Abstract
Structural analysis of nuclear receptor subfamily V orphan nuclear receptors suggests that ligand-independent mechanisms must regulate this subclass of receptors. Here, we report that steroidogenic factor 1 (SF-1) and liver receptor homolog 1 are repressed via posttranslational SUMO modification at conserved lysines within the hinge domain. Indeed, mutating these lysines or adding the SUMO isopeptidase SENP1 dramatically increased both native and Gal4-chimera receptor activities. The mechanism by which SUMO conjugation attenuates SF-1 activity was found to be largely histone deacetylase independent and was unaffected by the AF2 corepressor Dax1. Instead, our data suggest that SUMO-mediated repression involves direct interaction of the DEAD-box protein DP103 with sumoylated SF-1. Of potential E3-SUMO ligase candidates, PIASy and PIASxα strongly promoted SF-1 sumoylation, and addition of DP103 enhanced both PIAS-dependent receptor sumoylation and SF-1 relocalization to discrete nuclear bodies. Taken together, we propose that DEAD-box RNA helicases are directly coupled to transcriptional repression by protein sumoylation.
Steroidogenic factor 1 (SF-1) and liver receptor homolog 1 (LRH-1) are two closely related transcription factors belonging to the nuclear receptor subfamily V (NR5A) that contain a highly conserved DNA binding domain (DBD), a large hinge domain and a ligand binding domain (LBD) (Fig. 1A). Drosophila melanogaster Ftz-F1 is the founding member of this subfamily and interacts directly with the pair-rule gene product of Ftz to control parasegmentation at early embryonic stages (25). The mammalian orthologs SF-1 and LRH-1 are also critical in tissue development and organogenesis (19, 27, 33). During development, SF-1 is essential for male differentiation, adrenogonadal morphogenesis, and terminal differentiation of the ventromedial hypothalamus, and in the adult, this receptor regulates genes involved in steroid biosynthesis and endocrine signaling (34, 44). Although SF-1 null mice die at birth from adrenal failure, SF-1 heterozygous mice live. However, further analyses of these heterozygous mice show that despite seemingly adequate levels of SF-1, the amount of active SF-1 protein is insufficient to overcome defects in adrenal morphogenesis (2, 3). In humans, SF-1 haploinsufficiency is associated with severe adrenal disease and gonadal dysgenesis (1, 28). LRH-1 acts far earlier in development than SF-1, as evidenced by the embryonic lethality observed in LRH-1 null embryos (33). In vitro and in vivo analyses have implicated LRH-1 in bile acid homeostasis (13, 26), where a heterozygous phenotype has also emerged in the intestine (4). In addition, LRH-1 controls tissue conversion of androgens to estrogen by regulating aromatase gene expression (7, 17)
FIG. 1.
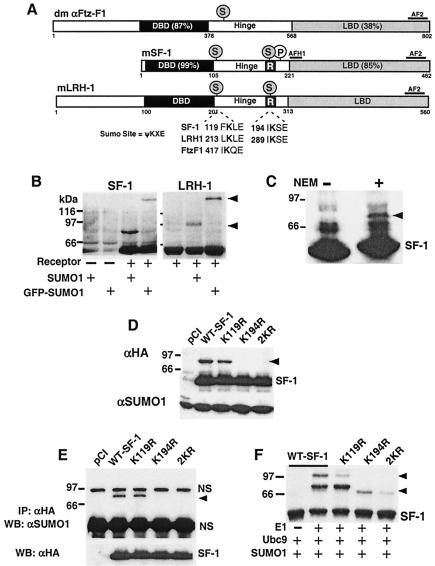
Subfamily V receptors are sumoylated in the hinge region. (A) A schematic of the domain structures and percentages of protein identity for Drosophila (dm) Ftz-F1 and mouse SF-1 and LRH-1 are shown, with SUMO sites (S) and phosphorylation sites (P) indicated. The repression domain is also shown (R, black square). (B) An anti-HA Western blot of COS-7 lysates is shown after transfection with HA-epitope tagged SF-1 or LRH-1 and SUMO1 or GFP-SUMO1. The slower-migrating forms of each receptor are indicated (arrowheads), and all lysates were prepared in the presence of NEM, an inhibitor of SUMO isopeptidases. (C) Western blots are shown for Y1 whole-cell lysates treated with (+) or without (−) 20 mM NEM. Protein was detected with an anti-SF-1 antibody. Upshifted SF-1 after NEM treatment is indicated by an arrowhead. (D) An anti-HA Western blot of COS-7 cells is shown for empty vector control (pCI), the HA-SF-1 wild type, and lysine mutants with sumoylated SF-1 (arrowhead) and nonsumoylated SF-1 (SF-1) indicated; SUMO1 was coexpressed in all conditions. A control immunoblot for SUMO1 is shown below. (E) An anti-SUMO1 Western blot of HA-immunoprecipitated lysates from COS-7 cells transfected with the wild type or lysine mutants of SF-1 is shown, with sumoylated SF-1 (arrowhead) and nonspecific bands (NS) indicated. A control immunoblot for HA-SF-1 expression is shown below. One microgram of each plasmid was added for all transfections. (F) In vitro sumoylation of the in vitro-transcribed and -translated 35S-labeled wild-type and lysine mutants of SF-1 (1 μl) was carried out as described in Materials and Methods. Unmodified SF-1 (SF-1) and sumoylated SF-1 (arrowheads) are indicated. α, anti; IP, immunoprecipitation; WB, Western blotting.
Despite the fact that the high-resolution crystal structure of LRH-1 revealed a large hydrophobic pocket within the LBD (38), natural ligands have yet to emerge for this subclass of receptors. As such, the question of how subfamily V receptors are regulated is unclear. In many cellular contexts, this subclass of receptors is active and presumably recruits coactivators in a ligand-independent manner. NR5A receptor activity depends on two distinct regions in the LBD, an activation function in helix 1 and the C-terminal AF2 domain (8, 20). In both SF-1 and LRH-1, a repression domain has been identified in the hinge region (32, 47). For SF-1, this domain is reported to interact with the DEAD-box RNA helicase DP103 (Ddx20 or Gemin-3) (49), although the precise mechanism of SF-1 repression by DP103 is unknown.
Phosphorylation and sumoylation are posttranslational modifications known to modulate nuclear receptors. Phosphorylation of SF-1 is proposed to increase receptor activity by stabilization of the LBD and enhanced cofactor recruitment (8, 11, 15). On the other hand, sumoylation of transcription factors, such as Elk-1, Lef1, and nearly all steroid nuclear receptors, results in their transcriptional repression (5, 18, 35, 39, 42, 50). Sumoylation occurs at canonical motifs of ψKXE, where ψ is a hydrophobic amino acid and K is the acceptor lysine for covalent attachment of the small ubiquitin-like modifier (SUMO). SF-1, LRH-1, and other invertebrate NR5 receptors are predicted to be sumoylated given the presence of a conserved IKSE or I/VKQE site in the hinge region (Fig. 1A). SUMO modification of proteins is analogous to ubiquitination, involving a three-step ATP-dependent reaction. Processed SUMO protein is loaded onto the heterodimeric E1 enzyme (SAE1/SAE2) and transferred from E1 to the sole E2 enzyme Ubc9, which then mediates SUMO conjugation to the protein substrate with aid from E3-SUMO ligases. Protein inhibitor of activated stats (PIAS) proteins comprise the largest of three identified E3-SUMO ligase classes (29). This protein conjugation is dynamic and easily reversed by Sentrin/SUMO-specific proteases (SENP/SUSP), which cleave SUMO from its substrate. However unlike ubiquitin conjugation, which primarily facilitates protein degradation, SUMO modification of transcription factors often results in transcriptional repression. Others have proposed that this repression involves direct recruitment of histone deacetylases (HDACs) (40, 51) or a relocalization of the SUMO-marked protein to promyelocytic leukemia protein (PML) nuclear bodies (9, 39).
