Abstract
Cancer remains one of the leading causes of death, albeit enormous efforts to cure the disease. To overcome the major challenges in cancer therapy, we need to have a better understanding of the tumour microenvironment (TME), as well as a more effective means to screen anti-cancer drug leads; both can be achieved using advanced technologies, including the emerging tumour-on-a-chip technology. Here, we review the recent development of the tumour-on-a-chip technology, which integrates microfluidics, microfabrication, tissue engineering and biomaterials research, and offers new opportunities for building and applying functional three-dimensional in vitro human tumour models for oncology research, immunotherapy studies and drug screening. In particular, tumour-on-a-chip microdevices allow well-controlled microscopic studies of the interaction among tumour cells, immune cells and cells in the TME, of which simple tissue cultures and animal models are not amenable to do. The challenges in developing the next-generation tumour-on-a-chip technology are also discussed.
Keywords: tumour-on-a-chip, microfluidics, tumour microenvironment, drug screening
1. Introduction
Cancer remains one of the leading causes of death in the USA and many other countries in the world, despite the extensive research and enormous efforts in drug discovery over the last few decades to cure the disease. This is partly due to the high cost of developing a new anti-cancer drug, as well as the need to better understand cancer development and the tumour microenvironment (TME), including the roles of inflammation, different effectors and suppressors of immune responses, the heterogeneity of tumour stroma, and the function of tumour vasculature. To make significant improvements in cancer therapy, it is necessary to develop more effective approaches to screen anti-cancer drug leads and to have a better understanding of TME using advanced technologies, including the organs-on-chips technology [1–5].
To date most cancer research and anti-cancer drug screening have been conducted using cell culture and animal models. While animal models of cancer can provide essential in vivo information of tumour growth and response to drug molecules, they could be very costly and the results may have very large variations among the animals used, thus it is difficult to obtain relevant statistics. Further, small animal models such as mouse models for cancer studies may not accurately represent what happens in humans [6]. On the other hand, two- and three-dimensional cell cultures have been widely used for screening anti-cancer drugs, and studying cell signalling, proliferation, migration and drug responses including altered protein/gene expression [7,8]. These in vitro models may use co-culturing of multiple cell types in hydrogel matrices and include patient-derived cells [9,10]. Although cell culture models are low cost, easy to handle and typically have high repeatability, they may not be able to mimic the microenvironment in an organ or an animal, thus are not suitable to study the effect of complex spatial organization and interaction of cells.
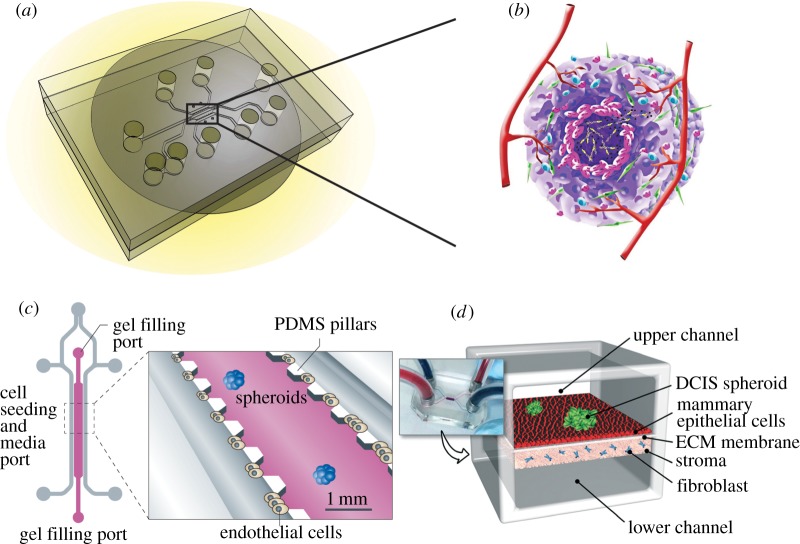
As an alternative to animal models and cell culture models to address the complex problem of cancer development and treatment, ‘tumour-on-a-chip’ technology has emerged recently as a new tool for cancer studies, providing a unique approach which integrates microfluidics, microfabrication, tissue engineering and biomaterials research, possessing the potential to significantly advance our understanding of cancer biology, allowing accelerated and cost-effective drug discovery [4,11]. As shown in figure 1, a tumour-on-a-chip system consists of a microfluidic device that has tissue culture, nutrient and small molecule supply and waste removal functions (figure 1a) [12]. Ideally, a three-dimensional tumour could grow on the chip with a complex tissue structure comprised of tumour cells, stromal cells and blood vessels either self-organized or spatially organized by design, mimicking some aspects of a tumour (figure 1b) [13]. Examples of first-generation tumour-on-a-chip systems include a chip in which lung cancer spheroids were embedded in micro-patterned three-dimensional matrices immediately contiguous to a microchannel lined with endothelial cells (figure 1c) [4], and a breast tumour-on-a-chip model comprised the upper and lower cell culture chambers separated by an ECM-derived membrane that mimics a basement membrane in vivo (figure 1d) [13]. Previous reviews in the literature on tumour-on-a-chip technology include the construction of three-dimensional tumour models [14–17], its applications to specific cancer studies such as metastasis [18,19], and its utilities in drug discovery [20,21].
Figure 1.
The concept of tumour-on-a-chip. (a) A microfluidic device that has tissue culture, nutrient and small molecule supply and waste removal functions for growing tumours on a chip. Adapted from [12]. (b) The ultimate goal is to grow a three-dimensional tumour on chip with a complex tissue structure consisting of tumour cells, stromal cells and blood vessels. (c) An example of tumour-on-a-chip in which lung cancer spheroids were embedded in micro-patterned three-dimensional matrices immediately contiguous to a microchannel lined with endothelial cells. Reprinted with permission from Macmillan Publishers Ltd. (d) The physiological microarchitecture is recapitulated in the breast-cancer-on-chip microdevice with two cell-culture chambers separated by an ECM-derived membrane. Adapted from [13] with permission from the Royal Society of Chemistry.
This review aims to showcase the recent developments of the tumour-on-a-chip technology to mimic TMEs for cancer biology studies and biomedical applications. In §2, three-dimensional in vitro tumour models established on microfluidic chips are reviewed. Specific microdevices mimicking various TMEs are elaborated in §3. In §4, examples of tumour-on-a-chip applications are discussed. The challenges in developing the next-generation tumour-on-a-chip technology are summarized in §5.
2. Three-dimensional in vitro tumour models on chip
To characterize and study the invasiveness and detailed cancer biology of different tumours, in vitro culture of cancer cells from a tumour sample is routinely used. A two-dimensional monolayer cell culture on thermoplastics is the gold standard for in vitro maintenance and multiplication of cells. Although two-dimensional cell cultures have been widely used in various cellular assays (e.g. migration and toxicity assays) to characterize the metastatic property and drug response of cancer cells, two-dimensional cultures cannot recapitulate the three-dimensional architecture of tissue's complexity, biophysical and biochemical property of extracellular matrix (ECM), and cell–cell interactions of human tumours [22–24]. Furthermore, cell cycle, cellular signalling and drug sensitivity can be different if cell culture is performed in a three-dimensional instead of a two-dimensional microenvironment [25–27]. In vivo three-dimensional models using animal xenografts are also popular but suffer from ethical concerns and are unable to mimic human-specific biology and physiology. In vitro three-dimensional tumour models are created by adapting several three-dimensional tissue engineering methods to construct cells into three-dimensional space and mimic the in vivo TME in the body (table 1). Among them, top-down methods use decellularized scaffolds and bottom-up methods use cells to build up tumour tissues for in vitro three-dimensional tumour models. In tumour-on-a-chip systems, three-dimensional tumour tissues are often cultured initially by established tissue culture protocols, then transferred onto the microfluidic chip for analysis. Thus, in vitro three-dimensional tumour models can be adopted in tumour biology research and the development of therapeutics for personalized medicine [28,29]. Next, we discuss common techniques for creating three-dimensional in vitro tumour models in detail (figure 2).
Table 1.
Comparison of in vitro tumour models.
| three-dimensional tumour models | processes | advantages | disadvantages |
|---|---|---|---|
| transwell assays | migration, invasion through ECM, transendothelial migration | recovery of motile cell population, easy to perform | no control over gradient, endpoint assay, inability to create multiplex gradient, no cell–cell interaction |
| tumour spheroids | mimicry of tumour mass in three-dimensional configuration | a micro-tumour with three-dimensional structure, necrotic core and nutrient transport property; perfusable with microfluidics | no vasculature on spheroids |
| ex vivo tumour section | direct in situ analysis on in vitro cultured tissue | retains primary tumour and stroma | require primary tumour tissue for every experiment |
| scaffold | solid extracellular support for three-dimensional cell culture | a characterized ECM structure for three-dimensional cell culture | difficult to uniformly distribute cells in scaffolds, difficult to perfuse the model |
| bioink three-dimensional printing | layer-by-layer construction of cells | printing multiple cell types and ECMs; high spatial precision | specific bioink formulation is needed for optimal cell survival |
| microfluidic microvascular model | patterning microscopic vascular capillary | perfusable model, microscopic observation for kinetics, incorporation of gradients | size limited to small tumours |
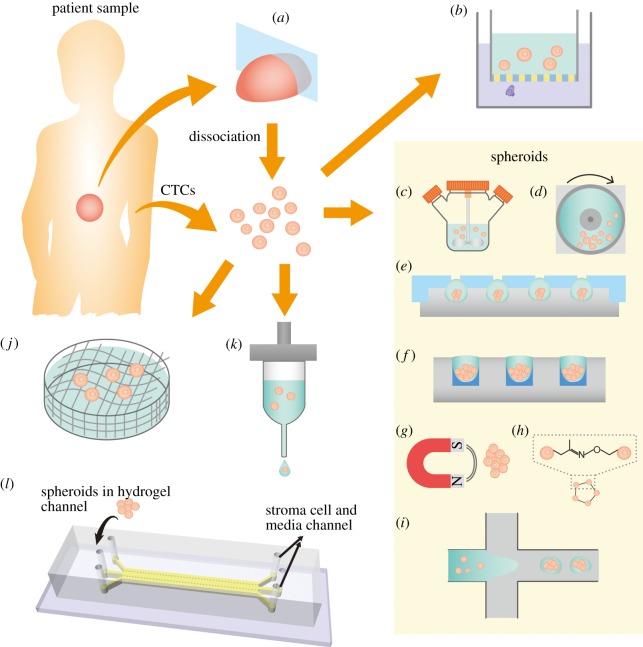
Figure 2.
Existing techniques to create three-dimensional in vitro tumour models. (a) An ex vivo tumour culture based on a tumour tissue section. (b) Single tumour cells embedded in hydrogel on transwell insert is one of the earliest three-dimensional models that can also characterize invasiveness. Tumour spheroids can be prepared from dissociated cells from tumour or circulating tumour cells (CTCs) by (c) the spinning mask method, (d) NASA microgravity apparatus, (e) the hanging drop method, (f) the liquid overlay method, (g) magnetic levitation after cells are incubated with magnetic nanoparticles, (h) bio-orthogonal chemistry and (i) microfluidic methods such as flow focusing, droplet microfluidics and digital microfluidics. Alternative to spheroids, three-dimensional tumour models can be fabricated by seeding cells in artificial three-dimensional matrices. (j) Cancer cells can be seeded in fabricated scaffolds. (k) Cell-embedded bioink can be printed as building blocks for tissues. (l) Microfluidics–microvascular model uses a microdevice to model a multiple tissue-type microenvironment. Adapted from [28,29].
2.1. Ex vivo tumour culture
Primary tumour tissues from biopsy or surgical resection can be embedded in ECM and cultured as an in vitro model [30–34] (figure 2a). The embedded tumour sections retain the tumour vasculature, nearby stroma and the heterogeneity of the tumour cells. Microfluidic technology can be combined with an ex vivo tumour section culture system for parallel drug sensitivity testing while maintaining continuous control over culture conditions [35].
2.2. Conventional transwell model
Transwell inserts (also known as Boyden chambers) are widely used to perform conventional migration, invasion and transendothelial migration assays, to assess the migration of cancer cells in combination with a chemical gradient. A transwell insert is composed of a polymeric porous membrane to allow cancer cells to migrate through the pores. A transwell insert is routinely used together with a multiwell plate with chemoattractants inside wells. In a migration assay, the ability of cancer cells to translocate through the pores is measured. The invasion assay further characterizes the migration of cancer cells through a three-dimensional ECM layer on the porous membrane. In addition, the transendothelial migration ability of cancer cells can be characterized by using a transwell insert with a confluent endothelial cell layer grown on top of the membrane [36] (figure 2b).
The transwell assay is usually performed as an endpoint assay because it is difficult to image the kinetic behaviour of cells migrating through the pores. Moreover, the steepness of the chemoattractant gradient established between the well and inside the transwell insert is difficult to control, making the transwell assay results semi-quantitative. However, transwell assays are quite suitable for more motile or invasive cell subpopulations as they can be recovered after the transwell assay [36].
2.3. Tumour spheroids
A tumour spheroid is derived from three-dimensional aggregations of cells under non-adherent cell culture conditions [37]. The tumour spheroid resembles a small tumour mass in its morphology, growth kinetics, nutrient transport and cell–cell as well as cell–matrix interactions. Thus, the tumour spheroid serves as an excellent in vitro three-dimensional tumour model [28,38].
Tumour spheroids can be generated by using single- or multiple-cell suspensions from permanent cell lines as well as dissociated cells from primary isolated tumour tissues and organotypic tissues [38]. Several common methods to generate tumour spheroids include suspension culture, hanging drop method, liquid overlay on non-adherent substrates, two-phase encapsulation and assembly by bio-conjugate chemistry or magnetic particles [28]. In the suspension culture, cells are placed in a spinner flask [39] (figure 2c) or a NASA microgravity vessel [40] (figure 2d) to promote spheroid formation by inducing aggregation. The suspension culture is advantageous in high-throughput production of spheroids, but the disadvantages are limited control over spheroid size and uniformity. The hanging drop method uses microtitre plate or microstructures to inversely hold a cell suspension droplet [41–47] (figure 2e). The cells aggregate under gravity and subsequently form spheroids inside the droplet. The hanging drop method is of moderate throughput, but it possesses better control over the size of the spheroid. Microfluidic perfusion networks in combination with hanging drop methods have been used for continuous spheroid culture and drug screening [44–47]. Alternative to the hanging droplet method, using the liquid overlay method, cell suspension is cultured on non-adherent substrates to produce spheroids [48–50] (figure 2f). The advantage of the liquid overlay method is its simplicity of operation, but the disadvantage lies in its poor control over spheroid size. Similarly, to produce tumour spheroids by avoiding cell adhesion to cultureware and inducing aggregation, an aqueous two-phase system can also compartmentalize cell suspension and produce spheroids without the concern of drying and possible inefficiency in chemical transport and toxicity of an oil phase [51–53]. Three-dimensional spheroids can also be formed by assembly of cells using bio-orthogonal chemistry [54] or incubation of cells with magnetic particles [55,56] (figure 2g,h).
