Abstract
The p53 tumor suppressor is the most commonly mutated gene in human cancers. The ability of p53 to induce cell cycle arrest, apoptosis, DNA repair, and other p53-dependent activities is well known; however, the mechanism by which p53 induces a specific activity over another is unclear. Here, we showed that stringent regulation of and by p53 family isoforms facilitates differential target gene expression and thus determines cell fate. Through the use of engineered deletion mutants, we found that activation domain 2 is required for induction of the proapoptotic target gene insulin-like growth factor binding protein 3 (IGFBP3) by p53 and that the basic domain inhibits induction of this gene by p53. Thus, for the first time we provide evidence that the basic domain of p53 is inhibitory in vivo as has been determined in vitro. We also showed that the in vivo inhibitory activity of the basic domain depends upon activation domain 1, such that combined deletion of activation domain 1 and the basic domain was required to alleviate the inhibition by the basic domain. Importantly, deletion of the inhibitory functional domains, namely N-terminal activation domain 1 and the C-terminal basic domain, is paralleled in nature. We found that the IGFBP3 promoter was activated by p53(ΔNΔBD), which mimics a naturally occurring N- and C-terminally truncated human p53 isoform, and by p53AS, a C-terminally truncated murine p53 isoform generated through alternative splicing, but not by full-length human or murine p53. In addition, we found that the C termini of p63 and p73 inhibit the induction of IGFBP3, such that C-terminally truncated p63 and p73 isoforms induce the expression of IGFBP3, whereas full-length ones cannot. We also demonstrated that IGFBP3 is an important effector of the apoptosis induced by N- and C-terminally truncated p53, such that knockdown of IGFBP3 by using an IGFBP3 neutralizing antibody or IGFBP3 small interfering RNA partially rescues the cell death induced by N- and C-terminally truncated p53. In addition, we identified that histone deacetylase activity, not p53 DNA binding ability, governs the regulation of IGFBP3 by full-length p53 family proteins, as inhibition of histone deacetylases restores the induction of IGFBP3 by exogenous full-length p53, p63, and p73 proteins. Furthermore, we found that activation of p53 or inhibition of histone deacetylases alone was not sufficient to induce IGFBP3; however, combined treatment endowed endogenous p53 with this activity. To better understand the significance of this regulation, we performed a microarray study and identified several target genes differentially regulated by full-length p53 and p53 lacking the N-terminal activation domain 1 and the C-terminal basic domain. Taken together, our data suggest a novel mechanism by which p53 family proteins differentially regulate gene expression and provide an insight for designing a combined therapy for cancer treatment.
The tumor suppressor p53 is the most commonly mutated gene in human cancers (43). After activation by cellular stresses, p53, a sequence-specific transcription factor, functions to transactivate genes that mediate cell cycle arrest, apoptosis, DNA repair, inhibition of angiogenesis and metastasis, and other p53-dependent activities (24, 30). The p53 protein contains several functional domains: activation domain 1 (AD1) within residues 1 to 42, activation domain 2 (AD2) within residues 43 to 63, the proline-rich domain (PRD) within residues 64 to 91, the sequence-specific DNA binding domain (DBD) within residues 100 to 300, the nuclear localization signal within residues 316 to 325, the tetramerization domain (TD) within residues 334 to 356, and the C-terminal basic domain (BD) within residues 364 to 393.
AD1 is important for transactivation; this domain contains residues that contact the basal transcriptional machinery (34). Previously, along with others we identified AD2 and characterized the requirement of AD2 for p53-dependent apoptosis (7, 8, 61, 70). In addition, along with others we have shown that the PRD is necessary for the induction of apoptosis and contributes to growth suppression (50, 60, 62, 67). The C-terminal BD has been subjected to extensive analysis. All evidence suggests that the BD is an important regulatory domain. Previous studies have shown that deletion of the BD and peptides or antibodies targeted to the BD increase p53-specific DNA binding activity in vitro (23, 25, 26, 55, 56). In addition, the function of the BD is altered through posttranslational modifications such as phosphorylation by casein kinase II or protein kinase C and acetylation by p300/CBP as well as through interactions with other proteins such as the calcium binding protein S100b and the DNA repair proteins XPB and XPD (reviewed in references 4 and 30).
Recently, the p63 and p73 proteins have been identified as p53 homologues (2, 28, 44, 59, 63). p53 family members share significant similarity at the amino acid level within the AD, the DBD, and the TD. Like p53, both p63 and p73 bind to the canonical p53-responsive element, transactivate p53 target gene expression, and induce apoptosis when overexpressed (reviewed in reference 64). Unlike p53, the genes encoding p63 and p73 are rarely mutated in human cancer (39, 65). Rather than displaying a propensity for tumor formation as in the p53 knockout mouse model (14), the p63 and p73 knockout animals demonstrate discrete developmental defects (39, 65). In addition, p63 and p73 undergo alternative splicing of their C termini, resulting in three p63 isoforms (α to γ) and seven p73 isoforms (α to η). These isoforms are transcribed from an upstream promoter as well as from a cryptic promoter within intron 3, called the TA and ΔN isoforms, respectively (reviewed in reference 64). Differential expression of these isoforms is believed to facilitate differential target gene regulation by p63 and p73.
With the realization that each p63 and p73 isoform functions in similar yet different manners, new light is shed on the functional importance of previously identified p53 isoforms. Although different mechanisms generate p53 isoforms, mouse and human p53 exist as both N- and C-terminally truncated forms (reviewed in reference 11). N-terminal truncation of human and mouse p53 yields ΔNp53 and Δ40p53, respectively. Human ΔNp53 is generated through alternative splicing of intron 2 with the subsequent use of the translational start site at codon 40 (21). Mouse Δ40p53 lacks the first 40 amino acids and is generated through the use of an internal translational start site at codon 41. C-terminal truncation of human and mouse p53 yields I9+p53 and p53AS and occurs through alternative splicing of introns 9 and 10, respectively (1, 18, 32). In addition, several studies strongly suggest that calpain and interaction with mismatched DNA induce both N- and C-terminal cleavage of p53 (31, 37, 40, 42, 45). To date, the biological functions of these p53 isoforms remain unclear.
Insulin-like growth factor binding protein 3 (IGFBP3) belongs to the IGFBP family. There are six known members (reviewed in reference 17). Previously, IGFBP3 was shown to be regulated by p53 (5). IGFBP3 contains eleven decamers of the p53-responsive element within the promoter (3) and two canonical p53-responsive elements, called box A and box B, within introns 1 and 2, respectively (5). Since IGFBP3 is the major serum IGFBP, many IGF-dependent functions have been described; however, recent evidence supports the IGF-independent functions of IGFBP3. Interestingly, IGF-independent functions oppose the IGF-dependent functions, such that IGFBP3 directly inhibits growth, induces apoptosis, and sensitizes cells to apoptosis (6, 22, 29, 53).
Here, we showed that p53 isoforms differentially regulate target gene expression. Through the use of engineered deletion mutants, we first found that p53 functional domains dictate target gene specificity. We found that AD2 is required for, and the basic domain inhibits, induction of IGFBP3 by p53. Subsequently, we found a common theme among the p53 family of transcription factors: the C terminus of p53 family isoforms inhibits transactivation of IGFBP3, such that C-terminally truncated p53, p63, and p73 isoforms induce the expression of IGFBP3, whereas full-length ones cannot. We showed that IGFBP3 is an important mediator of p53(ΔAD1ΔBD)-dependent apoptosis. We also determined that histone deacetylase (HDAC) activity governs the induction of IGFBP3 by full-length p53 family proteins, as the inhibition of HDACs restores the ability of exogenous full-length p53, p63, and p73 isoforms to induce IGFBP3. Interestingly, we found that the simultaneous stabilization of p53 through the DNA damage pathway and the inhibition of HDACs restore the ability of endogenous full-length p53 to induce IGFBP3. Our data suggest a mechanism by which p53 family isoforms differentially interact with regulatory proteins to facilitate a highly regulated gene expression profile.
MATERIALS AND METHODS
Plasmids and mutagenesis.
Human p53 proteins encoding full-length p53, p53(R249S), p53(ΔAD1ΔBD), p53(ΔAD1), p53(ΔBD), p53(AD1−ΔBD), p53(ΔAD2ΔBD), p53(AD2−ΔBD), and p53(ΔAD1AD2−ΔBD) were as previously described (69). All cDNAs, except p53(R249S) and p53(AD1−ΔBD), were N-terminally hemagglutinin (HA)-tagged. To generate p53(ΔC20), a human cDNA fragment that encodes residues 1 to 373 was amplified by the 5′ end primer 5HA (GATCGAATTCACCATGGGCTACCCATACGATGTTCCAGATTACGCTGAGGAGCCGCAGTCAGATCC) and the 3′ end primer C373 (AGAATTCTCACTTTTTGGACTTCAG). To generate p53(ΔC10), a human cDNA fragment that encodes residues 1 to 383 was amplified by the 5′ end primer 5HA and the 3′ end primer C383 (AGAATTCTCAGAGTTTTTTATGGCG). To generate ΔNp53, a human cDNA fragment that encodes residues 40 to 393 was amplified by the 5′ end primer ΔNp53 (AGAATTACCATGGATGATTTGATGCTGTCCCCGG) and the 3′ end primer C393 (AGAATTCTCAGTCTGAGTCAGGCCCTTCTGTC). To generate p53(ΔNΔBD), the 3′ end cDNA fragment starting from the StuI site in ΔNp53 was replaced with the corresponding cDNA fragment in p53(ΔBD). To generate I9+p53, a human cDNA fragment that encodes an alternative splice form of p53 was generated by using the 5′ end primer 5hp53 (AGAATTCACCATGGAGGAGCCGCAGTCAGATCCTA) and the consecutive 3′ end primers I9-up (TTGAAAGCTGGTCTGGTCCTGAAGGGTGAAATATTC) and I9-dn (AGAATTCTTAACAATTTTCTTTTTGAAAGCTGGTCTGGTC). Full-length murine p53 was generated by using 5′ end primer 5mp53 (AGAATTCACCATGACTGCCATGGAGGAGTC) and the 3′ end primer mC387 (TGAATTCTCAGTCTGAGTCAGGCCCCAC). p53(A135V), a murine cDNA that encodes a naturally occurring p53 DNA binding mutant was as previously described (38). To generate Δ40p53, a murine cDNA fragment that encodes residues 41 to 387 was amplified by using the 5′ end primer Δ40p53 (AGAATTCACCATGGACGATCTGTTGCTGCC) and the 3′ end primer mC387. To generate p53AS, a murine cDNA fragment that encodes an alternative splice form of p53 was generated by using the 5′ end primer 5mp53 and the consecutive 3′ end primers AS-up (TTGATCAAGGCTTGGAAGGCTCTAGGCTGGAGGCTGGAGTGAGCCCTGCT) and AS-dn (TGAATTCCTAGCAGTTTGGGCTTTCCTCCTTGATCAAGGCTTGGAAG).
cDNAs encoding murine p63 isoforms, p63α, ΔNp63α, p63γ, and ΔNp63γ were kindly given by C. Di Como (20). All p63 proteins were N-terminally Myc-tagged as previously described (20). All p63-expressing constructs were confirmed by DNA sequencing. cDNAs encoding human p73 isoforms, p73α, ΔNp73α, p73β, and ΔNp73β were as previously described (35).
The luciferase reporter constructs under the control of the p21 promoter, IGFBP3 box A, and IGFBP3 box B were as previously described (5, 41). Luciferase reporter constructs under the control of the IGFBP3 promoter, pGL2/−1165 IGFBP3 and pGL2/−705 IGFBP3, were kindly given by Xiao-Fan Wang. Deletion constructs of the IGFBP3 promoter in the luciferase reporter vector pGL2 were generated by PCR and confirmed by sequencing. To generate pGL2/−256 IGFBP3, a genomic fragment of the IGFBP3 promoter spanning nucleotides (nt) −256 to +72, with +1 being the transcriptional start site, was amplified by using the 5′ end primer −256 BP3 (AGGTACCTGGCCGGGCACACCTTG) and the 3′ end primer GLprimer2 (Promega). pGL2/−183 IGFBP3, pGL2/−116 IGFBP3, and pGL2/−91 IGFBP3 were amplified by using the 5′ end primers −183 BP3 (AGGTACCCGGGCGAGTCTCGAGCTG), −116 BP3 (AGGTACCCAGCCGTGCCTGCGCCGA), or −91 BP3 (AGGTACCCCTCCCAACCCCCACTCC) with the 3′ end primer GLprimer2.