Here we identify sumoylation as an important posttranslational regulatory mechanism for dampening the activity of subfamily V nuclear receptors. Potential mechanisms for sumoylation-mediated repression were investigated and found to involve a functional interaction between the receptor and the DEAD-box RNA helicase DP103.
MATERIALS AND METHODS
Plasmids.
Full-length mSUMO1 (101 amino acids [aa]) was PCR amplified from embryonic mouse hypothalamic-enriched cDNA with primers 5′-CTCGAGATGTCTGACCAGGAGGCAAAA-3′and 5′-TCTAGACTAAACCGTCGAGTGACCCCC-3′,TA-cloned into pCRII (Invitrogen), and subcloned into XhoI-XbaI pCI-neo. Processed His6-human SUMO1 (hSUMO1) (97 aa) was subcloned from His6-hSUMO1-pcDNA3 (F. Poulat) into pGEX4T1 at BamHI. Hemagglutinin (HA)-tagged murine SENP1 (mSENP1) was PCR cloned from mouse hypothalamic-enriched cDNA with primers 5′-CCGGAATTCATGTACCCATACGACGTACCAGATTACGCTAGCTTGGATGACACAGCTGATGGGGTG-3′and 5′-ACCTCTAGAGTCGACTCACAAGAGCTTCCGGTGGAG-3′into EcoRI-SalI of pCI-neo. HA-tagged mSF1 in pCI-neo, HA-tagged SF-1 S203A, and green fluorescent protein (GFP)-HA-SF-1 in pCMV were described previously (8). K119R, K194R, and 2KR mutants of HA-SF-1-pCI-neo and GFP-HA-SF-1 were created by PCR mutagenesis (Stratagene). All Gal4 constructs contained a HA epitope tag N-terminal to the Gal4 DBD. A C-terminal fragment containing the hinge-LBD (aa 105 to 462) of SF-1 and mutants was generated by PCR from HA-SF-1-pCI-neo with primers 5′-ACGCGTCCTTGAAGCAGCAGAAGAAAGCA-3′and 5′-AAGCTTTCAAGTCTGCTTGGCCTG-3′and subcloned 3′ to the HA-Gal4-DBD. A similar strategy was used to create all pGAL-LRH-1 constructs with the LRH-1 (aa 198 to 562) fused to Gal4. FLAG-mPIASxα was cloned from RIKEN clone 4921511I02 with primers 5′-CCGGAATTCATGGACTACAAAGACGACGACGACAAAGCGGATTTCGAGGAGTTG-3′and 5′-CCGCTCGAGTCACTGTTGCACAGTATCAGA-3′,and FLAG-mPIAS1 was cloned from mouse hypothalamic cDNA with primers 5′-CTCGAGATGGACTACAAAGACGACGACGACAAAGCGGACAGTGCGGAACTAAAG-3′and 5′-CCGCTCGAGTCAGTCCAATGAGATAATGTC-3′.PCR products were subcloned into pCI-neo, pBH4, and pGADT7. pVP16-PIAS1 and pVP16-PIASxα were generated by inserting FLAG-mPIAS1 and FLAG-mPIASxα PCR fragments downstream of the VP16 activation domain in a pVP16 vector (Clontech). The following constructs were generous gifts: T7 tagged-mPIASy pCMV (from R. Grosschedl), FLAG-mPIAS3 pCMV (from K. Shuai), full-length mDP103 pcDNA3 (from Y. Sadovsky), and C-terminal hDP103 pGEX (aa 414 to 824) and full-length 2FLAG-hDP103 pcDNA3 (from C. Glass).
Cell transfections, luciferase assays, and metabolic labeling.
COS-7 cells were plated at a density of 50,000 cells/ml/12-well plate or 1.5 × 106 cells/10-cm-diameter plate in medium (Dulbecco's modified Eagle's H21 with 4.5 g of glucose/liter with 10% calf serum and antibiotics) 18 h prior to transfection. Transfections were carried out by using FuGene 6 (Roche). For luciferase assays, cells were transfected with no more than 500 ng of total DNA per well and harvested 48 h after transfection (BD Pharmingen). All transfections were performed in triplicate and repeated at least twice. Results were normalized to β-galactosidase activity and expressed as relative luciferase units or activation, as indicated. For metabolic labeling, COS-7 cells were plated in full medium and transfected 18 h after plating. Cys/Met-deficient medium (Dulbecco's modified Eagle's H21 with 4.5 g of glucose/liter, 10% dialyzed fetal bovine serum, 2 mM glutamine, and antibiotics) was added to washed cells 48 h posttransfection, followed by 1 h of pulse-labeling with 350 μCi of [35S]Cys and [35S]Met (Redivue, AGQ0080; Amersham), washing, and incubation in full medium for relevant chase periods. Cells lysates were subjected to immunoprecipitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and autoradiography, and the signal was quantified by phosphorimaging.
Yeast interaction system.
An expression cassette containing full-length mouse SF-1 (no heterologous activation domain) was integrated in yeast strain YM4271 containing two integrated reporters, HIS and LacZ, driven by four tandem copies of the SF-1 response elements, using the manufacturer's protocols (Clontech). Full-length FLAG-tagged mPIAS1, mPIASxα, and mPIASy were subcloned into pGADT7 for transformation into yeast reporter strains. Transformants were plated on selective medium and analyzed on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) medium and by liquid β-galactosidase assays.
Western analysis, immunoprecipitation, and coimmunoprecipitation.
Cells were washed twice in cold phosphate-buffered saline (calcium and magnesium free); lysed in 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, and protease inhibitors (Roche); and precleared by centrifugation at 14,000 rpm with a Sorvall Microfuge for 30 min. When appropriate, all solutions contained 20 mM N-ethylmaleimide (NEM; Sigma) to inhibit SUMO isopeptidases. Protein concentrations were determined by the Bradford method (Pierce). Equal amounts of total protein were loaded for Western blot analysis. Wild-type and mutant receptors were affinity purified by using anti-HA affinity matrix (Covance/Babco) in lysis buffer (as described above), washed in a modified lysis buffer containing 300 mM KCl and 0.05% NP-40, and subjected to SDS-PAGE and Western blotting following incubation with primary antibodies (anti-HA, 1:2,000 [Covance/Babco]; anti-FLAGM2, 1:2,000 [Sigma]; anti-SUMO1, 1:500 [Zymed]) and a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:10,000; Bio-Rad). The signal was developed by chemiluminescence (ECL kit; Amersham). For coimmunoprecipitation of FLAG-hDP103 and sumoylated SF1, cells were transfected and lysed as for in vivo sumoylation in 10 mM NEM. Lysates were incubated with anti-FLAG M2 agarose beads (Sigma) in pull-down buffer (50 mM Tris HCl [pH 7.6], 150 mM NaCl, 0.5 mM EDTA, 0.01% NP-40, 2 mM NEM, protease inhibitors), and precipitates analyzed by Western blotting (anti-HA, 1:2,000 [Covance/Babco]; anti-hDP103, 1:2,000 [BD Biosciences]).
Recombinant protein expression, in vitro sumoylation assay, and GST pull downs.
Recombinant His6-hSUMO1 (aa 1 to 97) was expressed and purified by TALON chromatography (Clontech). Recombinant His6-hE1 (SAE1/SAE2) and His6-hUbc9 were obtained commercially (LAE Biotech). In vitro-transcribed and -translated 35S-SF-1 and variants thereof were produced (Promega) and incubated with 150 ng of E1, 750 ng of His6-Ubc9, and 900 ng of His6-SUMO1 in 50 mM Tris (pH 7.6), 5 mM MgCl2, 1 mM dithiothreitol, and 2.5 mM ATP at 37°C for 1.5 h, and the reaction was stopped by boiling in protein loading buffer. Samples were subjected to SDS-PAGE followed by autoradiography. Glutathione S-transferase (GST) pull-down assays were carried out with 35S-SF-1 or variants thereof and purified GST-C-terminal hDP103 as described previously (15, 21).