Recently, several microfluidic techniques have been developed to create tumour spheroids by either hydrodynamic trapping of cells in stagnation regions or in microwell structures [57–60], aggregating multiple cells in double-emulsions or hydrogel droplets [61–64], or aggregating cells on a digital microfluidic platform [65] (figure 2i). The advantages of generating spheroids by microfluidics include control over spheroid size with continuous perfusion, as well as real time and in situ observation of spheroid formation kinetics. However, spheroids produced in some microfluidic models are difficult to retrieve for off-chip analysis [57,63,66].
2.4. Three-dimensional cell culture in three-dimensional matrices
Tissue engineering methods have been adopted to create three-dimensional tumour models. A scaffold is a biocompatible and chemically stable extracellular support structure serving as an instructive support for cell attachment, growth and morphogenesis into tissues [67] (figure 2j). A porous scaffold can be made from decellularized tissues or from fabrication of several natural ECM proteins or biocompatible polymers, such as collagen, hyaluronic acid, silk protein, polyethylene glycol (PEG) and polylactic acid [68,69]. The scaffold is commonly prepared by freeze drying, electrospinning, phase separation and microscale macromolecular self-assembly [70–74].
Tumour cells cultured in scaffolds showed less sensitivity to chemotherapy and yield tumours with more invasive phenotypes [70,75–77]. While the porous scaffolds have the mechanical and chemical characteristic of ECM for three-dimensional tumour cell culture, the disadvantages include lack of vasculature structure in fabricated scaffolds that hinder perfusion for long-term culture, as well as poor control on cell placement positions inside the scaffold.
Alternative to scaffolds, a bottom-up approach using cells or few-cell spheroids as building blocks has emerged, inspired by the embryonic developmental processes [78,79] (figure 2k). Hydrogels as ECM support are embedded with cells or few cell spheroids as building blocks (also known as bioinks) [80,81]. Several natural polymers such as collagen, fibrin, Matrigel®, hyaluronan, chitosan, gelatin and alginate, as well as synthetic polymers such as PEG can be used to create property-controlled hydrogel matrices. The bioink containing multiple cell types and multiple ECMs can be printed at high density into large-scale tissues and organs through the layer-by-layer additive bioprinting. The cell positions in three dimensions can be automatically and precisely controlled using bioprinting to create multicellular tissue with vasculature mimicking the in vivo tissue hierarchy and the microenvironment [80,82,83]. Common bioprinting methods include inkjet printing [84,85], microextrusion printing [86,87], laser-induced transfer printing [88] and stereolithography [89,90].
2.5. Microfluidic tumour–microvascular model
The vasculature plays a pivotal role in tissue engineering and tumour biology [91]. Tissue engineering with vasculature is important for three-dimensional persistent tissue culture. Moreover, the growth and dissemination of cancer requires growth of new vasculatures for nutrient transport [92]. Many cell types in the vasculature such as endothelial cells interact with cancer and modulate the TME as well as the cancer phenotype [93]. Conventional transwell assays, tumour spheroids and scaffold approaches share the disadvantage of their inability to incorporate tumour–vasculature interactions in the culture. Using microfluidic technology, capillary lumen structures have been fabricated to mimic the microvasculature in tissues [94]. Common methods to create capillary lumen structures as microvasculatures include moulding the capillaries in hydrogels by needles or rods [95–98], by photoresists [99–101], by sacrificial carbohydrates [102] or creating lumens based on viscous fingering instabilities [103,104]. Alternatively, an endothelial vascular network as the microvasculature can be formed by endothelial sprouting in hydrogels [105–112], monolayer on ECM hydrogel [113–115] or on a porous membrane [116,117], and monolayer in microchannels [118,119] (figure 2l).
Creating a functional microvasculature network together with the three-dimensional tumour model is essential to recapitulate the TME in vitro. By using a microfluidic perfusable platform to co-culture vasculature and cancer cells, it allows better kinetic examination of important cancer progression stages such as angiogenesis, intravasation and extravasation in a controlled microenvironment [99,110,112,120,121]. Future challenges for microfluidic tumour–vasculature model include validation of the platform to clinical tumour tissues and increase complexity of the emulated microenvironment, such as chemical gradients and fluid flow at biologically relevant speed and rhythms. TMEs are complex and each component within often interacts and affects one another. Current efforts have focused on mimicking specific TME to answer different biological questions. A microfluidic platform with active control components such as microvalves and micropumps can be programmed to recapitulate multiplex physical and chemical gradients together with multiple cell types to better mimic the complex microenvironment of a tumour. However, the design and optimization of such platforms still pose great challenges, and their robustness and reproducibility are also major hurdles that need to be overcome.
3. Mimicking tumour microenvironment using microdevices
Cancer is a complex and heterogeneous metastatic disease modulated by genetic, epigenetic and cellular signalling influenced by its surrounding stroma. The cancer cells grow uncontrollably into a primary tumour and interact with the supportive and immune cells as well as the biochemical and biophysical components of ECM in the nearby stroma. Within the TME, three aspects are important: (i) hypoxia in the necrotic core of primary tumour tissue further drives metabolic shifts of cancer cells in the peri-necrotic niche; (ii) new vasculature growth is induced by the tumour and tumour-associated stroma for nutrients in the peri-vascular niche; (iii) cancer cells interact with stroma to evade the immune system and adopt invasive and migratory phenotypes to metastasize to distant tissues in the metastatic niche [93,122] (figure 3).
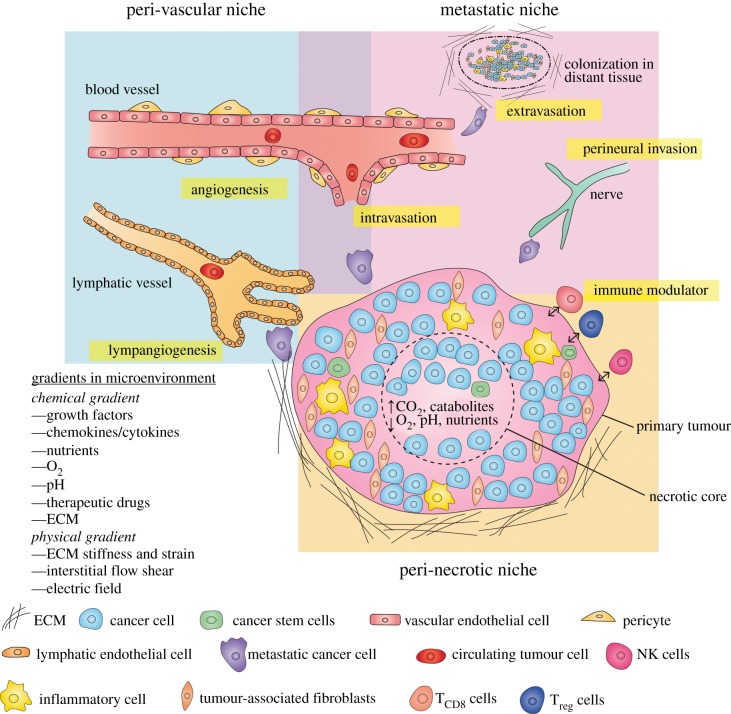
Figure 3.
A TME of solid tumour consists of the peri-vascular niche, metastatic niche and peri-necrotic niche. Hypoxia is a result of growth/nutrient imbalance induces metabolic reprogramming in the peri-necrotic niche. Tumour induces angiogenesis and lymphangiogenesis in an effort to gain more nutrients and access to the circulatory system in the peri-vascular niche. Cancer cells shed, invade, intravasate, extravasate to metastasize to distant tissues and create secondary tumours. The TME is complex and dynamic in cell–cell interactions and biophysical as well as biochemical interactions between the tumour and the stroma. Adapted from [123,124].
In the peri-necrotic niche, the metabolic state of cancer cells is reprogrammed under hypoxia and ischaemia due to an increase in the tumour mass. A necrotic microenvironment with dramatically low oxygen and nutrient concentrations as well as high acidity further induces the heterogeneity within the cancer cell population and promotes cancer cell survival in the harsh environment, as well as their metabolic resistance to many cancer therapeutics [125,126].
Within the peri-vascular niche, by cross-talking with stroma, the cancer cells also induce outgrowth of new vasculatures (angiogenesis) and new lymphatic vessels (lymphangiogenesis) for nutrient and gas transport to enable cell survival and proliferation [127,128]. However, the tumour vasculatures are often immature and leaky in comparison to the normal vasculature [129]. The peri-vascular niche also overlaps with the metastatic niche. New vasculature allows dissemination of cancer cells as they shed to circulating tumour cells, and among them tumour initiating cells can grow into secondary metastasis when seeded in distant tissues.
The metastatic niche must be developed for invasive cancer cells to shed from the primary tumour, invade through the basement membrane into the stroma, intravasate into nearby vascular or lymphatic vessels, travel and survive in the circulatory system, extravasate into a distant tissue site and form new micro-metastasis in new sites [123]. In some forms of cancer, cancer cells can also invade the nervous system during the process termed as peri-neural invasion, which is a contributor to tumour-related pain [130]. The complex sequential process that cancer cells undergo is also known as a metastatic cascade [131]. Recently, the theory of tumour initiating cells or cancer stem cells as a rare group of circulating tumour cells suggest that the microenvironment is important for cancer stem cells to seed in distant tissues and form new metastasis [123].
Microfluidic platforms allow recapitulation, manipulation and observation of cancer cell responses in TME on a chip. An in vitro model recapitulating the cancer cells as well as their microenvironment can enable more biomimetic and clinically relevant outcomes to accelerate our knowledge in tumour biology and improve cancer therapeutic development. In this section, we briefly review the microdevices developed in the past few decades to study different TMEs, including the peri-necrotic niche, peri-vascular niche and metastatic niche [122,124,132].
3.1. Peri-necrotic niche: modelling hypoxia and necrosis
In most tumour types, hypoxia is a mediator of tumour progression and therapeutic resistance [125]. As the primary tumour grows and its hyper-proliferating area increases, an imbalance between the hyper-proliferative cancer cell growth and nutrient as well as the gas supply from the vasculature causes ischaemia in the local tissue [125]. New vasculature to deliver more nutrients and gas is induced by the perivascular niche and in part by the hypoxia. However, the new vasculature is often abnormal and fails to rectify the nutrient deficit. The persistent hypoxia in the tumour has several effects including the selection of survival cancer cell genotypes, upregulation of pro-survival gene expressions, metabolic switches into anaerobic glycolysis, epithelial–mesenchymal transition (EMT) and therapeutic resistance [133]. Thus, creating an in vitro platform to recapitulate the hypoxia in an in vivo TME is very important.
In the conventional tissue culture laboratory, precise control over gas concentrations is challenging due to continuous oxygen diffusion into the culture medium in ambient air [126]. A CO2 incubator equipped with additional nitrogen gas mass flow controller can regulate the oxygen concentration within the incubator, but the oxygen gradient is still different in comparison to oxygen tension in the tissue. Alternatively, biochemical induction of key transcription factor of cellular hypoxia response such as hypoxia inducible factor (HIF) can be done to induce cellular signalling pathways in the hypoxia condition. The biochemical induction limits the spectrum of hypoxia study to dedicated signalling pathways [126].
Alternatively, with microfluidics, the gas permeability of the chip material provides the advantage of creating a hypoxic microenvironment to simulate the peri-necrotic niche. Poly(dimethylsiloxane) (PDMS) is a biocompatible silicone rubber with high gas permeability, and it has been a popular material for microfluidic chip fabrication by using soft lithography techniques [134,135]. A low oxygen environment or an oxygen gradient can be created by flowing different gases, gas-equilibrated liquids or oxygen scavengers in microfluidic networks [136–138]. Using poor gas-permeable thermoplastic as the microfluidic chip material or embedding a thin thermoplastic sheet can also improve the control over the gas environment inside the chip [138–140].
On two-dimensional microfluidic platforms, Zhang et al. [141] used SUM159 breast cancer cells to demonstrate increased migration in mesenchymal mode as well as production of lactate under hypoxic conditions. The acidic microenvironment derived from metabolic reprogramming is also a factor for cell migration. Neutralization of the environmental acidity can inhibit the migration velocity of cancer cells and simultaneously improve the efficiencies of therapeutics targeting HIF-1α, colony stimulating factor 1 receptor (CSF-1R) and C-C chemokine receptor type 4 (CCR4) [141]. These results demonstrate the importance of oxygen concentration as well as the pH level in the microenvironment to regulate the migration potential of cancer cells. Other two-dimensional microdevices can create stable oxygen gradients generated by oxygen scavengers, which become very useful to screen for cell survival and drug response under different oxygen concentrations [66,138,140,142] (figure 4a).
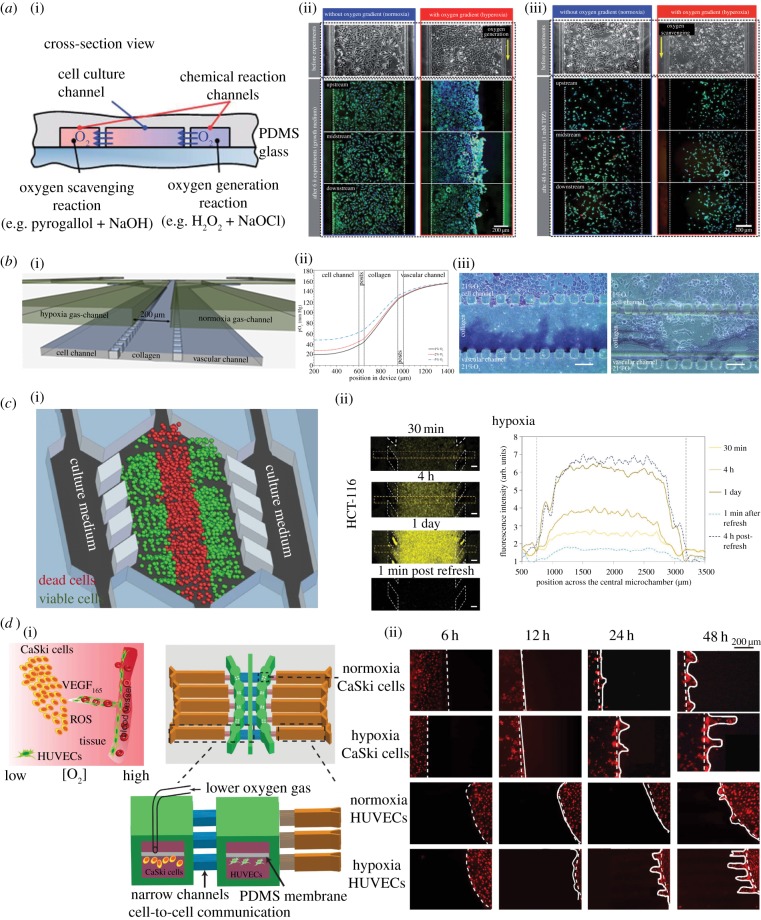
Figure 4.