To generate constructs expressing small interfering RNAs (siRNAs) targeting the IGFBP3 coding region, pSuper-si-IGFBP3-CR1 and pSuper-si-IGFBP3-CR2 64-nt oligonucleotides were annealed and cloned into pSuper. With the siRNA targeting region shown in bold, the oligonucleotides for pSuper-si-IGFBP3-CR1 were the forward oligonucleotide 5′-GATCCCCGAGCACAGATACCCAGAACTTCAAGAGAGTTCTGGGTATCTGTGCTCTTTTTGGAAA-3′ and the reverse oligonucleotide 5′-AGCTTTTCCAAAAAGAGCACAGATACCCAGAACTCTCTTGAAGTTCTGGGTATCTGTGCTCGGG-3′. The oligonucleotides for pSuper-si-IGFBP3-CR2 were the forward oligonucleotide 5′-GATCCCCGGGGAAGGAGGACGTGCACTTCAAGAGAGTGCACGTCCTCCTTCCCCTTTTTGGAAA-3′ and the reverse oligonucleotide 5′-AGCTTTTCCAAAAAGGGGAAGGAGGACGTGCACTCTCTTGAAGTGCACGTCCTCCTTCCCCGGG-3′.The oligonucleotides for the control siRNA targeting rat p53 were the forward oligonucleotide 5′-GATCCCCGAGCATTGCCCGGAGCTGCTTCAAGAGAGCAGCTCCGGGCAATGCTCCTTTTTGGAAA-3′ and the reverse oligonucleotide 5′-AGCTTTTCCAAAAAGAGCATTGCCCGGAGCTGCTCTCTTGAAGCAGCTCCGGGCAATGCTCCGGG-3′.
Cell lines.
The culture, transfection, and generation of MCF7 cell lines were performed as previously described (70). Individual clones were screened for the inducible expression of HA-tagged p53(ΔBD) by Western blot analysis with monoclonal antibodies against p53. Individual clones were screened for the inducible expression of Myc-tagged p63γ and Myc-tagged ΔNp63γ by Western blot analysis with monoclonal antibodies against Myc. The MCF7 cell lines p53-24, p53(ΔAD1ΔBD)-15, p63α-11, ΔNp63α-9, p73α-2, and p73β-31 were as previously described (13, 68, 69). EB and H1299 cells were maintained in Dulbecco's modified Eagle medium containing 10% fetal bovine serum.
Luciferase assay.
A dual luciferase reporter assay was used to determine the transcriptional activity of p53, engineered p53 mutants, and p53 family isoforms. The luciferase reporter constructs used were pGL2/p21-A (41), which is regulated by the p21 promoter with two p53-responsive elements; pGL2/IGFBP3-Box A (5), which contains the p53-responsive element within intron 1 of the IGFBP3 gene; pGL2/IGFBP3-Box B (5), which contains the p53-responsive element within intron 2 of the IGFBP3 gene; and pGL2/−1165 IGFBP3, pGL2/−705 IGFBP3, pGL2/−256 IGFBP3, pGL2/−183 IGFBP3, pGL2/−116 IGFBP3, and pGL2/−91 IGFBP3, which are regulated by the IGFBP3 promoter. In all, 1 μg of a luciferase reporter, 1 μg of pcDNA3 control vector or pcDNA3 vector that expresses p53 family isoforms, and 25 ng of Renilla luciferase assay vector pRL-CMV (Promega, Madison, Wis.) were cotransfected by using the calcium phosphate method into MCF7, H1299, or EB cells. A dual luciferase assay was performed in triplicate according to the manufacturer's instructions (Promega). The fold increase in relative luciferase activity is a product of the luciferase activity induced by p53 family isoforms divided by that induced by an empty pcDNA3 vector.
Western blot analysis.
Western blot analysis was performed as previously described (70) with anti-p53 monoclonal antibodies DO-1, PAb1801, PAb240, and PAb421; antiactin polyclonal antibody (Sigma); anti-p21 polyclonal antibody (C-19) (Santa Cruz); anti-HA polyclonal antibody (Y11) (Santa Cruz); anti-HA monoclonal antibody (HA.11) (Covance); and anti-Myc monoclonal antibody (9B11) (Cell Signaling). To analyze IGFBP3 protein, conditioned medium was collected from MCF7 cells induced or uninduced to express p53 and mutants for 72 h. The conditioned medium was concentrated by using a VivaSpin 6-ml concentrator (Vivascience), resuspended with 2× sodium dodecyl sulfate sample buffer, and boiled for 5 min. Western blot analysis was then performed as described by using anti-IGFBP3 polyclonal antibody (Upstate).
RNA isolation, Northern blot analysis, and reverse transcription-PCR (RT-PCR).
Total RNA was isolated by using Trizol reagents (Invitrogen). Northern blot analyses were performed as described (70). The p21, glyceraldehyde-3-phosphate dehydrogenase (GADPH), and aquaporin 3 probes were prepared as previously described (66, 70); the IGFBP3 cDNA probe was made from a 700-bp EcoRI-XhoI fragment. First-strand cDNA was synthesized by using iScript (Bio-Rad) according to the manufacturer's instructions. The level of the transcripts for IGFBP3 and GADPH were determined by PCR. The primers used to amplify GAPDH were as previously described (9). The primers used to amplify a 942-bp IGFBP3 cDNA fragment were the 5′ end primer 5-BP3-CR (AGAATTCTGTACTGTCGCCCCATCC) and 3′ end primer 3-BP3-CR (AGAATTCCGTCTACTTGCTCTGCATGC).
ChIP assay.
A chromatin immunoprecipitation (ChIP) assay was performed as previously described (36). After 24 h of induction (+) or no induction (−) of full-length p53 or p53(ΔAD1ΔBD) in MCF7 cells, chromatin was cross-linked in 1% formaldehyde in phosphate-buffered saline (PBS), and nuclei were extracted. Chromatin was sonicated to yield 500- to 1,000-bp DNA fragments and immunoprecipitated with a mixture of anti-HA polyclonal antibody (Y11) (Santa Cruz) and anti-p53 PAb1801 monoclonal antibody. After reverse cross-linking and phenol-chloroform extraction, DNA fragments bound by p53 were purified over a QIAGEN column. PCR was performed to visualize the enriched DNA fragments. Primers designed to amplify the 11 decamers of the p53-responsive element within the IGFBP3 promoter were the 5′ end primer −282BP3 (TGCTGAGGTGGCCTGGAGT) and the 3′ end primer +51BP3 (TCCAGGCAGGAAGCGGCTGATC). Primers that amplify the 5′ p53-responsive element within the p21 promoter were as previously described (36).
Trypan blue dye exclusion assay.
MCF7-p53-24 or MCF7-p53(ΔAD1ΔBD)-15 cells were seeded at a density of 2 × 104 cells/well in 24-well plates in the presence or absence of tetracycline. At 12 h after plating, anti-rabbit polyclonal antibody (Sigma) or anti-IGFBP3 polyclonal antibody (Upstate) was added to the cell culture supernatant at a dilution of 1:250. At 24 h after antibody addition, both floating cells in the medium and live cells on the plate were collected and concentrated by centrifugation. After staining with trypan blue (Sigma) for 10 min, both live (unstained) and dead (stained) cells were counted in a hemocytometer. The percentage of dead cells was calculated as the number of dead cells divided by the total number of cells counted.
Proliferation assays.
A CellTiter96 nonradioactive cell proliferation assay (Promega) was used to perform an MTT [3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide] assay. MCF7-p53(ΔAD1ΔBD)-15 cells were seeded at a density of 5 × 103 cells/well in 96-well plates in the presence and absence of tetracycline. To use IGFBP3 siRNA to characterize the contribution of IGFBP3 during MCF7-p53(ΔAD1ΔBD)-15-mediated cell death, after adherence to the plate, cells were transfected with 50 ng (per 96-well plate) of either a control siRNA expression vector targeting rat p53 or a mix of siRNA expression vectors targeting two locations within the coding region of IGFBP3 by using FuGENE 6 transfection reagent (Roche). At 44 h after plating, cells were labeled with the dye solution for 4 h and permeabilized for at least 1 h, and the absorbance at 570 nm was read by a microplate reader. To use IGFBP3 neutralizing antibody, at 18 h after plating, anti-rabbit polyclonal antibody (Sigma) or anti-IGFBP3 polyclonal antibody (Upstate) was added to the cell culture supernatant at a dilution of 1:250. At 72 h after plating, samples were processed as above, and absorbance at 570 nm was determined. P values were determined by a Student's t test.
A CellTiter-Glo luminescent cell viability assay (Promega) was used to determine the cellular ATP content. MCF7-p53(ΔAD1ΔBD)-15 cells were transfected with pSuper empty vector or a mix of siRNA expression vectors targeting two locations within the coding region of IGFBP3 by using FuGENE 6 transfection reagent (Roche). At 24 h after transfection, cells were seeded at a density of 5 × 103/well in 96-well plates in the presence and absence of tetracycline. At 36 h after plating, the CellTiter-Glo luminescent cell viability assay (Promega) was performed according to the manufacturer's instructions. The relative luciferase unit is expressed as a percentage of the product of luciferase activity induced by ATP content in the presence of p53 divided by that induced in the absence of p53. P values were determined by a Student's t test.
Drug treatments.
To verify that cisplatin stabilizes endogenous p53 and that trichostatin A (TSA) increases acetylation of histones, MCF7 cells were treated with 50 μM cisplatin (Sigma) dissolved in H2O and 100 ng of TSA (Upstate) per ml dissolved in 75% ethanol. To determine the effect upon IGFBP3 expression, MCF7 cells were treated with 50 μM cisplatin and 100 ng of TSA per ml for 12 h. To determine whether exogenous p53, p63α, or p73α induces IGFBP3 in the presence of TSA, MCF7 cells were induced or uninduced to express p53, p63α, or p73α for 12 h prior to treatment with 100 ng of TSA per ml for another 12 h.
Affymetrix gene chip analysis.
Total RNA from MCF7 cells induced and uninduced to express full-length p53 and p53(ΔAD1ΔBD) was isolated, labeled, and hybridized to an Affymetrix gene chip (U133 plus 2.0).
RESULTS
IGFBP3 is induced by p53(ΔAD1ΔBD) but not by full-length p53.