Chromatin immunoprecipitation assay.
HeLa luciferase reporter (Stratagene) cells containing an integrated promoter-reporter of five Gal4 binding sites fused to the luciferase gene were electroporated with pCI-Neo and HA-tagged pGal-SF-1 constructs (4 μg). The method used follows that described in reference 46, with PCR conditions of 25 cycles at 95°C for 30 s, 53°C for 1 min, and 72°C for 1 min and by using primers described previously (40) to amplify a 5′ 330-bp region of luciferase cDNA.
Nuclear localization and immunohistochemistry.
COS-7 cells were plated at 6,000 cells/well in four-well chamber slides (Lab-Tek) and transfected in duplicate 24 h later (total DNA, 0.5 μg/well). At 48 h posttransfection, cells were fixed in 4% paraformaldehyde, permeabilized in phosphate-buffered saline containing 0.3% Triton X-100, incubated with primary antibodies (rabbit anti-T7, 1:300 [ICL]; mouse anti-FLAGM2, 1:5,000 [Sigma]; mouse anti-SF2/ASF, 1:1,000 [Zymed]; goat anti-Sp100, 1:50 [Zymed]; mouse anti-PML [PG-M3], 1:75 [Santa Cruz]) followed by secondary antibodies (Cy-3 goat anti-rabbit, 1:1,000; Cy3-donkey anti-mouse 1:1,000 [Molecular Probes]; Texas Red rabbit anti-goat, 1:500 [Vector]), and imaged on a Zeiss LSM510 confocal microscope.
RESULTS
Subfamily V receptors are sumoylated in the hinge region.
Although sumoylation is known to repress steroid receptor activity, this modification has not been investigated for so-called orphan nuclear receptors, which can function in a ligand-independent manner. In a modified one-hybrid yeast screen for SF-1 protein partners, we identified Ubc9 or the E2 SUMO conjugating enzyme as a strong interacting protein (data not shown). We next sought to determine whether SF-1 and LRH-1 could be sumoylated. Indeed, sequence analysis of all vertebrate species of SF-1 and LRH-1 revealed two highly conserved canonical sumoylation motifs at the N- and C-terminal hinge regions, while insect Ftz-F1 variants contained one site in the N-terminal hinge region (Fig. 1A).
Sumoylation of both SF-1 and LRH-1 was demonstrated in a cellular system, as evidenced by slower-migrating bands after coexpression of receptor with either SUMO1 or GFP-SUMO1 (Fig. 1B). In addition, a similar slower-migrating SF-1 species was detected in NEM-treated lysates made from both Y1 and αT3 cells (Fig. 1C and data not shown), suggesting that endogenous SF-1 is sumoylated. Further analysis revealed that Lys194 served as the major acceptor lysine for SF-1 sumoylation, as evidenced by the loss of the slower-migrating band with the single mutation K194R and double mutation (K119R and K194R, referred to as 2KR) but not with K119R (Fig. 1D). Our results for SF-1 are similar to those of other recent reports (6, 22). The identity of these slower-migrating SF-1 species as sumoylated receptors was confirmed by immunoprecipitation of HA epitope-tagged SF-1, followed by Western blotting with an anti-SUMO1 antibody (Fig. 1E), and as predicted, no sumoylated species were observed with K194R or 2KR mutant proteins. These results were confirmed in an in vitro sumoylation assay, with Lys194 identified as a major site and Lys119 presumed to be a minor sumoylation site (Fig. 1F). Amounts of sumoylated SF-1 diminish in both the K194R and 2KR mutants; the faint residual upshifted band observed in the 2KR variant imply that a minor third site can be sumoylated in vitro. Taken together, we conclude that subfamily V receptors are sumoylated in vivo and in vitro.
Sumoylation of SF-1 attenuates transcriptional activity.
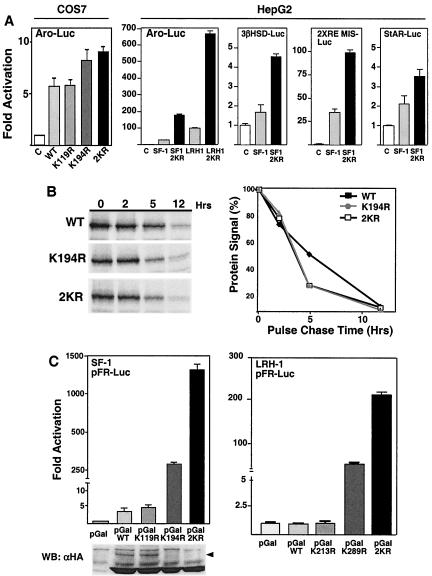
Previous studies identified a regulatory domain which when mutated led to increased receptor activity; this domain contained the major sumoylation site for SF-1 and LRH-1 (Fig. 1A) (32, 47). Consistent with these reports, we found increased activity of NR5A promoter reporters with either SF-1 or LRH-1 sumoylation mutants (Fig. 2A). Increased receptor activity observed with both the K194R and 2KR receptor mutants was not due to increased protein stability, as judged by results from pulse-chase metabolic labeling experiments (Fig. 2B). Gal4-SF-1/LRH-1 fusion receptors containing the full hinge and LBD also showed a dramatic increase in activity following mutation of the sumoylation acceptor sites. Strikingly, the single mutant K194R was at least 70-fold more active than the wild type, and mutation of both sumoylation sites (2KR) resulted in greater than 300-fold activation (Fig. 2C, left panel). While K119R exhibited comparable activation to that of the wild type, the double mutant at both Lys119 and Lys194 showed remarkable synergism; this is consistent with Lys119 as a minor site. Similar to native receptors, Gal4-SF-1 and Gal4-K119R are efficiently sumoylated, whereas Gal4-K194R and Gal4-2KR exhibit no detectable sumoylation (Fig. 2C, left lower panel). Nearly identical results were observed for Gal4-LRH-1 constructs, where double mutation of K213R and K289R in the hinge region led to strong receptor activation (Fig. 2C, right panel).
FIG. 2.
Sumoylation represses SF-1 transcriptional activity. (A) Transcriptional activity of the wild type and lysine mutants of SF-1 (50 ng) on the aromatase-luciferase reporter (Aro-Luc, 500 ng) is shown for both COS-7 (no SUMO1 added) and HepG2 cells (50 ng of SUMO1 added). Other promoter-luciferase reporters used in HepG2 cells were the 3β-hydroxysteroid dehydrogenase promoter (3βHSD Luc, −153/+2 bp), a synthetic promoter containing tandem SF-1 response elements from the mouse Müllerian inhibiting substance promoter (2XRE MIS), and the StAR promoter (StAR Luc, −966/+1); 250 ng of each promoter was used. (B) The stability of wild-type (WT) and lysine mutant (K194R or 2KR) SF-1 proteins in COS-7 cells was determined after metabolic labeling, followed by a chase for 0, 2, 5, and 12 h. An autoradiogram of immunoprecipitated HA proteins from whole-cell lysates is shown, with phosphorimaging data graphed as the percentage of labeled protein remaining after each chase period; levels of protein at time zero were taken to be 100%. (C) Transcriptional activity is shown for the Gal4-SF-1 wild type (pGalWT, aa 105 to 462, 25 ng) or Gal4-SF-1 lysine mutants (pGalK119R, pGalK194R, and pGal2KR; 25 ng) on the Gal4-luciferase reporter (pFR-Luc, 200 ng; Stratagene) in COS-7 cells (left panel). Anti-HA Western blotting shows expression levels of the Gal4-SF-1 WT or KR mutants, with slower-migrating forms of sumoylated Gal4-SF-1 protein indicated (arrowhead). Transcriptional activities of the Gal4-LRH-1 wild type (pGalWT, aa 198 to 560, 25 ng) and lysine mutants (pGalK213R, pGalK289R, and pGal2KR) are shown (right panel). All luciferase activity is expressed as activation over parent vectors: pCI-neo (C) for panels in A and pM (pGal) for panels in C. Hrs, hours; WB, Western blotting.
