Abstract
The adaptive attachment of marine mussels to a wide range of substrates in a high-energy, saline environment has been explored for decades and is a significant driver of bioinspired wet adhesion research. Mussel attachment relies on a fibrous holdfast known as the byssus, which is made by a specialized appendage called the foot. Multiple adhesive and structural proteins are rapidly synthesized, secreted and moulded by the foot into holdfast threads. About 10 well-characterized proteins, namely the mussel foot proteins (Mfps), the preCols and the thread matrix proteins, are reported as representing the bulk of these structures. To explore how robust this proposition is, we sequenced the transcriptome of the glandular tissues that produce and secrete the various holdfast components using next-generation sequencing methods. Surprisingly, we found around 15 highly expressed genes that have not previously been characterized, but bear key similarities to the previously defined mussel foot proteins, suggesting additional contribution to byssal function. We verified the validity of these transcripts by polymerase chain reaction, cloning and Sanger sequencing as well as confirming their presence as proteins in the byssus. These newly identified proteins greatly expand the palette of mussel holdfast biochemistry and provide new targets for investigation into bioinspired wet adhesion.
Keywords: transcriptomics, biomaterials, mussel adhesion, load-bearing proteins
1. Introduction
Along with barnacles [1], sandcastle worms [2] and sea stars [3], marine mussels are among the pre-eminent model systems of bioadhesion [4–7] and have been explored extensively to develop bioinspired water-compatible adhesives for medical and industrial applications [5,8]. Moreover, mussel adhesion is scrutinized for clues to undermine and prevent adhesion, given the prohibitive economic and environmental costs associated with biofouling [9,10]. Mussels offer distinct advantages as a model system for bioadhesion research because (i) processing speed and extra-organismal secretion of the adhesive allow for easy, non-invasive collection of relatively large quantities of unadulterated sample, (ii) mussels are readily available along most temperate coastlines and amenable to mariculture, and (iii) the protein-based nature of the adhesive allows characterization by standard biochemical and molecular techniques.
Mytilus californianus (Conrad, 1837) inhabits the rocky intertidal zones of the temperate Eastern Pacific region. Intense wave action and extreme tidal exposure make this a particularly harsh environment [11]. However, M. californianus can populate and thrive in this ecological niche owing in large part to its holdfast byssus. The proteinaceous byssus consists of a stem rooted in the soft tissue within the mussel. From the stem radiate numerous load-bearing collagenous fibres that terminate in porous spatulate adhesive plaques chemically adhered to the rocky substrate [12]. It is the entire byssal system that ultimately enables mussels to remain firmly anchored in place. Interfacial adhesion is an essential component [13], but the energy dissipative architecture of plaque [14] and thread [15] greatly dampen the load seen at the actual plaque–substrate interface.
Byssal threads are made one at a time by rapid injection moulding and protein self-assembly in the ventral pedal groove (figure 1). The pedal groove is lined with diverse glandular tissues that secrete the byssus-forming proteins [16–18]. At the tip of the foot, the cup-like distal depression is surrounded by the phenol gland, which secretes the interfacial and plaque-forming proteins: Mytilus californianus foot proteins (Mcfp) -2, -3, -4, -5, -6 [8]. Mcfp-3 and Mcfp-5 are deposited at the interface and chemically bind to the substratum [19]. Mcfp-6 is present at the interface to maintain a reducing environment important for adhesion [20]. These interfacial proteins interact with Mcfp-2, a major component of the porous network within the plaque [21]. Mcfp-4 bridges Mcfp-2 to the collagenous protein fibres that splay into the plaque from the thread [22]. The collagen gland runs the length of the pedal groove and secretes a unique family of collagens called preCols (variants -D, -P, -NG). The preCols self-assemble to form a graded fibre of preCol species to impart varied mechanical properties along its length [23]. Thread matrix proteins (TMPs) are also integrated into the thread to bridge collagen fibres laterally [24]. The accessory gland, a thin gland which extends along both lips of the pedal groove and around the distal depression, secretes a protective coating—or biological varnish—covering the entire thread and plaque. A major coating protein, Mcfp-1, has been shown to possess wear-resistant properties [25]. Many amino acids in the Mfps are extensively post-translationally modified and include hydroxyarginine, phosphoserine, hydroxyproline and dihydroxyproline, but the amino acid modification 3,4-dihydroxyphenylalanine (dopa) is the hallmark of many of the Mfps [8]. This multi-functional modification is important to cohesive and adhesive plaque interactions via metal coordination, covalent cross-linking, H-bonding and π-cation interactions, which all play integral roles in the formation and maturation of the robust holdfast.
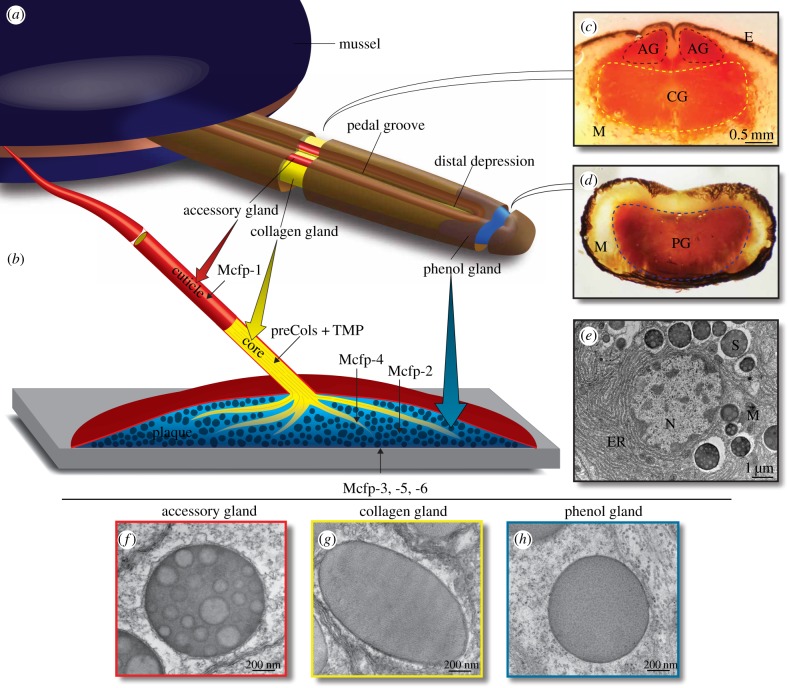
Figure 1.
The mussel foot fabricates the byssal threads, plaques and coating by injection moulding. (a) A diagram of a mussel with foot extended. For clarity, the foot is drawn with the distal depression and pedal groove up, although during plaque deposition these features face the substratum. Three distinct glands inside the foot: the accessory gland (red), collagen gland (yellow) and phenol gland (blue) secrete proteinaceous components which self-assemble in the pedal groove and distal depression to form the byssus structures, the cuticle, collagen core and plaque, respectively. (b) A single byssus element showing the porous plaque (blue) attached to the substrate (grey). The collagen thread (yellow) integrates into the plaque at the distal end and anchors inside the mussel at the other end. Both the thread and plaque are protected by a hard cuticle (red). The protein components of each structure are indicated. (c) Transverse cross-section of foot from the centre as indicated by the artificial opening in the foot diagram. Dashed borders indicate the accessory gland (AG) and collagen gland (CG) and represent the location of tissue sampling for RNA isolation. Muscle (M) and pigmented epithelium (E) are also indicated. The section is chemically stained for dopa (Arnow stain), a major modification in Mfp-1, highlighting the location of the accessory gland with respect to the collagen gland. (d) Transverse cross-section of the foot tip as indicated by the artificial opening in the foot diagram. This section is also stained for dopa, a prevalent modification in Mfp-3 and -5 in the phenol gland (PG). (e) Transmission electron micrograph of a portion of an accessory gland cell showing the extensive rough endoplasmic reticulum (ER), cuticle secretion granules (S) and for reference the nucleus (N), and mitochondria (M). Transmission electron micrographs highlight the secretory vesicles within the accessory (f), collagen (g) and phenol (h) glands.
The maximum adhesion energy of the adhesive Mfps was measured to be approximately 15 mJ m−2 [26]. However, energy to failure for the native thread and plaque was nearly 10 000 times greater [27]. One explanation for this functional discrepancy is that plaque architecture and rupturing reversible sacrificial bonds exposing hidden polymer lengths are dissipating the applied force. However, it is also plausible that the analysis of plaque components is incomplete and that significant additional but unknown bio-macromolecular components exist.