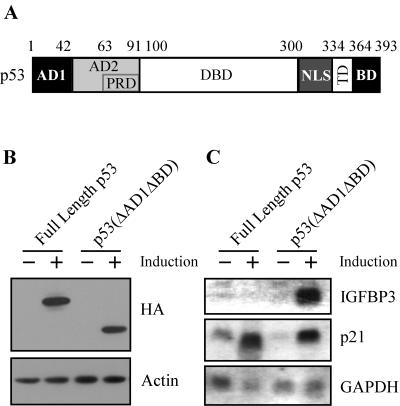
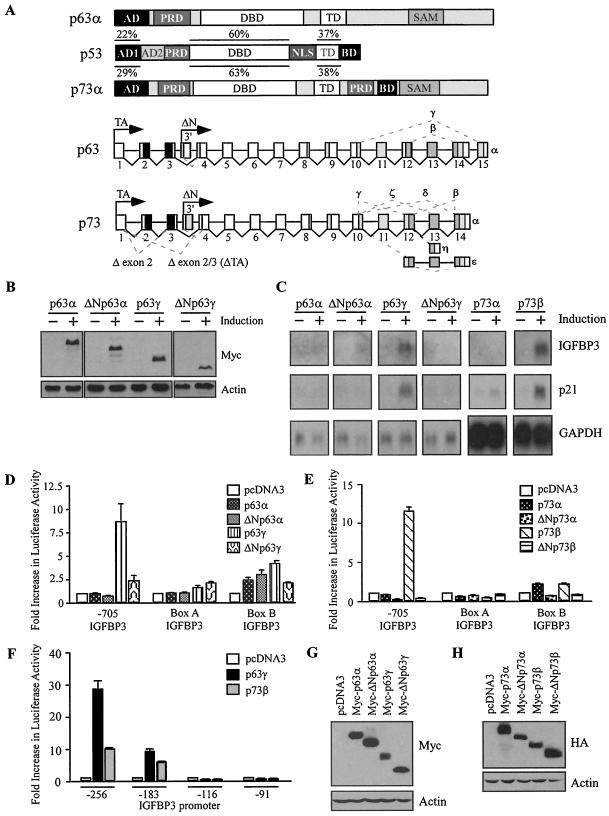
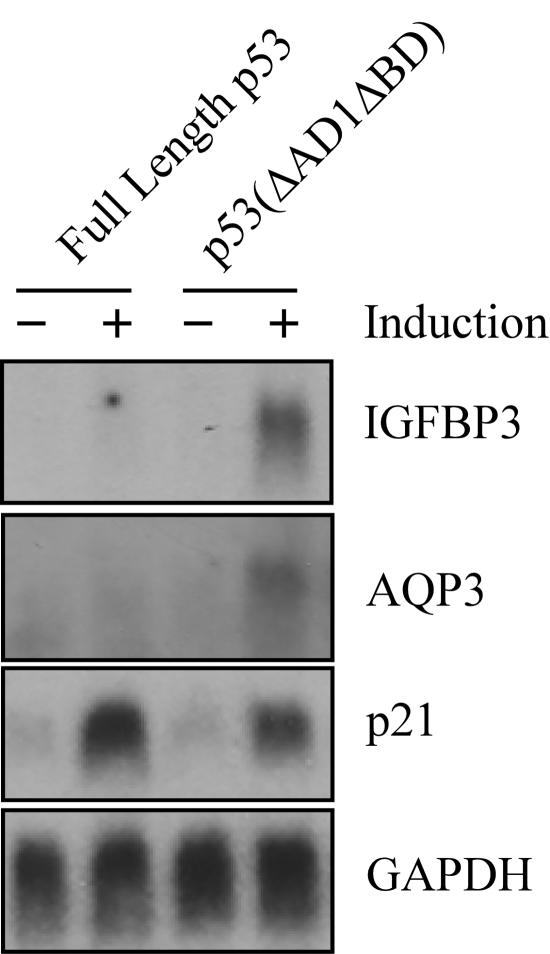
p53 contains several functional domains (Fig. 1A). During the characterization of these domains by using stable inducible cell lines, we have previously reported that p53(ΔAD1ΔBD), which lacks the N-terminal AD1 and the C-terminal BD, is more potent than full-length p53 to induce apoptosis in MCF7 breast adenocarcinoma cells (69). We chose MCF7-HA-p53-24 and MCF7-HA-p53(ΔAD1ΔBD)-15 cell lines because the proteins expressed by these lines are equivalent, although they are expressed at levels twofold higher than endogenous p53 levels induced by cisplatin (Fig. 1B and data not shown). While the BD has been characterized as a negative regulatory domain in vitro (23, 25, 56), to date, no one has been able to demonstrate the negative activity of this domain in vivo. We reasoned that the study of p53(ΔAD1ΔBD) might shed insight into how the BD functions as a negative regulator. Interestingly, we found that IGFBP3, a proapoptotic target gene, was induced by p53(ΔAD1ΔBD) but not by full-length p53, as analyzed by Northern blotting (Fig. 1C). To demonstrate that both full-length p53 and p53(ΔAD1ΔBD) are transcriptionally active, we analyzed the expression of p21. Both full-length p53 and p53(ΔAD1ΔBD) were able to induce p21. GAPDH was used as a loading control. These data suggest that AD1 and the BD inhibit the ability of full-length p53 to induce IGFBP3.
FIG. 1.
IGFBP3 expression is induced by p53(ΔAD1ΔBD), but not by full-length p53. (A) Schematic representation of p53 functional domains: AD1, AD2, PRD, sequence-specific DBD, the nuclear localization signal (NLS), TD, and the C-terminal BD. Numbers indicate amino acid residues. (B) The level of p53 and actin was assayed by Western blot analysis by using anti-HA monoclonal and antiactin polyclonal antibodies, respectively, in MCF7-HA-p53-24 and MCF7-HA-p53(ΔAD1ΔBD)-15 cells in the absence (−) or presence (+) of p53 or p53(ΔAD1ΔBD) for 24 h. (C) A Northern blot was prepared by using total RNAs isolated from MCF7-HA-p53-24 and MCF7-HA-p53(ΔAD1ΔBD)-15 cells in the absence (−) or presence (+) of p53 or p53(ΔAD1ΔBD) for 24 h. The blot was sequentially probed with cDNAs derived from IGFBP3, p21, and GAPDH genes.
IGFBP3 was shown to be regulated by p53 in EB-1 cells, a colorectal carcinoma cell line that expresses exogenous p53 under control of the metallothionein promoter (5). Because we identified a unique situation where p53(ΔAD1ΔBD), but not full-length p53, induced IGFBP3, we wanted to characterize the mechanism regulating the ability of full-length p53 to induce IGFBP3 in MCF7 cells. Interestingly, O-glycosylation masks the PAb421 epitope of p53 in EB-1 cells and the resulting O-glycosylated p53 is activated for DNA binding in vitro (57).
The IGFBP3 promoter is activated by p53(ΔAD1ΔBD) but not by full-length p53.
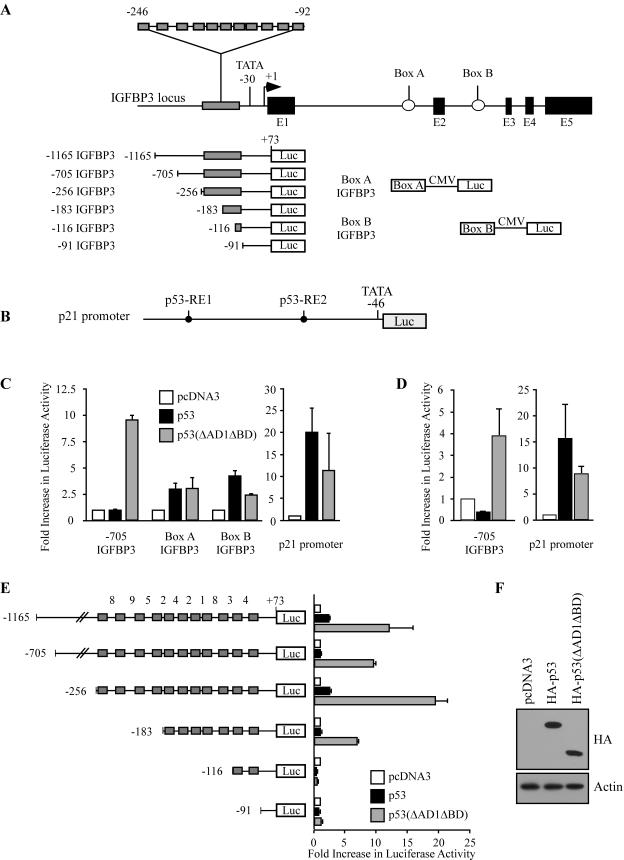
The canonical p53-responsive element contains two decamers [RRRC(A/T)(A/T)GYYY] separated by a spacer of 0 to 13 bp, where R represents purine and Y represents pyrimidine (15). IGFBP3 is a unique p53 target gene in that it contains 11 tandem decamers of the p53-responsive element within the proximal promoter, with each decamer separated by ≤9 bp (3). Thus, the IGFBP3 promoter potentially contains 10 tandem canonical p53-responsive elements spanning the region from nt −246 to −92, with +1 as the transcriptional start site. In addition, IGFBP3 contains two canonical p53-responsive elements, called box A and box B, within introns 1 and 2, respectively (5) (Fig. 2A).
FIG. 2.
Activation of the IGFBP3 promoter by p53(ΔAD1ΔBD) requires all 11 decamers of the p53-responsive element. (A) Schematic representation of the IGFBP3 locus and luciferase reporter constructs. CMV, cytomegalovirus. (B) Schematic representation of the p21 luciferase reporter construct. (C) The IGFBP3 promoter is activated by p53(ΔAD1ΔBD) but not by full-length p53 in MCF7 cells. MCF7 cells were cotransfected with 1 μg of the luciferase reporter under control of the IGFBP3 promoter, box A, or box B and 1 μg of empty pcDNA3 or pcDNA3 vector expressing HA-p53 or HA-p53(ΔAD1ΔBD). (D) The IGFBP3 promoter is activated by p53(ΔAD1ΔBD) but not by full-length p53 in EB cells. The experiment was performed as described above except that EB cells were used. (E) The 11 decamers of the p53-responsive element function as a unit. MCF7 cells were cotransfected with 1 μg of a luciferase reporter under control of various lengths of the IGFBP3 promoter and 1 μg of empty pcDNA3 or pcDNA3 vector expressing HA-p53 or HA-p53(ΔAD1ΔBD). The number between each decamer indicates the number of base pairs of separation. (F) HA-p53 and HA-p53(ΔAD1ΔBD) are expressed at equal levels. MCF7 cells were transfected with 5 μg of empty pcDNA3 or pcDNA3 vector expressing HA-p53 or HA-p53(ΔAD1ΔBD). HA-p53 and HA-p53(ΔAD1ΔBD) were detected with anti-HA monoclonal antibody. Actin was detected with antiactin polyclonal antibody.
To determine which regions within the IGFBP3 gene are responsive to p53(ΔAD1ΔBD), we analyzed the ability of transiently overexpressed HA-p53(ΔAD1ΔBD) and full-length HA-p53 to activate the IGFBP3 promoter as well as box A and box B in MCF7 cells by using a dual luciferase reporter assay. We found that the −1165 and −705 IGFBP3 reporter constructs (Fig. 2A), which contain the IGFBP3 promoter from nt −1165 to +73 and from nt −705 to +73, respectively, were activated by p53(ΔAD1ΔBD) but not by full-length p53 (Fig. 2C and E). Surprisingly, reporters under the control of box A or box B linked to a minimal cytomegalovirus promoter (Fig. 2A) were only weakly activated by p53(ΔAD1ΔBD) and full-length p53 (Fig. 2C). The luciferase reporter under the control of the p21 promoter with two canonical p53-responsive elements (Fig. 2B) was activated by both full-length p53 and p53(ΔAD1ΔBD), albeit to a lesser extent by the latter (Fig. 2C). Equal levels of p53 protein are expressed by HA-p53 and HA-p53(ΔAD1ΔBD) as detected by Western blot analysis (Fig. 2F). These data suggest that the p53-responsive elements within the IGFBP3 promoter are primarily responsible for the induction of IGFBP3 by p53(ΔAD1ΔBD).
To determine whether our observation was cell type specific, we tested the ability of full-length p53 and p53(ΔAD1ΔBD) to activate the IGFBP3 promoter in EB colon and H1299 non-small cell lung carcinoma cells. We found the same pattern of activation in EB and H1299 cells: the IGFBP3 promoter was activated by p53(ΔAD1ΔBD) but not by full-length p53 (Fig. 2D and data not shown). Again, the p21 promoter was activated by both p53(ΔAD1ΔBD) and full-length p53 (Fig. 2D and data not shown).