Microdevices to model the peri-necrotic niche. (a) (i) The cross-sectional view of oxygen gradient generating microfluidic chip by Chen et al. (ii) A549 cell culture for 6 h with and without oxygen gradient (hyperoxia). Hyperoxia induced cell death is visible on the right side of the channel. (iii) A549 cell culture under 4 h 1 mM hypoxia-dependent anti-cancer drug Tirapazamine (TPZ) with and without oxygen gradient. Increased efficacy of TPZ is seen on the left side. Adapted from [66] with permission of The Royal Society of Chemistry. (b) (i) A microfluidic chip by Acosta et al. to create oxygen gradient by diffusion of gas between two gas supply channels. (ii) The predicted oxygen concentration gradient at steady state within the cross-section of microfluidic device. (iii) Increase invasion into collagen hydrogel by PANC-1 cancer cells under hypoxia condition. Reproduced from [143], with permission of AIP Publishing. (c) (i) A microdevice by Ayuso et al. to provide nutrient and oxygen gradient across it. (ii) Increased hypoxia in HCT-116 colon cancer cells imaged by hypoxia-sensitive dye [144]. (d) (i) Schematic diagram of an integrated microfluidic oxygen gradient generator by Lin et al. (ii) Cell migration of CaSki cancer cells and HUVEC endothelial cells under 5% O2 concentration for 2 days. Adapted from [145].
The response of cancer cells to hypoxic environments in three dimensions can also be examined by cell embedded hydrogel models. Xu et al. [146] demonstrated that the proliferation and invasion of glioblastoma U87MG cells under hypoxia conditions. By flowing normoxia gas in one control channel and hypoxia gas in another near the PANC-1 pancreatic adenocarcinoma cells, Acosta et al. [143] showed that hypoxia generated a more aggressive phenotype invading into the collagen gel (figure 4b).
In addition, microfluidic platforms have been used to examine kinetic formation of a necrotic core of a three-dimensional cell embedded hydrogel tumour model [144,147]. Ayuso et al. developed a three-dimensional cell embedded hydrogel system to observe the kinetic formation of necrotic cores in HCT-116 colon cancer cell model as well as U-251MG glioblastoma cell model over a 6-day period. Furthermore, real-time dynamic changes of oxygen and glucose concentrations, cell proliferation, apoptosis, reactive oxygen species formation and drug response can all be studied in situ on chip [144] (figure 4c). Co-culture multiple cell types with oxygen control is also possible with microfluidic platforms. Lin et al. [145] demonstrated that both cell migration and VEGF165 and HIF-1α were upregulated in CaSki cervical cancer cells under hypoxia conditions (figure 4d). A similar response was also observed with U87 glioblastoma cells in an alginate hydrogel [148]. Expressions of VEGF provide evidence that cancer cells under hypoxic environments are stimulated to induce angiogenesis and that there can be cellular signalling cross-talk between the peri-necrotic niche and peri-vascular niche. One imperative future direction is to develop more complex microdevices to recapitulate multiple microenvironments for detailed kinetic analysis of signalling crosstalks between various microenvironments, such as elucidation of the interdependency of necrosis and neo-angiogenesis in the cross-talk of the peri-necrotic niche and peri-vascular niche. Within the peri-necrotic niche, it has been challenging to validate the in vitro necrotic tumour model to tumour lysis and to incorporate stroma to investigate tumour–stromal cell interaction. Tumour lysis is the rapid death of large populations of cells that causes sudden metabolic disturbances, leading to tumour lysis syndrome (TLS). TLS contributes to high mortality of cancer. It can happen spontaneously due to tumour necrosis or can be initiated from anti-cancer therapies [149]. Although three-dimensional tumour spheroids on chip developed recently exhibit necrotic cores as a micro-tumour model [11], validation of this micro-tumour model with a human tumour and its tumour lysis kinetics remain a challenge. Moreover, incorporation of chemical gradients and co-culturing stroma cells such as fibroblasts, macrophages and natural killer cells to observe how necrosis contributes to stroma remodelling and chronic inflammation remains a difficult task [150]. Novel microdevice design integrated with biosensors and active flow control components such as microvalves is necessary to address these technical challenges.
3.2. Peri-vascular niche: modelling angiogenesis and quiescence of cancer cells
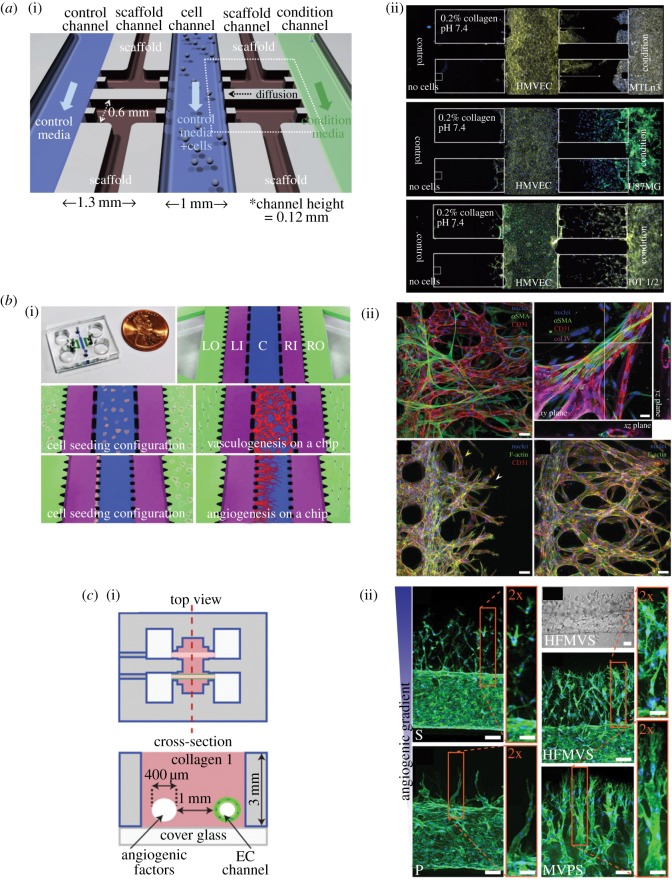
As a tumour grows and demands more nutrients for proliferation and survival, the tumour attracts neovascularization of blood vessels and lymphatic vessels through angiogenesis and lymphangiogenesis [127,128]. It has also been suggested that endothelial cells in peri-vascular niche can regulate quiescence of cancer cells as well as emergence after latency [151]. Using microfluidic technologies and tissue engineering, in vitro platforms with tumour and vasculature interactions can be developed and used to improve contemporary anti-angiogenic therapy. Several hydrogel microdevices focused specifically on angiogenesis induction by cancer cells in the co-culture configuration. Chung et al. [152] showed sprouting of endothelial cells into collagen hydrogel by VEGF gradient as well as by MTLn3 rat mammary adenocarcinoma cells (figure 5a). Cross et al. [99] also demonstrated formation and lumen structure and invasion of hydrogel of human umbilical vein endothelial cells when co-cultured with an oral squamous cell carcinoma cell line, OSCC3. Patra et al. [153] showed that when co-culturing HUVEC cells with HepG2 hepatocellular carcinoma cells in tumour spheroids, HUVEC cells migrated outwards to the proliferative edge and formed lumen-like structures under stimulation of pro-angiogenic factors. Liu et al. [154] used a three-dimensional hydrogel microfluidic device to study angiogenesis induction by salivary gland adenoid cystic carcinoma and oral squamous cell carcinoma cells. Both cell lines can induce strong angiogenesis and the angiogenesis can be inhibited under anti-angiogenic therapy. Aside from studying angiogenetic sprouting, Kim et al. [110] demonstrated that a perfusable microvascular network could be created on chip as a vasculogenesis model (figure 5b). Instead of generating microvascular networks in a hydrogel, Bischel et al. [104] and Nguyen et al. [96] reported methods to pattern endothelia in a capillary lumen structure as a model of an artificial blood vessel and angiogenesis assay (figure 5c).
Figure 5.
Microdevices to model the peri-vascular niche. (a) (i) Schematic for a three-dimensional microfluidic chip with scaffold channel for hydrogel patterning and flow channels to pattern different cell types for study of angiogenesis and invasion. (ii) Angiogenesis of endothelial cells in the middle towards MTLn3 cancer cells and invasion of cancer cells towards the vasculature. Adapted from [152] with permission of The Royal Society of Chemistry. (b) (i) Microfluidic chip design for creation of microvascular network and angiogenic sprouting. (ii) (top) Immunofluorescence staining of a fully functional microvascular network with endothelial cells, pericytes, cancer cells and leucocytes; (bottom) immunofluorescence staining of the angiogenic sprouting model. Adapted from [110] with permission of The Royal Society of Chemistry. (c) (i) Device schematic of a microdevice holding three-dimensional hydrogel with microchannels. (ii) Angiogenic sprouting of endothelium towards different angiogenic factors [96].
In the peri-vascular niche, aside from signalling between cancer cells and endothelial cells, other cell–cell interactions and physiochemical factors in the stroma also influence the angiogenesis. Using a multi-culture microdevice, Theberge et al. [155] demonstrated that the microenvironment would change when macrophages interacted with endothelial cells and fibroblasts. In the presence of macrophage with fibroblast and endothelial cells, expressions of several pro-angiogenic factors such as HGF, VEGF, interleukin-8 and anti-angiogenic factor matrix metalloproteinase-12 all increased. Angiogenesis is promoted but the endothelial tubules are abnormal due to the presence of other anti-angiogenic factors that are also secreted by macrophages. This observation supports our current knowledge that stroma cells in the microenvironment are also important in regulating the leaky vasculature cancer phenotype. These reported investigations demonstrate that microfluidic platforms offer new opportunities to recapitulate all the microenvironment components in vivo to yield physiologically and clinically relevant results in an in vitro assay.
Aside from the biochemical factors and cell–cell interaction in the stroma that can affect angiogenesis, it has been found that interstitial flow and shear stress also regulate the sprouting of microvasculatures. The advantage of microfluidic models over other conventional three-dimensional tumour models is their capability to create a perfusable vasculature with precise control with flow manipulations. Song & Munn [156] showed that both interstitial flow and VEGF gradient regulate the angiogenic sprouting and vascular dilation on a tumour-microvasculature-on-chip. Song et al. [157] further demonstrated that interstitial flow enhanced anastomosis, achieving perfusion by connecting multiple vascular sprouts.
In addition, the shear stress acting on endothelial cells can also regulate barrier function and induce expression of pro-angiogenic factors, such as VEGF [158]. Buchanan reported increased secretion of pro-angiogenic factors when endothelial cells were co-cultured with MDA-MB-231 breast cancer cells [159,160]. However, higher shear stress (10 dyne cm−2) applied on endothelial cells may increase perfusion and decrease secretion of several pro-angiogenic factors, as well as downregulate HIF-1α. These results indicate that interstitial flow, biochemical factors and cell–cell interactions all contribute to the regulation of angiogenesis in the TME. Using microfluidics, a perfusable and controllable platform supporting kinetic analysis of multiple cell co-culture is a promising approach to understand the pivotal roles of each factor and their interactions in regulating tumour angiogenesis. Testing the effectiveness and response to novel anti-angiogenic therapeutic tools using the tumour-on-a-chip platforms could provide detailed kinetic analysis and clinical relevant results.
The key challenge of adopting the peri-vascular niche is to incorporate multiplex chemical, physical and gas gradients (oxygen and nitric oxide) to elucidate its interplay with the peri-necrotic niche. The interdependency between necrosis and neo-angiogenesis is essential for understanding the growth of solid tumour and remodelling of the TME [161]. To identify the essential features in recreating an in vitro perivascular niche, a high-throughput microdevice is required to study microvasculature functions under different combinations of chemical and physical factors. The factors include but are not limited to pro-angiogenic growth factors, stiffness of stroma, shear stress of interstitial flow and concentration gradients of oxygen and nitric oxide.
3.3. Metastatic niche: modelling tumour–stroma interaction and metastasis
In a metastatic niche, cancer cells adapt into invasive and migratory phenotypes, shed from the primary tumour, intravasate, extravasate and colonize distant microenvironments through the metastasis cascade. Many microfluidic devices have been developed to inspect each process in the metastatic cascade.
First, cancer cells must locally invade into nearby stroma. Chung et al. [152] demonstrated the invasion of MtLn3, U87MG and 10 T 1/2 cancer cells into collagen hydrogels. In microdevices, by taking advantage of the laminar flow and limited mass transport at the microscale, stable chemical gradients can be established to investigate chemotactic invasion of three-dimensional cancer models that is difficult to achieve by conventional macroscale methods. Liu et al. [162] studied how MCF breast cancer cells embedded in the basement membrane extract hydrogel are guided by epidermal growth factor (EGF) to invade the matrix. Multiplex chemical gradients can also be easily established in a three-dimensional microfluidic model. Kim et al. [163] showed stromal cell derived factor-1α (SDF-1α) and EGF cooperatively modulated the migration of MDA-MB-231 cells.
Microfabricated porous microdevices can also be used to select and examine migratory cancer cells from tumour spheroids guided by EGF gradients similar to that in a conventional transwell assay. Using such devices, Kuo et al. [164,165] found decreased EpCAM expression in migratory cells, suggesting that the cells underwent the EMT and gained invasive properties.
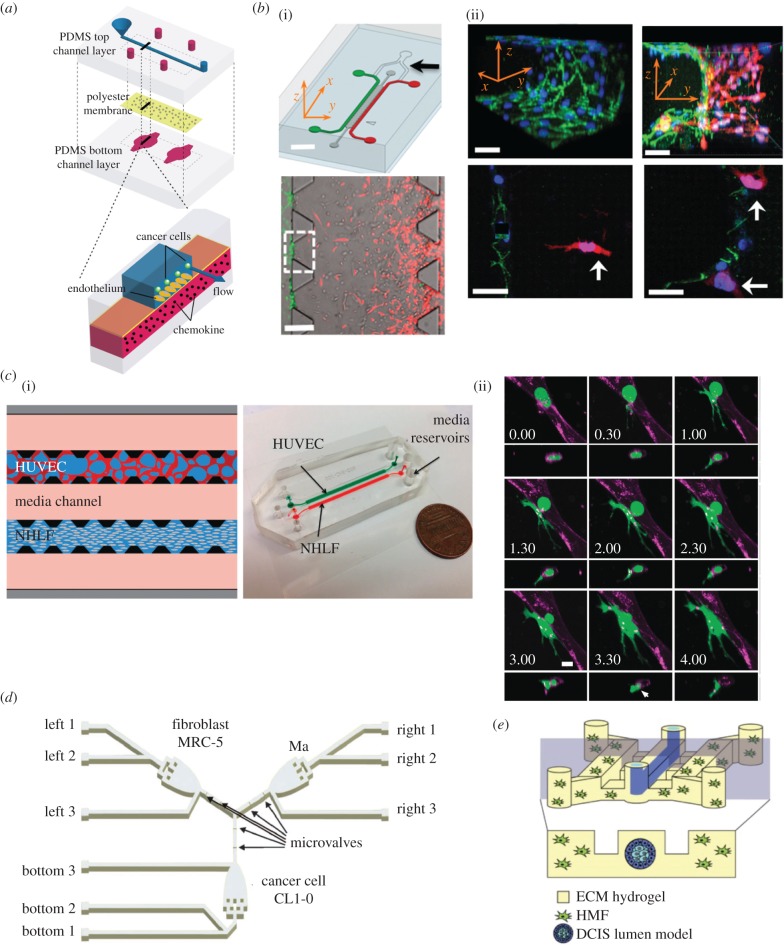
The second stage for metastasis is for cancer cells to adhere to endothelium and intravasate into the circulatory system. Song et al. [116] developed a microfluidic platform to culture uniform endothelium on a porous membrane to allow chemical transport and study how MDA-MB-231 cells adhere to the endothelium through CXCL12-CXCR4-dependent signalling (figure 6a). Zervantonakis et al. created a microfluidic tumour-ECM hydrogel–vasculature interface model to study how HT1080 fibrosarcoma cells interacted with the endothelial monolayer [166] (figure 6b). While the fibrosarcoma cells have the ability to intravasate across the endothelium, when macrophages are present at the endothelium, macrophages can secrete TNF-α and increase endothelial permeability. As a result, the fibrosarcoma intravasation through the endothelium is increased. Such three-dimensional microfluidic models combined with high-resolution microscopy enable real-time observation of cancer metastasis kinetics and further capture important parameters determining the microenvironment. Using a similar approach, Lee et al. [169] demonstrated that TNF-α also promoted the intravasation of MDA-MB-231 cells.