Next-generation sequencing (NGS), particularly RNA transcriptome sequencing, is effective in characterizing swathes of novel proteins, and has been showcased in recent characterizations of various biomaterials and bioadhesives [28–31]. The ability to sequence the full pool of mRNA transcripts in a sample is universally advantageous for any biomolecular investigation, but there is added value in the case of biomaterials, in that these are often so heavily processed and cross-linked that little of value is extractable from mature materials. Transcriptomics offers a welcome alternative to traditional protein characterization by partial sequencing and degenerate cloning, and opens doors that have long been closed.
Owing to the highly cross-linked nature of the mature mussel plaques and depending on the unknown degree of pre-secretory processing, it is conceivable that some Mfps are not available/amenable to extraction and purification. We approach the protein make-up of the byssus from a transcriptomics standpoint, using NGS to survey the entire mRNA transcript pool of each gland. As these tissues are specialized to rapidly produce and secrete massive amounts of byssus-forming proteins, it is reasonable to predict abundant quantities of the mRNA transcripts corresponding to the known Mfps and potentially novel Mfps as well.
2. Material and methods
2.1. Transcriptome generation
Live Mytilus californianus specimens were collected from the Goleta, CA, pier (coordinates 34.413574, −119.828492) and kept in an open seawater system until dissected. The feet were excised from shucked mussels, and then sliced into successive thin transverse cross-sections from the foot tip (distal end) to the base (proximal end). Each slice was laid flat and precise gland dissection was accomplished under a dissecting microscope. The collagen gland and accessory glands were isolated from a slice from the centre of the foot length and carefully isolated from the surrounding muscle, and pigmented epithelium (figure 1a,c). The phenol gland was similarly isolated from slices near the foot tip, just distal to the distal depression (figure 1a,d). Single isolated tissue samples (approx. 60 mg) from each gland region were collected and flash frozen in liquid nitrogen. The RNA was purified using a Purelink RNA isolation kit (ThermoFisher Scientific, Waltham, MA) following the manufacturer's protocol after homogenization with a mortar and pestle under liquid nitrogen. RNA quality was assessed on a TapeStation 2200 (Agilent Technologies, Santa Clara, CA) and quantified by a Cubit 2.0 Fluorometer. The mRNA was purified using Dynabeads® Oligo (dT)25 (ThermoFisher Scientific) and the RNA library was prepared using the TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA) and sequenced on a NextSeq 500 (Illumina) running at either 100 or 150 cycles.
2.2. Transcriptome assembly
Bioinformatic analyses were performed using a locally installed instance of the Galaxy bioinformatics platform [32]. Low-quality reads and adapter sequences were removed using the read processing tool Trimmomatic [33]. The trimmed reads were assembled into mRNA isotigs using the Trinity software package [34]. The software RSEM [35] was used to map the trimmed reads onto the assembled transcripts to estimate the transcription levels of each isotig. The transcriptome assemblies were organized based on their transcript abundance and the most abundant transcripts (FPKM > 500) were analysed and annotated.
2.3. Polymerase chain reaction validation
Selected transcriptome isotig sequences were verified by traditional cloning experiments. Primer pairs were designed to anneal 5′ of the start codon and 3′ of the stop codon for the putative transcripts of interest (electronic supplementary material, table S1). Following polymerase chain reaction (PCR) amplification, the products were cloned into the Escherichia coli vector pCR4 using a TOPO-TA cloning kit (Invitrogen, Carlsbad, CA), transformed into chemically competent E. coli (Invitrogen), and screened by growing on Luria–Bertani agar with kanamycin. Plasmid DNA was isolated from positive clones and sequenced by Sanger sequencing (Genewiz, Newbury Park, CA). These sequences were deposited into GenBank under accessions: KY627765–KY627780.
2.4. Matrix-assisted laser desorption ionization mass spectrometry
Live mussels were artificially induced to secrete plaque-forming proteins by injecting the base of the foot with 0.56 M potassium chloride to stimulate the pedal ganglion, as described previously [36]. The resulting induced plaques were then collected from the distal depression and placed immediately in 5% acetic acid, 6 M guanidine hydrochloride. This crude extract was directly separated by reverse phase chromatography on a C18 column (Brownlee OD-300, 7 µm, 250 × 4.6 mm) using a linear gradient from buffer A (99.9% water, 0.1% trifluoroacetic acid) to buffer B (95% acetonitrile, 5% water, 0.1% trifluoroacetic acid) over the course of 60 min. The protein elution profile was monitored at an absorbance of 280 nm. Individual peak fractions were analysed by matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) following mixing with a saturated solution of α-cyano-4-hydroxycinnamic acid in a 50 : 50 solution of water : acetonitrile, with 0.1% trifluoroacetic acid.
3. Results
3.1. Transcriptome generation and analysis
The three gland tissues were carefully dissected under a microscope; the phenol gland (plaque formation) from the distal portion of the foot; the collagen gland (thread core formation) and the accessory gland (cuticle formation) from the middle of the foot (figure 1). It is noteworthy that significant cross-contamination between the last two glands (collagen and accessory) was expected, as the demarcation between these two glandular regions is fuzzy. High-quality RNA was obtained from each tissue with RNA integrity numbers of 9.3, 9.5 and 9.3, respectively. Each transcriptome was assembled from the millions of reads into thousands of transcripts (isotigs); read and assembly statistics are shown in table 1.
Table 1.
Transcriptome preparation and assembly of mussel foot secretory glands.
| phenol gland | collagen gland | accessory gland | |
|---|---|---|---|
| RNA purification | |||
| RNA integrity number | 9.3 | 9.5 | 9.3 |
| Illumina sequencing | |||
| read length | 2 × 100 | 2 × 100 | 2 × 150 |
| number of untrimmed reads | 56 650 426 | 43 293 488 | 87 720 502 |
| number of trimmed reads | 54 764 254 | 40 746 558 | 60 426 226 |
| assembly | |||
| min. isotig length | 201 | 201 | 201 |
| max. isotig length | 9916 | 8373 | 10 084 |
| mean isotig length | 541.95 | 464.18 | 513.25 |
| standard deviation of isotig length | 562.28 | 439.1 | 522.61 |
| median isotig length | 339 | 314 | 337 |
| N50 isotig length | 701 | 520 | 611 |
| number of isotigs | 44 774 | 39 426 | 59 030 |
| number of isotigs ≥1 kb | 5162 | 3020 | 5728 |
| number of isotigs in N50 | 8804 | 8895 | 12 369 |
| number of bases in all isotigs | 24 265 373 | 18 300 884 | 30 297 309 |
| number of bases in isotigs ≥1 kb | 9 104 355 | 5 044 646 | 10 047 098 |
| GC content of isotigs | 33.33% | 34.00% | 34.11% |
| RSEM | |||
| # transcripts with FPKM > 500 | 171 | 187 | 240 |
| % total transcripts with FPKM > 500 | 0.38 | 0.47 | 0.41 |
| % cumulative FPKM of transcripts with FPKM > 500 | 89.80 | 86.30 | 76.30 |
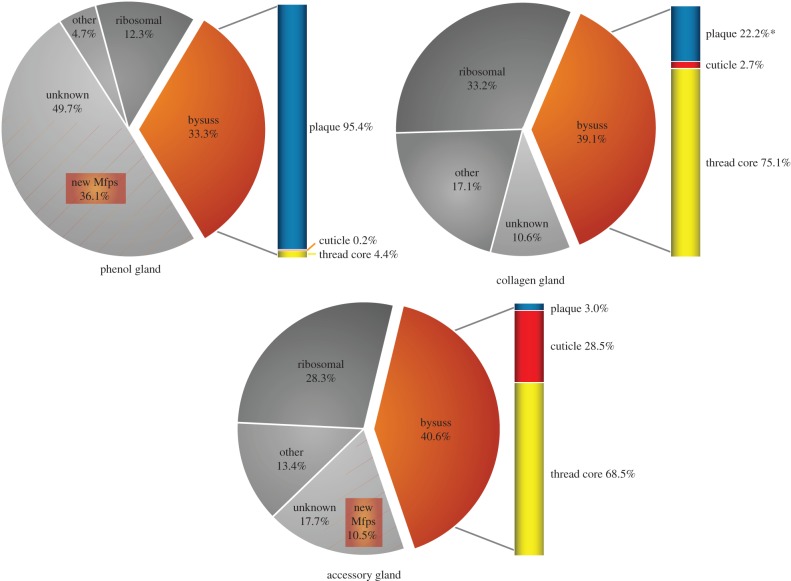
Illumina reads were then mapped back to the assembled isotigs to estimate abundance. Each dataset was sorted by descending abundance in terms of fragments per kilobase per million fragments mapped (FPKM). The most abundant isotigs (FPKM > 500) represent only approximately 0.5% of the total isotigs, but in terms of expression abundance (cumulative FPKM) they constitute 89.8%, 86.3% and 76.3% of the total transcripts in the phenol, collagen and accessory glands, respectively. These top transcripts were manually classified into the following groupings: byssus-associated proteins (Mfps, preCols and TMP), ribosomal proteins, unknown transcripts (no database hits) and other proteins (e.g. mitochondrial-associated proteins, housekeeping proteins). Cumulative FPKM percentages for each classification show that a significant portion of the transcripts in each gland are byssus-associated proteins (33–40%), as well as ribosome-associated proteins (12–33%) (figure 2). Transcripts classified as ‘unknown’ (NCBI BLAST E-value > 1 × 10−10) are also significantly represented in the transcriptomes, particularly in the phenol gland (49.7%). The byssus proteins were further categorized per their localization in the byssus, showing strong representation of expected secretory products from each gland; for example, the phenol gland secretes mostly Mfps known to be associated with the plaque (Mcfp-2, -3, -4, -5, -6), the collagen gland has high representation of collagen and TMP, and so on. Table 2 shows an abbreviated summary of the most abundant transcripts (having removed the significant amount of ribosome-associated protein transcripts, contaminating rRNA, protein replicates and unknown transcripts lacking a clear open reading frame), yielding a general view of the major protein products of the gland tissue.
