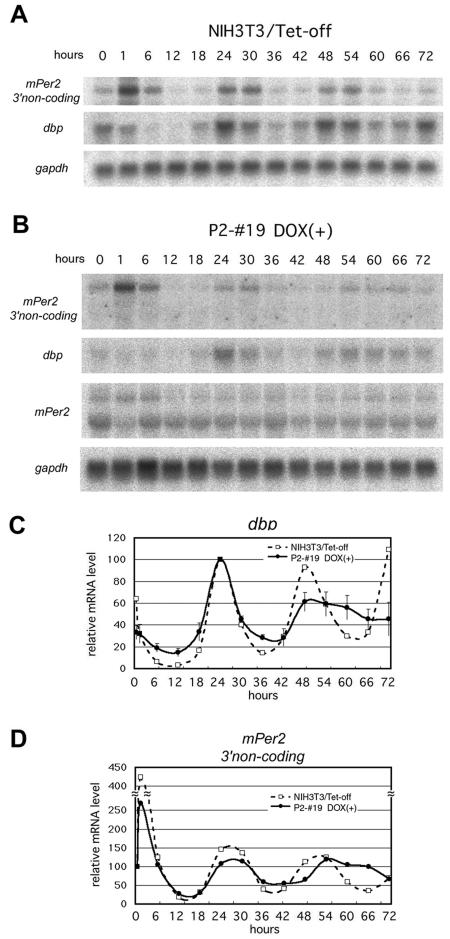
As shown in Fig. 3B, our data indicate that the cycle of mPer2 mRNA expression is not indispensable for the oscillation of the circadian feedback loop for at least two cycles, as shown in the Dox+ P2-#19 line. What, then, is the role of cyclic transcription of mPer2? To answer this question, we compared the endogenous mPer2 and dbp mRNA accumulation rhythms in the Tet-Off single stable transfected NIH 3T3 cell line (NIH 3T3/Tet-Off) and in the P2-#19 Tet-Off/mPer2 double stable cell line under Dox+ conditions for 3 days after the serum shock. In NIH 3T3/Tet-Off cells, the robust rhythm of mPer2 and dbp mRNA accumulation persisted for 3 days after the serum shock without dampening (Fig. 7A). On the other hand, with a doxycycline-treated P2-#19 line, the circadian endogenous mPer2 and dbp mRNA accumulation rhythm was observed until the second day, as shown in Fig. 3B, but no apparent rhythm was detected on the third day (Fig. 7B). These results support the hypothesis that cyclic transcriptional regulation of mPer2 is essential to sustain the oscillation of the circadian clock in mammalian cultured fibroblasts.
FIG. 7.
Constitutive mPer2 mRNA expression results in rapid dampening of circadian dbp and endogenous mPer2 mRNA expression rhythm. (A) The pTet-Off vector single stably transfected NIH 3T3 cell line exhibited robust circadian oscillation of mPer2 and dbp mRNA expression without apparent dampening for 3 days. (B) The P2-#19 Tet-Off/mPer2 double stable cell line cultured with doxycycline [Dox(+)] also showed circadian endogenous mPer2 (represented as 3′ noncoding) and dbp mRNA expression after serum shock for 2 days. However, amplitude was rapidly dampened, and no apparent rhythm was observed during the third day. (C and D) Expression profiles of dbp (C) and endogenous mPer2 (D) quantified from panels A and B. Open squares with broken lines represent pTet-Off single stably transfected NIH 3T3 cells, and solid circles with solid lines represent the P2-#19 cell line under Dox+ conditions. Error bars represent means ± standard errors of the mean. (n = 3).
DISCUSSION
Although a considerable number of studies about the mPer genes have been done, the role of mPer genes in circadian rhythms has not been addressed. In the present study, we address the role of the mPer2 gene, the indispensable component of mammalian circadian rhythms, and its products in the mammalian circadian oscillatory loop by establishing doxycycline-regulatable mPer2 expression in NIH 3T3 cell lines as the model of mammalian circadian system. The Tet-Off-induced mPER proteins are functionally active for suppressing CLOCK/BMAL1-mediated transcription and metabolized and phosphorylated in cell lines similar to that seen in vivo. In this cell line, the Tet-Off-induced constitutive high levels, but not low levels, of overexpression of mPer2 mRNA and mPER2 proteins severely impair serum shock-induced cyclic circadian clock gene expression, which indicates that mPer2 is the crucial molecule for determining the state of the circadian rhythms. The impairment of circadian clock oscillation in Tet-Off-induced mPer2 overexpression in cells is the first demonstration that the cyclicity of mPer2 gene product may involve regulating the mammalian circadian clock.