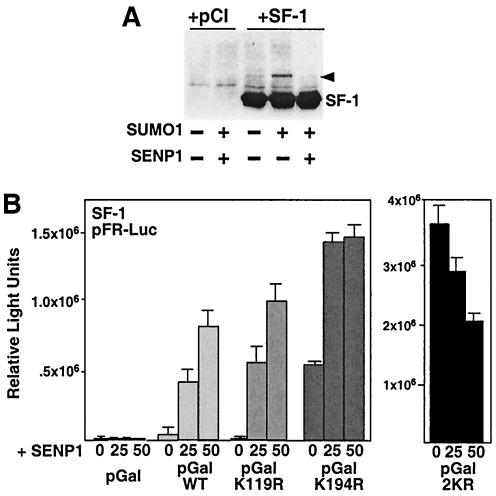
To confirm that receptor sumoylation served to repress SF-1 activity, we asked whether removing the SUMO conjugate from SF-1 with the SUMO isopeptidase SENP1 would yield similar results, as observed with the SF-1 lysine mutants. Indeed, coexpression of SENP1 with SF-1 and SUMO1 resulted in a marked attenuation of sumoylated SF-1 (Fig. 3A). Furthermore, activities of both the wild type and the K119R mutant were enhanced after the addition of small amounts of SENP1 expression vector (25 ng), reaching levels observed with the K194R mutant (Fig. 3B, left panel). Addition of SENP1 failed to activate the 2KR variant, providing further evidence that Lys119 and Lys194 are the sites of sumoylation (Fig. 3B, right panel). Collectively, our data suggest that Lys194 plays a dominant role in mediating repression of SF-1 via sumoylation and that receptor sumoylation represents a major silencing mechanism.
FIG. 3.
Adding SENP1 increases activity of wild-type SF-1 but not the 2KR sumoylation mutant. (A) A Western blot is shown for COS-7 cells transfected with empty vector (pCI) or wild-type SF-1 (1 μg of each) in the presence or absence of SUMO1 and SENP1 (1 μg of each), with sumoylated SF-1 (arrowhead) and nonsumoylated SF-1 (SF-1) indicated. (B) Effects of increasing amounts of SENP1 (0, 25, and 50 ng) are shown for transcriptional activity of the Gal4-SF-1 wild type (pGalWT) and Gal4-SF-1 lysine mutants (pGalK119R, pGalK194R, and pGal2KR) on the pFR-Luc Gal4 reporter in COS-7 cells. Luciferase activity is expressed in relative luciferase units. Amounts of transfected plasmids are identical to those used for Fig. 2C. +, with; −, without; WT, wild type
A DEAD-box protein mediates repression via SF-1 sumoylation.
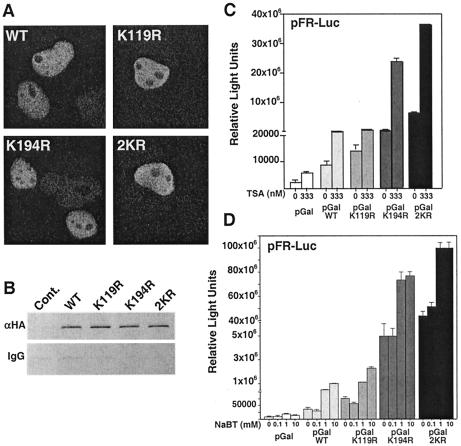
The mechanisms by which protein sumoylation leads to transcriptional repression are diverse. Recent literature suggests that repression by sumoylation involves (i) nuclear relocalization with a concomitant decrease of promoter occupancy or (ii) direct recruitment of HDACs. Therefore, we asked whether sumoylation mutants differ in their subnuclear localization. Both GFP-wild type and GFP-SUMO mutants yielded nearly identical patterns of nuclear localization (Fig. 4A). Consistent with these results, no apparent differences were noted in the promoter occupancy of Gal4-wild type compared to the K194R mutant as judged by chromatin immunoprecipitation (ChIP) results with a HeLa cell line containing a stably integrated Gal4 reporter (Fig. 4B). We next asked whether SF-1 sumoylation promotes recruitment of HDACs by using the class I and II HDAC inhibitors, trichostatin A (TSA) and sodium butyrate (NaBT). If HDAC recruitment is essential for SUMO-mediated repression, mutating the major sumoylation sites within SF-1 should prevent derepression by TSA or NaBT. Instead, addition of TSA or NaBT led to a dramatic increase in the activity of all receptor variants (Fig. 4C and D). Our results differ from those recently shown for Elk-1, where loss of sumoylation eliminates TSA sensitivity (51), and thus, we suggest that repression of SF-1 via sumoylation is largely HDAC independent.
FIG. 4.
SF-1 sumoylation mutants exhibit wild-type localization, promoter occupancy, and sensitivity to HDAC inhibitors. (A) Nuclear localization is shown for transfected GFP-SF-1 wild type (WT) and lysine mutants (K119R, K194R, and 2KR) in COS-7 cells; 100 ng of each plasmid was used and resulted in expression of GFP-SF-1 in 15% of all cells. (B) ChIP assays are shown for the control vector, N-terminal HA-tagged Gal4-SF-1 (WT), and Gal4 lysine mutants in HeLa cells containing integrated Gal4 response elements fused to luciferase with anti-HA or control immunoglobulin G (IgG). TSA (C) and NaBT (D) effects on transcriptional activity of the Gal4-SF-1 wild type (pGalWT) and lysine mutants (pGalK119R, pGalK194R, and pGal2KR) in COS-7 cells. TSA (0 or 333 nM) or NaBT (0, 0.1, 1, or 10 mM) was added to cells at 12 h posttransfection and incubated for 24 h. Cont., control; αHA, anti-HA.
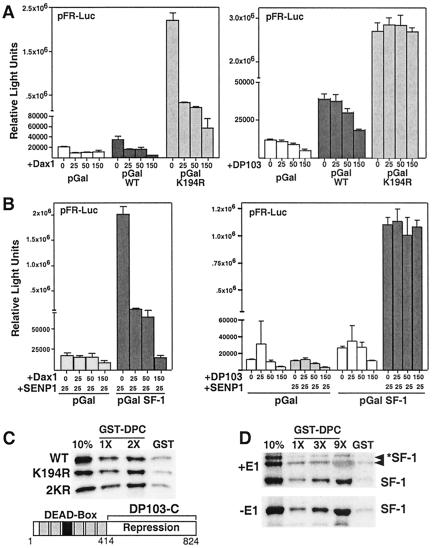
For subfamily V, two types of repressors have been identified. The first includes the orphan nuclear receptors Dax1 and SHP, which interfere with the AF2 in the LBD. The second is the RNA helicase DEAD-box protein DP103 (32). Indeed, while Dax1 was able to repress the Gal4-K194R mutant as effectively as Gal4-wild type (Fig. 5A, left panel), DP103 was ineffective at repressing the Gal4-K194R and 2KR mutants (Fig. 5A, right panel, and data not shown). Moreover, addition of SENP1 failed to abolish Dax1-mediated repression of SF-1 (Fig. 5B, left panel). In contrast, addition of SENP1 completely eliminated DP103-mediated repression of Gal4-SF-1 (Fig. 5B, right panel). Our work contrasts a recent report showing no difference between DP103-mediated repression in wild-type and K194R (22). This discrepancy may reflect a difference in cell types or the significantly greater amounts of DP103 used compared to experiments shown here. Nonetheless, our data agree with those reported by Ou and colleagues showing Lys194 to be essential for DP103 repression of SF-1 (32).