Figure 2.
Cumulative transcript abundance by functional classification shows the dominance of byssal proteins in each transcriptome. Transcriptome summary for the phenol gland (a), the collagen gland (b) and the accessory gland (c). The sum of FPKM values for each transcript classification is expressed as a percentage of the total transcripts considered. Only transcripts with FPKM > 500 are represented in the graphs; however, these transcripts represent approximately 75–90% of the cumulative transcript abundance (table 1). Bysuss transcripts encode well-known components of the functional byssus structure (e.g. Mfps, preCols). Ribosomal transcripts encode ribosomal-associated proteins. ‘Unknown’ transcripts are those that do not have significant blast hits in the NCBI database (E-value > 1 × 10−10). ‘Other’ transcripts constitute everything else—i.e. positive blast hits for characterized proteins (e.g. miscellaneous housekeeping proteins, mitochondrial proteins, actin, etc.). The byssus transcripts are further broken down by localization to the byssal structure and the percentages are represented in the bar to the right of each graph. Many of the unknown transcripts in the phenol gland and accessory gland are candidate novel mussel foot proteins destined for the plaque and cuticle, respectively, and their abundance is demarcated by the area shaded by the orange lines.
Table 2.
Abbreviated summary of the most abundant protein transcripts by gland (FPKM > 500)a. Novel Mcfps (red), ubiquitous byssal transcripts (orange), ubiquitous transcripts (yellow).
| phenol gland |
collagen gland | accessory gland | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| no. | transcript ID | FPKM | annotation | no. | transcript ID | FPKM | annotation | no. | transcript ID | FPKM | annotation |
| 1 | c16253_g1_i2 | 202344 | Mcfp-7p v1 | 1 | c15274_g3_i1 | 34809 | thread matrix protein | 1 | c15006_g1_i1 | 62158 | Mcfp-1 |
| 2 | c16566_g1_i1 | 181351 | Mcfp-3 | 4 | c15428_g2_i1 | 23710 | byssus glycosyl-hydrolase-like protein | 2 | c12737_g1_i1 | 54485 | preCol-NG |
| 3 | c16253_g1_i3 | 100298 | Mcfp-8p | 5 | c15431_g1_i1 | 20321 | Mcfp-6 | 3 | c12602_g1_i1 | 43047 | preCol-D |
| 4 | c25234_g2_i1 | 67476 | Mcfp-5 | 12 | c13832_g1_i2 | 14614 | preCol-NG | 10 | c21940_g1_i1 | 17743 | preCol-P |
| 7 | c12339_g1_i2 | 50307 | Mcfp-9p | 16 | c13842_g1_i1 | 12085 | preCol-D | 11 | c10092_g1_i1 | 16342 | Mcfp-15p |
| 8 | c15788_g1_i2 | 49635 | Mcfp-6 | 46 | c13944_g2_i1 | 3923 | Mcfp-1 | 12 | c13615_g1_i1 | 15067 | proximal thread matrix protein |
| 10 | c16355_g1_i1 | 38388 | Mcfp-10p | 57 | c11464_g1_i1 | 3054 | ferritin | 18 | c12213_g2_i1 | 8904 | Mcfp-16p |
| 11 | c14767_g3_i1 | 36244 | Mcfp-2 | 72 | c4969_g1_i2 | 2691 | preCol-P | 19 | c12177_g1_i1 | 7633 | Mcfp-17p |
| 14 | c16285_g1_i1 | 31890 | Mcfp-11p | 82 | c15051_g2_i1 | 2394 | ubiquitin | 20 | c10298_g1_i1 | 6996 | ferritin |
| 18 | c17523_g1_i3 | 21901 | Mcfp-12p | 96 | c14448_g1_i1 | 2065 | Qm-like protein | 21 | c7297_g1_i1 | 6700 | Mcfp-18p |
| 20 | c14769_g2_i1 | 20094 | Mcfp-13p | 125 | c14069_g1_i1 | 1233 | ADP/ATP carrier protein | 30 | c14420_g2_i1 | 5754 | Mcfp-6 |
| 31 | c14593_g2_i1 | 6547 | preCol-D | 130 | c14595_g1_i2 | 1017 | translationally controlled tumour protein | 47 | c7837_g1_i1 | 3414 | thread matrix protein |
| 44 | c16361_g1_i1 | 3419 | Mcfp-4 | 132 | c12387_g1_i1 | 990 | laminin | 73 | c11070_g1_i1 | 2935 | ubiquitin |
| 48 | c12727_g1_i1 | 2902 | Mcfp-14p | 133 | c12234_g1_i1 | 988 | nucleoside diphosphate kinase A | 81 | c13002_g1_i1 | 2869 | translationally controlled tumour protein |
| 49 | c4587_g1_i1 | 2583 | ferritin | 142 | c15318_g1_i1 | 781 | HSP90 | 85 | c10653_g1_i1 | 2780 | nucleoside diphosphate kinase |
| 66 | c14916_g1_i1 | 2243 | preCol-NG | 145 | c15481_g1_i3 | 694 | cysteine peptidase | 87 | c14408_g1_i1 | 2752 | byssus glycosyl-hydrolase-like protein |
| 81 | c17081_g2_i2 | 2148 | cysteine peptidase | 150 | c15498_g1_i3 | 666 | tubulin | 89 | c14993_g2_i2 | 2685 | CD109 antigen like |
| 100 | c2677_g1_i1 | 1873 | translationally controlled tumour protein | 151 | c12430_g1_i1 | 660 | cyclophilin | 103 | c11948_g1_i1 | 2387 | RACK1 |
| 113 | c11248_g1_i1 | 1684 | myticusin | 157 | c12246_g1_i1 | 622 | Mcfp-3 | 135 | c35090_g1_i1 | 1736 | ADP/ATP carrier protein |
| 120 | c14097_g1_i1 | 1543 | RACK1 | 165 | c11529_g1_i1 | 547 | peptidyl-prolyl cis-trans-isomerase | 143 | c14194_g1_i2 | 1562 | tubulin |
| 123 | c14926_g1_i1 | 1508 | peptidyl-prolyl cis-trans-isomerase | 149 | c14497_g2_i1 | 1453 | peptidyl-prolyl cis-trans- isomerase | ||||
| 127 | c14273_g1_i1 | 1457 | ubiquitin | 161 | c10909_g1_i1 | 1212 | apextrin-like protein | ||||
| 129 | c12717_g1_i1 | 1433 | thymosin | 165 | c11834_g1_i1 | 1150 | HSP90 | ||||
| 131 | c16041_g1_i1 | 1392 | p63 | 180 | c10390_g1_i1 | 992 | lysozyme | ||||
| 140 | c10847_g1_i1 | 1247 | laminin | 181 | c7263_g1_i1 | 966 | cytochrome c | ||||
| 152 | c17536_g1_i1 | 947 | tubulin | 187 | c14497_g1_i2 | 871 | vigilin-like | ||||
| 159 | c10384_g1_i1 | 750 | ADP/ATP carrier protein | 189 | c13637_g1_i1 | 859 | heat shock protein-70 | ||||
| 169 | c17374_g1_i1 | 651 | vigilin-like protein | 191 | c12280_g1_i1 | 849 | arginine kinase | ||||
| 177 | c28964_g1_i1 | 595 | Mcfp-1 | 192 | c14066_g4_i1 | 847 | cysteine peptidase | ||||
| 181 | c15527_g1_i1 | 554 | nucleoside diphosphate kinase | 195 | c14595_g1_i1 | 817 | procollagen-proline dioxygenase | ||||
| 197 | c7713_g1_i1 | 809 | transcription factor BTF3 | ||||||||
| 198 | c14213_g1_i2 | 774 | actin | ||||||||
| 220 | c10009_g1_i1 | 667 | tropomyosin | ||||||||
| 226 | c11241_g1_i1 | 635 | glyceraldehyde-3-phosphate dehydrogenase | ||||||||
| 233 | c8853_g2_i1 | 605 | Sec61 gamma | ||||||||
| 236 | c28684_g1_i1 | 598 | cyclophilin | ||||||||
| 245 | c12384_g1_i1 | 528 | adenosylhomocysteinase A-like | ||||||||
| 246 | c9332_g1_i1 | 526 | translocon-associated protein subunit beta | ||||||||
aFor simplicity, the table excludes ribosomal-associated protein transcripts, contaminating rRNA, protein replicates and unknown transcripts without clear open reading frames; see the electronic supplementary material for complete table.