Because IGFPB3 is unique in that the promoter contains 11 tandem decamers of the p53-responsive element, we wanted to determine which decamers were necessary or sufficient for activation by p53(ΔAD1ΔBD). Thus, we generated four new reporter constructs: −256 IGFBP3, −183 IGFBP3, −116 IGFBP3, and −91 IGFBP3 (Fig. 2A). We found that the −256 IGFBP3 reporter, which contains all 11 decamers, was strongly activated by p53(ΔAD1ΔBD) (Fig. 2E). Surprisingly, we found that the 11 decamers function as a unit. Deletion of four upstream decamers inhibited activation of the −183 IGFBP3 reporter by p53(ΔAD1ΔBD) (Fig. 2E). In addition, the presence of only two decamers, recapitulating the canonical p53-responsive element, was not sufficient for activation of the −116 IGFBP3 reporter by p53(ΔAD1ΔBD) (Fig. 2E). Similarly, no activity was detected when all 11 decamers were deleted as in the −91 IGFBP3 reporter (Fig. 2E). Consistent with the above data, full-length p53 did not strongly activate any of the reporter constructs (Fig. 2E). Taken together, these data and results obtained by Northern blot analysis (Fig. 1C) indicate that p53(ΔAD1ΔBD) but not full-length p53 is capable of transactivating the IGFBP3 gene.
Activation of the IGFBP3 promoter by p53 requires the presence of AD2 and deletion of the BD.
The mechanism by which p53 induces a specific activity over another remains unclear in p53 research. We reasoned that p53 functional domains may play a role in target gene selection. To this end, we used deletion and point mutation of p53 to analyze the requirements of p53 functional domains for activation of the IGFBP3 promoter.
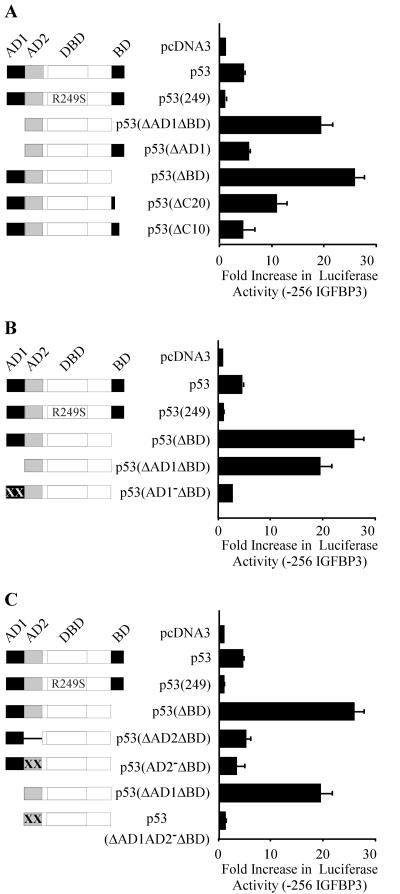
Because p53(ΔAD1ΔBD) lacks two functional domains, we first wanted to determine which domain is inhibitory. We found that deletion of only AD1 resulted in activity similar to full-length p53 (Fig. 3A); however, deletion of only the BD enabled strong activation of the IGFBP3 promoter (Fig. 3A). We then wanted to identify the inhibitory residues within the BD. We found that p53(ΔC20), which lacks the C-terminal 20 amino acids, strongly activated the IGFBP3 promoter, albeit less than p53(ΔBD) (Fig. 3A). p53(ΔC10), which lacks the C-terminal 10 amino acids, had activity similar to that of full-length p53 (Fig. 3A). p53(R249S), a tumor-derived DNA binding mutant, served as a negative control and did not activate the IGFBP3 promoter. p53 protein was expressed by all p53 constructs used in these studies (data not shown).
FIG. 3.
Activation of the IGFBP3 promoter by p53 requires the presence of AD2 and deletion of the BD. Schematic representations of p53 and mutants are shown at left. Graphs (right) show increases (n-fold) in activation of the −256 IGFBP3 reporter by p53 and mutants. MCF7 cells were cotransfected with 1 μg of luciferase reporter under control of the IGFBP3 promoter and 1 μg of empty pcDNA3 or pcDNA3 vector expressing p53 and various mutants. (A) The BD inhibits p53 activation of the IGFBP3 promoter. (B) AD1 is dispensable, but an inactive AD1 is inhibitory for p53 activation of the IGFBP3 promoter. (C) AD2 is required for activation of the IGFBP3 promoter.
Next, we wanted to characterize the N-terminal requirement. We reasoned that AD1 is not required because p53(ΔAD1ΔBD) strongly activates the IGFBP3 promoter. To test this, we used a double point mutation in AD1 (Gln22-Ser23), resulting in a well-defined AD1-deficient mutant (AD1−) that is incapable of interacting with the transcriptional machinery (34). We found that p53(AD1−ΔBD), which contains the nonfunctional AD1 and lacks the BD, was not able to activate the IGFBP3 promoter, whereas p53(ΔBD), which lacks only the BD, was active (Fig. 3B). Thus, AD1 is dispensable, but an inactive AD1 is inhibitory. This finding suggests that the conformation of p53 might be important for the induction of IGFBP3.
Because AD1 is dispensable, we wanted to characterize the role of AD2. We have previously found that AD2 is important for the induction of apoptosis (70). Interestingly, we found that deletion of AD2 (ΔAD2) or mutation of AD2 (AD2−) abolished the activity of p53(ΔBD) at the IGFBP3 promoter (Fig. 3C). Further strengthening the importance of AD2 was the finding that mutation of AD2 (AD2−) abolished the activity of p53(ΔAD1ΔBD) at the IGFBP3 promoter (Fig. 3C). Taken together, these data suggest that a functional AD2 is required for activation of the IGFBP3 promoter.
The activity of naturally occurring p53 isoforms correlates with the activity of our engineered p53 mutants.
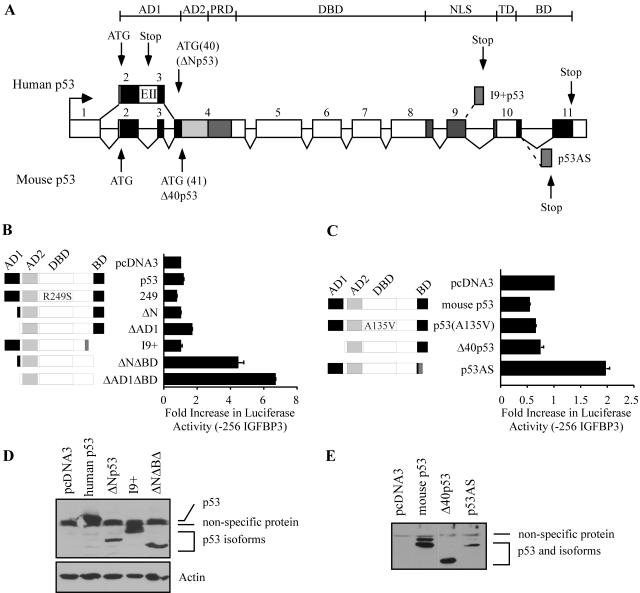
p53 has been reported to exist as both individual and combined N- and C-terminally truncated isoforms (Fig. 4A). To determine whether the behavior of our engineered p53 mutants represents the behavior of naturally occurring p53 isoforms, we generated untagged constructs expressing human and mouse p53 isoforms that have been reported to exist in the literature, namely human ΔNp53, mouse Δ40p53, human I9+p53, human p53(ΔNΔBD), and mouse p53AS (Fig. 4A). Similar to p53(ΔAD1) which lacks the first 42 amino acids, human ΔNp53 lacks the first 39 amino acids. ΔNp53 is generated through alternative splicing of intron 2. The resulting exon contains three stop codons; thus, the translational start site of ΔNp53 is at codon 40 of full-length p53 (21). Mouse Δ40p53 lacks the first 40 amino acids and is generated through the use of an internal translational start site at codon 41 (reviewed in reference 11). As expected, the activity of ΔNp53, Δ40p53, and p53(ΔAD1) was analogous, with all constructs having similar levels of activity at the IGFBP3 promoter compared to full-length human and mouse p53 (Fig. 4B and C). I9+p53 is generated through alternative splicing of intron 9 and results in the substitution of 62 C-terminal amino acids by 10 unique amino acids (18). Because alternative splicing abolishes the TD, I9+p53 should be transcriptionally inactive. As expected, I9+p53 did not activate the IGFBP3 promoter (Fig. 4B). C-terminal cleavage of p53, detectable by the lack of anti-p53 PAb421 epitope located within amino acids 371 to 380, occurs through interaction with single-stranded DNA (ssDNA) or cleavage by calpain (42, 45). We reasoned that p53(ΔBD), which lacks amino acids 364 to 393, mimics a C-terminally cleaved p53. It is also likely that a p53 protein exists that is alternatively spliced at the N terminus and is subsequently truncated at the C terminus through interaction with ssDNA or calpain. Thus, we generated p53(ΔNΔBD). As expected, the activities of p53(ΔNΔBD) and p53(ΔAD1ΔBD) were analogous, with both constructs strongly activating the IGFBP3 promoter (Fig. 4B). In addition, we found that mouse p53AS activated the IGFBP3 promoter, whereas full-length mouse p53 did not (Fig. 4C). p53AS is generated through alternative splicing of intron 10 and results in the substitution of 26 C-terminal amino acids by 17 unique amino acids. Mouse p53(A135V), a naturally occurring DNA binding mutant, was inactive and served as a negative control (Fig. 4C). p53 protein was expressed by these constructs as detected by Western blot analysis (Fig. 4D and E). Taken together, these data suggest that naturally occurring p53 isoforms differentially regulate IGFBP3. Unfortunately, we were not able to detect endogenous levels of naturally occurring isoforms by Western blot analysis. However, we cannot rule out the possibility that undetectable levels exist and may possess significant function.
FIG. 4.
The activity of naturally occurring p53 isoforms correlates with the activity of engineered mutants. (A) Schematic representation of human (top) and murine (bottom) p53 isoforms. Numbers indicate exons. Dotted lines indicate alternative splicing. Human ΔNp53 is generated through alternative splicing of intron 2. The alternative exon (EII) contains three stop codons; thus, translation begins at codon 40. Additionally, human ΔNp53 is generated through the use of an internal translational start site at codon 40. Mouse Δ40p53 is generated through the use of an internal translational start site at codon 41. Human I9+p53 and mouse p53AS are generated through alternative splicing of introns 9 and 10, respectively. (B) Naturally occurring human p53 isoforms activate the IGFBP3 promoter. Shown is a schematic representation of human p53 isoforms (left). MCF7 cells were cotransfected with 1 μg of luciferase reporter under control of the IGFBP3 promoter and 1 μg of empty pcDNA3 or pcDNA3 vector expressing p53 and various mutants (right). (C) Naturally occurring mouse p53 isoforms activate the IGFBP3 promoter. The schematic representation and graphs are analogous to those described for panel B. (D and E) p53 isoforms are expressed from various p53 constructs. MCF7 cells were transfected with empty pcDNA3 or pcDNA3 vector expressing various human p53 isoforms. H1299 cells were transfected with empty pcDNA3 or pcDNA3 vector expressing various mouse p53 isoforms. p53 was detected with anti-p53 monoclonal antibodies. Actin was detected with antiactin polyclonal antibody.
IGFBP3 expression is induced by C-terminally truncated p63 and p73 isoforms, p63γ and p73β, but not by full-length ones.
Since the C terminus of p53 plays a critical role in regulating IGFBP3, we reasoned that the C termini of p63 and p73 might also affect the regulation of IGFBP3. Both p63 and p73 undergo alternative splicing of their C termini, resulting in three p63 isoforms (α to γ) and seven p73 isoforms (α to η) (Fig. 5A). These isoforms, called the TA and ΔN isoforms, are transcribed from the upstream promoter as well as from a cryptic promoter within intron 3, respectively. Thus, many naturally occurring p63 and p73 proteins are generated through alternative splicing and the use of two transcriptional start sites.
FIG. 5.