Using this model of constitutive expression of mPer2 genes under some adequate conditions, we found that mPER2 protein accumulation in these cells showed clear circadian oscillation, even in the presence of constitutive mPer2 mRNA expression. This finding suggests that posttranscriptional regulation of mPer2 plays an important role in generating the mPER2 accumulation cycle and following cycling of circadian feedback loops in mammals. This is similar to the role of Drosophila per, since cycling at the protein level without accompanying mRNA cycling was also reported (9, 11, 26, 30). Although the mechanism that generates the mPER2 protein cycle in the absence of the apparent mRNA accumulation cycle is still unclear at present, our finding using MG132 suggests that the proteasome-mediated proteolysis step has an important role in the generation. Ubiquitin-proteasome-mediated degradation is speculated to be important in Drosophila, in which the mutation of Drosophila slimb, an F-box ubiquitin ligase of the PER protein, is known to induce the constant accumulation of PER protein and behavioral arrhythmicity (12, 14). Alternatively, it has been known that phosphorylation of mPER proteins contributes to the protein instability, although the expression of casein kinase I ε and/or casein kinase I δ is constant throughout the day (17, 23).
Theoretically, all processes from synthesis to degradation of mPER2 proteins probably underly their cyclic turnover. The regulation of protein synthesis possibly contributes to generation of the protein rhythms, since all processes from synthesis to degradation of mPER2 proteins can theoretically underly their cyclic turnover. Recently, it was reported that Nocturnin regulates polyadenylation length of mRNA (5). Although Nocturnin is likely to contribute to the mRNA stability, the polyadenylation tail of mRNA was closely related to the regulation of translation because the polyadenylation tail is essential for activation of the translation initiation factors as well as mRNA stability. To regulate translation initiation, the cytoplasmic polyadenylation element sequence in the 3′ untranslated region of mRNA plays an important role, and the consensus sequence is interestingly conserved in mPer1 and mPer2. Moreover, the β-globin 3′ untranslated region used in our mPer2 expression construct to make Tet-Off/mPer2 cell lines also includes cytoplasmic polyadenylation element consensus sequence (data not shown). Therefore, this kind of translational regulation may also contribute to the generation of the mPER2 cycle of the Tet-Off/mPer2 cell lines.
The above-mentioned data strongly suggest that the protein cycle of mPER2 is essential for oscillation of the circadian feedback loops in mammalian cells. This is not to say that mRNA oscillations do not play a role under normal circumstances, but they may be indispensable for maintaining the rhythms since there is a severe dampening of amplitude of clock outputs without mRNA cycling.
Acknowledgments
We thank M. Yasuda and M. Sakaguchi for technical assistance.
This work was supported in part by grants from the Special Coordination Funds for Scientific Research and by grants-in-aid for science from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Ministry of Health, Welfare and Labor; SRF; and the Cosmetology Research Foundation.
REFERENCES
- 1.Akashi, M., and E. Nishida. 2000. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 14:645-649. [PMC free article] [PubMed] [Google Scholar]
- 2.Akashi, M., Y. Tsuchiya, and E. Nishida. 2002. Control of intracellular dynamics of mammalian period proteins by casein kinase I ε (CKIε) and CKIδ in cultured cells. Mol. Cell. Biol. 22:1693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allada, R., P. Emery, J. S. Takahashi, and M. Rosbash. 2001. Stopping time: the genetics of fly and mouse circadian clocks. Annu. Rev. Neurosci. 24:1091-1119. [DOI] [PubMed] [Google Scholar]
- 4.Bae, K., X. Jin, E. S. Maywood, M. H. Hastings, S. M. Reppert, and D. R. Weaver. 2001. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30:525-536. [DOI] [PubMed] [Google Scholar]
- 5.Baggs, J. E., and C. B. Green. 2003. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13:189-198. [DOI] [PubMed] [Google Scholar]
- 6.Balsalobre, A., F. Damiola, and U. Schibler. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93:929-937. [DOI] [PubMed] [Google Scholar]