3.2. Novel mussel foot protein transcripts
The most abundant unknown transcripts for each gland were further scrutinized for those possessing clear open reading frames and predicted signal peptides [37], suggesting plausible accurate transcript assembly, accurate start codon assignment and secretory destination (table 3). In addition to their apparent secretory fate, many of these novel transcripts boast adherence to pervasive Mfp themes, in particular elevated pIs of approximately 8.5–10.5 and distinct amino acid compositional bias for glycine, tyrosine (precursor to dopa), lysine, arginine, serine and histidine, and strikingly deficient in hydrophobic and acidic amino acids. Here we suggest that many, if not all, of these proteins play a significant role in the structure and function of the byssus, and, in this spirit, putative Mytilus californianus foot proteins 7–19 (Mcfp-7p, Mcfp-8p, etc.) are assigned names that follow the previously well-characterized Mfps. These novel transcripts make up a significant portion of the transcriptome for the phenol gland (36%) and the accessory gland (10%); incorporating these transcripts yields an estimated byssus-associated transcript portion of 69% and 58%, respectively (figure 2). No novel transcripts were observed among the top 50 in the collagen gland. All the reported novel Mcfp-p sequences were validated using PCR amplification, cloning and Sanger sequencing. Generally, there was minimal disparity between the sequences obtained by NGS and traditional sequencing, and in several cases the two methods yielded identical data (table 3). Mcfp-10p, however, was only 73% identical between these methods, but this discrepancy is attributed to an assembly artefact reporting two tandem repeat domains instead of the actual three evidenced by traditional sequencing. Sanger-generated sequences are presented as the actual, because assembly and sequencing errors are a major concern in NGS, and PCR primers were designed to anneal outside the open reading frame, giving no presupposed sequencing bias. Notably, assembling repetitive and low complexity sequences is difficult and prone to error [38]. The full amino acid sequences of the novel phenol gland proteins, Mcfp-7p–15p, are divided into two groups: the small proteins, less than 15 kDa (figure 3), and the larger proteins, more than 25 kDa (figure 4). The full sequences from the accessory gland Mcfp-16p–19p are shown in figure 5.
Table 3.
Putative, novel highly abundant mussel foot proteins.
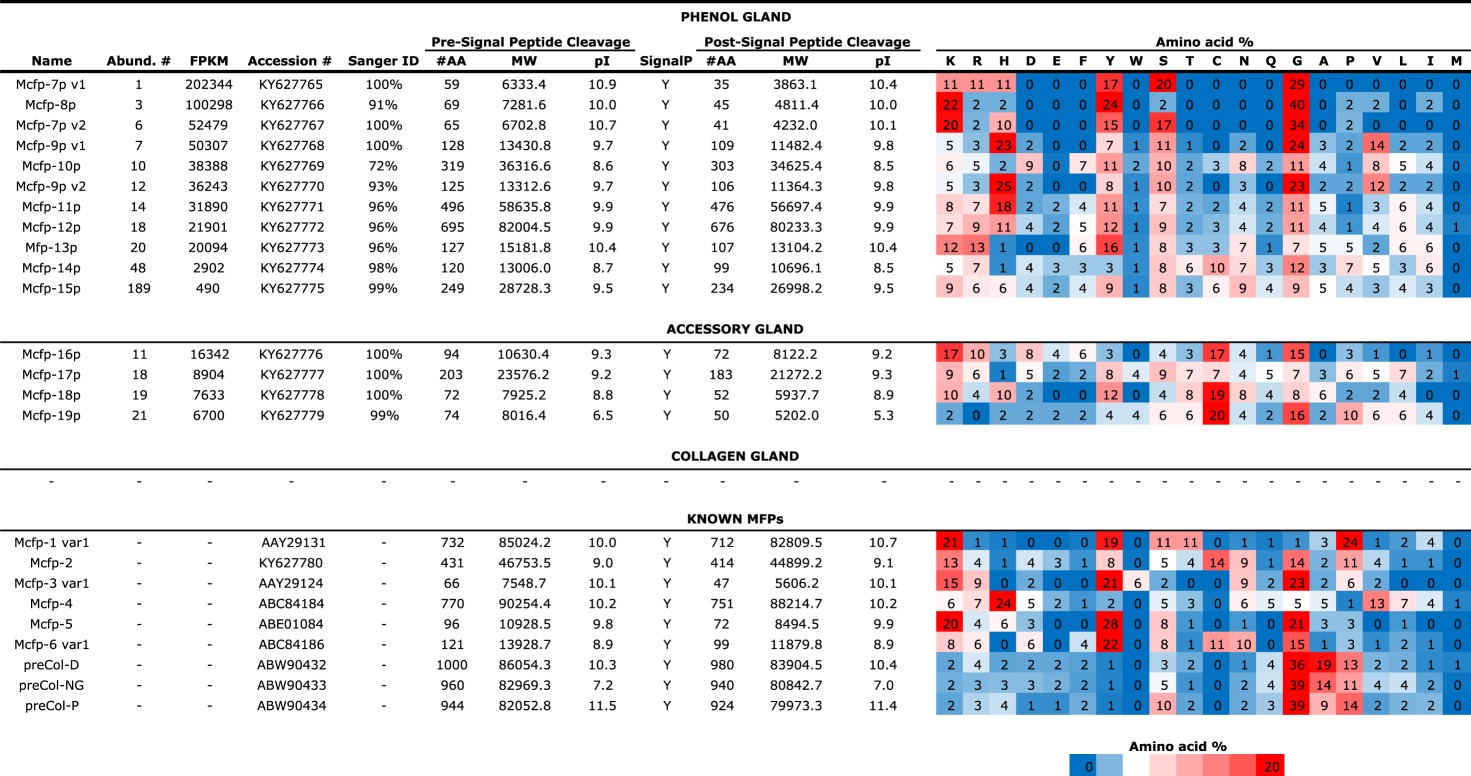 |
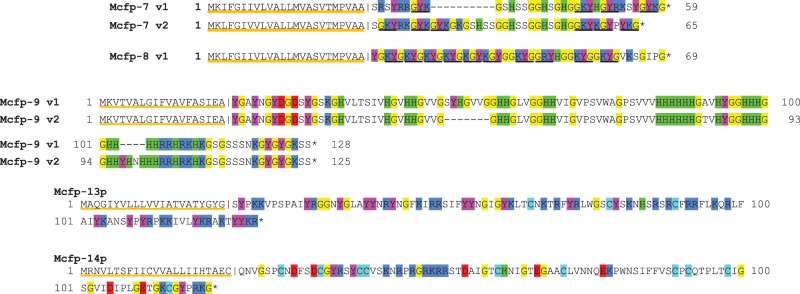
Figure 3.
Small new putative mussel foot proteins, Mcfp-7, -8, -9, -13, -14, from the phenol gland. Full-length primary sequences are shown, predicted signal peptides for each protein are underlined orange, and the cleavage site is marked with a vertical line. Key amino acids of the post-signal cleavage portions are highlighted: arginine and lysine (dark blue), tyrosine (magenta), glycine (yellow), histidine (green), and aspartate and glutamate (red). Mcfp-7 has a central serine and histidine-rich domain flanked by domains rich in KYG triplets (underlined in black). There are two variants of Mcfp-7, with the first being an approximate shorter version of the latter. Mcfp-9 also has two variants, which only differ by two short insertion/deletion segments.