The C termini of p63 and p73 facilitate differential regulation of IGFBP3. (A) Schematic representation of p53 family functional domains and isoforms. SAM, sterile-α-motif domain; TA, isoforms transcribed from the upstream promoter; and ΔN, isoforms transcribed from the cryptic promoter within intron 3. Dotted lines indicate alternative splicing. Percent identity is indicated. (B) The levels of p63 and actin were assayed by Western blot analysis by using anti-Myc monoclonal and antiactin polyclonal antibodies, respectively, in MCF7-Myc-p63α-11, MCF7-Myc-ΔNp63α-9, MCF7-Myc-p63γ-19, and MCF7-Myc-ΔNp63γ-18 cells in the absence (−) or presence (+) of p63 for 24 h. (C) IGFBP3 is induced by p63γ and p73β but not by p63α, ΔNp63α, ΔNp63γ, or p73α. A Northern blot was prepared by using total RNAs isolated from cells uninduced (−) or induced (+) to express various p63 or p73 isoforms for 24 h. The blot was sequentially probed with cDNAs derived from IGFBP3, p21, and GAPDH genes. (D and E) p63γ and p73β activate the IGFBP3 promoter. MCF7 cells were cotransfected with 1 μg of luciferase reporter under control of the IGFBP3 promoter, box A, or box B and 1 μg of empty pcDNA3 or pcDNA3 vector expressing various p63 (D) or p73 (E) isoforms. (F) The 11 decamers function as a unit in response to p63γ and p73β. (G and H) Proteins are expressed from various p63 and p73 constructs. MCF7 cells were transfected with 5 μg of empty pcDNA3 or pcDNA3 vector expressing Myc-tagged p63 isoforms or HA-tagged p73 isoforms. Levels of p63, p73, and actin were determined by Western blotting with anti-Myc monoclonal antibody, anti-HA monoclonal antibody, and antiactin polyclonal antibody, respectively.
To determine whether the N and/or the C termini of p63 and p73 modulate the regulation of IGFBP3, previously characterized stable MCF7 cell lines were analyzed for their ability to regulate IGFBP3 as follows: for cell lines inducibly expressing the Myc-tagged p63 isoforms, p63α (clone 11), ΔNp63α (clone 9), p63γ (clone 19), or ΔNp63γ (clone 18) was used; for cell lines expressing the HA-tagged p73 isoforms, p73α (clone 2) or p73β (clone 31) was used. Equal levels of p63 are expressed by the representative clones as detected by Western blot analysis (Fig. 5B). Interestingly, we found that IGFBP3 was strongly induced by the C-terminally truncated isoforms p63γ and p73β but not by the full-length or the ΔN isoforms, as detected by Northern blotting (Fig. 5C). We also analyzed the ability of the p63 and p73 isoforms to regulate the well-characterized p53 target gene, p21. Similar to the regulation of IGFBP3 by p63 and p73 isoforms, the C-terminally truncated isoforms p63γ and p73β strongly induce p21. In addition, p21 is induced by full-length p73α, albeit weakly. Thus, a pattern emerges for p53 family regulation of IGFBP3: the C terminus of the p53 family is inhibitory for the induction of IGFBP3.
p63γ and p73β activate the IGFBP3 promoter.
To determine which regions within the IGFBP3 gene are responsive to p63γ and p73β, we analyzed the ability of various p63 and p73 isoforms to activate the IGFBP3 promoter as well as box A and box B in MCF7 cells by using a dual luciferase reporter assay. We found that the −705 IGFBP3 reporter was activated by p63γ but not by p63α, ΔNp63α, or ΔNp63γ (Fig. 5D). None of the p63 isoforms analyzed strongly activated box A or box B (Fig. 5D). Similarly, the −705 IGFBP3 reporter was activated by p73β but not by p73α, ΔNp73α, or ΔNp73β. None of the p73 isoforms analyzed strongly activated box A or box B (Fig. 5E). Next, we wanted to determine which decamers within the IGFBP3 promoter were necessary or sufficient for activation by p63γ and p73β. We found that all 11 decamers were required for activation of the IGFBP3 promoter. Deletion of four upstream decamers, as with the −183 IGFBP3 construct, severely inhibited p63γ and p73β activity at the IGFBP3 promoter (Fig. 5F). No activity was detected when 9 or all 11 decamers were deleted, as with the −116 IGFBP3 or −91 IGFBP3 construct, respectively (Fig. 5F). Thus, we found that both p63γ and p73β behave similarly to p53(ΔAD1ΔBD). Protein was expressed by these p63 and p73 constructs as detected by Western blotting (Fig. 5G and 5H). Taken together, these data and results obtained by Northern blot analysis (Fig. 5C) indicate that p63γ and p73β, but not the α or ΔN isoforms, utilize the IGFBP3 promoter to induce IGFBP3.
IGFBP3 is an important effector of p53(ΔAD1ΔBD)-dependent cell death.
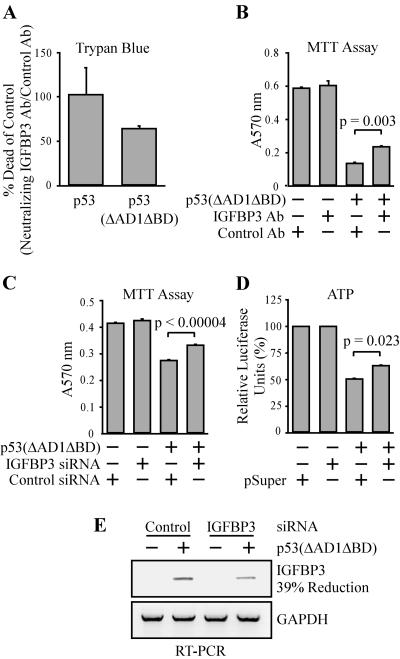
Because p53(ΔAD1ΔBD) is more potent than full-length p53 to induce apoptosis in MCF7 cells (69) and IGFBP3 is induced by p53(ΔAD1ΔBD) but not by full-length p53 (Fig. 1C), we wanted to characterize the contribution of IGFBP3 in p53- and p53(ΔAD1ΔBD)-dependent apoptosis. To do this, we performed a trypan blue dye exclusion assay and found that cell death induced by p53(ΔAD1ΔBD) was reduced by 36% with the addition of the IGFBP3 neutralizing antibody to the cell culture supernatant compared to the cells incubated with the rabbit control antibody (Fig. 6A). Cell death induced by full-length p53 was not reduced by incubation with the IGFBP3 antibody (Fig. 6A). In addition, we performed an MTT assay and found that incubation with the IGFBP3 neutralizing antibody increases the survival of cells expressing p53(ΔAD1ΔBD) in comparison to cells incubated with the rabbit control antibody (Fig. 6B). To confirm these data, we measured cell viability in the presence of p53(ΔAD1ΔBD) with and without knockdown of IGFBP3 by siRNA. We used transient transfection of vector-expressed siRNA hairpins that target two locations within the IGFBP3 coding region. Using the MTT assay, we found that transient knockdown of IGFBP3 with siRNA increases the viability of cells expressing p53(ΔAD1ΔBD) in comparison to cells transfected with control siRNA targeting rat p53 (Fig. 6C). In addition, we measured cellular ATP as an indication of cell viability and found that siRNA targeting IGFBP3 increases the viability of cells expressing p53(ΔAD1ΔBD) compared to cells transfected with empty pSuper vector (Fig. 6D). To demonstrate the extent of the transient knockdown of induced IGFBP3 levels, RT-PCR was performed on p53(ΔAD1ΔBD)-15 cells transiently transfected with a mix of the IGFBP3-siRNA expression vectors pSuper-si-IGFBP3-CR-1 and pSuper-si-IGFBP3-CR-2 or a control siRNA expression vector targeting rat p53 and were subsequently induced or uninduced to express p53(ΔAD1ΔBD). A 39% reduction of induced IGFBP3 levels was detected by RT-PCR (Fig. 6E). Although we cannot completely abolish induced IGFBP3 levels by using transient transfection of pSuper-si-IGFBP3-CR-1 and pSuper-si-IGFBP3-CR-2, we do detect a modest but reproducible reduction of cell death induced by p53(ΔAD1ΔBD) in the presence of IGFBP3 siRNA (Fig. 6C and D). We suspect that the reduction of cell death would be greater given more efficient IGFBP3 knockdown. Taken together, these data suggest that IGFBP3 is an important effector of p53-dependent apoptosis.
FIG. 6.
IGFBP3 is an effector of p53(ΔAD1ΔBD)-mediated apoptosis. (A and B) Neutralizing IGFBP3 antibody reduces cell death induced by p53(ΔAD1ΔBD) but not by full-length p53. Results are shown of a trypan blue dye exclusion assay of MCF7-p53-24 and MCF7-p53(ΔAD1ΔBD)-15 cells and MTT assay of MCF7-p53(ΔAD1ΔBD)-15 cells induced or uninduced to express the p53 protein in the presence of anti-IGFBP3 neutralizing antibody or anti-rabbit control antibody. Experiments were performed as described in Materials and Methods. (C and D) Knockdown of IGFBP3 by siRNA enhances viability of cells induced to express MCF7-p53(ΔAD1ΔBD)-15. Shown are the results of an MTT assay and an ATP luminescence assay of MCF7-p53(ΔAD1ΔBD)-15 cells induced or uninduced to express the p53 protein transiently transfected with vector-expressed siRNA against IGFBP3 (mix of two siRNAs targeting the coding region of IGFBP3) or with control siRNA against rat p53 or empty pSuper vector. Experiments were performed as described in Materials and Methods. (E) A 39% reduction of the p53(ΔAD1ΔBD)-induced IGFBP3 mRNA level by IGFBP3 siRNA. Levels of the transcripts for IGFBP3 and GAPDH with transient transfection of vectors expressing siRNA against IGFBP3 (mix of two siRNAs targeting the coding region of IGFBP3) or control siRNA against rat p53 were determined by RT-PCR with 28 cycles.
Full-length p53 and p53(ΔAD1ΔBD) bind to the IGFBP3 promoter in vivo.
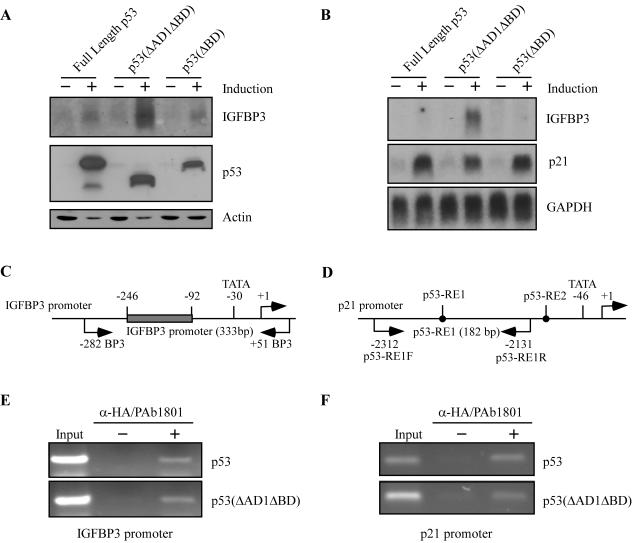
To gain insight into how the BD regulates p53 target gene selection, we wanted to characterize the mechanism that enables p53(ΔAD1ΔBD), but not full-length p53, to induce IGFBP3. First, we wanted to characterize the induction of IGFBP3 by p53(ΔBD), because deletion of the BD renders p53 capable of activating the IGFBP3 promoter (Fig. 3A). Thus, we generated several stable MCF7 cell lines inducibly expressing HA-tagged p53(ΔBD) in the tetracycline-repressible system. A representative cell line, clone 3, was selected for presentation. Western blot analysis showed that the level of p53(ΔBD) protein expressed in this line was comparable to that of full-length p53 and p53(ΔAD1ΔBD) in M7-p53-24 and M7-p53(ΔAD1ΔBD)-15 cell lines, respectively (Fig. 7A). To demonstrate that p53(ΔBD) is transcriptionally active, we analyzed the expression of p21. Like full-length p53 and p53(ΔAD1ΔBD), p53(ΔBD) was able to induce p21 (Fig. 7B). Surprisingly, we found that p53(ΔBD) did not significantly induce IGFBP3, as determined by Northern blot analysis (Fig. 7B). Although unexpected, our result is not unprecedented as deletion of the BD has been shown to activate DNA binding in vitro (25); however, to date deletion of the BD has not been shown to enhance the expression of a target gene. Western blot analysis further confirmed that p53(ΔBD) was not capable of inducing IGFBP3, since secreted IGFBP3 was substantially increased by p53(ΔAD1ΔBD) but not by full-length p53 or p53(ΔBD) (Fig. 7A). Although we do detect a slight increase in the secreted IGFBP3 protein level when full-length p53 or p53(ΔBD) is expressed, a corresponding increase in IGFBP3 mRNA level is not detected. Thus, we attribute the modest induction of IGFBP3 protein by full-length p53 and p53(ΔBD) to posttranslational stabilization of the IGFPB3 protein that may be effected by another p53 target gene. As hinted by the study of AD1, where AD1 is dispensable but an inactive AD1 is inhibitory (Fig. 3B), these data indicate that the conformation of the p53 protein is important for the regulation of IGFBP3.