FIG. 5.
The DEAD-box protein DP103 mediates SF-1 repression by sumoylation at Lys194 and binds to sumoylated SF-1. (A) Repression of SF-1 by Dax1 and DP103. Increasing amounts of mDax1 or mDP103 (0, 25, 50, or 150 ng) were cotransfected in COS-7 cells with Gal4-SF-1 (pGalWT) or the pGalK194R (25 ng each) on the Gal4-luciferase reporter (pFR-Luc, 250 ng). (B) Effect of SENP1 on repression by Dax1 and mDP103. Increasing amounts of Dax1 or DP103 (as in panel A) were cotransfected with control vector (pGal) or the Gal4-SF-1 wild type (pGal-SF-1), with or without SENP1 (25 ng), on the Gal4-luciferase reporter (as in panel A). (C) GST pull-down assays show binding of the 35S-labeled SF-1 wild type (WT) and lysine mutants (K194R and 2KR) to increasing amounts of GST-hDP103 C-terminal aa 414 to 824 (GST-DPC) (1× and 2× indicate relative amounts used). The 10% input and GST controls are indicated. A schematic of human DP103 shows the unique C-terminal region and conserved helicase domain motifs (gray rectangles), including the signature DEAD-box motif (black rectangle). (D) GST pull-down assays show binding of in vitro sumoylated 35S-labeled SF-1 (+E1, *SF-1, upper panel) to increasing amounts of GST-hDP103 C-terminal (1×, 3×, and 9×) and to a nonsumoylated SF-1 control made in reactions lacking E1 enzyme (−E1, SF-1, lower panel).
To test the hypothesis that sumoylation at Lys194 allows DP103 to function as a repressor, interaction between DP103 and sumoylated SF-1 was explored by direct binding assays. As shown previously, only the C-terminal half of DP103 interacts with SF-1 (32). Mutation of Lys194 and/or Lys119 did not result in an appreciable loss of binding, suggesting that Lys194 is not the sole determinant for DP103 interaction with SF-1 (Fig. 5C). Furthermore, DP103 is able to interact efficiently with in vitro sumoylated forms of SF-1 (Fig. 5D). These results provide evidence that the DEAD-box protein DP103 interacts with sumoylated SF-1 and directly participates in receptor repression.
DP103 promotes PIAS-dependent sumoylation and subnuclear relocalization of SF-1.
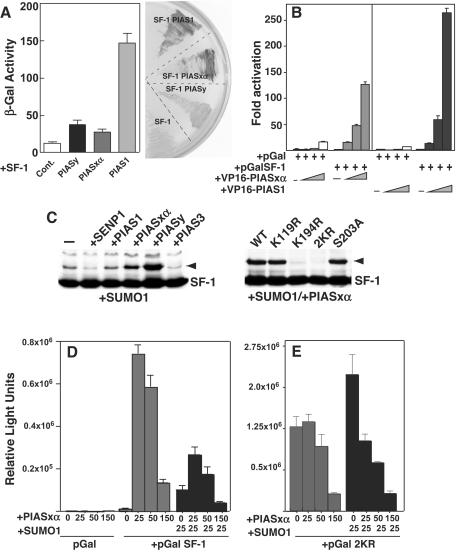
To further explore how DP103 may affect SF-1 activity, we first defined the optimal E3-SUMO ligase in vivo. One of the defining characteristics of an E3-SUMO ligase is its ability to interact with and promote sumoylation of a given substrate. In both the yeast and mammalian two-hybrid assays, SF-1 interacted strongly with PIAS1 and less well with PIASxα and PIASy (Fig. 6A and B). However, despite this strong interaction, PIAS1 does not serve as an efficient E3-SUMO ligase for SF-1 in vivo. In a survey of four PIAS members, only PIASxα and PIASy promoted SF-1 sumoylation in a dose-dependent manner; this effect was not observed for PIAS1 or PIAS3 (Fig. 6C, left panel). In contrast to results from the in vitro assay, overexpression of PIAS proteins in vivo does not reveal detectable sumoylation at noncanonical sites, as evidenced by the 2KR mutant (Fig. 6C, right panel, and data not shown). Interestingly, mutating the major phosphorylation site of SF-1 adjacent to Lys194 (S203A) had no effect on receptor sumoylation (Fig. 6C, right panel). Next, the functional effects of overexpressing PIAS proteins on wild-type and 2KR receptors were determined. Consistent with PIAS-dependent activation of other nuclear receptors (24), we observed an initial activation phase, followed by repression when PIASxα is added to the wild-type receptor (Fig. 6D). Addition of SUMO1 further enhanced receptor repression, suggesting that increased sumoylation does silence SF-1 activity. In contrast, increased repression was not observed with the double mutant 2KR (Fig. 6E). The global repression observed with increasing amounts of SUMO1 added to either wild-type or mutant receptors most likely reflects the multiple nuclear substrates affected by the sumoylation machinery, including corepressors and coactivators (23).
FIG. 6.
PIASxα and PIASy are E3-SUMO ligases for SF-1. (A) Interactions between full-length SF-1 and Gal4AD-PIASxα, PIAS1, -PIASy fusion proteins are shown in yeast expressing SF-1, driving SF-1 response elements fused to LacZ (bar graph) and when grown on X-Gal medium. The bottom sector (SF-1) shows β-galactosidase (β-Gal) activity resulting from yeast expressing SF-1 (SF-1) and the empty vector pGADT7. Other sectors show activity in strains with SF-1 and transformed PIAS fusion proteins, as indicated. Cont., control. (B) A mammalian two-hybrid system showing transcriptional activity of wild-type pGal4-SF-1 with increasing concentrations of VP16-PIAS fusion proteins (25, 50, or 150 ng). The empty vector control is shown (pGal). (C) Western blots for COS-7 cells cotransfected with wild-type HA-SF-1, SUMO1, and with individual PIAS proteins or SENP1 are shown (left panel). The right panel shows a Western blot for HA-SF-1 (WT) and lysine and S203A SF-1 mutants after the addition of PIASxα and SUMO1; 1 μg of each plasmid was added. Sumoylated SF-1 (arrowhead) and nonsumoylated SF-1 (SF-1) are indicated. (D) The transcriptional activity of pGal-SF-1 (25 ng) is shown after increasing amounts of PIASxα were cotransfected in COS-7 cells (25, 50, or 150 ng) in the absence or presence of SUMO1 (25 ng) with the Gal4-luciferase reporter (pFR-Luc, 250 ng). The empty vector control is shown (pGal). (E) Activity of the double sumoylation mutant of SF-1 (pGal-2KR) is shown after increasing amounts of PIASxα, as described for panel C.