Figure 6.
Microdevices to model the metastatic niche. (a) A microfluidic vasculature with region-specific activation of endothelium for cancer cell adhesion analysis [116]. (b) (i) A microfluidic tumour–vascular interface model. (ii) Invasion of the endothelium by fibrosarcoma cells [166]. (c) (i) A microfluidic microvascular network platform. (ii) The extravasation dynamics of MDA-MB-231 cells. Adapted from [109] with permission of The Royal Society of Chemistry. (d) A microdevice to study how the paracrine signalling between macrophage, lung adenocarcinoma cells, and myofibroblasts can affect the invasiveness of the cancer. Adapted from [167] with permission of The Royal Society of Chemistry. (e) The tubeless lumen model to study invasive transition of MCF10aDCIS ductal carcinoma by mammary fibroblast [168].
The intravasated cancer cells enter the blood vessel and become circulating tumour cells (CTCs) that travel throughout the body in the circulatory system. The CTCs have been a very active topic for the role in metastasis and the clinical potential as a diagnostic and prognostic tool [170]. While the amount of CTCs is very low in peripheral blood, it is hypothesized that cancer stem cells or tumour initiating cells can seed in distant tissues and grow into secondary tumours [171]. Many microfluidic platforms have been developed for capture and analysis of CTCs, more dedicated articles can be found in the literature [172–174].
At distant sites, the circulating tumour cells need to extravasate through the endothelium and settle in the new microenvironment. Zhang et al. [119] demonstrated that chemokine CXCL12 could stimulate salivary gland adenoid cystic carcinoma cells to extravasate through HUVEC endothelium. The stimulated extravasation can also be inhibited by CXCR4 antagonist AMD3100. Chen et al. [109] employed a microvascular network in hydrogel and loaded MDA-MB-231, HT-1080 and MCF-10A cells by perfusion (figure 6c). The extravasation events (transendothelial migration) of the cells from the microvascular network into hydrogel can be tracked via time-lapsed microscopy. Interestingly, different cancer cell subpopulations exhibit different migration capabilities. Trapped cells as well as clustered cells showed much higher rate of migration into the ECM. Activation of tumour integrins β1 was found to be necessary for both extravasation and bone marrow colonization using the microvascular network microdevice [175]. Several microfluidic models also employed the ECM hydrogel–endothelium monolayer interface models commonly used in intravasation to study cancer extravasation by seeding cells in different microfluidic channels [112,176,177].
In the metastatic niche, other stromal cells and biophysical components also influence cancer cells' invasiveness. Multiple cell co-culture microdevices have been developed to study the effect of cell–cell interactions such as autocrine and paracrine signalling on the invasiveness of the cancer cells. Small vesicles containing nucleic acids and proteins (termed exosomes) may be the carriers to carry signalling molecules between the cancer and stromal cells [178]. Hsu et al. [167] developed a two-dimensional three-chamber PDMS microfluidic chip with microvalve control to selectively flow the conditioned media of fibroblast, macrophages and CL1-0 lung cancer cells to investigate how paracrine signalling from tumour stroma affected cancer cell invasiveness (figure 6d). Lung cancer cells release TGF-β1 to transform fibroblasts into myofibroblasts and in return promote the migration speed of cancer cells. However, macrophages can immunomodulate the myofibroblasts and the cancer cell migration speed decreases in the macrophage-pretreated and myofibroblast conditioned media. Interestingly, instead of pretreatment, direct combination of macrophage conditioned medium and myofibroblast conditioned medium resulted in a nearly threefold increase of lung cancer cells' migration speed [167]. Similar to other tri-culture microfluidic models, these results imply that the responses of cancer cells influenced under multiple factors can be quite complex and diverse [155,167].
Multiplex three-dimensional co-culture microdevices also serve as useful tools to investigate how the stroma interacts and modulates cancer cells. A breast cancer-on-a-chip device developed by Choi et al. [13] recapitulates the mammary duct and stroma as well as tumour spheroid in one microdevice model. Jeong et al. [179] used multiple hydrogel chambers embedded with tumour spheroids and fibroblasts to show that cancer associated fibroblasts promote cancer cell proliferation and drug resistance. Liu et al. [180] developed a four-chamber co-culture microdevice to simulate the microenvironment of bladder cancer with T24 cancer cells, macrophages, fibroblasts and HUVECs embedded in hydrogel. The bladder cancer cells grew into reticular structures and stromal cell phenotype changed despite the lack of three-dimensional tissue hierarchy in the system. Bischel et al. [168] patterned a three-dimensional lumen structure in a microdevice by viscous fingering method and successfully verified that the invasion of ductal carcinoma in situ (DCIS) of breast cancer cells was induced by mammary fibroblasts (figure 6e). By using second harmonic imaging, increased collagen modifications were found near the invasive region, suggesting that the ECM was remodelled by invasive cancer cells.
Aside from cell–cell interaction in the metastatic niche, physical factors such as interstitial flow and mechanical stimulation can regulate invasiveness of cancer cells. Polacheck et al. [181] developed a microfluidic culture chip to apply a stable interstitial flow to MDA-MB-231 cells embedded in a collagen hydrogel. Cancer cells at different densities responded to interstitial flows differently. At low cell density, cells migrated with the interstitial flow and the migration was dependent on CCR7 signalling. When CCR7 signalling was blocked, the migration directionality was reversed. Jeon et al. [112] demonstrated that the presence of interstitial flow in a microvascular network reduced the extravasation of cancer cells and decreased the permeability of vasculature. By applying cyclic tensile strain on myofibroblasts in a PDMS microdevice, Huang et al. [182] showed that tensile strain reduced the ability of the myofibroblast to accelerate cancer cell migration. The effect of cyclic tensile strain is also modulated by IL-1β secreted by other cells in the stroma, which implies the complicated interaction between cancer cells and different stromal cell types in the microenvironment. The stiffness of the ECM and stromal cells can also regulate the invasiveness of cancer cells [183,184]. Finally, transepithelial potential differences in tissues can generate physiological electric field and guide the migration of cancer cells through electrotaxis [185–187]. In short, many metastatic niche studies verified that invasive cancer cells could interact with and modulate the biophysical and biochemical properties of the stroma, as well as all the cellular components in the complex microenvironment. Similar to the challenges in mimicking TMEs, incorporating multiplex chemical, physical, and cell factors in a metastatic niche is critical in order to create a reliable in vitro micro-tumour model and investigate how each component contributes to the modulation of the metastatic cascade.
4. Applications of tumour-on-a-chip technology
The development and application of tumour-on-a-chip technology has the potential to address many important biological questions by replicating major aspects of the tumour structure, microenvironment and tumour biology. For example, a tumour-on-a-chip system may allow us to study the complexity of cancer growth and progression in a controlled fashion, capture and analyse spatio-temporal dynamics of tumour cells interacting with stromal cells, immune cells and other cells in the blood, and perform high-resolution imaging to understand some of the molecular and cellular mechanisms of tumour growth and metastasis. Tumour-on-a-chip approaches may allow the use of patients' own tumour cells to determine how they respond to anti-cancer drug or immunotherapy and to better predict cancer aggressiveness, achieving the best possible clinical outcome by extending the survival rate and reducing the chances of relapses and emergence of drug-resistant tumours. Although the technology is still in its early stages, the current designs of microfluidic tumour-on-a-chip systems have already shown promise in growing simple three-dimensional tumours and having good control over the TME. Some of the applications include multiplexed drug screening, transport of nanoparticles, transcription analysis, proteomic analysis and metabolic changes in cells.
4.1. Multiplexed drug screening
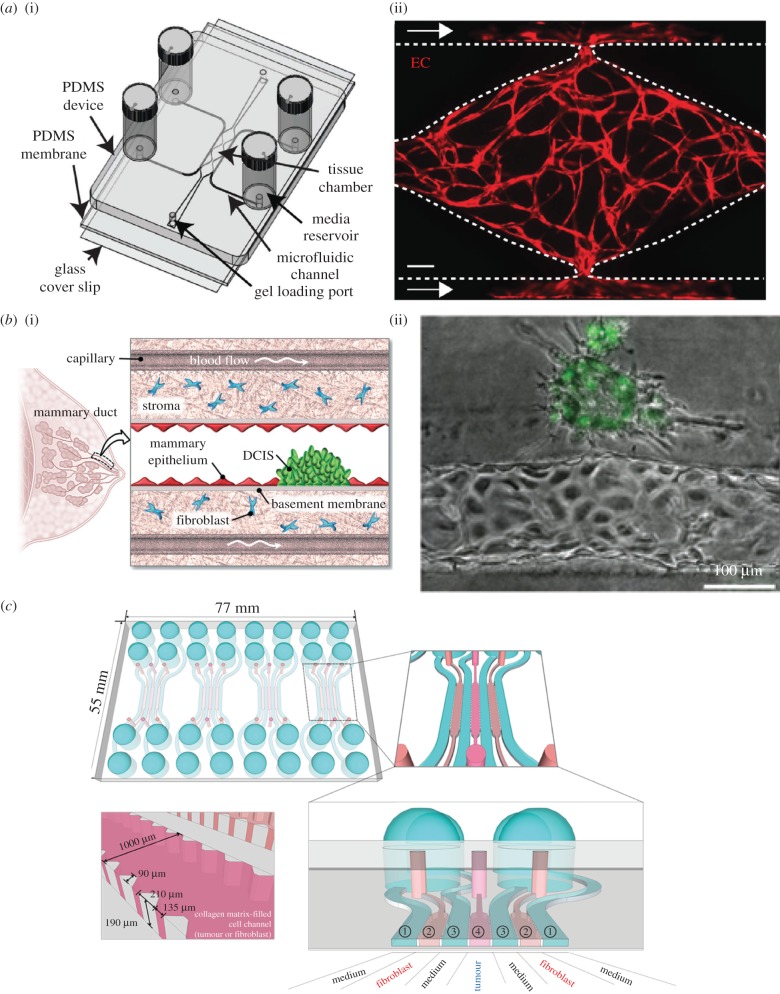
Conventional pre-clinical drug screening is expensive and time-consuming, and requires large number of cells. Recent advances in microfluidics technology have enabled cost-effective high-throughput screening. Aside from having a lower cost and faster processing speed, microfluidic chips require a much smaller sample volume. Furthermore, these chips can be customized to monitor the effects of anti-cancer drugs on any number of parameters, including cell migration [188]. Specifically, Zhang et al. developed a microfluidic device with 3120 different microchambers in which cell density was varied throughout the chambers, and the average migration velocity and the percentage of migrating cells were quantified. This device can create chemical gradients of multiple anti-tumour drugs and generate multiplicates of sample data on a single chip to specifically monitor mesenchymal migration and survival of tumour cells upon exposure to drugs that inhibit cell migration, including axitinib [189]. In a study by Sobrino et al. [124], vascularized microtumours were created on a PDMS membrane to study the effects of vascular targeting agents, such as apatinib and linifanib (figure 7a). A key drawback to this approach is the absorption of the agents in question by the PDMS membrane. Further work is needed to determine the effects of the partition coefficients of various types of drugs in different types of microfluidic platforms. Choi et al. explored the TME as a crucial regulator of tumour progression by designing a microchip with two microchannels surrounding a basement membrane with epithelial and stromal cells to simulate pre-invasive breast cancer lesions. Tumour spheroids were cultured on top of the epithelial cell layer [13] (figure 7b). This model replicates the three-dimensional microarchitecture in vivo and enables simulation of physiological delivery of intravenously administered paclitaxel by continuous flow through the lower microchannel [13]. This device can be scaled up easily for multiplexed screening of drug molecules based on their efficacy and safety, and the platform is flexible enough to be used for models of other types of cancer.
Figure 7.
Microdevices for tumour-on-a-chip studies. (a) Establishment of vascularized micro-organs (VMOs). (i) A schematic depicts the microfluidic platform of the VMO, which consists of a thick layer of PDMS with patterned tissue chambers and microfluidic channels, bonded on top of thin PDMS membrane and a glass coverslip. The cell–ECM suspension is injected through the gel-loading ports at either end of the tissue chamber. The four media reservoirs are attached to the inlets and outlets of the microfluidic channels. (ii) A depiction of a representative tissue chamber at day 7 with a fully developed vascular network. Transduced endothelial cells, shown in red, are migrating out and fusing with microfluidic channels. This platform is used later in the study to establish a human colon cancer micro-tumour. Adapted from [124] with permission from Nature Publishing Group. (b) A human breast cancer-on-a-chip. (i) A depiction of DCIS (carcinoma) in a mammary duct, with basement membrane, epithelium, stroma and capillary blood flow. Adapted from [13] with permission from the Royal Society of Chemistry. (ii) An image showing the interaction between breast cancer cells and an artificial microvessel embedded in a microfluidic tumour-on-a-chip device. Adapted from [98] with permission from the American Association for Cancer Research. (c) Design of microfluidic chip for tumour spheroid–fibroblast co-culture. This chip is used for three-dimensional co-culture of human colorectal cancer cells and fibroblasts. There are four units per chip and seven channels per unit for loading with either cells or media. The bottom-left shows in detail a cell-loading channel. Adapted from [179] with permission.