FIG. 7.
IGFBP3 is induced by p53(ΔAD1ΔBD) but not by full-length p53 or p53(ΔBD). (A) Secreted IGFBP3 is induced by p53(ΔAD1ΔBD). Cell extracts and concentrated conditioned medium were prepared from cells uninduced (−) or induced (+) to express p53, p53(ΔAD1ΔBD), or p53(ΔBD). Levels of p53, actin, and secreted IGFBP3 in p53-24, p53(ΔAD1ΔBD)-15, and p53(ΔBD)-3 cell lines were determined by Western blotting with anti-p53 monoclonal antibodies, antiactin polyclonal antibody, and anti-IGFBP3 polyclonal antibody, respectively. (B) p53(ΔAD1ΔBD) transactivates the gene encoding IGFBP3. A Northern blot was prepared by using total RNAs isolated from cells uninduced (−) or induced (+) to express p53, p53(ΔAD1ΔBD), or p53(ΔBD) for 24 h. The blot was sequentially probed with cDNAs derived from IGFBP3, p21, and GAPDH genes. (C and D) Schematic representation of the IGFBP3 and p21 promoters with the location of the transcriptional start site, the TATA box, p53-responsive elements, and primers used for ChIP assays. (E) p53 and p53(ΔAD1ΔBD) bind the IGFBP3 promoter in vivo. A ChIP assay was performed as described in Materials and Methods. (F) p53 and p53(ΔAD1ΔBD) bind the p53-responsive element within the p21 promoter in vivo.
To determine whether p53 DNA binding is the mechanism governing the regulation of IGFBP3 by p53, we performed ChIP assays. After a 24-h induction of full-length p53 or p53(ΔAD1ΔBD), chromatin was cross-linked, sonicated, and immunoprecipitated with a mixture of anti-HA polyclonal and anti-p53 PAb1801 monoclonal antibodies. To visualize the enriched DNA fragments, PCR was performed to amplify the region of the IGFBP3 promoter spanning the 11 decamers as well as the upstream p53-responsive element within the p21 promoter (Fig. 7C and D). Chromatin prepared without induction of full-length p53 or p53(ΔAD1ΔBD) was used as a negative control. We found that both full-length p53 and p53(ΔAD1ΔBD) bound to the IGFBP3 promoter (Fig. 7E); however, only p53(ΔAD1ΔBD) induces IGFBP3 (Fig. 1C). No enrichment of the DNA fragment containing the IGFBP3 promoter was detected in the samples not induced to express full-length p53 or p53(ΔAD1ΔBD). As expected, both full-length p53 and p53(ΔAD1ΔBD) bound to the p21 promoter (Fig. 7F). Thus, p53 DNA binding is not the mechanism by which IGFBP3 is induced by p53(ΔAD1ΔBD) but not by full-length p53.
HDAC activity inhibits the ability of full-length p53 family isoforms to induce IGFBP3.
Because full-length p53 binds the IGFBP3 promoter in vivo, we reasoned that under certain circumstances, full-length p53 would become competent to induce IGFBP3. Such circumstances might include the addition of an activating posttranslational modification, removal of an inhibitory posttranslational modification, dissociation from an inhibitory p53-associated protein, or a combination of these effects. To this end, we used MCF7 cells to characterize the regulation of IGFBP3 by endogenous p53 activated by the DNA damaging agent cisplatin in the presence and absence of the HDAC inhibitor TSA.
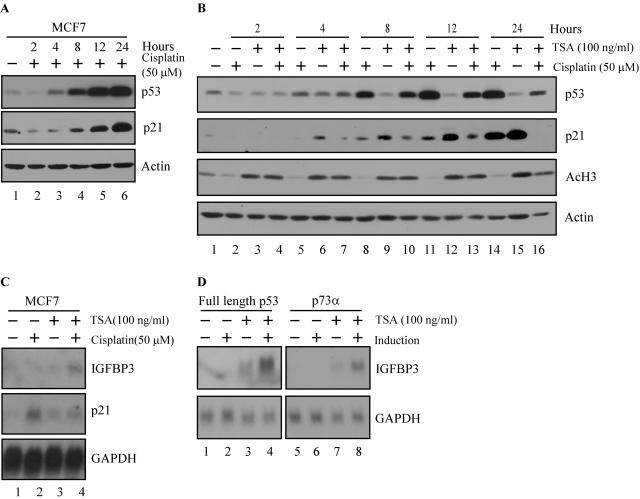
Western blotting showed that endogenous p53 protein was stabilized within 4 h upon DNA damage with 50 μM cisplatin, and the stabilization continued throughout 24 h (Fig. 8A). p21 protein was detected as early as 8 h after cisplatin treatment and increased throughout 24 h (Fig. 8A). HDAC inhibition, evidenced by increased levels of acetylated histone H3, was detected within 2 h upon treatment with 100 ng of TSA per ml, and the inhibition continued throughout 24 h (Fig. 8B). Because MCF7 cells did not tolerate a combined treatment of cisplatin and TSA for more than 12 h, we chose this time point for future analyses. We found that activation of p53 by cisplatin or inhibition of HDAC activity by TSA was not sufficient to induce IGFBP3, as detected by Northern blotting (Fig. 8C). However, the combined treatment with cisplatin and TSA enabled endogenous full-length p53 to adopt the conformation necessary to induce IGFBP3 (Fig. 8C). In contrast, p21 was induced upon DNA damage, but its expression was inhibited by the combined treatment (Fig. 8B and C).
FIG. 8.
HDAC activity inhibits the induction of IGFBP3 by full-length p53 family isoforms. (A) Time course of p53 stabilization by the DNA damage agent cisplatin. MCF7 cells were treated with 50 μM cisplatin, and cell extracts were collected at the time points indicated. p53, p21, and actin levels were determined by Western blotting by using anti-p53 monoclonal antibodies, anti-p21 polyclonal antibody, and antiactin polyclonal antibody, respectively. (B) Time course of p53 stabilization with cisplatin and/or HDAC inhibition with TSA. MCF7 cells were treated with 50 μM cisplatin and/or 100 ng of TSA per ml, and cell extracts were collected at the time points indicated. p53, acetylated histone H3, p21, and actin levels were determined by Western blotting by using anti-p53 monoclonal antibodies, anti-acetylated histone H3 polyclonal antibody, anti-p21 polyclonal antibody, and antiactin polyclonal antibody, respectively. (C) Inhibition of HDACs enables induction of IGFBP3 by endogenous p53. A Northern blot was prepared by using total RNAs isolated from MCF7 cells treated as indicated for 12 h. The blot was sequentially probed with cDNAs derived from IGFBP3, p21, and GAPDH genes. (D) Inhibition of HDACs restores the ability of exogenous p53 to induce IGFBP3. A Northern blot was prepared by using total RNAs isolated from cells uninduced (−) or induced (+) to express p53 or p73α in the presence or absence of 100 ng of TSA per ml. The blot was sequentially probed with cDNAs derived from IGFBP3 and GAPDH genes.
To verify our observation that inhibition of HDAC activity restores the ability of endogenous p53 to induce IGFBP3, we tested the ability of exogenous full-length p53 and p73α to induce IGFBP3 in the presence of TSA. As noted previously, IGFBP3 was not induced by full-length p53 or p73α (Fig. 1C, 5C, and 8D). However, TSA treatment restored the ability of exogenous full-length p53 and p73α to induce IGFBP3 in the M7-p53-24 and M7-p73α-2 cell lines, respectively (Fig. 8D). Similarly, TSA treatment rendered full-length p63α competent to induce IGFBP3 (data not shown). We note that a slight increase in IGFBP3 mRNA was detected in the presence of TSA alone in M7-p53-24 and M7-p73α-2 cell lines (Fig. 8D). It is possible that a small amount of p53 or p73 was expressed in the uninduced condition, leading to the modest induction of IGFBP3 in the presence of TSA. Taken together, these data demonstrate that HDACs inhibit the ability of full-length p53 family proteins to induce IGFBP3 and that this inhibition can be overcome by combination treatments that simultaneously activate p53 and inhibit HDACs.
Differential target gene regulation by full-length p53 and p53(ΔAD1ΔBD).
p53 family isoforms differentially regulate IGFBP3. Specifically, AD1 and the C-terminal BD inhibit induction of IGFBP3 by p53. Importantly, this inhibition can be overcome with inhibition of HDACs. To better understand the significance of this regulation, we wanted to determine whether other target genes are also under similar control by p53 isoforms. Thus, we performed Affymetrix gene chip analysis by using MCF7 cells induced and uninduced to express full-length p53 or p53(ΔAD1ΔBD). We identified several common target genes such as p21, FDXR, and others that were induced by full-length p53 and p53(ΔAD1ΔBD) (Table 1). We also identified several differentially regulated target genes that were induced by p53(ΔAD1ΔBD) but not by full-length p53 and vice versa (Table 1). Importantly, Northern blot analysis confirmed that the water and glycerol transporter aquaporin 3 (AQP3) is regulated similarly to IGFBP3, such that AQP3 is induced by p53(ΔAD1ΔBD) but not by full-length p53 (Fig. 9). Although AQP3 has not been linked to apoptosis to date, we cannot rule out the possibility that the increased expression of AQP3 may be proapoptotic by altering the water balance of the MCF7 cells.
TABLE 1.
Differential target gene regulation by full-length p53 and p53 (ΔAD1ΔBD)
| Target gene | GenBank accession no. | Increase (fold) in expression of:
|
|
|---|---|---|---|
| Full-length p53 | p53 (ΔAD1ΔBD) | ||
| Common target gene | |||
| p21 | NM_000389.1 | 5.0 | 3.5 |
| FDXR | NM_004110.2 | 5.1 | 3.2 |
| Fas/Apo1 | X83493.1 | 4.1 | 2.5 |
| GADD45 | NM_001924.2 | 3.6 | 2.5 |
| CYFIP2 | AL161999.1 | 2.3 | 2.0 |
| TP53INP1 | AW341649 | 3.2 | 3.4 |
| TUBB | NM_001069.1 | 3.1 | 3.9 |
| CAV2 | NM_001233.1 | 3.4 | 3.8 |
| Palladin | NM_016081.1 | 2.3 | 3.0 |
| Differentially regulated gene | |||
| AQP3 | AB001325 | 3.2 | |
| EVA1 | AF275945.1 | 4.6 | |
| MYO 10 | NM_012334.1 | 2.2 | |
| Novel gene | AI188104 | 3.2 | |
| ANXA4 | NM_001153.2 | 2.4 | |
| MLF2 | NM_005439.1 | 2.1 | |
| POMZP3 | NM_012230.1 | 3.0 | |
FIG. 9.
Differential target gene regulation by full-length p53 and p53(ΔAD1ΔBD). A Northern blot was prepared by using total RNAs isolated from cells uninduced (−) or induced (+) to express p53 or p53(ΔAD1ΔBD) for 24 h. The blot was sequentially probed with cDNAs derived from AQP3, IGFBP3, p21, and GAPDH genes.