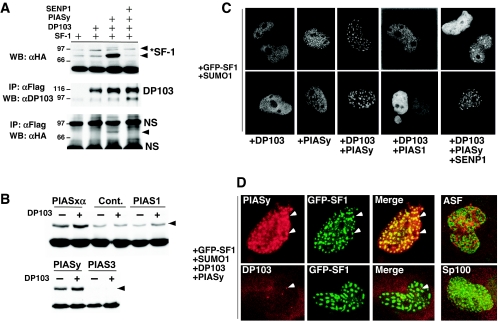
To determine how sumoylation affects interaction between DP103 and SF-1, the levels of receptor sumoylation were driven by the optimal E3-SUMO ligase PIASy. DP103 interacted with SF-1 in the presence of PIASy but not under basal levels of sumoylation or after addition of SENP1 (Fig. 7A). Surprisingly, DP103 enhanced PIAS-mediated sumoylation (two- to threefold) for all PIAS proteins, except PIAS3 (Fig. 7B). No significant increase in sumoylation was observed with DP103 alone (control). Whether this effect arises from increased ligase activity of PIAS proteins or by protecting sumoylated SF-1 from desumoylation remains to be determined. Finally, we asked whether DP103 would alter the subnuclear localization of SF-1. Although our previous results suggested that the nuclear pattern of SF-1 does not change under basal levels of sumoylation, a dramatic relocalization of GFP-SF-1 was revealed when DP103 was coexpressed with PIASy and SUMO1; two representative cells with prominent nuclear bodies are shown (Fig. 7C). Addition of SUMO1, PIASy, or DP103 alone or a combination of DP103 plus PIAS1, PIASxα, or PIAS3 showed no SF-1 relocalization (Fig. 7C; data not shown). However, we noted the presence of fine GFP-SF-1 foci in some cells with PIASy alone (Fig. 7C). The ability of DP103 and PIASy to shuttle SF-1 to discrete nuclear bodies does not apparently require SF-1 sumoylation, as evidenced by a speckled pattern after the addition of SENP1 or with the K119R, K194R, and 2KR GFP-SF-1 mutants (Fig. 7C and data not shown). Further analysis revealed colocalization of GFP-SF-1 with PIASy but not with DP103, which localizes to Cajal bodies or gems (Fig. 7D). These GFP-SF-1 nuclear bodies appear distinct from endogenous splicing speckles, as shown by the nonoverlapping patterns between GFP-SF-1 and splicing factor 2 (SF2)ASF. Moreover, these foci do not resemble PML nuclear bodies (PML-NBs), given that we failed to detect obvious PML-NBs in COS-7 cells under our culture conditions with two markers, Sp100 and PML (Fig. 7D and data not shown). Collectively, our data suggest that DP103 promotes PIAS-mediated sumoylation and, together with PIASy, relocalizes SF-1 to discrete nuclear foci. Whether these foci are functionally significant remains to be determined; however, their formation correlates well with optimal receptor sumoylation, suggesting a functional complex between SF-1, PIASy, and DP103.
FIG. 7.
DP103 interacts with sumoylated SF-1 in vivo and promotes PIASy-mediated SF-1 relocalization into nuclear bodies. (Α) Coimmunoprecipitation of sumoylated SF-1 from COS-7 cells transfected with wild-type HA-SF-1, SUMO1, and combinations of FLAG-DP103, T7-PIASy, and SENP1 (1 μg of each) with anti-FLAG-M2 agarose beads. Western blots (WB) for HA-SF-1 (3% input lysate, upper panel) and immunoprecipitated (IP) DP103 (10% IP protein, middle panel) are shown with sumoylated SF-1 indicated (*SF-1). SF-1 (black arrowheads) and nonspecific bands (NS) are indicated in an anti-HA (αHA) Western blot of IP DP103 protein (lower panel). αFlag, anti-Flag; αDP103, anti-DP103. (B) Western blots are shown of COS-7 cells cotransfected with wild-type HA-SF-1, SUMO1, and individual PIAS proteins or empty vector (Cont.) with (+) or without (−) DP103 (1 μg). Sumoylated SF-1 (arrowhead) and nonsumoylated SF-1 (SF-1) are indicated. (C) Nuclear localization of GFP-SF-1 transfected into COS-7 cells is shown with different combinations of FLAG-DP103, T7-PIASy, FLAG-PIAS1, and SENP1, as indicated. All cells were transfected with SUMO1 (100 ng). (D) Subnuclear signals are shown for wild-type GFP-SF-1 (green), and indirect immunofluorescence is shown for T7-PIASy (red) or FLAG-hDP103 (red). Colocalization of GFP-SF-1 and T7-PIASy signals are shown in the merged figure (upper panels), and the endogenous DP103 signals (lower panels) are indicated (arrowheads). Staining for endogenous SF2/ASF (marker for splicing speckles) or Sp100 (marker for PML-NBs) is shown (red). Note that no positive staining is observed for endogenous Sp100. In all conditions, cells were transfected with 100 ng (each) of GFP-SF-1, PIASy, hDP103, and SUMO1.
DISCUSSION
In this study, we report that subfamily V nuclear receptors are sumoylated at evolutionarily conserved sites. As established for other transcription factors, SUMO modification of SF-1 and LRH-1 significantly attenuates transcriptional activity. Mutating the acceptor lysines in both SF-1 and LRH-1 resulted in a more active receptor, and at least in the Gal4 context, the relative increase is reminiscent of ligand-dependent receptor activation. Thus, for subfamily V receptors, the extent of sumoylation represents one mechanism to both regulate and restrain receptor activity. Our data also suggest that sumoylation of the so-called repression domain in SF-1/LRH-1 marks the receptor for repression by the DEAD-box protein DP103. Moreover, this ATPase/RNA helicase was found to enhance PIAS-dependent receptor sumoylation and to promote PIASy-dependent shuttling of SF-1 to discrete nuclear bodies or foci. Subnuclear relocalization of SF-1 correlated strongly with conditions that promote extensive receptor sumoylation, suggesting that physical interactions between SF-1, DP103, and PIASy are linked to transcriptional repression.
Repression of SF-1 via sumoylation.
In contrast to the ubiquitously expressed E1 and E2 sumoylation enzymes, most of the known E3-SUMO ligases exhibit restricted expression patterns and therefore may direct tissue-specific sumoylation of protein substrates (48). In considering SF-1 sumoylation, three E3-SUMO ligases (PIASxα, PIASy, and PIAS1) are all highly expressed in the adult testes (14, 48), where SF-1 regulates multiple genes. SF-1 is also needed for male sexual differentiation (37, 45), and it is possible that sumoylation of SF-1 is sexually dimorphic during development. Thus, silencing of male-specific genes in the ovary can be partially explained by lowered levels of SF-1 or by the actions of Dax1 (30, 41) but may also involve sumoylation. Interestingly, other factors that function in sexual differentiation, namely Sox9 and WT-1, contain sumoylation sites, and the combinatorial effects of sumoylation may ensure gene silencing in the female. Finally, it is worth considering the in vivo ratio of nonsumoylated to sumoylated receptors. In this regard, SF-1 haploinsufficiency (2, 28) may stem from inadequate SF-1 activity due to a reduction of protein levels coupled with extensive receptor sumoylation.
Currently, our studies are limited to a loss-of-function analysis. Attempts to provide SUMO1 in cis to SF-1, as shown for other proteins (18, 50), have failed due to the precise excision of SUMO1 in COS-7 cells (L. A. Lebedeva and H. A. Ingraham, unpublished data). Whether SF-1 or LRH-1 sumoylation confers any structural changes to the DBD, hinge, or LBD remains unclear; however, results from our ChIP analysis suggest that sumoylation does not alter the apparent DNA binding of a heterologous DBD. Moreover, given that Dax1-mediated repression of K194R SF-1 mutant is intact, we suggest that no gross conformational changes occur in the LBD of a sumoylation-defective receptor. Further structural analyses are needed and will require an appropriate SUMO-SF-1 chimera or SUMO stably conjugated to SF-1/LRH-1. Although our findings point to a functional role for Lys194 and Lys289 in SF-1 and LRH-1, respectively, the role of the minor sumoylation sites at Lys119 or Lys213 (Fig. 1A) is less apparent. Despite the fact that disumoylated SF-1 is only observed in vivo under conditions that promote efficient sumoylation, our functional analyses show that both the minor and major sumoylation sites act in concert to dampen receptor activity. In this regard, it remains to be established whether an ordered sumoylation of SF-1/LRH-1 occurs.
Recent studies report interdependency between sumoylation and phosphorylation. Mitogen-activated protein kinase-mediated phosphorylation of Elk-1 greatly reduced sumoylation at adjacent lysines and led to increased transcriptional activity (50), and phosphorylation of heat shock factor 1 is a prerequisite for stress-induced sumoylation (16). Currently, we find no apparent relationship between phosphorylation of Ser203 and sumoylation of SF-1. Indeed, the phospho-deficient S203A mutant was efficiently sumoylated, and all SF-1 SUMO mutants showed equivalent levels of phosphorylated Ser203 in SF-1 (M. B. Lee, unpublished data). However, it remains possible that the rate and extent of either phosphorylation or sumoylation are altered following modification of the Lys194 or Ser203, respectively.