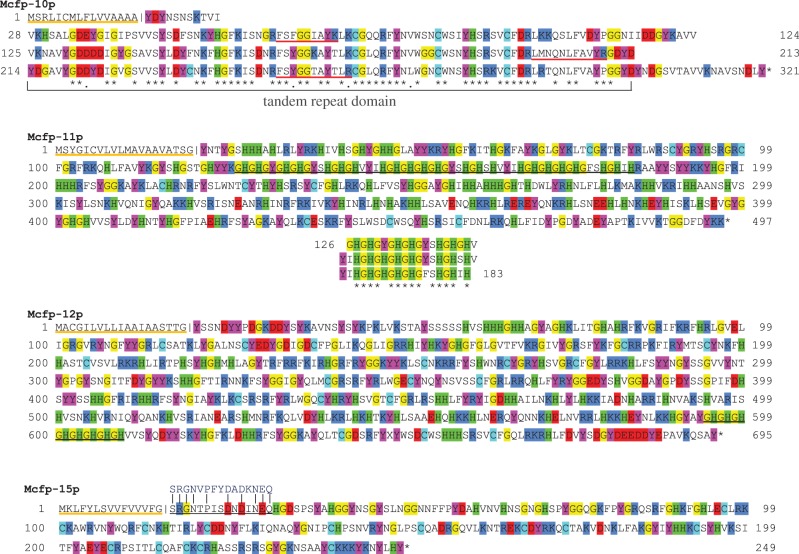
Figure 4.
Large new putative mussel foot proteins, Mcfp-10, -11, -12 and -15, from the phenol gland. The predicted signal peptides for each protein are underlined orange, and the cleavage site is marked with a vertical line. Key amino acids of the post-signal cleavage portions are highlighted: arginine and lysine (dark blue), tyrosine (magenta), glycine (yellow), histidine (green), aspartate and glutamate (red), and cysteine (light blue). Mcfp-10p is arranged to show the three tandem repeat domains of approximately 90 amino acids; identities are marked by asterisks. Mcfp-11p is a histidine-rich protein with a particular -[GH]- repeat domain (57 amino acids long, underlined in black); this segment is further broken down as three tandem repeats below the sequence. Mcfp-12 is also a histidine-rich protein containing a -[GH]- repeat but only 16 amino acids long (underlined in black). Mcfp-15p shows the sequence determined by Edman degradation (underlined in black) from the closely related species Mytilus edulis (written above in blue).
Figure 5.
Mcfp-16, -17, -18 and -19 primary amino acid sequences from the accessory gland. The predicted signal peptides for each protein are underlined orange, and the cleavage site is marked with a vertical line. Key amino acids of the post-signal cleavage portions are highlighted: arginine and lysine (dark blue), tyrosine (magenta), glycine (yellow), histidine (green), aspartate and glutamate (red), and cysteine (light blue).
3.3. Protein-level evidence of novel Mcfp-p proteins
Several of the presented novel Mcfp-p proteins have been tentatively detected by mass spectrometry. KCl-induced plaque proteins were chromatographically separated and the resulting fractions were analysed by MALDI-MS to reveal several peaks that match the predicted masses of the mature Mcfp-p proteins (figure 6), namely Mcfp-7p, -8p, -10p, -13p and -14p. Furthermore, we isolated Mcfp-10p and obtained partial sequence of tryptic peptides by liquid chromatography-tandem mass spectrometry (LC-MS/MS) unequivocally verifying the natural production of this protein (electronic supplementary material, figure S1).
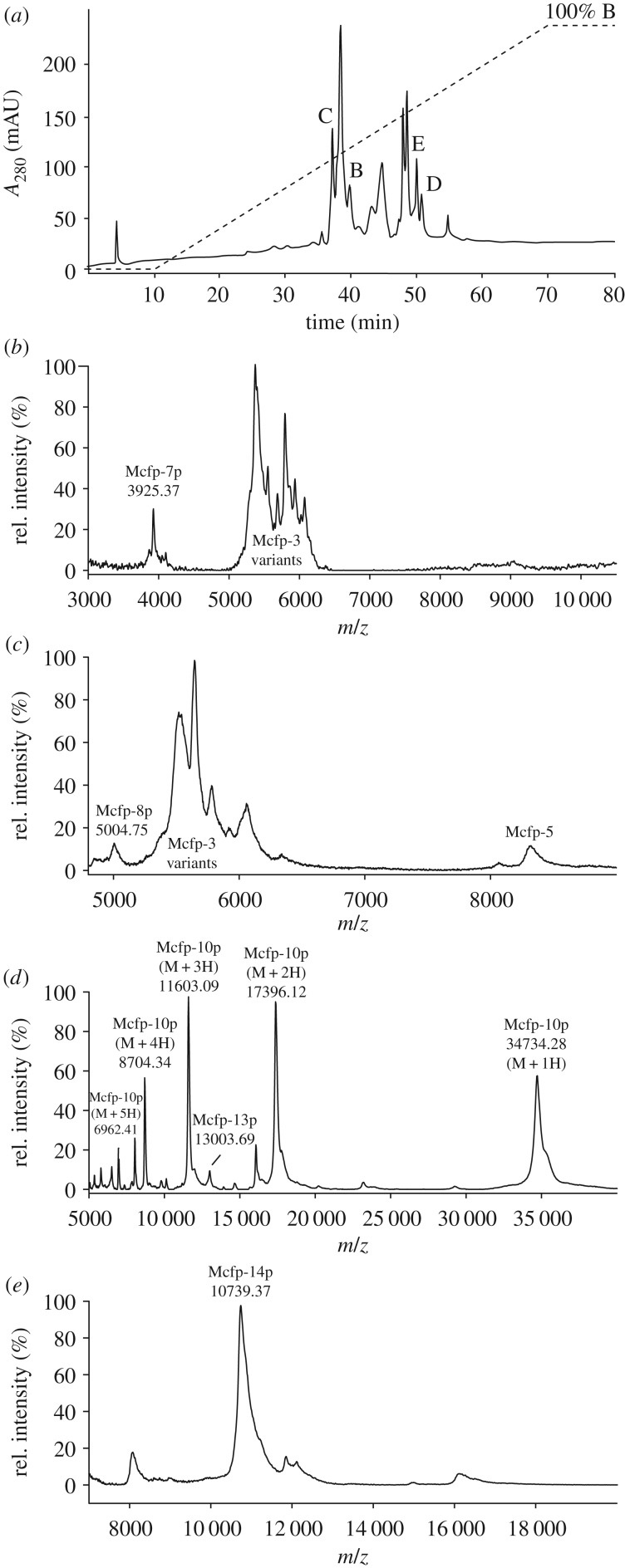
Figure 6.
MALDI-MS of induced M. californianus plaques indicate the presence of Mcfp-p proteins. (a) HPLC chromatogram of the fractionation of acetic acid–guanidine-solubilized byssus proteins. Protein elution was monitored at an absorbance of 280 nm (solid line); the elution gradient is also shown (dashed line). HPLC yielded various peaks consistent with predicted masses of the novel Mcfp-p sequences as shown by MALDI-MS, specifically Mcfp-7 (b), Mcfp-8p (c), Mcfp-10p (d), Mcfp-13p (d) and Mcfp-14p (e).
With respect to Mcfp-15p, we previously isolated an abundant 28 kDa protein from the plaques of Mytilus galloprovincialis. The N-terminal sequence was deduced by Edman degradation to be SRGNVPFYDADKNEQ. Efforts to obtain the full sequence via degenerate molecular sequencing techniques were unproductive at the time; however, the full sequence was easily identified in the transcriptome data from this partial sequence as Mcfp-15p (figure 4). Indeed, the sequence matches the predicted mature N-terminus and has a similar molecular mass of 27 kDa (table 2). Additionally, this transcript has a significant high FPKM value of 490 (just below our arbitrary cut-off of 500, but still in the upper 0.5%), which suggests a high level of actual protein production.
4. Discussion
To cope with the continually changing lift and drag forces around them, mussels have evolved the capacity to quickly and continuously deposit plaques and threads to maintain the integrity of their byssus. Micrographs of the cells responsible for byssus production illustrate the overwhelming amount of rough endoplasmic reticulum and secretory granules in the gland tissue [39,40], and the transcriptomes of these glands corroborate the dominant presence of transcripts for byssus proteins as well as their synthesis machinery (ribosomal proteins).
An important consideration in interpreting the collated data is that the expression levels reported here are qualitative and not strictly quantitative; the exact hierarchy of transcript abundance should not be taken as absolute, but as a strong indication of the dominant species. This assumption is corroborated by the agreement between the abundant transcripts of a particular gland and previously characterized byssal precursors. Another relevant assumption is that transcript abundance is a good indication of protein abundance, which is not always true, particularly in cases of translational control [41]. However, as we are focused on the most abundant transcripts of specialized secretory gland tissues, this assumption is reasonable.