- 7.Bunger, M. K., L. D. Wilsbacher, S. M. Moran, C. Clendenin, L. A. Radcliffe, J. B. Hogenesch, M. C. Simon, J. S. Takahashi, and C. A. Bradfield. 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camacho, F., M. Cilio, Y. Guo, D. M. Virshup, K. Patel, O. Khorkova, S. Styren, B. Morse, Z. Yao, and G. A. Keesler. 2001. Human casein kinase Iδ phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 489:159-165. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, Y., and P. E. Hardin. 1998. Drosophila photoreceptors contain an autonomous circadian oscillator that can function without period mRNA cycling. J. Neurosci. 18:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunlap, J. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 11.Frisch, B., P. E. Hardin, M. J. Hamblen-Coyle, M. Rosbash, and J. C. Hall. 1994. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron 12:555-570. [DOI] [PubMed] [Google Scholar]
- 12.Grima, B., A. Lamouroux, E. Chelot, C. Papin, B. Limboug-Bouchon, and F. Rouyer. 2002. The F-box protein Slimb controls the levels of clock proteins Period and Timeless. Nature 420:178-182. [DOI] [PubMed] [Google Scholar]
- 13.Hardin, P. E., J. C. Hall, and M. Rosbash. 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343:536-540. [DOI] [PubMed] [Google Scholar]
- 14.Ko, H. W., J. Jiang, and I. Edery. 2002. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420:673-678. [DOI] [PubMed] [Google Scholar]
- 15.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193-205. [DOI] [PubMed] [Google Scholar]
- 16.Lee, C., J. P. Etchegaray, F. R. Cagampang, A. S. Loudon, and S. M. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107:855-867. [DOI] [PubMed] [Google Scholar]
- 17.Lowrey, P. L., K. Shimomura, M. P. Antoch, S. Yamazaki, P. D. Zemenides, M. R. Ralph, M. Menaker, and J. S. Takahashi. 2000. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pando, M. P., D. Morse, N. Cermakian, and P. Sassone-Corsi. 2002. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell 110:107-117. [DOI] [PubMed] [Google Scholar]
- 19.Preitner, N., F. Damiola, L. Lopez-Molina, J. Zakany, D. Duboule, U. Albrecht, and U. Schibler. 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251-260. [DOI] [PubMed] [Google Scholar]
- 20.Reppert, S. M., and D. R. Weaver. 2002. Coordination of circadian timing in mammals. Nature 418:935-941. [DOI] [PubMed] [Google Scholar]
- 21.Ripperger, J. A., L. P. Shearman, S. M. Reppert, and U. Schibler. 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14:679-689. [PMC free article] [PubMed] [Google Scholar]
- 22.Shigeyoshi, Y., E. Meyer-Bernstein, K. Yagita, W. Fu, Y. Chen, T. Takumi, P. Schotland, A. Sehgal, and H. Okamura. 2002. Restoration of circadian behavioural rhythms in a period null Drosophila mutant (per01) by mammalian period homologues mPer1 and mPer2. Genes Cells 7:163-171. [DOI] [PubMed] [Google Scholar]
- 23.Toh, K. L., C. R. Jones, Y. He, E. J. Eide, W. A. Hinz, D. M. Virshup, L. J. Ptacek, and Y.-H. Fu. 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291:1040-1043. [DOI] [PubMed] [Google Scholar]
- 24.van der Horst, G. T. J., M. Muijtjens, K. Kobayashi, R. Takano, S. Kanno, M. Takao, J. de Wit, A. Verkerk, A. P. M. Eker, D. von Leenen, R. Buijs, D. Bootsma, J. H. J. Hoeijmakers, and A. Yasui. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627-630. [DOI] [PubMed] [Google Scholar]
- 25.Vitaterna, M. H., D. P. King, A.-M. Chang, J. M. Kornhauser, P. L. Lowrey, J. D. McDonald, W. F. Dove, L. H. Pinto, F. W. Turek, and J. S. Takahashi. 1994. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vosshall, L. B., and M. W. Young. 1995. Circadian rhythms in Drosophila can be driven by period expression in a restricted group of central brain cells. Neuron 15:345-360. [DOI] [PubMed] [Google Scholar]
- 27.Yagita, K., F. Tamanini, G. T. J. van der Horst, and H. Okamura. 2001. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292:278-281. [DOI] [PubMed] [Google Scholar]
- 28.Yagita, K., F. Tamanini, M. Yasuda, J. H. J. Hoeijmakers, G. T. J. van der Horst, and H. Okamura. 2002. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitination of the mPER2 clock protein. EMBO J. 21:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi, S., S. Mitsui, L. Yan, K. Yagita, S. Miyake, and H. Okamura. 2000. Role of DBP in the circadian oscillatory mechanism. Mol. Cell. Biol. 20:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, Z., and A. Sehgal. 2001. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29:453-467. [DOI] [PubMed] [Google Scholar]
- 31.Young, M. W. 1998. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu. Rev. Biochem. 67:135-152. [DOI] [PubMed] [Google Scholar]
- 32.Young, M. W., and S. A. Kay. 2001. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2:702-715. [DOI] [PubMed] [Google Scholar]
- 33.Zheng, B., U. Albrecht., K. Kaasik, M. Sage, W. Lu, S. Vaishnav, Q. Li, Z. S. Sun, G. Eichele, A. Bradley, and C. C. Lee. 2001. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105:683-694. [DOI] [PubMed] [Google Scholar]