DEAD-box proteins and transcriptional repression.
Historically, DEAD-box (Ddx) RNA helicases are associated with splicing, in part because they were initially identified as protein components of the spliceosome (43). However, other functions for Ddx family members have been noted, and there is mounting evidence that they function to silence transcription factors, including nuclear receptors, Egr1 to 4, and the Ets-like repressor, METS (12, 21, 36, 49). Additionally, GRTH (Ddx25), which is expressed in the testes, is reported to attenuate expression of SF-1 target genes, including steroidogenic enzymes (10). For DP103 and another DEAD-box protein, DP97, the repression domain has been mapped to the C-terminal region and does not require the N-terminal ATPase/helicase domain characteristic of this gene family (21, 36). Attenuation and silencing of transcription are multilayered and multidimensional. So how may Ddx proteins and sumoylation lead to transcriptional repression? Recruitment of HDACs upon protein sumoylation, or by Ddx proteins, offers the most plausible explanation and is consistent with prior literature. Indeed, DP103 interacts with the N-terminal repression domain of METS and promotes HDAC recruitment (21). However, our data imply that repression through DP103 is TSA and NaBT insensitive and suggest that repression by Ddx proteins must involve additional mechanisms other than recruitment of class I or II HDACs. In considering other mechanisms, it is possible that DP103 protects SF-1 from desumoylation. This hypothesis is consistent with the observations that DP103 increased PIAS-dependent SF-1 sumoylation and that additional SENP1 eliminates repression by DP103. The interaction between DP103 and SF-1 remains to be mapped and is likely to involve multiple interfaces based on our finding that Lys194 and/or sumoylation at Lys119/Lys194 is not the sole determinant of this interaction. Another possible scenario is that DP103 represses SF-1 by facilitating PIASy-mediated relocalization of SF-1. However, we noted that sumoylation is dispensable for movement of SF-1 to nuclear bodies; this observation is reminiscent of PIASy-dependent relocalization of both wild-type and sumoylation-defective Lef1 into nuclear bodies that partially overlap with PML-NBs (39). Thus, while sumoylation is not required for subnuclear relocalization of SF-1 (or Lef1), conditions that promote optimal sumoylation do correlate with altered nuclear distribution of SF-1.
Given that DEAD-box proteins are present in both splicing and translational complexes (31), repression may be coupled to transcript processing or translational control. However, studies to date, including ours, have yet to identify a function for the RNA helicase (unwindase) and RNA binding motifs in repression. Indeed, the N-terminal portion of DP103 is dispensable for interaction and repression of SF-1 and METS (21, 49) and for relocalization of SF-1 to nuclear bodies (our unpublished data). Further in vitro and in vivo experiments aimed at delineating the precise role of sumoylation in DEAD-box-mediated transcriptional repression will be of interest.
Acknowledgments
We thank D. Morgan for helpful discussions and F. Poulat for sharing unpublished data regarding the PIAS1/SF-1 interaction and for His6-hSUMO1-pcDNA3. We also thank D. Pearce, R. Grosschedl, K. Shuai, Y. Sadovsky, and C. Glass for reagents. We especially thank B. Panning and C. de la Cruz for discussion and reagents for immunocytochemistry experiments.
Support for this work was funded in part by a National University of Singapore Fellowship to M.B.L., an NSF Predoctoral fellowship award to L.A.L, a Susan B. Komen award to M.S., an NRSA fellowship to S.A.W., an AHA Beginning Grant in Aid to M.D., and an RO1-NIH-NIDDK grant to H.A.I.
REFERENCES
- 1.Achermann, J. C., J. J. Meeks, and J. L. Jameson. 2001. Phenotypic spectrum of mutations in DAX-1 and SF-1. Mol. Cell. Endocrinol. 185:17-25. [DOI] [PubMed] [Google Scholar]
- 2.Bland, M. L., R. C. Fowkes, and H. A. Ingraham. 2004. Differential requirement for steroidogenic factor-1 gene dosage in adrenal development versus endocrine function. Mol. Endocrinol. 18:941-952. [DOI] [PubMed] [Google Scholar]
- 3.Bland, M. L., C. A. Jamieson, S. F. Akana, S. R. Bornstein, G. Eisenhofer, M. F. Dallman, and H. A. Ingraham. 2000. Haploinsufficiency of steroidogenic factor-1 in mice disrupts adrenal development leading to an impaired stress response. Proc. Natl. Acad. Sci. USA 97:14488-14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botrugno, O. A., E. Fayard, J. S. Annicotte, C. Haby, T. Brennan, O. Wendling, T. Tanaka, T. Kodama, W. Thomas, J. Auwerx, and K. Schoonjans. 2004. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol. Cell 15:499-509. [DOI] [PubMed] [Google Scholar]
- 5.Chauchereau, A., L. Amazit, M. Quesne, A. Guiochon-Mantel, and E. Milgrom. 2003. Sumoylation of the progesterone receptor and of the steroid receptor coactivator SRC-1. J. Biol. Chem. 278:12335-12343. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W. Y., W. C. Lee, N. C. Hsu, F. Huang, and B. C. Chung. 2004. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 279:38730-38735. [DOI] [PubMed] [Google Scholar]
- 7.Clyne, C. D., A. Kovacic, C. J. Speed, J. Zhou, V. Pezzi, and E. R. Simpson. 2004. Regulation of aromatase expression by the nuclear receptor LRH-1 in adipose tissue. Mol. Cell. Endocrinol. 215:39-44. [DOI] [PubMed] [Google Scholar]
- 8.Desclozeaux, M., I. N. Krylova, F. Horn, R. J. Fletterick, and H. A. Ingraham. 2002. Phosphorylation and intramolecular stabilization of the ligand binding domain in the nuclear receptor steroidogenic factor 1. Mol. Cell. Biol. 22:7193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobreva, G., J. Dambacher, and R. Grosschedl. 2003. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 17:3048-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dufau, M. L., C. Tsai-Morris, P. Tang, and A. Khanum. 2001. Regulation of steroidogenic enzymes and a novel testicular RNA helicase. J. Steroid Biochem. Mol. Biol. 76:187-197. [DOI] [PubMed] [Google Scholar]
- 11.Fowkes, R. C., M. Desclozeaux, M. V. Patel, S. J. Aylwin, P. King, H. A. Ingraham, and J. M. Burrin. 2003. Steroidogenic factor-1 and the gonadotrope-specific element enhance basal and pituitary adenylate cyclase-activating polypeptide-stimulated transcription of the human glycoprotein hormone alpha-subunit gene in gonadotropes. Mol. Endocrinol. 17:2177-2188. [DOI] [PubMed] [Google Scholar]
- 12.Gillian, A. L., and J. Svaren. 2003. The Ddx20/DP103 dead Box protein represses transcriptional activation by Egr2/Krox-20. J. Biol. Chem. 279:9056-9063. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, P. R. Maloney, T. M. Willson, and S. A. Kliewer. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517-526. [DOI] [PubMed] [Google Scholar]
- 14.Gross, M., B. Liu, J. Tan, F. S. French, M. Carey, and K. Shuai. 2001. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene 20:3880-3887. [DOI] [PubMed] [Google Scholar]
- 15.Hammer, G. D., I. Krylova, Y. Zhang, B. D. Darimont, K. Simpson, N. L. Weigel, and H. A. Ingraham. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3:521-526. [DOI] [PubMed] [Google Scholar]