4.2. Transport and delivery of nanoparticles
Microfluidic systems can be used to evaluate nanoparticle transport in vitro and optimize nanoparticle designs by selecting the right size, shape and surface chemistry, such that the nanoparticle systems identified would have higher rates of success in drug delivery or in vivo imaging, thereby reducing the number of costly animal studies [190]. Recently, Albanese et al. designed a tumour-on-a-chip microfluidic model to study how nanoparticles were transported in the three-dimensional tumour spheroid. They showed that flow rate affected the accumulation of the nanoparticles in the in vitro spheroid model [11]. Kwak et al. [191] developed a tumour-microenvironment-on-a-chip model to recapitulate the complex transport of drugs and nanoparticles within a three-dimensional model of breast cancer and endothelial cells. They could quantify the effects of nanoparticle size on extravasation and interstitial diffusion. There was a significant decrease in both parameters between the 100 and 200 nm nanoparticles [191]. Bagley et al. demonstrated the use of plasmonic nanoantennae to enhance transport into a model of ovarian cancer via heat generation. They also used temperature-controlled microfluidic devices to measure diffusion of the nanoparticles in vitro [192]. The use of microfluidic devices to aid in the rapid development of translatable nanoparticles for TME studies is a very active and promising area of research.
4.3. Analysis of transcription
Using droplets in a microfluidic platform is an effective way to conduct transcription analysis on the level of single cells. Zhang et al. [193] developed a microfluidic device for performing single copy RT-PCR (reverse transcription polymerase chain reaction) using agarose droplets, which contained both sample and RT-PCR reagents. The platform was validated by showing significant differences in expression of the EpCAM cancer biomarker gene between different types of cancer cells [193]. Microfluidic droplets were also used in a separate study by Hayes et al. [194] to evaluate ECM gene expression levels in patient samples of colorectal cancer in order to find a potential correlation between differential expression and metastatic potential. A study by Jang et al. [195] demonstrated that a droplet-based model of microtumours can be used effectively to analyse the gene expression of markers related to the EMT. Developing high-throughput single-cell analytical techniques and using patient samples to find correlates or clusters of genes of interest has the potential to greatly expand the number of therapeutic targets currently known to us.
4.4. Proteomic analysis
Quantitative analysis of the cancer proteome has the potential to have a tremendous impact not only on molecular diagnostic technology, but also on discovering novel therapeutic targets. In a study by Sun et al. [196] a microfluidic cytometry imaging system was developed that is capable of quantitative, single-cell proteomic analysis in both cultured cell lines and patient samples, using as little as 1000 cells. Its clinical application was demonstrated by analysing four proteins within the mTOR signalling pathway using human brain tumour samples, and comparing the results to that using well-established clinical immunohistochemical (IHC) protocols. The IHC findings corroborated the single-cell analysis in all but one case [196]. In a study by Jeong et al. [179], human colorectal cancer cells were co-cultured with fibroblasts on a PDMS microfluidic chip, which was then used to quantify the level of proteins involved in angiogenesis, apoptosis and cell motility (figure 7c). On a larger scale, Xu et al. [197] designed a biomimetic multi-organ microfluidic chip to assess changes in the expression levels of CXCR4, RANKL and other markers in the various ‘distant organs’ after tumour cell invasion.
4.5. Analysis of metabolites and energy metabolism
Cancer cells tend to continuously multiply without cell cycle check, thus understanding the mechanism of cancer cell energy metabolism is critical to both basic cancer research and cancer therapeutics. Microdevices are well suited to study tumour cell energy metabolism by controlling both the oxygen supply and nutrient depletion to the cells. Xu et al. [198] designed a three-dimensional microfluidic chip to study the energy metabolism in tumour-associated fibroblasts and bladder tumour cells, specifically measuring lactic acid concentration and mitochondrial-related gene expressions. Culture media were perfused through the microfluidic channels which contained fibroblasts or bladder tumour cells or both. The conditioned media of co-cultured cells had the highest lactate concentration, suggesting that the aerobic glycolysis increased under the co-culturing condition. This microfluidic platform provided a unique non-contact co-culture condition to investigate energy metabolisms between different cell types. Zhu et al. [199] also examined cancer cell metabolism using a microfluidic tumour–endothelial cell co-culture system. Lactic acid and mitochondrial protein levels were measured and found to increase in the co-culture group. Similar approaches can also be adopted to screen for drug resistance, or investigate energy metabolism in regenerative medicine [200,201].
As cancer cells that release high levels of lactate correlate to increased metastasis [202], a microdevice has been developed to perform single-cell analysis of metabolites. Mongersun et al. [203] developed a droplet microfluidic platform that quantifies the lactate release rate down to single-cell resolution. The PDMS/glass microfluidic chip with the flow focusing design produced droplets containing single cancer cells and allowed real-time monitoring of lactate release within each individual droplet [203]. Performing metabolic analysis on single populations of tumour cells can yield significant insights into the mechanisms of tumour heterogeneity and energy metabolism reprogramming, both are important for predicting cancer metastatic potential as well as drug resistance [204].
5. Conclusion and future perspectives
To fully realize the potential of tumour-on-a-chip approaches, a number of key questions must be addressed. For example, how to model the mechanism of intravasation and extravasation using such a system; how to allow tumour-associated tissues to mature on a chip with respect to self-organization, if there is a minimum number of components needed to construct a tumour-on-a-chip system that allows a tumour to grow on a chip. Clearly, different approaches need to be developed to quantitatively analyse tumour–matrix interactions (including matrix remodelling and growth factors) in order to understand the enhanced permeability and retention effect as well as the phenotype of dormancy. It is also very important to reflect the heterogeneity and evolution present in the tumour by using a tumour-on-a-chip system. As a control, we need to have both cancerous and normal tissues grown on a chip in order to compare different features. It is also possible to use tumour-on-a-chip approaches to study tumour–immune response as to how bacteria and viruses trigger oncogenesis.
Tumour-on-a-chip systems may have different designs and complexities, depending on the medical relevance and biological question(s) to be addressed. It is necessary to avoid constructing oversimplified or overcomplicated systems, and have sufficient complexity driven by need. Accordingly, a tumour-on-a-chip system may include one or more of the following considerations: (i) structural features including three-dimensional tumour constructs and microfluidics designs; (ii) biomechanical and kinematic parameters such as matrix stiffness and anisotropy, cell adhesion and flow conditions; (iii) cell types and sources, including patient cells, cell lines, stromal cells, stem cells and progenitor cells; (iv) cell metabolism, culture media transport, waste removal and cytotoxicity; (v) physiological levels of concentration and concentration gradients of circulating factors. To accurately capture important features of a tumour, it may also be necessary to consider metastatic sites, recapitulation of cancer–immune cell interactions, and integrate real-time, on-chip monitoring of relevant biophysical and biochemical parameters. While preformed scaffolds for tumour structure and organization have certain advantages, the self-organized tumour structure through evolution of cell–cell interaction may provide a better model for tumour-on-a-chip platforms. It is likely that different tumour-on-a-chip systems with different features and complexities are needed for different cancers and/or to address different questions.
The emerging tumour-on-a-chip technology has the potential to transform the fields of oncology and cancer biology. However, there are roadblocks in technology development, including design, optimization, analysis and validation. Consistency of device properties (such as its biocompatibility, fit for purpose, ease of handling and mechanical properties) relies on material choices. Most devices have been built on PDMS-based substrates, which have been outstanding for studies on biological mechanisms, but have severe limitations when used with hydrophobic drugs. Other mouldable and printable surrogates must be explored to overcome this limitation such as off-stoichiometry thiol-enes [205], epoxy resin [206] and perfluorinated polymers [207]. Systematic manipulation and automation of the physical and chemical parameters within the microfluidic device will require integration of microdevice printing experts with polymer chemists and material scientists. Material choice and user operability are chief concerns when considering the scalability of the device and good manufacturing practice development. Further, it is important to establish the shelf-life (longevity post-manufacturing and pre-utility) and sustainability (e.g. duration of cell culture and waste removal in the system) of tumour-on-a-chip devices.
Although the potential of tumour-on-a-chip systems as cancer research tools has been demonstrated through proof-of-concept reports, major challenges for translating the technology to clinical practice remain, including the validation of device functionalities by comparing with well-established in vivo tumour models, and the correlation of the results obtained using tumour-on-a-chip systems with clinical tumour tissues. The tumour-on-a-chip system has the unique advantages of precisely manipulating the physical and chemical factors in the TME, co-culturing stromal cells with cancer cells, providing an optical window for real-time observation of molecular and sub-cellular processes through microscopy, and integrating with biosensors for quantitation [208]. Tumour-on-a-chip systems can be superior to animal xenograft models concerning physiochemical differences, biological variation, cost, and ease of statistical analysis. To fully realize the potential of the tumour-on-a-chip technology, it is essential for researchers in biomedical engineering, material science, biophysics, cell biology and oncology to make concerted efforts in designing and optimizing tumour-on-a-chip systems for cancer research, drug discovery and in translating the technology to clinical use [209].
Acknowledgements
G.B. thank Drs Kaiming Ye, Kam Leong, Philip LeDuc, Bahareh Behkam, Utkan Demirci, Gretchen Mahler, Shelly Peyton, Peter Searson, Shay Soker and Brian Wamhoff for helpful discussions.
Data accessibility
This article has no additional data.
Authors' contributions
G.B., A.Q.S., H.F.T. and A.T. wrote the review article together.
Competing interests
We declare we have no competing interests.
Funding
This work was supported in part by the Cancer Prevention and Research Institute of Texas (RR140081 to G.B.). H.F.T. and A.Q.S. acknowledge financial support from the OIST Graduate University with subsidy funding from the Cabinet Office, Government of Japan. H.F.T. was in addition supported by a JSPS Research Fellowship for Young Scientists (DC1) and a JSPS Grant in Aid for JSPS Fellows (JP1700362).
References
- 1.Alemany M, Semino C. 2014. Bioengineering 3D environments for cancer models. Adv. Drug Deliv. Rev. 79–80, 40–49. ( 10.1016/j.addr.2014.06.004) [DOI] [PubMed] [Google Scholar]
- 2.Wu MM, Swartz MA. 2014. Modeling tumor microenvironments in vitro. J. Biomech. Eng. 136, 021011 ( 10.1115/1.4026447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hielscher AC, Gerecht S. 2012. Engineering approaches for investigating tumor angiogenesis: exploiting the role of the extracellular matrix. Cancer Res. 72, 6089–6096. ( 10.1158/0008-5472.CAN-12-2773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esch EW, Bahinski A, Huh D. 2015. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discovery 14, 248–260. ( 10.1038/nrd4539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatia SN, Ingber D. 2014. Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772. ( 10.1038/nbt.2989) [DOI] [PubMed] [Google Scholar]
- 6.Begley CG, Ellis LM. 2012. Drug development: raise standards for preclinical cancer research. Nature 483, 531–533. ( 10.1038/483531a) [DOI] [PubMed] [Google Scholar]
- 7.Ehsan SM, Welch-Reardon KM, Waterman ML, Hughes CC, George SC. 2014. A three-dimensional in vitro model of tumor cell intravasation. Integr. Biol. 6, 603–610. ( 10.1039/c3ib40170g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fong EL, et al. 2013. Modeling Ewing sarcoma tumors in vitro with 3D scaffolds. Proc. Natl Acad. Sci. USA 110, 6500–6505. ( 10.1073/pnas.1221403110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokol ES, Miller DH, Breggia A, Spencer KC, Arendt LM, Gupta PB. 2016. Growth of human breast tissues from patient cells in 3D hydrogel scaffolds. Breast Cancer Res. 18, 19 ( 10.1186/s13058-016-0677-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efremov AN, Stanganello E, Welle A, Scholpp S, Levkin PA. 2013. Micropatterned superhydrophobic structures for the simultaneous culture of multiple cell types and the study of cell–cell communication. Biomaterials 34, 1757–1763. ( 10.1016/j.biomaterials.2012.11.034) [DOI] [PubMed] [Google Scholar]
- 11.Albanese A, Lam AK, Sykes EA, Rocheleau JV, Chan WC. 2013. Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat. Commun. 4, 2718 ( 10.1038/ncomms3718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozcelikkale A, Moon HR, Linnes M, Han B. 2017. In vitro microfluidic models of tumor microenvironment to screen transport of drugs and nanoparticles. WIREs Nanomed. Nanobiotechnol. e1460. ( 10.1002/wnan.1460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi Y, et al. 2015. A microengineered pathophysiological model of early-stage breast cancer. Lab. Chip 15, 3350–3357. ( 10.1039/c5lc00514k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutmacher DW, et al. 2009. Translating tissue engineering technology platforms into cancer research. J. Cell. Mol. Med. 13, 1417–1427. ( 10.1111/j.1582-4934.2009.00853.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvalho MR, Lima D, Reis RL, Correlo VM, Oliveira JM. 2015. Evaluating biomaterial- and microfluidic-based 3D tumor models. Trends Biotechnol. 33, 667–678. ( 10.1016/j.tibtech.2015.09.009) [DOI] [PubMed] [Google Scholar]