DISCUSSION
How p53 differentially regulates target gene expression and thus chooses cell fate is complex. Previous studies have shown that the level of p53 (8, 48), the sequence of p53-responsive elements (47, 58), the proapoptotic AD2 (61, 70), the PRD (50, 69), and p53-associated proteins, such as ASPP1 and ASPP2 (33, 52), play an important role in this process. In this study, we have identified a common theme among the p53 family proteins: the C terminus is inhibitory, such that C-terminally truncated p53, p63, and p73 isoforms induce the expression of IGFBP3, an important effector of apoptosis, whereas full-length isoforms cannot. Thus, for the first time, we provide evidence that the BD of p53 is inhibitory in vivo as has been described in vitro. We have also shown that the in vivo inhibitory activity of the BD depends upon AD1, such that alleviation of inhibition by the BD requires deletion of AD1. We attribute the inhibition to HDAC activity. Inhibition of HDAC activity restores the ability of endogenous and exogenous full-length p53, p63, and p73 isoforms to induce IGFBP3. Moreover, inhibition of HDAC activity further enhances induction of IGFBP3 by C-terminally truncated p53 family isoforms (data not shown). We also identified several target genes differentially regulated by p53(ΔAD1ΔBD) and full-length p53, emphasizing the importance of p53 isoforms and functional domains for target gene selection. In summary, we have found that differential target gene selection requires the coordination of p53 family isoforms with the promoter environment.
p53 family isoforms differentially regulate target gene expression.
Our studies indicate that coordinated and integrated signals are required for the induction of the proapoptotic target gene IGFBP3 by the p53 family. We showed that p53 functional domains regulate the ability of p53 to activate the IGFBP3 promoter, such that AD2 is required and AD1 and the BD are inhibitory (Fig. 3). Interestingly, deletion of the inhibitory functional domains, namely the N-terminal AD1 and the C-terminal BD, is paralleled in nature. N-terminal truncation of p53 has been reported to occur (i) through alternative splicing of human p53 to yield ΔNp53/p47 (21); (ii) through the use of the internal translational start site at codon 40 and 41 in human and mouse p53, yielding ΔNp53 and Δ40p53, respectively (11, 12); (iii) through cleavage by calpain to yield a 42-kDa human and 41-kDa mouse p53 (31, 45); (iv) through interaction with mismatched double-stranded DNA to yield mouse p40(ΔN) (40); and (v) through interaction with an N- and C-terminally truncated p53 protein to yield mouse p50(ΔN23) (42). C-terminal truncation of p53 has been reported to occur (i) through alternative splicing to yield human I9+p53 and murine p53AS (1, 18); (ii) through cleavage by calpain (45); and (iii) through interaction with the 3′ end of ssDNA to yield murine p50(ΔC) and p40(ΔC) (42). Combined N- and C-terminal truncation has been reported to occur through interaction with mismatched DNA to yield p35, which is reported to have intrinsic protease ability (42), and through cleavage by calpain to yield a 33-kDa murine p53 (45). We postulate that combined N- and C-terminal truncation could occur sequentially. Alternative splicing or internal translational initiation at codon 40 generates ΔNp53, which is subsequently truncated at the C terminus through interaction with ssDNA or calpain. In the case of mouse p53, internal translational initiation of the p53AS transcript at codon 41 generates Δ40p53AS, which is analogous to p53(ΔAD1ΔBD). We found that both ΔNp53 and Δ40p53 behave similarly to p53(ΔAD1) (Fig. 4B and C). I9+p53 lacks the TD and, as expected, is transcriptionally inactive (Fig. 4B). Interestingly, full-length mouse p53 and p53AS behave similarly to full-length p53 and human p53(ΔBD): mouse p53AS activates the IGFBP3 promoter, whereas full-length mouse p53 cannot (Fig. 4C). In addition, p53(ΔNΔBD), which mimics a sequentially truncated p53 isoform, is extremely active at the IGFBP3 promoter (Fig. 4B). Thus, for the first time, we showed that p53 isoforms do have different functions. In addition, using Affymetrix gene chip analysis, we identified several target genes differentially regulated by full-length p53 and p53(ΔAD1ΔBD) (Table 1).
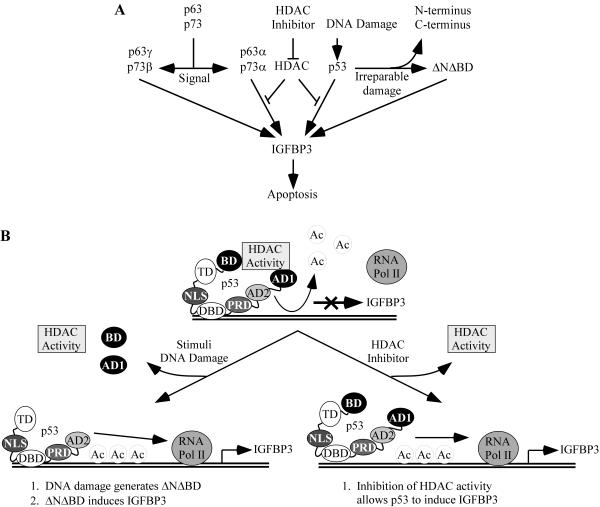
Based on these data, we suggest a model according to which specific environmental stimuli trigger the generation of p53 isoforms that will differentially regulate target gene expression (Fig. 10A). Genotoxic stress induces DNA damage. The DNA damage pathway activates and stabilizes full-length p53, which in turn induces p53-dependent cell cycle arrest through induction of p21. The arrest allows a cell time to repair its damaged DNA. However, if the damage is extensive, free 3′ ends of the damaged DNA could induce N- and/or C-terminal cleavage of ΔNp53 and full-length p53 to yield p53(ΔNΔBD). Since p53(ΔNΔBD) is resistant to negative regulation at the IGFBP3 promoter, IGFBP3 is induced. IGFBP3, along with other proapoptotic target genes, shifts the balance from p53-dependent cell cycle arrest to p53-dependent apoptosis. Thus, we propose that certain stimuli can alter the expression pattern of specific p53 isoforms and, subsequently, the expression pattern of p53 target genes (Fig. 10A). In support of this model, DNA damage has been shown to heighten the apoptotic response without affecting p53 levels (8). In addition, a recent study demonstrated that DNA damage-induced p53-dependent neuronal cell death was prevented upon calpain inhibition (54) and thus highlights the potential functional importance of calpain-dependent p53 isoforms.
FIG. 10.
A model of apoptosis for p53 family isoforms. Ac, acetyl.
Further strengthening the validity of our model is the evidence we found that p63 and p73 isoforms function in an analogous manner to p53 isoforms: the p63 and p73 C-terminally truncated isoforms, p63γ and p73β, but not full-length isoforms, induce the expression of IGFBP3 (Fig. 5C). Thus, the question arises, How are these isoforms differentially regulated? Since the regulation of alternative splicing has not been thoroughly studied, we propose a model where certain signals would activate alternative splicing to preferentially upregulate p63γ or p73β, which would carry out a specific transcriptional program, namely the induction of IGFBP3 and other proapoptotic target genes, and subsequently apoptosis (Fig. 10A). Therefore, future work needs to be done to characterize the regulation of alternative splicing.
Previously, we have shown that the ΔN isoforms of p63 and p73 contain a unique AD (13, 35) that differs from the AD in the TA isoforms. The AD in TAp63 and TAp73 shares 22 and 29% identity, respectively, to AD1 in p53 (Fig. 5A). Interestingly, we have shown that the p53 AD1 is inhibitory and AD2 is required for activation of the IGFBP3 promoter (Fig. 3B and C). Similarly, we found that the AD in the TA but not the ΔN isoforms is active in inducing IGFBP3 (Fig. 5C). Although the AD present in the TA isoforms is highly similar to AD1 in p53, perhaps the AD of TA possesses a wider range of functions, such that the critical residues or tertiary structure important for AD2 function are also contained within the TA AD. The fact that the ΔN isoforms cannot induce IGFBP3 suggests a functional difference between the ADs within the ΔN and TA isoforms.
The promoter environment contributes to differential target gene selection.
Our studies indicate that the promoter sequence in terms of the structure and number of the p53-responsive elements and the promoter topology in terms of the associated proteins are important for target gene selection. The structure of the p53-responsive element within the IGFBP3 promoter is unique in that it contains 11 tandem decamers of the p53-responsive element. We found that the 11 decamers function as a unit, such that all are required for full activation of IGFBP3 by p53(ΔAD1ΔBD), p63γ, and p73β (Fig. 2E and 5F). Since only two decamers constitute the canonical p53-responsive element, these data suggest that the unique arrangement is important for the differential regulation of IGFBP3 by p53 family isoforms. Similar to IGFBP3, p53-induced gene 3 (PIG3) is a proapoptotic target gene and contains a unique p53-responsive element (46). p53 activates the PIG3 promoter through a pentanucleotide microsatellite sequence (10). Interestingly, the pentanucleotide sequence is polymorphic, and increasing numbers of repeats confer greater responsiveness to p53 (10). To further support the importance of the responsive element, studies show that p53-responsive elements located within cell cycle arrest genes are more highly activated by p53 than the ones located in proapoptotic genes (47). It has been postulated that the spacing between the decamers is critical. Responsive elements found within cell cycle arrest genes do not contain interspersed sequences between the decamers, but the ones within proapoptotic genes do (47, 58).
Our studies also demonstrate that the coordination of p53 family isoforms with the promoter environment is important for transcriptional activation of IGFBP3. We found that full-length p53 and p53(ΔAD1ΔBD) bind to the IGFBP3 promoter in vivo, as determined by the ChIP assay (Fig. 7E); however, only p53(ΔAD1ΔBD) is capable of inducing IGFBP3 (Fig. 1C). We found that inhibition of HDAC activity restores the ability of exogenous full-length p53 to induce IGFBP3 (Fig. 8D). In addition, we found that p63 and p73 isoforms behave in a similar fashion: HDAC activity inhibits the induction of IGFBP3 by full-length p63 and p73 isoforms, p63α and p73α, but not by C-terminally truncated isoforms, p63γ and p73β (Fig. 8D and data not shown). Our data suggest a model according to which full-length p53 and p53(ΔAD1ΔBD) bind to the IGFBP3 promoter, but full-length p53 cannot recruit the basal transcriptional machinery due to its association with HDAC activity (Fig. 10B). Although the acetylation status of p53 has been shown to modulate its own activity (27), our model focuses on the ability of p53 to interact either directly or indirectly with HDACs that target the chromatin but not full-length p53, as p53(ΔAD1ΔBD), which is extremely active in inducing IGFBP3, lacks acetylation sites. In support of our model, recent studies have demonstrated that p53 target gene expression requires more than just p53 binding: p53 first binds to the p21 promoter and recruits p300, which then acetylates the p21 proximal promoter and cooperates with p53 to induce p21 (16, 36).
In our model, p53 family isoforms differentially interact with regulatory proteins to facilitate target gene expression. Differential protein interactions may depend upon the conformation of the p53 protein or specific epitopes, either gained or lost. In support of the importance of conformation, many studies have identified p53 mutants that can induce apoptosis but not cell cycle arrest and vice versa (19, 49, 51). For example, mutant p53(121F) is a more potent inducer of transcription-dependent apoptosis than full-length p53 (51). Although the structure of p53(121F) was not determined by crystallography, because full-length p53 is a fluid molecule and will not crystallize, we speculate that the conformation of p53(121F) is different from that of the full-length p53. Thus, p53(121F) may escape regulation by certain proteins, potentially HDACs or protein complexes with HDAC activity, to enable the induction of proapoptotic target genes. Interestingly, p53 has been shown to interact with mSin3A, a component of the transcriptional repression complex, as well as HDAC-1, HDAC-2, and HDAC-3 (27, 71). While mSin3A interacts with the PRD of p53 (71), the physical interaction between p53 and HDAC-1, HDAC-2, and HDAC-3 has not been mapped. Thus, it is possible that the HDACs interact with the N and C termini of p53, and thus the IGFBP3 promoter bound by p53(ΔAD1ΔBD) escapes this negative regulation. While many questions still remain and many mechanisms are yet to be discovered, one point becomes clear: the ability of the p53 family to induce the expression of proapoptotic target genes is under stringent regulation.