In addition to providing gland-specific information about known byssal proteins, the transcriptomes also revealed many putative new mussel foot proteins with no database homologues. Detected together with known Mfps, these putative proteins exhibit characteristics reminiscent of the established mussel foot proteins, but also bolster and highlight significant emerging themes in mussel adhesion. Because transcriptomic assembly is particularly prone to sequencing errors and cannot solely be relied upon for de novo sequencing of novel genes, we corroborated these novel Mcfp-p transcripts using traditional PCR and cloning, thus confirming their existence and lending plausibility to the sequences as translated products.
Two new variants known as Mcfp-7 and Mcfp-8 are only 35–45 amino acids long following signal peptide cleavage. They have an extremely biased composition, being rich in glycine, lysine and tyrosine (dopa), which are frequently grouped as KYG triplets. Mcfp-7 variants 1 and 2 and Mcfp-8 have 4, 6 and 10 KYG triplets, respectively. Indeed, two-thirds of the Mcfp-8 sequence consists of KYG repeats. Mcfp-7 has KYG triplets at both the N- and C-termini that separate a central histidine–serine–glycine domain. Lysine and dopa are critical residues for the interfacial adhesion of Mcfp-3 and 5 and, in fact, the C-terminal portion of Mcfp-5 is almost exclusively composed of these three residues and shows the highest adhesion compared with other domains in Mcfp-5 [42]. As was shown with synthetic siderophores modelled after the abundant YK sequences in Mfp-5, primary amines in the YK pairs synergistically displace hydrated cations from aluminosilicate rock surfaces, thereby triggering catechol–surface interactions with underlying metal oxides [43]. Fortuitously, just as the key role of lysine–dopa synergy in adhesion became apparent, we found Mcfp-8, which consists almost exclusively of these two residues with the addition of flexible glycine. The MALDI-MS peak at 3925 Da (figure 6a) potentially represents Mcfp-7p variant 2, with the addition of four hydroxyl groups, plausibly tyrosine to dopa modifications. Similarly, the peak at 5004 Da could reasonably represent Mcfp-8 with 12 hydroxylations (figure 6b), as it turns out 12 is the exact number of available sites (11 Tyr, 1 Arg) if we consider the same degree of modification as Mcfp-5.
Adhesion experiments with Mcfp-5-based peptide sequences also showed that chain length was important in enabling peptides to bridge between two surfaces [42]. Full-length Mfp-5 was excellent at providing bridging adhesion, but shorter homologues were not. At approximately half the length of Mcfp-3 and -5, Mcfp-7p and -8p may be adapted as non-bridging surface primers with which other larger bridging Mcfps interact.
Mcfp-9p has two variants, both approximately 100 residues in length and 23–25 mol% histidine—the highest of all the Mfps. Along with the other small Mfps these histidine-rich proteins may play an adhesive role at the substrate interface, diversifying functional groups and potentially coordinating surface-bound transition metals, such as Ni, Co, Cu and Zn [44]. Mcfp-9p histidine–metal ion coordination could also function as sacrificial bonds for self-healing and energy dissipation as shown in the mussel thread preCOLs [15,45], and/or in cohesion between the thread and the plaque mediated by Mcfp-4 [46]. One conspicuous feature of Mcfp-9p is the grouping of histidines into blocks, in one case six consecutive histidine residues, immediately reminiscent of the classic protein purification tag [44], but there are a few cases of natural polyhistidine in proteins of various functions including metal ion transporters, bacterial chaperonins and antimicrobial peptides [47].
Mcfp-10p is 303 residues in length and contains three tandem repeat domains of approximately 90 amino acids each. The function of these domains is unclear and there is no equivalent homologue in the database. The C-terminal portion of the YGH-rich protein 3 (accession no. ALA16022), identified in the Mytilus coruscus transcriptome, is 84% identical to one of the Mcfp-10 repeats, but that constitutes only a small portion of that large 500 residue protein, which as its name suggests is rich in tyrosine, glycine and histidine. Mcfp-10, however, is only 2 mol% histidine compared with 10% in the YGH-rich protein. Furthermore, the M. coruscus YGH-rich protein was only identified in a transcriptomic assembly and has not been vetted by cloning. It is, nonetheless, relevant that the Mcfp-10 repeat domain has been observed in other mussel byssus precursors. Mcfp-10p was eluted from a C18 high-performance liquid chromatography (HPLC) column and easily ionized by MALDI-MS, ms yielding a clean spectrum with clear representation of the multiply charged ions (M + 1H through M + 5H; figure 6c).
Mcfp-11p and Mcfp-12p are the two largest novel proteins reported here at 55 kDa and 80 kDa, respectively. The high mol% of histidine (17 and 11%) is notable and makes them most similar to the N-terminus of Mcfp-4 that is proposed to interact with the terminal ends of the collagen fibres through metal-mediated cross-links. These two proteins along with Mcfp-9 showcase histidine to be much more prevalent in the plaque than previously thought. While histidine could be playing a similar role to N-terminal Mcfp-4 in metal-mediated cohesive cross-linking [22], its physiologically relevant pKa also makes it a prime candidate as a coacervation intermediate in the fluid to solid transition apparent in plaque processing [48].
Mcfp-13p has a largely non-repetitive sequence rich in tyrosine, arginine and lysine, and various hydrophilic residues, and is poor in hydrophobic residues; this is reminiscent of Mcfp-3 and Mcfp-5. The distinguishable difference is the molecular weight of the three proteins (5, 8 and 13 kDa for Mcfp-3, -5 and -13p, respectively), again highlighting an apparent tendency to vary chain lengths. It is not unreasonable to propose an interfacial adhesive role for the protein.
Mcfp-14p is 10 mol% Cys and approximately 10.6 kDa, and could play a role in redox poise similar to the cysteine-rich Mcfp-6 [49]. Maintenance of a reducing environment safeguards against premature oxidation of dopa to the quinone, and thereby preserves dopa-mediated interactions for adhesion and metal coordination.
Mfp-15p has been confirmed as an extractable protein in byssal plaques (M. edulis) and exhibits nitrotetrazolium blue (NBT)-positive staining (assay for dopa), a mass of approximately 29 kDa, and an N-terminal amino acid sequence by Edman degradation. We found the corresponding homologue in our database, the N-terminal peptide sequence was not perfectly conserved but the protein mass is within 10% and the N-terminus has high homology, and the Tyr content is high (available for dopa conversion). Like many of the other sequences presented, its function is not readily apparent, but we do know that it is present as an extractable protein in the plaque.
Only a couple of novel genes including Mcfp-16-19p were identified from the accessory gland: all are relatively small proteins, 5–20 kDa, and fairly rich in cysteine, 7–20%. To date, Mcfp-1 is the only known protein in the thread cuticle, although cytochemical analyses suggest the presence of others; for example, the matrix and the embedded granules of byssal cuticle are differentially susceptible to enzymatic degradation [40]. Mcfp-1's high dopa content, affinity for Fe and localization of Fe–dopa coordination complexes in the granules leave little doubt that Mcfp-1 is a prominent granular component. This leaves the possibility that Mcfp-16-19p could be a matrix component. Cysteinyl–dopa cross-links are a known cohesive component of the mussel plaque and are associated with the cysteine-rich Mcfp-6 [46], and Mcfp-16-19p could be performing a similar role in forming a robust cuticle matrix. The redox potential of cysteine also allows it to maintain the reducing environment at the plaque–substrate interface essential for the function/maintenance of dopa [20]. Dopa is not, however, exclusive to the plaque but a key feature throughout the byssus and a functionality present in almost all the Mfps. As such Mcfp-6 might have a more universal role throughout the byssus in regulating the oxidation of dopa to dopaquinone, as such we observed that Mcfp-6 transcripts are more ubiquitous in the byssus than previously thought. An emerging interpretation is that redox regulation is complex within the byssus and probably plays a dynamic role in helping regulate the development and maturation of the byssus.
Naturally, the discovery of so many novel Mfps provokes curiosity about why they escaped previous isolation and characterization efforts. There are several plausible explanations: first, the tanned leathery mature form of byssus makes for poor protein extractability; add to this that protein extractability from even the induced plaque exudates is less than approximately 50%. Second, some of the new and traditional Mfps share similar molecular weights, making a mixture of the two appear as a cluster of protein variants in gel electrophoresis and mass spectrometry; for example, Mcfp-9p (11.5 kDa) and Mcfp-6 (11.6 kDa), and Mcfp-12p (80 kDa) and Mcfp-4 (90 kDa). Small proteins like Mcfp-7p and -8p could have easily been overlooked (below the detection range) or attributed to degradation products. Mass spectrometry has the caveat of being empirically biased towards ‘fly-able’ ions and can hardly be relied on for a full representation of protein species in a sample. Likewise, partial peptide fragment sequencing can also be extremely biased based on protease susceptibility as well as fly-ability. For these and other reasons, it is not unfathomable these and other mussel foot proteins have been overlooked.