- 16.Peela N, et al. 2017. Advanced biomaterials and microengineering technologies to recapitulate the stepwise process of cancer metastasis. Biomaterials 133, 176–207. ( 10.1016/j.biomaterials.2017.04.017) [DOI] [PubMed] [Google Scholar]
- 17.Young EW. 2013. Cells, tissues, and organs on chips: challenges and opportunities for the cancer tumor microenvironment. Integr. Biol. 5, 1096–1109. ( 10.1039/c3ib40076j) [DOI] [PubMed] [Google Scholar]
- 18.Portillo-Lara R, Annabi N. 2016. Microengineered cancer-on-a-chip platforms to study the metastatic microenvironment. Lab. Chip 16, 4063–4081. ( 10.1039/c6lc00718j) [DOI] [PubMed] [Google Scholar]
- 19.Lee E, Song HG, Chen CS. 2016. Biomimetic on-a-chip platforms for studying cancer metastasis. Curr. Opin. Chem. Eng. 11, 20–27. ( 10.1016/j.coche.2015.12.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghaemmaghami AM, Hancock MJ, Harrington H, Kaji H, Khademhosseini A. 2012. Biomimetic tissues on a chip for drug discovery. Drug Discov. Today 17, 173–181. ( 10.1016/j.drudis.2011.10.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashaninejad N, Nikmaneshi MR, Moghadas H, Oskouei AK, Rismanian M, Barisam M, Saidi MS, Firoozabadi B. 2016. Organ-tumor-on-a-chip for chemosensitivity assay: a critical review. Micromach. Basel 7, 130 ( 10.3390/mi7080130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedl P, Wolf K. 2003. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 3, 362–374. ( 10.1038/nrc1075) [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez L Lara, Schneider IC. 2013. Directed cell migration in multi-cue environments. Integr. Biol. 5, 1306–1323. ( 10.1039/c3ib40137e) [DOI] [PubMed] [Google Scholar]
- 24.Cortese B, Palama IE, D'Amone S, Gigli G. 2014. Influence of electrotaxis on cell behaviour. Integr. Biol. 6, 817–830. ( 10.1039/c4ib00142g) [DOI] [PubMed] [Google Scholar]
- 25.Stock K, et al. 2016. Capturing tumor complexity in vitro: comparative analysis of 2D and 3D tumor models for drug discovery. Sci. Rep. 6, 28951 ( 10.1038/srep28951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riedl A, et al. 2017. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6 K signaling and drug responses. J. Cell Sci. 130, 203–218. ( 10.1242/jcs.188102) [DOI] [PubMed] [Google Scholar]
- 27.Howes AL, Richardson RD, Finlay D, Vuori K. 2014. 3-Dimensional culture systems for anti-cancer compound profiling and high-throughput screening reveal increases in EGFR inhibitor-mediated cytotoxicity compared to monolayer culture systems. PLoS ONE 9, e108283 ( 10.1371/journal.pone.0108283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benien P, Swami A. 2014. 3D tumor models: history, advances and future perspectives. Future Oncol. 10, 1311–1327. ( 10.2217/fon.13.274) [DOI] [PubMed] [Google Scholar]
- 29.Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. 2016. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 4, 12 ( 10.3389/fbioe.2016.00012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller BE, Mcinerney D, Jackson D, Miller FR. 1986. Metabolic cooperation between mouse mammary-tumor subpopulations in 3-dimensional collagen gel cultures. Cancer Res. 46, 89–93. [PubMed] [Google Scholar]
- 31.Dark GG, Hill SA, Prise VE, Tozer GM, Pettit GR, Chaplin DJ. 1997. Combretastatin A-4, an agent that displays potent and selective toxicity toward tumor vasculature. Cancer Res. 57, 1829–1834. [PubMed] [Google Scholar]
- 32.Yamada A, Tsuchida T, Kato T, Kawamoto K. 2002. Ultrastructural analysis of brain tumors using collagen gel culture. Brain Tumor Pathol. 19, 11–14. ( 10.1007/BF02482450) [DOI] [PubMed] [Google Scholar]
- 33.Naipal KA, et al. 2016. Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer 16, 78 ( 10.1186/s12885-016-2119-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies E, et al. 2015. Capturing complex tumour biology in vitro: histological and molecular characterisation of precision cut slices. Sci. Rep. 5, 17187 ( 10.1038/srep17187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang TC, Mikheev AM, Huynh W, Monnat RJ, Rostomily RC, Folch A. 2014. Parallel microfluidic chemosensitivity testing on individual slice cultures. Lab. Chip 14, 4540–4551. ( 10.1039/c4lc00642a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller WA, Luscinskas FW. 2008. Assays of transendothelial migration in vitro. Methods Enzymol. 443, 155–176. ( 10.1016/S0076-6879(08)02009-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta G, Hsiao AY, Ingram M, Luker GD, Takayama S. 2012. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Control Release 164, 192–204. ( 10.1016/j.jconrel.2012.04.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. 2010. Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 148, 3–15. ( 10.1016/j.jbiotec.2010.01.012) [DOI] [PubMed] [Google Scholar]
- 39.Sutherland RM, McCredie JA, Inch WR. 1971. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J. Natl. Cancer Inst. 46, 113–120. [PubMed] [Google Scholar]
- 40.Ingram M, Techy GB, Saroufeem R, Yazan O, Narayan KS, Goodwin TJ, Spaulding GF. 1997. Three-dimensional growth patterns of various human tumor cell lines in simulated microgravity of a NASA bioreactor. In Vitro Cell. Dev. Biol. Anim. 33, 459–466. ( 10.1007/s11626-997-0064-8) [DOI] [PubMed] [Google Scholar]
- 41.Kelm JM, Timmins NE, Brown CJ, Fussenegger M, Nielsen LK. 2003. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 83, 173–180. ( 10.1002/bit.10655) [DOI] [PubMed] [Google Scholar]
- 42.Hsiao AY, Tung YC, Kuo CH, Mosadegh B, Bedenis R, Pienta KJ, Takayama S. 2012. Micro-ring structures stabilize microdroplets to enable long term spheroid culture in 384 hanging drop array plates. Biomed. Microdevices 14, 313–323. ( 10.1007/s10544-011-9608-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S. 2011. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 136, 473–478. ( 10.1039/c0an00609b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frey O, Misun PM, Fluri DA, Hengstler JG, Hierlemann A. 2014. Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nat. Commun. 5, 4250 ( 10.1038/ncomms5250) [DOI] [PubMed] [Google Scholar]
- 45.Kim JY, Fluri DA, Kelm JM, Hierlemann A, Frey O. 2015. 96-well format-based microfluidic platform for parallel interconnection of multiple multicellular spheroids. J. Lab Autom. 20, 274–282. ( 10.1177/2211068214564056) [DOI] [PubMed] [Google Scholar]
- 46.Kim JY, et al. 2015. 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. J. Biotechnol. 205, 24–35. ( 10.1016/j.jbiotec.2015.01.003) [DOI] [PubMed] [Google Scholar]
- 47.Yazdi S Rismani, Shadmani A, Burgel SC, Misun PM, Hierlemann A, Frey O. 2015. Adding the ‘heart’ to hanging drop networks for microphysiological multi-tissue experiments. Lab. Chip 15, 4138–4147. ( 10.1039/c5lc01000d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuhas JM, Li AP, Martinez AO, Ladman AJ. 1977. A simplified method for production and growth of multicellular tumor spheroids. Cancer Res. 37, 3639–3643. [PubMed] [Google Scholar]
- 49.Santini MT, Rainaldi G. 1999. Three-dimensional spheroid model in tumor biology. Pathobiology 67, 148–157. ( 10.1159/000028065) [DOI] [PubMed] [Google Scholar]
- 50.Ivascu A, Kubbies M. 2006. Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J. Biomol. Screen. 11, 922–932. ( 10.1177/1087057106292763) [DOI] [PubMed] [Google Scholar]
- 51.Lemmo S, Atefi E, Luker GD, Tavana H. 2014. Optimization of aqueous biphasic tumor spheroid microtechnology for anti-cancer drug testing in 3D culture. Cell. Mol. Bioeng. 7, 344–354. ( 10.1007/s12195-014-0349-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han C, Takayama S, Park J. 2015. Formation and manipulation of cell spheroids using a density adjusted PEG/DEX aqueous two phase system. Sci. Rep. 5, 11891 ( 10.1038/srep11891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ham SL, Joshi R, Luker GD, Tavana H. 2016. Engineered breast cancer cell spheroids reproduce biologic properties of solid tumors. Adv. Healthc Mater. 5, 2788–2798. ( 10.1002/adhm.201600644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Brien PJ, Luo W, Rogozhnikov D, Chen J, Yousaf MN. 2015. Spheroid and tissue assembly via click chemistry in microfluidic flow. Bioconjug. Chem. 26, 1939–1949. ( 10.1021/acs.bioconjchem.5b00376) [DOI] [PubMed] [Google Scholar]
- 55.Tseng H, et al. 2015. A spheroid toxicity assay using magnetic 3D bioprinting and real-time mobile device-based imaging. Sci. Rep. 5, 13987 ( 10.1038/srep13987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin RZ, Chu WC, Chiang CC, Lai CH, Chang HY. 2008. Magnetic reconstruction of three-dimensional tissues from multicellular spheroids. Tissue Eng. Part C Methods 14, 197–205. ( 10.1089/ten.tec.2008.0061) [DOI] [PubMed] [Google Scholar]
- 57.Wu LY, Di Carlo D, Lee LP. 2008. Microfluidic self-assembly of tumor spheroids for anticancer drug discovery. Biomed. Microdevices 10, 197–202. ( 10.1007/s10544-007-9125-8) [DOI] [PubMed] [Google Scholar]
- 58.Hsiao AY, Torisawa YS, Tung YC, Sud S, Taichman RS, Pienta KJ, Takayama S. 2009. Microfluidic system for formation of PC-3 prostate cancer co-culture spheroids. Biomaterials 30, 3020–3027. ( 10.1016/j.biomaterials.2009.02.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu CY, Tseng SY, Yang SM, Hsu L, Liu CH, Chang HY. 2014. A microfluidic chip with a U-shaped microstructure array for multicellular spheroid formation, culturing and analysis. Biofabrication 6, 015009 ( 10.1088/1758-5082/6/1/015009) [DOI] [PubMed] [Google Scholar]
- 60.Liu W, Tian C, Yan M, Zhao L, Ma C, Li T, Xu J, Wang J. 2016. Heterotypic 3D tumor culture in a reusable platform using pneumatic microfluidics. Lab. Chip 16, 4106–4120. ( 10.1039/c6lc00996d) [DOI] [PubMed] [Google Scholar]
- 61.Chan HF, Zhang Y, Ho YP, Chiu YL, Jung Y, Leong KW. 2013. Rapid formation of multicellular spheroids in double-emulsion droplets with controllable microenvironment. Sci. Rep. 3, 3462 ( 10.1038/srep03462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cui X, Liu Y, Hartanto Y, Bi J, Dai S, Zhang H. 2016. Multicellular spheroids formation and recovery in microfluidics-generated thermoresponsive microgel droplets. Colloid Interface Sci. Commun. 14, 4–7. ( 10.1016/j.colcom.2016.09.001) [DOI] [Google Scholar]
- 63.Sabhachandani P, Motwani V, Cohen N, Sarkar S, Torchilin V, Konry T. 2016. Generation and functional assessment of 3D multicellular spheroids in droplet based microfluidics platform. Lab. Chip 16, 497–505. ( 10.1039/c5lc01139f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McMillan KS, McCluskey AG, Sorensen A, Boyd M, Zagnoni M. 2016. Emulsion technologies for multicellular tumour spheroid radiation assays. Analyst 141, 100–110. ( 10.1039/c5an01382h) [DOI] [PubMed] [Google Scholar]
- 65.Aijian AP, Garrell RL. 2015. Digital microfluidics for automated hanging drop cell spheroid culture. J. Lab Autom. 20, 283–295. ( 10.1177/2211068214562002) [DOI] [PubMed] [Google Scholar]
- 66.Chen YA, King AD, Shih HC, Peng CC, Wu CY, Liao WH, Tung YC. 2011. Generation of oxygen gradients in microfluidic devices for cell culture using spatially confined chemical reactions. Lab. Chip 11, 3626–3633. ( 10.1039/c1lc20325h) [DOI] [PubMed] [Google Scholar]
- 67.Hollister SJ. 2005. Porous scaffold design for tissue engineering. Nat. Mater. 4, 518–524. ( 10.1038/nmat1421) [DOI] [PubMed] [Google Scholar]
- 68.Nyga A, Cheema U, Loizidou M. 2011. 3D tumour models: novel in vitro approaches to cancer studies. J. Cell Commun. Signal. 5, 239–248. ( 10.1007/s12079-011-0132-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drury JL, Mooney DJ. 2003. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials 24, 4337–4351. ( 10.1016/S0142-9612(03)00340-5) [DOI] [PubMed] [Google Scholar]
- 70.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. 2007. Engineering tumors with 3D scaffolds. Nat. Methods 4, 855–860. ( 10.1038/nmeth1085) [DOI] [PubMed] [Google Scholar]
- 71.Li WJ, Laurencin CT, Caterson EJ, Tuan RS, Ko FK. 2002. Electrospun nanofibrous structure: a novel scaffold for tissue engineering. J. Biomed. Mater. Res. 60, 613–621. ( 10.1002/jbm.10167) [DOI] [PubMed] [Google Scholar]
- 72.Wu X, et al. 2010. Preparation of aligned porous gelatin scaffolds by unidirectional freeze-drying method. Acta Biomater. 6, 1167–1177. ( 10.1016/j.actbio.2009.08.041) [DOI] [PubMed] [Google Scholar]
- 73.Lin JY, Lin WJ, Hong WH, Hung WC, Nowotarski SH, Gouveia SM, Cristo I, Lin KH. 2011. Morphology and organization of tissue cells in 3D microenvironment of monodisperse foam scaffolds. Soft Matter 7, 10 010–10 016. ( 10.1039/c1sm05371j) [DOI] [Google Scholar]
- 74.Lu TL, Li YH, Chen T. 2013. Techniques for fabrication and construction of three-dimensional scaffolds for tissue engineering. Int. J. Nanomed. 8, 337–350. ( 10.2147/Ijn.S38635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun YS, Peng SW, Lin KH, Cheng JY. 2012. Electrotaxis of lung cancer cells in ordered three-dimensional scaffolds. Biomicrofluidics 6, 014102 ( 10.1063/1.3671399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu WD, Zhang L, Wu CL, Liu ZG, Lei GY, Liu J, Gao W, Hu YR. 2014. Development of an acellular tumor extracellular matrix as a three-dimensional scaffold for tumor engineering. PLoS ONE 9, e0103672 ( 10.1371/journal.pone.0103672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dunne LW, Huang Z, Meng WX, Fan XJ, Zhang NY, Zhang QX, An ZG. 2014. Human decellularized adipose tissue scaffold as a model for breast cancer cell growth and drug treatments. Biomaterials 35, 4940–4949. ( 10.1016/j.biomaterials.2014.03.003) [DOI] [PubMed] [Google Scholar]
- 78.Mironov V, Visconti RP, Kasyanov V, Forgacs G, Drake CJ, Markwald RR. 2009. Organ printing: tissue spheroids as building blocks. Biomaterials 30, 2164–2174. ( 10.1016/j.biomaterials.2008.12.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tibbitt MW, Anseth KS. 2009. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 103, 655–663. ( 10.1002/bit.22361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knowlton S, Onal S, Yu CH, Zhao JJ, Tasoglu S. 2015. Bioprinting for cancer research. Trends Biotechnol. 33, 504–513. ( 10.1016/j.tibtech.2015.06.007) [DOI] [PubMed] [Google Scholar]