- 16.Hietakangas, V., J. K. Ahlskog, A. M. Jakobsson, M. Hellesuo, N. M. Sahlberg, C. I. Holmberg, A. Mikhailov, J. J. Palvimo, L. Pirkkala, and L. Sistonen. 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 23:2953-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinshelwood, M. M., J. J. Repa, J. M. Shelton, J. A. Richardson, D. J. Mangelsdorf, and C. R. Mendelson. 2003. Expression of LRH-1 and SF-1 in the mouse ovary: localization in different cell types correlates with differing function. Mol. Cell. Endocrinol. 207:39-45. [DOI] [PubMed] [Google Scholar]
- 18.Holmstrom, S., M. E. Van Antwerp, and J. A. Iniguez-Lluhi. 2003. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc. Natl. Acad. Sci. USA 100:15758-15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingraham, H. A., D. S. Lala, Y. Ikeda, X. Luo, W. H. Shen, M. W. Nachtigal, R. Abbud, J. H. Nilson, and K. L. Parker. 1994. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 8:2302-2312. [DOI] [PubMed] [Google Scholar]
- 20.Ito, M., Y. Park, J. Weck, K. E. Mayo, and J. L. Jameson. 2000. Synergistic activation of the inhibin alpha-promoter by steroidogenic factor-1 and cyclic adenosine 3′,5′-monophosphate. Mol. Endocrinol. 14:66-81. [DOI] [PubMed] [Google Scholar]
- 21.Klappacher, G. W., V. V. Lunyak, D. B. Sykes, D. Sawka-Verhelle, J. Sage, G. Brard, S. D. Ngo, D. Gangadharan, T. Jacks, M. P. Kamps, D. W. Rose, M. G. Rosenfeld, and C. K. Glass. 2002. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell 109:169-180. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu, T., H. Mizusaki, T. Mukai, H. Ogawa, D. Baba, M. Shirakawa, S. Hatakeyama, K. I. Nakayama, H. Yamamoto, A. Kikuchi, and K. I. Morohashi. 2004. Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4BP/SF-1 regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol. 18:2451-2462. [DOI] [PubMed] [Google Scholar]
- 23.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J. Biol. Chem. 277:30283-30288. [DOI] [PubMed] [Google Scholar]
- 24.Kotaja, N., M. Vihinen, J. J. Palvimo, and O. A. Janne. 2002. Androgen receptor-interacting protein 3 and other PIAS proteins cooperate with glucocorticoid receptor-interacting protein 1 in steroid receptor-dependent signaling. J. Biol. Chem. 277:17781-17788. [DOI] [PubMed] [Google Scholar]
- 25.Lavorgna, G., H. Ueda, J. Clos, and C. Wu. 1991. FTZ-F1, a steroid hormone receptor-like protein implicated in the activation of fushi tarazu. Science 252:848-851. [DOI] [PubMed] [Google Scholar]
- 26.Lu, T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6:507-515. [DOI] [PubMed] [Google Scholar]
- 27.Luo, X., Y. Ikeda, and K. L. Parker. 1994. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 77:481-490. [DOI] [PubMed] [Google Scholar]
- 28.Mallet, D., P. Bretones, L. Michel-Calemard, F. Dijoud, M. David, and Y. Morel. 2004. Gonadal dysgenesis without adrenal insufficiency in a 46, XY patient heterozygous for the nonsense C16X mutation: a case of SF1 haploinsufficiency. J. Clin. Endocrinol. Metab. 89:4829-4832. [DOI] [PubMed] [Google Scholar]
- 29.Melchior, F., M. Schergaut, and A. Pichler. 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28:612-618. [DOI] [PubMed] [Google Scholar]
- 30.Nachtigal, M. W., Y. Hirokawa, D. L. Enyeart-VanHouten, J. N. Flanagan, G. D. Hammer, and H. A. Ingraham. 1998. Wilms' tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell 93:445-454. [DOI] [PubMed] [Google Scholar]
- 31.Nelson, P. T., A. G. Hatzigeorgiou, and Z. Mourelatos. 2004. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 10:387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou, Q., J. F. Mouillet, X. Yan, C. Dorn, P. A. Crawford, and Y. Sadovsky. 2001. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 15:69-79. [DOI] [PubMed] [Google Scholar]
- 33.Pare, J. F., D. Malenfant, C. Courtemanche, M. Jacob-Wagner, S. Roy, D. Allard, and L. Belanger. 2004. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis, and regulated by a DR4 element. J. Biol. Chem. 279:21206-21216. [DOI] [PubMed] [Google Scholar]
- 34.Parker, K. L., and B. P. Schimmer. 1997. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr. Rev. 18:361-377. [DOI] [PubMed] [Google Scholar]
- 35.Poukka, H., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2000. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. USA 97:14145-14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajendran, R. R., A. C. Nye, J. Frasor, R. D. Balsara, P. G. Martini, and B. S. Katzenellenbogen. 2003. Regulation of nuclear receptor transcriptional activity by a novel DEAD box RNA helicase (DP97). J. Biol. Chem. 278:4628-4638. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, L. M., J. Shen, and H. A. Ingraham. 1999. New solutions to an ancient riddle: defining the differences between Adam and Eve. Am. J. Hum. Genet. 65:933-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sablin, E. P., I. N. Krylova, R. J. Fletterick, and H. A. Ingraham. 2003. Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Mol. Cell 11:1575-1585. [DOI] [PubMed] [Google Scholar]
- 39.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiio, Y., and R. N. Eisenman. 2003. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100:13225-13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swain, A., V. Narvaez, P. Burgoyne, G. Camerino, and R. Lovell-Badge. 1998. Dax1 antagonizes Sry action in mammalian sex determination. Nature 391:761-767. [DOI] [PubMed] [Google Scholar]
- 42.Tallec, L. P., O. Kirsh, M. C. Lecomte, S. Viengchareun, M. C. Zennaro, A. Dejean, and M. Lombes. 2003. Protein inhibitor of activated signal transducer and activator of transcription 1 interacts with the N-terminal domain of mineralocorticoid receptor and represses its transcriptional activity: implication of small ubiquitin-related modifier 1 modification. Mol. Endocrinol. 17:2529-2542. [DOI] [PubMed] [Google Scholar]
- 43.Tanner, N. K., and P. Linder. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251-262. [DOI] [PubMed] [Google Scholar]
- 44.Tran, P. V., M. B. Lee, O. Marin, B. Xu, K. R. Jones, L. F. Reichardt, J. R. Rubenstein, and H. A. Ingraham. 2003. Requirement of the orphan nuclear receptor SF-1 in terminal differentiation of ventromedial hypothalamic neurons. Mol. Cell. Neurosci. 22:441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vilain, E. 2000. Genetics of sexual development. Annu. Rev. Sex Res. 11:1-25. [PubMed] [Google Scholar]
- 46.Wu, W. S., S. Vallian, E. Seto, W. M. Yang, D. Edmondson, S. Roth, and K. S. Chang. 2001. The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol. Cell. Biol. 21:2259-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, P. L., S. F. Shan, Y. Y. Kong, Y. H. Xie, and Y. Wang. 2003. Characterization of a strong repression domain in the hinge region of orphan nuclear receptor hB1F/hLRH-1. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35:909-916. [PubMed] [Google Scholar]
- 48.Yan, W., H. Santti, O. A. Janne, J. J. Palvimo, and J. Toppari. 2003. Expression of the E3 SUMO-1 ligases PIASx and PIAS1 during spermatogenesis in the rat. Gene Expr. Patterns 3:301-308. [DOI] [PubMed] [Google Scholar]
- 49.Yan, X., J. F. Mouillet, Q. Ou, and Y. Sadovsky. 2003. A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol. Cell. Biol. 23:414-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, S. H., E. Jaffray, R. T. Hay, and A. D. Sharrocks. 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 12:63-74. [DOI] [PubMed] [Google Scholar]
- 51.Yang, S. H., and A. D. Sharrocks. 2004. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13:611-617. [DOI] [PubMed] [Google Scholar]