Several recent studies also used transcriptomics and mass spectrometry to investigate novel mussel byssus proteins in the mussels Dreissena polymopha [50], Mytilus coruscus [51] and Perna viridis [28]. With the exception of Mcfp-10p (discussed above), we did not find significant sequence homology between the novel byssus sequences reported for these other species and the abundant transcripts from Mytilus californianus foot glands, although some sequences from these various studies seem to follow the general theme of glycine-rich basic proteins with particular bias towards characteristic amino acids (tyrosine, cysteine, lysine, arginine and/or serine). The high glycine content coupled with the observation that many of the Mfps are intrinsically disordered [17] could suggest that the amino acid composition is the key unifying element between some of these various novel Mfps. The lack of primary sequence homology does not necessarily negate the possibility of similar functional roles.
Significantly, we did not find any transcripts for highly acidic proteins. Coacervates offer an advantageous avenue for byssus processing and delivery, but, by definition, complex coacervates are electrostatically stabilized liquid–liquid phase separations usually involving a polycation and a polyanion [52]. Many of the Mfps are potential polycation candidates in such systems but the polyanion protein counterparts have not materialized even in these transcriptomic searches, leaving us to speculate on alternative polyanion possibilities, such as polysaccharides [25], or heavily phosphorylated proteins, as in the Phragmatopoma coacervate system [53].
5. Conclusion
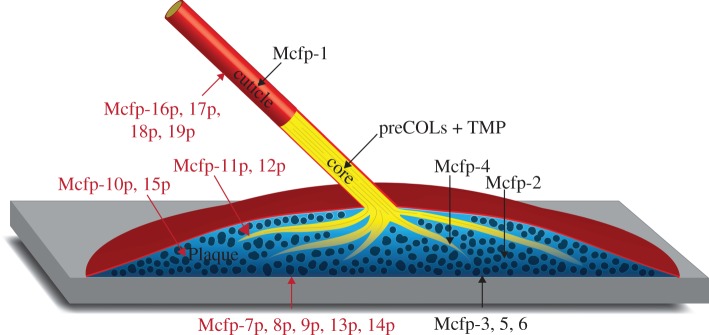
Our NGS data illustrate how transcriptomic analysis can offer significant insights into the composition of protein-based biomaterials, and are particularly advantageous in heavily cross-linked materials. Here, we have nearly doubled the suite of mussel foot proteins thought to play functional roles in the mussel byssus, and speculate on their potential localization (figure 7). It is, however, unclear whether the entire array of Mfps is required for proper plaque function or a subset of the Mfp suite is used to match specific environmental conditions. The knowledge of these new Mfps sets the groundwork for future biochemical investigations to build a more complete model of byssus structure and function in this premier system of bioadhesion.
Figure 7.
Putative locations of the new mussel foot proteins in the plaque (red). Mcfp-7, -8, -9, -13, -14 at the substrate interface. Mcfp-10, -15 in the bulk of the plaque. Mcfp-11, -12 at the collagen interface. Finally, Mcfp-16, -17, -18, -19 in the cuticle coating.
Supplementary Material
Acknowledgements
We thank Sabrina Pankey for help with transcriptomic sample preparation and Todd Oakley for generous use of his bioinformatic server.
Data accessibility
The transcriptome assembly projects are available under DDBJ/EMBL/GenBank accessions: GFII00000000 (phenol gland), GFIH00000000 (collagen gland) and GFIB00000000 (accessory gland). The raw sequencing reads are available from the NCBI Sequence Read Archive (SRA) under accessions: SRR5275489, SRR5275488 and SRR5275487, respectively. Novel mussel foot protein sequences are available under GenBank accessions: KY627765-KY627779. Additional data supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
D.G.D. and J.H.W. conceived the research, analysed the data and wrote the manuscript. D.G.D., A.F., S.S. and J.M.E. planned and executed the experiments, and processed the data. All authors edited and revised the manuscript.
Competing interests
We declare no competing interests.
Funding
This research was supported by the Materials Research Science and Engineering Centers Program of the National Science Foundation under award DMR 1121053 and the National Institutes of Health under grant no. R01-DE018468. We acknowledge support from the Center for Scientific Computing from the CNSI, MRL: an NSF MRSEC DMR-1121053 and NSF CNS-0960316.
References
- 1.Kamino K. 2013. Mini-review: barnacle adhesives and adhesion. Biofouling 29, 735–749. ( 10.1080/08927014.2013.800863) [DOI] [PubMed] [Google Scholar]
- 2.Stewart RJ. 2004. The tube cement of Phragmatopoma californica: a solid foam. J. Exp. Biol. 207, 4727–4734. ( 10.1242/jeb.01330) [DOI] [PubMed] [Google Scholar]
- 3.Hennebert E, Wattiez R, Demeuldre M, Ladurner P, Hwang DS, Waite JH, Flammang P. 2014. Sea star tenacity mediated by a protein that fragments, then aggregates. Proc. Natl Acad. Sci. USA 111, 6317–6322. ( 10.1073/pnas.1400089111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waite JH. 1987. Nature's underwater adhesive specialist. Int. J. Adhes. Adhes. 7, 9–14. ( 10.1016/0143-7496(87)90048-0) [DOI] [Google Scholar]
- 5.Li L, Zeng H. 2016. Marine mussel adhesion and bio-inspired wet adhesives. Biotribology 5, 44–51. ( 10.1016/j.biotri.2015.09.004) [DOI] [Google Scholar]
- 6.Brubaker CE, Messersmith PB. 2012. The present and future of biologically inspired adhesive interfaces and materials. Langmuir 28, 2200–2205. ( 10.1021/la300044v) [DOI] [PubMed] [Google Scholar]
- 7.Stewart RJ, Ransom TC, Hlady V. 2011. Natural underwater adhesives. J. Polym. Sci. B Polym. Phys. 49, 757–771. ( 10.1002/polb.22256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee BP, Messersmith PB, Israelachvili JN, Waite JH. 2011. Mussel-inspired adhesives and coatings. Annu. Rev. Mater. Res. 41, 99–132. ( 10.1146/annurev-matsci-062910-100429) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callow ME, Callow JA. 2002. Marine biofouling: a sticky problem. Biologist 49, 1–5. [PubMed] [Google Scholar]
- 10.Fitridge I, Dempster T, Guenther J, de Nys R. 2012. The impact and control of biofouling in marine aquaculture: a review. Biofouling 28, 649–669. ( 10.1080/08927014.2012.700478) [DOI] [PubMed] [Google Scholar]
- 11.Denny M. 1995. Predicting physical disturbance: mechanistic approaches to the study of survivorship on wave-swept shores. Ecol. Monogr. 65, 371–418. ( 10.2307/2963496) [DOI] [Google Scholar]
- 12.Tamarin A, Lewis P, Askey J. 1976. The structure and formation of the byssus attachment plaque in Mytilus. J. Morphol. 149, 199–221. ( 10.1002/jmor.1051490205) [DOI] [PubMed] [Google Scholar]
- 13.Sone ED. 2016. Interfacial phenomena in marine and freshwater mussel adhesion. In Biological adhesives (ed. Smith AM.), pp. 129–151. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 14.Filippidi E, DeMartini DG, Malo de Molina P, Danner EW, Kim J, Helgeson ME, Waite JH, Valentine MT. 2015. The microscopic network structure of mussel (Mytilus) adhesive plaques. J. R. Soc. Interface 12, 20150827 ( 10.1098/rsif.2015.0827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waite JH, Qin XX, Coyne KJ. 1998. The peculiar collagens of mussel byssus. Matrix. Biol. 17, 93–106. ( 10.1016/S0945-053X(98)90023-3) [DOI] [PubMed] [Google Scholar]
- 16.Tamarin A, Keller PJ. 1972. An ultrastructural study of the byssal thread forming system in Mytilus. J. Ultra. Res. 40, 401–416. ( 10.1016/S0022-5320(72)90110-4) [DOI] [PubMed] [Google Scholar]
- 17.Waite JH. 2017. Mussel adhesion-essential footwork. J. Exp. Biol. 220, 517–530. ( 10.1242/jeb.134056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Priemel T, Degtyar E, Dean MN, Harrington MJ. 2017. Rapid self-assembly of complex biomolecular architectures during mussel byssus biofabrication. Nat. Commun. 8, 1–12. ( 10.1038/ncomms14539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Q, Danner E, Waite JH, Israelachvili JN, Zeng H, Hwang DS. 2013. Adhesion of mussel foot proteins to different substrate surfaces. J. R. Soc. Interface 10, 20120759 ( 10.1098/rsif.2012.0759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicklisch SC, Das TS, Martinez Rodriguez NR, Waite JH, Israelachvili JN. 2013. Antioxidant efficacy and adhesion rescue by a recombinant mussel foot protein-6. Biotechnol. Prog. 29, 1587–1593. ( 10.1002/btpr.1810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rzepecki LM, Hansen KM, Waite JH. 1992. Characterization of a cystine-rich polyphenolic protein family from the blue mussel Mytilus edulis. Biol. Bull. 183, 123–137. ( 10.2307/1542413) [DOI] [PubMed] [Google Scholar]