- 81.Murphy SV, Atala A. 2014. 3D bioprinting of tissues and organs. Nat. Biotechnol. 32, 773–785. ( 10.1038/nbt.2958) [DOI] [PubMed] [Google Scholar]
- 82.Zhan YS, Duchamp M, Oklu R, Ellisen LW, Langer R, Khademhosseini A. 2016. Bioprinting the cancer microenvironment. ACS Biomater. Sci. Eng. 2, 1710–1721. ( 10.1021/acsbiomaterials.6b00246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA. 2016. Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl Acad. Sci. USA 113, 3179–3184. ( 10.1073/pnas.1521342113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Binder KW, Allen AJ, Yoo JJ, Atala A. 2011. Drop-on-demand inkjet bioprinting: a primer. Gene Therapy Regulation 6, 33–49. ( 10.1142/s1568558611000258) [DOI] [Google Scholar]
- 85.Nakamura M, et al. 2005. Biocompatible inkjet printing technique for designed seeding of individual living cells. Tissue Eng. 11, 1658–1666. ( 10.1089/ten.2005.11.1658) [DOI] [PubMed] [Google Scholar]
- 86.Ozbolat IT, Hospodiuk M. 2016. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials 76, 321–343. ( 10.1016/j.biomaterials.2015.10.076) [DOI] [PubMed] [Google Scholar]
- 87.Khalil S, Sun W. 2007. Biopolymer deposition for freeform fabrication of hydrogel tissue constructs. Mater. Sci. Eng.: C 27, 469–478. ( 10.1016/j.msec.2006.05.023) [DOI] [Google Scholar]
- 88.Guillotin B, et al. 2010. Laser assisted bioprinting of engineered tissue with high cell density and microscale organization. Biomaterials 31, 7250–7256. ( 10.1016/j.biomaterials.2010.05.055) [DOI] [PubMed] [Google Scholar]
- 89.Dhariwala B, Hunt E, Boland T. 2004. Rapid prototyping of tissue-engineering constructs, using photopolymerizable hydrogels and stereolithography. Tissue Eng. 10, 1316–1322. ( 10.1089/1076327042500256) [DOI] [PubMed] [Google Scholar]
- 90.Tsang VL, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN. 2007. Fabrication of 3D hepatic tissues by additive photopatterning of cellular hydrogels. FASEB J. 21, 790–801. ( 10.1096/fj.06-7117com) [DOI] [PubMed] [Google Scholar]
- 91.van der Meer AD, Poot AA, Duits MH, Feijen J, Vermes I. 2009. Microfluidic technology in vascular research. J. Biomed. Biotechnol. 2009, 823148 ( 10.1155/2009/823148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chaffer CL, Weinberg RA. 2011. A perspective on cancer cell metastasis. Science 331, 1559–1564. ( 10.1126/science.1203543) [DOI] [PubMed] [Google Scholar]
- 93.Quail DF, Joyce JA. 2013. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437. ( 10.1038/nm.3394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hasan A, Paul A, Vrana NE, Zhao X, Memic A, Hwang YS, Dokmeci MR, Khademhosseini A. 2014. Microfluidic techniques for development of 3D vascularized tissue. Biomaterials 35, 7308–7325. ( 10.1016/j.biomaterials.2014.04.091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chrobak KM, Potter DR, Tien J. 2006. Formation of perfused, functional microvascular tubes in vitro. Microvasc. Res. 71, 185–196. ( 10.1016/j.mvr.2006.02.005) [DOI] [PubMed] [Google Scholar]
- 96.Nguyen DH, Stapleton SC, Yang MT, Cha SS, Choi CK, Galie PA, Chen CS. 2013. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc. Natl Acad. Sci. USA 110, 6712–6717. ( 10.1073/pnas.1221526110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tourovskaia A, Fauver M, Kramer G, Simonson S, Neumann T. 2014. Tissue-engineered microenvironment systems for modeling human vasculature. Exp. Biol. Med. 239, 1264–1271. ( 10.1177/1535370214539228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong AD, Searson PC. 2014. Live-cell imaging of invasion and intravasation in an artificial microvessel platform. Cancer Res. 74, 4937–4945. ( 10.1158/0008-5472.CAN-14-1042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cross VL, Zheng Y, Choi NW, Verbridge SS, Sutermaster BA, Bonassar LJ, Fischbach C, Stroock AD. 2010. Dense type I collagen matrices that support cellular remodeling and microfabrication for studies of tumor angiogenesis and vasculogenesis in vitro. Biomaterials 31, 8596–8607. ( 10.1016/j.biomaterials.2010.07.072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng Y, et al. 2012. In vitro microvessels for the study of angiogenesis and thrombosis. Proc. Natl Acad. Sci. USA 109, 9342–9347. ( 10.1073/pnas.1201240109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morgan JP, et al. 2013. Formation of microvascular networks in vitro. Nat. Protoc. 8, 1820–1836. ( 10.1038/nprot.2013.110) [DOI] [PubMed] [Google Scholar]
- 102.Miller JS, et al. 2012. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 11, 768–774. ( 10.1038/nmat3357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bischel LL, Lee SH, Beebe DJ. 2012. A practical method for patterning lumens through ECM hydrogels via viscous finger patterning. J. Lab Autom. 17, 96–103. ( 10.1177/2211068211426694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bischel LL, Young EWK, Mader BR, Beebe DJ. 2013. Tubeless microfluidic angiogenesis assay with three-dimensional endothelial-lined microvessels. Biomaterials 34, 1471–1477. ( 10.1016/j.biomaterials.2012.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim J, Chung M, Kim S, Jo DH, Kim JH, Jeon NL. 2015. Engineering of a biomimetic pericyte-covered 3D microvascular network. PLoS ONE 10, e0133880 ( 10.1371/journal.pone.0133880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hsu YH, Moya ML, Hughes CC, George SC, Lee AP. 2013. A microfluidic platform for generating large-scale nearly identical human microphysiological vascularized tissue arrays. Lab. Chip 13, 2990–2998. ( 10.1039/c3lc50424g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yeon JH, Ryu HR, Chung M, Hu QP, Jeon NL. 2012. In vitro formation and characterization of a perfusable three-dimensional tubular capillary network in microfluidic devices. Lab. Chip 12, 2815–2822. ( 10.1039/c2lc40131b) [DOI] [PubMed] [Google Scholar]
- 108.Carrion B, Huang CP, Ghajar CM, Kachgal S, Kniazeva E, Jeon NL, Putnam AJ. 2010. Recreating the perivascular niche ex vivo using a microfluidic approach. Biotechnol. Bioeng. 107, 1020–1028. ( 10.1002/bit.22891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen MB, Whisler JA, Jeon JS, Kamm RD. 2013. Mechanisms of tumor cell extravasation in an in vitro microvascular network platform. Integr. Biol. 5, 1262–1271. ( 10.1039/c3ib40149a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim S, Lee H, Chung M, Jeon NL. 2013. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab. Chip 13, 1489–1500. ( 10.1039/c3lc41320a) [DOI] [PubMed] [Google Scholar]
- 111.Jeon JS, Bersini S, Whisler JA, Chen MB, Dubini G, Charest JL, Moretti M, Kamm RD. 2014. Generation of 3D functional microvascular networks with human mesenchymal stem cells in microfluidic systems. Integr. Biol. 6, 555–563. ( 10.1039/c3ib40267c) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD. 2015. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc. Natl Acad. Sci. USA 112, 214–219. ( 10.1073/pnas.1417115112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chan JM, Zervantonakis IK, Rimchala T, Polacheck WJ, Whisler J, Kamm RD. 2012. Engineering of in vitro 3D capillary beds by self-directed angiogenic sprouting. PLoS ONE 7, e50582 ( 10.1371/journal.pone.0050582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shin Y, Jeon JS, Han S, Jung GS, Shin S, Lee SH, Sudo R, Kamm RD, Chung S. 2011. In vitro 3D collective sprouting angiogenesis under orchestrated ANG-1 and VEGF gradients. Lab. Chip 11, 2175–2181. ( 10.1039/c1lc20039a) [DOI] [PubMed] [Google Scholar]
- 115.Jeong GS, Han S, Shin Y, Kwon GH, Kamm RD, Lee SH, Chung S. 2011. Sprouting angiogenesis under a chemical gradient regulated by interactions with an endothelial monolayer in a microfluidic platform. Anal. Chem. 83, 8454–8459. ( 10.1021/ac202170e) [DOI] [PubMed] [Google Scholar]
- 116.Song JW, Cavnar SP, Walker AC, Luker KE, Gupta M, Tung YC, Luker GD, Takayama S. 2009. Microfluidic endothelium for studying the intravascular adhesion of metastatic breast cancer cells. PLoS ONE 4, e5756 ( 10.1371/journal.pone.0005756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Srigunapalan S, Lam C, Wheeler AR, Simmons CA. 2011. A microfluidic membrane device to mimic critical components of the vascular microenvironment. Biomicrofluidics 5, 13409 ( 10.1063/1.3530598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Prabhakarpandian B, Shen MC, Pant K, Kiani MF. 2011. Microfluidic devices for modeling cell–cell and particle–cell interactions in the microvasculature. Microvasc. Res. 82, 210–220. ( 10.1016/j.mvr.2011.06.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Q, Liu T, Qin J. 2012. A microfluidic-based device for study of transendothelial invasion of tumor aggregates in realtime. Lab. Chip 12, 2837–2842. ( 10.1039/c2lc00030j) [DOI] [PubMed] [Google Scholar]
- 120.Pfisterer L, Korff T. 2016. Spheroid-based in vitro angiogenesis model. Methods Mol. Biol. 1430, 167–177. ( 10.1007/978-1-4939-3628-1_11) [DOI] [PubMed] [Google Scholar]
- 121.Moya M, Tran D, George SC. 2013. An integrated in vitro model of perfused tumor and cardiac tissue. Stem Cell Res. Ther. 4(Suppl. 1), S15 ( 10.1186/scrt376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Allen M, Louise Jones J. 2011. Jekyll and Hyde: the role of the microenvironment on the progression of cancer. J. Pathol. 223, 162–176. ( 10.1002/path.2803) [DOI] [PubMed] [Google Scholar]
- 123.Psaila B, Lyden D. 2009. The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer 9, 285–293. ( 10.1038/nrc2621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sobrino A, et al. 2016. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 6, 31589 ( 10.1038/srep31589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brown JM, Wilson WR. 2004. Exploiting tumour hypoxia in cancer treatment. Nat. Rev. Cancer 4, 437–447. ( 10.1038/nrc1367) [DOI] [PubMed] [Google Scholar]
- 126.Byrne MB, Leslie MT, Gaskins HR, Kenis PJ. 2014. Methods to study the tumor microenvironment under controlled oxygen conditions. Trends Biotechnol. 32, 556–563. ( 10.1016/j.tibtech.2014.09.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stacker SA, Williams SP, Karnezis T, Shayan R, Fox SB, Achen MG. 2014. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat. Rev. Cancer 14, 159–172. ( 10.1038/nrc3677) [DOI] [PubMed] [Google Scholar]
- 128.Hillen F, Griffioen AW. 2007. Tumour vascularization: sprouting angiogenesis and beyond. Cancer Metastasis Rev. 26, 489–502. ( 10.1007/s10555-007-9094-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Azzi S, Hebda JK, Gavard J. 2013. Vascular permeability and drug delivery in cancers. Front. Oncol. 3, 211 ( 10.3389/fonc.2013.00211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bapat AA, Hostetter G, Von Hoff DD, Han H. 2011. Perineural invasion and associated pain in pancreatic cancer. Nat. Rev. Cancer 11, 695–707. ( 10.1038/nrc3131) [DOI] [PubMed] [Google Scholar]
- 131.Valastyan S, Weinberg RA. 2011. Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292. ( 10.1016/j.cell.2011.09.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ma H, Xu H, Qin J. 2013. Biomimetic tumor microenvironment on a microfluidic platform. Biomicrofluidics 7, 11501 ( 10.1063/1.4774070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Martin JD, Fukumura D, Duda DG, Boucher Y, Jain RK. 2016. Reengineering the tumor microenvironment to alleviate hypoxia and overcome cancer heterogeneity. Cold Spring Harb. Perspect. Med. 6, a027094 ( 10.1101/cshperspect.a027094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cox ME, Dunn B. 1986. Oxygen diffusion in poly(dimethyl siloxane) using fluorescence quenching. I. Measurement technique and analysis. J. Polym. Sci. A Polym. Chem. 24, 621–636. ( 10.1002/pola.1986.080240405) [DOI] [Google Scholar]
- 135.Merkel TC, Bondar VI, Nagai K, Freeman BD, Pinnau I. 2000. Gas sorption, diffusion, and permeation in poly(dimethylsiloxane). J. Polym. Sci. B Polym. Phys. 38, 415–434. ( 10.1002/%28Sici%291099-0488%2820000201%2938:3%3C415::Aid-Polb8%3E3.0.Co;2-Z) [DOI] [Google Scholar]
- 136.Thomas PC, Raghavan SR, Forry SP. 2011. Regulating oxygen levels in a microfluidic device. Anal. Chem. 83, 8821–8824. ( 10.1021/ac202300g) [DOI] [PubMed] [Google Scholar]
- 137.Lo JF, Sinkala E, Eddington DT. 2010. Oxygen gradients for open well cellular cultures via microfluidic substrates. Lab. Chip 10, 2394–2401. ( 10.1039/c004660d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Peng CC, Liao WH, Chen YH, Wu CY, Tung YC. 2013. A microfluidic cell culture array with various oxygen tensions. Lab. Chip 13, 3239–3245. ( 10.1039/c3lc50388g) [DOI] [PubMed] [Google Scholar]
- 139.Mehta G, Lee J, Cha W, Tung YC, Linderman JJ, Takayama S. 2009. Hard top soft bottom microfluidic devices for cell culture and chemical analysis. Anal. Chem. 81, 3714–3722. ( 10.1021/ac802178u) [DOI] [PubMed] [Google Scholar]
- 140.Chang CW, Cheng YJ, Tu M, Chen YH, Peng CC, Liao WH, Tung YC. 2014. A polydimethylsiloxane–polycarbonate hybrid microfluidic device capable of generating perpendicular chemical and oxygen gradients for cell culture studies. Lab. Chip 14, 3762–3772. ( 10.1039/c4lc00732h) [DOI] [PubMed] [Google Scholar]
- 141.Zhang Y, Wen J, Zhou L, Qin L. 2015. Utilizing a high-throughput microfluidic platform to study hypoxia-driven mesenchymal-mode cell migration. Integr. Biol. 7, 672–680. ( 10.1039/c5ib00059a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang L, Liu W, Wang Y, Wang JC, Tu Q, Liu R, Wang J. 2013. Construction of oxygen and chemical concentration gradients in a single microfluidic device for studying tumor cell–drug interactions in a dynamic hypoxia microenvironment. Lab. Chip 13, 695–705. ( 10.1039/c2lc40661f) [DOI] [PubMed] [Google Scholar]
- 143.Acosta MA, Jiang X, Huang PK, Cutler KB, Grant CS, Walker GM, Gamcsik MP. 2014. A microfluidic device to study cancer metastasis under chronic and intermittent hypoxia. Biomicrofluidics 8, 054117 ( 10.1063/1.4898788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ayuso JM, et al. 2016. Development and characterization of a microfluidic model of the tumour microenvironment. Sci. Rep. 6, 36086 ( 10.1038/srep36086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lin X, Chen Q, Liu W, Zhang J, Wang S, Lin Z, Lin JM. 2015. Oxygen-induced cell migration and on-line monitoring biomarkers modulation of cervical cancers on a microfluidic system. Sci. Rep. 5, 9643 ( 10.1038/srep09643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xu H, et al. 2015. Activation of hypoxia signaling induces phenotypic transformation of glioma cells: implications for bevacizumab antiangiogenic therapy. Oncotarget 6, 11 882–11 893. ( 10.18632/oncotarget.3592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Funamoto K, Zervantonakis IK, Liu Y, Ochs CJ, Kim C, Kamm RD. 2012. A novel microfluidic platform for high-resolution imaging of a three-dimensional cell culture under a controlled hypoxic environment. Lab. Chip 12, 4855–4863. ( 10.1039/c2lc40306d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Verbridge SS, Choi NW, Zheng Y, Brooks DJ, Stroock AD, Fischbach C. 2010. Oxygen-controlled three-dimensional cultures to analyze tumor angiogenesis. Tissue Eng. Part A 16, 2133–2141. ( 10.1089/ten.TEA.2009.0670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Feld J, Mehta H, Burkes RL. 2000. Acute spontaneous tumor lysis syndrome in adenocarcinoma of the lung: a case report. Am. J. Clin. Oncol. 23, 491–493. ( 10.1097/00000421-200010000-00012) [DOI] [PubMed] [Google Scholar]