- 22.Zhao H, Waite JH. 2006. Proteins in load-bearing junctions: the histidine-rich metal-binding protein of mussel byssus. Biochemistry 45, 14223–14231. ( 10.1021/bi061677n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun C, Waite JH. 2005. Mapping chemical gradients within and along a fibrous structural tissue, mussel byssal threads. J. Biol. Chem. 280, 39332–39336. ( 10.1074/jbc.M508674200) [DOI] [PubMed] [Google Scholar]
- 24.Sagert J, Waite JH. 2009. Hyperunstable matrix proteins in the byssus of Mytilus galloprovincialis. J. Exp. Biol. 212, 2224–2236. ( 10.1242/jeb.029686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller DR, Das S, Huang K-Y, Han S, Israelachvili JN, Waite JH. 2015. Mussel coating protein-derived complex coacervates mitigate frictional surface damage. ACS Biomater. Sci. Eng. 1, 1121–1128. ( 10.1021/acsbiomaterials.5b00252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danner EW, Kan Y, Hammer MU, Israelachvili JN, Waite JH. 2012. Adhesion of mussel foot protein Mefp-5 to mica: an underwater superglue. Biochemistry 51, 6511–6518. ( 10.1021/bi3002538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desmond KW, Zacchia NA, Waite JH, Valentine MT. 2015. Dynamics of mussel plaque detachment. Soft Matter. 11, 6832–6839. ( 10.1039/c5sm01072a) [DOI] [PubMed] [Google Scholar]
- 28.Guerette PA, et al. 2013. Accelerating the design of biomimetic materials by integrating RNA-seq with proteomics and materials science. Nat. Biotechnol. 31, 908–915. ( 10.1038/nbt.2671) [DOI] [PubMed] [Google Scholar]
- 29.Guerette PA, Hoon S, Ding D, Amini S, Masic A, Ravi V, Venkatesh B, Weaver JC, Miserez A. 2014. Nanoconfined β-sheets mechanically reinforce the supra-biomolecular network of robust squid sucker ring teeth. ACS Nano. 8, 7170–7179. ( 10.1021/nn502149u) [DOI] [PubMed] [Google Scholar]
- 30.So CR, et al. 2016. Sequence basis of barnacle cement nanostructure is defined by proteins with silk homology. Sci. Rep. 6, 36219 ( 10.1038/srep36219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues M, Ostermann T, Kremeser L, Lindner H, Beisel C, Berezikov E, Hobmayer B, Ladurner P. 2016. Profiling of adhesive-related genes in the freshwater cnidarian Hydra magnipapillata by transcriptomics and proteomics. Biofouling. 32, 1115–1129. ( 10.1080/08927014.2016.1233325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afgan Eet al. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10. ( 10.1093/nar/gkw343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 ( 10.1186/1471-2105-12-323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu J, Wei W, Danner E, Ashley RK, Israelachvili JN, Waite JH. 2011. Mussel protein adhesion depends on interprotein thiol-mediated redox modulation. Nat. Chem. Biol. 7, 588–590. ( 10.1038/nchembio.630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petersen TN, Brunak S, Heijne von G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. ( 10.1038/nmeth.1701) [DOI] [PubMed] [Google Scholar]
- 38.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. 2010. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26, 493–500. ( 10.1093/bioinformatics/btp692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuccarello LV. 1980. The collagen gland of Mytilus galloprovincialis: an ultrastructural and cytochemical study on secretory granules. J. Ultra. Res. 73, 135–147. ( 10.1016/S0022-5320(80)90119-7) [DOI] [PubMed] [Google Scholar]
- 40.Zuccarello LV. 1981. Ultrastructural and cytochemical study on the enzyme gland of the foot of a mollusc. Tissue Cell 13, 701–713. ( 10.1016/S0040-8166(81)80007-9) [DOI] [PubMed] [Google Scholar]
- 41.Vogel C, Marcotte EM. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232. ( 10.1038/nrg3185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei W, Yu J, Gebbie MA, Tan Y, Martinez Rodriguez NR, Israelachvili JN, Waite JH. 2015. Bridging adhesion of mussel-inspired peptides: role of charge, chain length, and surface type. Langmuir 31, 1105–1112. ( 10.1021/la504316q) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maier GP, Rapp MV, Waite JH, Israelachvili JN, Butler A. 2015. Adaptive synergy between catechol and lysine promotes wet adhesion by surface salt displacement. Science 349, 628–632. ( 10.1126/science.aab0556) [DOI] [PubMed] [Google Scholar]
- 44.Schmidt S, Reinecke A, Wojcik F, Pussak D, Hartmann L, Harrington MJ. 2014. Metal-mediated molecular self-healing in histidine-rich mussel peptides. Biomacromolecules 15, 1644–1652. ( 10.1021/bm500017u) [DOI] [PubMed] [Google Scholar]
- 45.Harrington MJ, Gupta HS, Fratzl P, Waite JH. 2009. Collagen insulated from tensile damage by domains that unfold reversibly: in situ X-ray investigation of mechanical yield and damage repair in the mussel byssus. J. Struct. Biol. 167, 47–54. ( 10.1016/j.jsb.2009.03.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao H, Waite JH. 2006. Linking adhesive and structural proteins in the attachment plaque of Mytilus californianus. J. Biol. Chem. 281, 26 150–26 158. ( 10.1074/jbc.M604357200) [DOI] [PubMed] [Google Scholar]
- 47.Rowinska-Zyrek M, Witkowska D, Potocki S, Remelli M, Kozlowski H. 2013. His-rich sequences—is plagiarism from nature a good idea? New J. Chem. 37, 58–70. ( 10.1039/C2NJ40558J) [DOI] [Google Scholar]
- 48.Tan Y, Hoon S, Guerette PA, Wei W, Ghadban A, Hao C, Miserez A, Waite JH. 2015. Infiltration of chitin by protein coacervates defines the squid beak mechanical gradient. Nat. Chem. Biol. 11, 488–495. ( 10.1038/nchembio.1833) [DOI] [PubMed] [Google Scholar]
- 49.Nicklisch SCT, Spahn JE, Zhou H, Gruian CM, Waite JH. 2016. Redox capacity of an extracellular matrix protein associated with adhesion in Mytilus californianus. Biochemistry 55, 2022–2030. ( 10.1021/acs.biochem.6b00044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gantayet A, Rees DJ, Sone ED. 2014. Novel proteins identified in the insoluble byssal matrix of the freshwater zebra mussel. Mar. Biotechnol. 16, 144–155. ( 10.1007/s10126-013-9537-9) [DOI] [PubMed] [Google Scholar]
- 51.Qin C-L, Pan Q-D, Qi Q, Fan M-H, Sun J-J, Li N-N, Liao Z. 2016. In-depth proteomic analysis of the byssus from marine mussel Mytilus coruscus. J. Proteom. 144, 87–98. ( 10.1016/j.jprot.2016.06.014) [DOI] [PubMed] [Google Scholar]
- 52.Hwang DS, Zeng H, Srivastava A, Krogstad DV, Tirrell M, Israelachvili JN, Waite JH. 2010. Viscosity and interfacial properties in a mussel-inspired adhesive coacervate. Soft Matter. 6, 3232–3236. ( 10.1039/C002632H) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao H, Sun C, Stewart RJ, Waite JH. 2005. Cement proteins of the tube-building polychaete Phragmatopoma californica. J. Biol. Chem. 280, 42938–42944. ( 10.1074/jbc.M508457200) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome assembly projects are available under DDBJ/EMBL/GenBank accessions: GFII00000000 (phenol gland), GFIH00000000 (collagen gland) and GFIB00000000 (accessory gland). The raw sequencing reads are available from the NCBI Sequence Read Archive (SRA) under accessions: SRR5275489, SRR5275488 and SRR5275487, respectively. Novel mussel foot protein sequences are available under GenBank accessions: KY627765-KY627779. Additional data supporting this article have been uploaded as part of the electronic supplementary material.