- 150.Christakou AE, Ohlin M, Onfelt B, Wiklund M. 2015. Ultrasonic three-dimensional on-chip cell culture for dynamic studies of tumor immune surveillance by natural killer cells. Lab. Chip 15, 3222–3231. ( 10.1039/c5lc00436e) [DOI] [PubMed] [Google Scholar]
- 151.Ghajar CM, et al. 2013. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 15, 807–817. ( 10.1038/ncb2767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. 2009. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab. Chip 9, 269–275. ( 10.1039/b807585a) [DOI] [PubMed] [Google Scholar]
- 153.Patra B, Peng YS, Peng CC, Liao WH, Chen YA, Lin KH, Tung YC, Lee CH. 2014. Migration and vascular lumen formation of endothelial cells in cancer cell spheroids of various sizes. Biomicrofluidics 8, 052109 ( 10.1063/1.4895568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu LL, et al. 2016. Biomimetic tumor-induced angiogenesis and anti-angiogenic therapy in a microfluidic model. RSC Adv. 6, 35 248–35 256. ( 10.1039/c6ra05645h) [DOI] [Google Scholar]
- 155.Theberge AB, Yu JQ, Young EWK, Ricke WA, Bushman W, Beebe DJ. 2015. Microfluidic multiculture assay to analyze biomolecular signaling in angiogenesis. Anal. Chem. 87, 3239–3246. ( 10.1021/ac503700f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Song JW, Munn LL. 2011. Fluid forces control endothelial sprouting. Proc. Natl Acad. Sci. USA 108, 15 342–15 347. ( 10.1073/pnas.1105316108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Song JW, Bazou D, Munn LL. 2012. Anastomosis of endothelial sprouts forms new vessels in a tissue analogue of angiogenesis. Integr. Biol. 4, 857–862. ( 10.1039/c2ib20061a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Galie PA, Nguyen DH, Choi CK, Cohen DM, Janmey PA, Chen CS. 2014. Fluid shear stress threshold regulates angiogenic sprouting. Proc. Natl Acad. Sci. USA 111, 7968–7973. ( 10.1073/pnas.1310842111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Buchanan CF, Verbridge SS, Vlachos PP, Rylander MN. 2014. Flow shear stress regulates endothelial barrier function and expression of angiogenic factors in a 3D microfluidic tumor vascular model. Cell Adhes. Migr. 8, 517–524. ( 10.4161/19336918.2014.970001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Buchanan CF, Voigt EE, Szot CS, Freeman JW, Vlachos PP, Rylander MN. 2014. Three-dimensional microfluidic collagen hydrogels for investigating flow-mediated tumor-endothelial signaling and vascular organization. Tissue Eng. Part C Methods 20, 64–75. ( 10.1089/ten.TEC.2012.0731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Duffy JP, Eibl G, Reber HA, Hines OJ. 2003. Influence of hypoxia and neoangiogenesis on the growth of pancreatic cancer. Mol. Cancer 2, 12 ( 10.1186/1476-4598-2-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu T, Li C, Li H, Zeng S, Qin J, Lin B. 2009. A microfluidic device for characterizing the invasion of cancer cells in 3-D matrix. Electrophoresis 30, 4285–4291. ( 10.1002/elps.200900289) [DOI] [PubMed] [Google Scholar]
- 163.Kim BJ, Hannanta-Anan P, Chau M, Kim YS, Swartz MA, Wu MM. 2013. Cooperative roles of SDF-1 alpha and EGF gradients on tumor cell migration revealed by a robust 3D microfluidic model. PLoS ONE 8, e68422 ( 10.1371/journal.pone.0068422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kuo CT, Chiang CL, Chang CH, Liu HK, Huang GS, Huang RY, Lee H, Huang CS, Wo AM. 2014. Modeling of cancer metastasis and drug resistance via biomimetic nano-cilia and microfluidics. Biomaterials 35, 1562–1571. ( 10.1016/j.biomaterials.2013.11.008) [DOI] [PubMed] [Google Scholar]
- 165.Kuo CT, et al. 2014. A spatiotemporally defined in vitro microenvironment for controllable signal delivery and drug screening. Analyst 139, 4846–4854. ( 10.1039/c4an00936c) [DOI] [PubMed] [Google Scholar]
- 166.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. 2012. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl Acad. Sci. USA 109, 13 515–13 520. ( 10.1073/pnas.1210182109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hsu TH, Kao YL, Lin WL, Xiao JL, Kuo PL, Wu CW, Liao WY, Lee CH. 2012. The migration speed of cancer cells influenced by macrophages and myofibroblasts co-cultured in a microfluidic chip. Integr. Biol. 4, 177–182. ( 10.1039/c2ib00112h) [DOI] [PubMed] [Google Scholar]
- 168.Bischel LL, Beebe DJ, Sung KE. 2015. Microfluidic model of ductal carcinoma in situ with 3D, organotypic structure. BMC Cancer 15, 12 ( 10.1186/s12885-015-1007-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee H, Park W, Ryu H, Jeon NL. 2014. A microfluidic platform for quantitative analysis of cancer angiogenesis and intravasation. Biomicrofluidics 8, 054102 ( 10.1063/1.4894595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Alix-Panabieres C, Pantel K. 2014. Challenges in circulating tumour cell research. Nat. Rev. Cancer 14, 623–631. ( 10.1038/nrc3820) [DOI] [PubMed] [Google Scholar]
- 171.Aceto N, Toner M, Maheswaran S, Haber DA. 2015. En route to metastasis: circulating tumor cell clusters and epithelial-to-mesenchymal transition. Trends Cancer 1, 44–52. ( 10.1016/j.trecan.2015.07.006) [DOI] [PubMed] [Google Scholar]
- 172.Myung JH, Hong S. 2015. Microfluidic devices to enrich and isolate circulating tumor cells. Lab. Chip 15, 4500–4511. ( 10.1039/c5lc00947b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dong Y, et al. 2013. Microfluidics and circulating tumor cells. J. Mol. Diagn. 15, 149–157. ( 10.1016/j.jmoldx.2012.09.004) [DOI] [PubMed] [Google Scholar]
- 174.Qian W, Zhang Y, Chen W. 2015. Capturing cancer: emerging microfluidic technologies for the capture and characterization of circulating tumor cells. Small 11, 3850–3872. ( 10.1002/smll.201403658) [DOI] [PubMed] [Google Scholar]
- 175.Chen MB, Lamar JM, Li R, Hynes RO, Kamm RD. 2016. Elucidation of the roles of tumor integrin beta 1 in the extravasation stage of the metastasis cascade. Cancer Res. 76, 2513–2524. ( 10.1158/0008-5472.Can-15-1325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jeon JS, Zervantonakis IK, Chung S, Kamm RD, Charest JL. 2013. In vitro model of tumor cell extravasation. PLoS ONE 8, e56910 ( 10.1371/journal.pone.0056910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, Moretti M, Kamm RD. 2014. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 35, 2454–2461. ( 10.1016/j.biomaterials.2013.11.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kahlert C, Kalluri R. 2013. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 91, 431–437. ( 10.1007/s00109-013-1020-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jeong SY, Lee JH, Shin Y, Chung S, Kuh HJ. 2016. Co-culture of tumor spheroids and fibroblasts in a collagen matrix-incorporated microfluidic chip mimics reciprocal activation in solid tumor microenvironment. PLoS ONE 11, e0159013 ( 10.1371/journal.pone.0159013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liu PF, et al. 2015. A bladder cancer microenvironment simulation system based on a microfluidic co-culture model. Oncotarget 6, 37 695–37 705. ( 10.18632/oncotarget.6070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Polacheck WJ, Charest JL, Kamm RD. 2011. Interstitial flow influences direction of tumor cell migration through competing mechanisms. Proc. Natl Acad. Sci. USA 108, 11 115–11 120. ( 10.1073/pnas.1103581108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang JW, Pan HJ, Yao WY, Tsao YW, Liao WY, Wu CW, Tung YC, Lee CH. 2013. Interaction between lung cancer cell and myofibroblast influenced by cyclic tensile strain. Lab. Chip 13, 1114–1120. ( 10.1039/c2lc41050h) [DOI] [PubMed] [Google Scholar]
- 183.Pathak A, Kumar S. 2012. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc. Natl Acad. Sci. USA 109, 10 334–10 339. ( 10.1073/pnas.1118073109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fenner J, Stacer AC, Winterroth F, Johnson TD, Luker KE, Luker GD. 2014. Macroscopic stiffness of breast tumors predicts metastasis. Sci. Rep. 4, 5512 ( 10.1038/srep05512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Djamgoz MB.A., Mycielska M, Madeja Z, Fraser SP, Korohoda W. 2001. Directional movement of rat prostate cancer cells in direct-current electric field: involvement of voltage-gated Na+ channel activity. J. Cell Sci. 114, 2697–2705. [DOI] [PubMed] [Google Scholar]
- 186.Tsai HF, Huang CW, Chang HF, Chen JJW, Lee CH, Cheng JY. 2013. Evaluation of EGFR and RTK signaling in the electrotaxis of lung adenocarcinoma cells under direct-current electric field stimulation. PLoS ONE 8, e73418 ( 10.1371/journal.pone.0073418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.McCaig CD, Song B, Rajnicek AM. 2009. Electrical dimensions in cell science. J. Cell Sci. 122, 4267–4276. ( 10.1242/jcs.023564) [DOI] [PubMed] [Google Scholar]
- 188.Mi S, Du Z, Xu Y, Wu Z, Qian X, Zhang M, Sun W. 2016. Microfluidic co-culture system for cancer migratory analysis and anti-metastatic drugs screening. Sci. Rep. 6, 35544 ( 10.1038/srep35544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang Y, Zhang W, Qin L. 2014. Mesenchymal-mode migration assay and antimetastatic drug screening with high-throughput microfluidic channel networks. Angew. Chem. Int. Ed. Engl. 53, 2344–2348. ( 10.1002/anie.201309885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Valencia PM, Farokhzad OC, Karnik R, Langer R. 2012. Microfluidic technologies for accelerating the clinical translation of nanoparticles. Nat. Nanotechnol. 7, 623–629. ( 10.1038/nnano.2012.168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kwak B, Ozcelikkale A, Shin CS, Park K, Han B. 2014. Simulation of complex transport of nanoparticles around a tumor using tumor-microenvironment-on-chip. J. Control Release 194, 157–167. ( 10.1016/j.jconrel.2014.08.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bagley AF, Scherz-Shouval R, Galie PA, Zhang AQ, Wyckoff J, Whitesell L, Chen CS, Lindquist S, Bhatia SN. 2015. Endothelial thermotolerance impairs nanoparticle transport in tumors. Cancer Res. 75, 3255–3267. ( 10.1158/0008-5472.CAN-15-0325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang H, Jenkins G, Zou Y, Zhu Z, Yang CJ. 2012. Massively parallel single-molecule and single-cell emulsion reverse transcription polymerase chain reaction using agarose droplet microfluidics. Anal. Chem. 84, 3599–3606. ( 10.1021/ac2033084) [DOI] [PubMed] [Google Scholar]
- 194.Hayes CJ, Dowling CM, Dwane S, McCumiskey ME, Tormey SM, Anne Merrigan B, Coffey JC, Kiely PA, Dalton TM. 2016. Extracellular matrix gene expression profiling using microfluidics for colorectal carcinoma stratification. Biomicrofluidics 10, 054124 ( 10.1063/1.4966245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jang M, Koh I, Lee SJ, Cheong JH, Kim P. 2017. Droplet-based microtumor model to assess cell–ECM interactions and drug resistance of gastric cancer cells. Sci. Rep. 7, 41541 ( 10.1038/srep41541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sun J, et al. 2010. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Res. 70, 6128–6138. ( 10.1158/0008-5472.CAN-10-0076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Xu Z, et al. 2016. Design and construction of a multi-organ microfluidic chip mimicking the in vivo microenvironment of lung cancer metastasis. ACS Appl. Mater. Interfaces 8, 25 840–25 847. ( 10.1021/acsami.6b08746) [DOI] [PubMed] [Google Scholar]
- 198.Xu XD, Shao SX, Cao YW, Yang XC, Shi HQ, Wang YL, Xue SY, Wang XS, Niu HT. 2015. The study of energy metabolism in bladder cancer cells in co-culture conditions using a microfluidic chip. Int. J. Clin. Exp. Med. 8, 12 327–12 336. [PMC free article] [PubMed] [Google Scholar]
- 199.Zhu G, Wang D, Li S, Yang X, Cao Y, Wang Y, Niu H. 2016. Acute effect of lactic acid on tumor-endothelial cell metabolic coupling in the tumor microenvironment. Oncol. Lett. 12, 3478–3484. ( 10.3892/ol.2016.5047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Martinez-Outschoorn UE, et al. 2011. Understanding the metabolic basis of drug resistance: therapeutic induction of the Warburg effect kills cancer cells. Cell Cycle 10, 2521–2528. ( 10.4161/cc.10.15.16584) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. ( 10.1126/science.1160809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dhup S, Dadhich RK, Porporato PE, Sonveaux P. 2012. Multiple biological activities of lactic acid in cancer: influences on tumor growth, angiogenesis and metastasis. Curr. Pharm. Des 18, 1319–1330. ( 10.2174/138161212799504902) [DOI] [PubMed] [Google Scholar]
- 203.Mongersun A, Smeenk I, Pratx G, Asuri P, Abbyad P. 2016. Droplet microfluidic platform for the determination of single-cell lactate release. Anal. Chem. 88, 3257–3263. ( 10.1021/acs.analchem.5b04681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tellez-Gabriel M, Ory B, Lamoureux F, Heymann MF, Heymann D. 2016. Tumour heterogeneity: the key advantages of single-cell analysis. Int. J. Mol. Sci.. 17, 2142 ( 10.3390/ijms17122142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Carlborg CF, Haraldsson T, Oberg K, Malkoch M, van der Wijngaart W. 2011. Beyond PDMS: off-stoichiometry thiol-ene (OSTE) based soft lithography for rapid prototyping of microfluidic devices. Lab. Chip 11, 3136–3147. ( 10.1039/c1lc20388f) [DOI] [PubMed] [Google Scholar]
- 206.Hashimoto M, Langer R, Kohane DS. 2013. Benchtop fabrication of microfluidic systems based on curable polymers with improved solvent compatibility. Lab. Chip 13, 252–259. ( 10.1039/c2lc40888k) [DOI] [PubMed] [Google Scholar]
- 207.Begolo S, Colas G, Viovy JL, Malaquin L. 2011. New family of fluorinated polymer chips for droplet and organic solvent microfluidics. Lab. Chip 11, 508–512. ( 10.1039/c0lc00356e) [DOI] [PubMed] [Google Scholar]
- 208.Wu SH, Lee KL, Chiou A, Cheng X, Wei PK. 2013. Optofluidic platform for real-time monitoring of live cell secretory activities using Fano resonance in gold nanoslits. Small 9, 3532–3540. ( 10.1002/smll.201203125) [DOI] [PubMed] [Google Scholar]
- 209.Jeanbart L, Swartz MA. 2015. Engineering opportunities in cancer immunotherapy. Proc. Natl Acad. Sci. USA 112, 14 467–14 472. ( 10.1073/pnas.1508516112) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.