Therapeutic implications.
Restoration of p53 functions, especially p53-dependent apoptosis, is an attractive cancer therapeutic strategy. To exploit p53-dependent apoptosis for the destruction of cancer cells, many studies have focused on strategies that preferentially activate p53 to induce proapoptotic target genes. Here, we found that treatment of MCF7 cells with the DNA damage agent cisplatin was sufficient to stabilize p53 levels with the subsequent induction of p21; however, cisplatin alone was not sufficient to enable p53 induction of IGFBP3, an important effector of apoptosis. However, combined treatment with cisplatin and the HDAC inhibitor TSA enabled IGFBP3 expression (Fig. 8C). Thus, our data presented here shed new insight into how we can modulate p53 activity to induce apoptosis in tumor cells, which can be explored for the design of combined therapies for cancer treatment.
Acknowledgments
We thank Charles Di Como for p63 cDNA constructs, Xiao-Fan Wang (Duke University) for IGFBP3 promoter constructs, and Jiandong Chen (University of South Florida) for EB and EB-1 cells.
This work was supported in part by the National Cancer Institute of the National Institutes of Health (grants CA076069, CA081237, and CA102188) and the Department of Defense Breast Cancer Research Program (under award number W81XWH-04-0349).
The opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the U.S. Army or the Department of Defense.
REFERENCES
- 1.Arai, N., D. Nomura, K. Yokota, D. Wolf, E. Brill, O. Shohat, and V. Rotter. 1986. Immunologically distinct p53 molecules generated by alternative splicing. Mol. Cell. Biol. 6:3232-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Augustin, M., C. Bamberger, D. Paul, and H. Schmale. 1998. Cloning and chromosomal mapping of the human p53-related KET gene to chromosome 3q27 and its murine homolog Ket to mouse chromosome 16. Mamm. Genome 9:899-902. [DOI] [PubMed] [Google Scholar]
- 3.Bourdon, J. C., V. Deguin-Chambon, J. C. Lelong, P. Dessen, P. May, B. Debuire, and E. May. 1997. Further characterisation of the p53 responsive element—identification of new candidate genes for trans-activation by p53. Oncogene 14:85-94. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, C. L., and W. Gu. 2003. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15:164-171. [DOI] [PubMed] [Google Scholar]
- 5.Buckbinder, L., R. Talbott, S. Velasco-Miguel, I. Takenaka, B. Faha, B. R. Seizinger, and N. Kley. 1995. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature 377:646-649. [DOI] [PubMed] [Google Scholar]
- 6.Butt, A. J., S. M. Firth, M. A. King, and R. C. Baxter. 2000. Insulin-like growth factor-binding protein-3 modulates expression of Bax and Bcl-2 and potentiates p53-independent radiation-induced apoptosis in human breast cancer cells. J. Biol. Chem. 275:39174-39181. [DOI] [PubMed] [Google Scholar]
- 7.Candau, R., D. M. Scolnick, P. Darpino, C. Y. Ying, T. D. Halazonetis, and S. L. Berger. 1997. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene 15:807-816. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., L. J. Ko, L. Jayaraman, and C. Prives. 1996. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 10:2438-2451. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., Y. Zheng, J. Zhu, J. Jiang, and J. Wang. 2001. p73 is transcriptionally regulated by DNA damage, p53, and p73. Oncogene 20:769-774. [DOI] [PubMed] [Google Scholar]
- 10.Contente, A., A. Dittmer, M. C. Koch, J. Roth, and M. Dobbelstein. 2002. A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat. Genet. 30:315-320. [DOI] [PubMed] [Google Scholar]
- 11.Courtois, S., C. C. de Fromentel, and P. Hainaut. 2004. p53 protein variants: structural and functional similarities with p63 and p73 isoforms. Oncogene 23:631-638. [DOI] [PubMed] [Google Scholar]
- 12.Courtois, S., G. Verhaegh, S. North, M. G. Luciani, P. Lassus, U. Hibner, M. Oren, and P. Hainaut. 2002. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene 21:6722-6728. [DOI] [PubMed] [Google Scholar]
- 13.Dohn, M., S. Zhang, and X. Chen. 2001. p63α and ΔNp63α can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20:3193-3205. [DOI] [PubMed] [Google Scholar]
- 14.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, Jr., J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 15.el-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 1:45-49. [DOI] [PubMed] [Google Scholar]
- 16.Espinosa, J. M., and B. M. Emerson. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 8:57-69. [DOI] [PubMed] [Google Scholar]
- 17.Firth, S. M., and R. C. Baxter. 2002. Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev. 23:824-854. [DOI] [PubMed] [Google Scholar]
- 18.Flaman, J. M., F. Waridel, A. Estreicher, A. Vannier, J. M. Limacher, D. Gilbert, R. Iggo, and T. Frebourg. 1996. The human tumour suppressor gene p53 is alternatively spliced in normal cells. Oncogene 12:813-818. [PubMed] [Google Scholar]
- 19.Friedlander, P., Y. Haupt, C. Prives, and M. Oren. 1996. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Mol. Cell. Biol. 16:4961-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaiddon, C., M. Lokshin, J. Ahn, T. Zhang, and C. Prives. 2001. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21:1874-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh, A., D. Stewart, and G. Matlashewski. 2004. Regulation of human p53 activity and cell localization by alternative splicing. Mol. Cell. Biol. 24:7987-7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gucev, Z. S., Y. Oh, K. M. Kelley, and R. G. Rosenfeld. 1996. Insulin-like growth factor binding protein 3 mediates retinoic acid- and transforming growth factor beta2-induced growth inhibition in human breast cancer cells. Cancer Res. 56:1545-1550. [PubMed] [Google Scholar]
- 23.Halazonetis, T. D., L. J. Davis, and A. N. Kandil. 1993. Wild-type p53 adopts a “mutant”-like conformation when bound to DNA. EMBO J. 12:1021-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harms, K., S. Nozell, and X. Chen. 2004. The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 61:822-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hupp, T. R., D. W. Meek, C. A. Midgley, and D. P. Lane. 1992. Regulation of the specific DNA binding function of p53. Cell 71:875-886. [DOI] [PubMed] [Google Scholar]
- 26.Jayaraman, J., and C. Prives. 1995. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell 81:1021-1029. [DOI] [PubMed] [Google Scholar]
- 27.Juan, L. J., W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin, and C. W. Wu. 2000. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275:20436-20443. [DOI] [PubMed] [Google Scholar]
- 28.Kaghad, M., H. Bonnet, A. Yang, L. Creancier, J. C. Biscan, A. Valent, A. Minty, P. Chalon, J. M. Lelias, X. Dumont, P. Ferrara, F. McKeon, and D. Caput. 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90:809-819. [DOI] [PubMed] [Google Scholar]
- 29.Kim, H. S., A. R. Ingermann, J. Tsubaki, S. M. Twigg, G. E. Walker, and Y. Oh. 2004. Insulin-like growth factor-binding protein 3 induces caspase-dependent apoptosis through a death receptor-mediated pathway in MCF-7 human breast cancer cells. Cancer Res. 64:2229-2237. [DOI] [PubMed] [Google Scholar]
- 30.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 31.Kubbutat, M. H., and K. H. Vousden. 1997. Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol. Cell. Biol. 17:460-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulesz-Martin, M. F., B. Lisafeld, H. Huang, N. D. Kisiel, and L. Lee. 1994. Endogenous p53 protein generated from wild-type alternatively spliced p53 RNA in mouse epidermal cells. Mol. Cell. Biol. 14:1698-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane, D. 2001. How cells choose to die. Nature 414:25, 27. [DOI] [PubMed] [Google Scholar]
- 34.Lin, J., J. Chen, B. Elenbaas, and A. J. Levine. 1994. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 8:1235-1246. [DOI] [PubMed] [Google Scholar]
- 35.Liu, G., S. Nozell, H. Xiao, and X. Chen. 2004. ΔNp73β is active in transactivation and growth suppression. Mol. Cell. Biol. 24:487-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, G., T. Xia, and X. Chen. 2003. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J. Biol. Chem. 278:17557-17565. [DOI] [PubMed] [Google Scholar]
- 37.Mee, T., A. L. Okorokov, S. Metcalfe, and J. Milner. 1999. Proteolytic cleavage of p53 mutants in response to mismatched DNA. Br. J. Cancer 81:212-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michalovitz, D., O. Halevy, and M. Oren. 1990. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell 62:671-680. [DOI] [PubMed] [Google Scholar]
- 39.Mills, A. A., B. Zheng, X. J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708-713. [DOI] [PubMed] [Google Scholar]
- 40.Molinari, M., A. L. Okorokov, and J. Milner. 1996. Interaction with damaged DNA induces selective proteolytic cleavage of p53 to yield 40 kDa and 35 kDa fragments competent for sequence-specific DNA binding. Oncogene 13:2077-2086. [PubMed] [Google Scholar]
- 41.Nozell, S., Y. Wu, K. McNaughton, G. Liu, A. Willis, J. C. Paik, and X. Chen. 2003. Characterization of p73 functional domains necessary for transactivation and growth suppression. Oncogene 22:4333-4347. [DOI] [PubMed] [Google Scholar]
- 42.Okorokov, A. L., F. Ponchel, and J. Milner. 1997. Induced N- and C-terminal cleavage of p53: a core fragment of p53, generated by interaction with damaged DNA, promotes cleavage of the N terminus of full-length p53, whereas ssDNA induces C-terminal cleavage of p53. EMBO J. 16:6008-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olivier, M., R. Eeles, M. Hollstein, M. A. Khan, C. C. Harris, and P. Hainaut. 2002. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 19:607-614. [DOI] [PubMed] [Google Scholar]
- 44.Osada, M., M. Ohba, C. Kawahara, C. Ishioka, R. Kanamaru, I. Katoh, Y. Ikawa, Y. Nimura, A. Nakagawara, M. Obinata, and S. Ikawa. 1998. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 4:839-843. [DOI] [PubMed] [Google Scholar]
- 45.Pariat, M., S. Carillo, M. Molinari, C. Salvat, L. Debussche, L. Bracco, J. Milner, and M. Piechaczyk. 1997. Proteolysis by calpains: a possible contribution to degradation of p53. Mol. Cell. Biol. 17:2806-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polyak, K., Y. Xia, J. L. Zweier, K. W. Kinzler, and B. Vogelstein. 1997. A model for p53-induced apoptosis. Nature 389:300-305. [DOI] [PubMed] [Google Scholar]
- 47.Qian, H., T. Wang, L. Naumovski, C. D. Lopez, and R. K. Brachmann. 2002. Groups of p53 target genes involved in specific p53 downstream effects cluster into different classes of DNA binding sites. Oncogene 21:7901-7911. [DOI] [PubMed] [Google Scholar]
- 48.Ronen, D., D. Schwartz, Y. Teitz, N. Goldfinger, and V. Rotter. 1996. Induction of HL-60 cells to undergo apoptosis is determined by high levels of wild-type p53 protein whereas differentiation of the cells is mediated by lower p53 levels. Cell Growth Differ. 7:21-30. [PubMed] [Google Scholar]
- 49.Ryan, K. M., and K. H. Vousden. 1998. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol. Cell. Biol. 18:3692-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakamuro, D., P. Sabbatini, E. White, and G. C. Prendergast. 1997. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene 15:887-898. [DOI] [PubMed] [Google Scholar]
- 51.Saller, E., E. Tom, M. Brunori, M. Otter, A. Estreicher, D. H. Mack, and R. Iggo. 1999. Increased apoptosis induction by 121F mutant p53. EMBO J. 18:4424-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samuels-Lev, Y., D. J. O'Connor, D. Bergamaschi, G. Trigiante, J. K. Hsieh, S. Zhong, I. Campargue, L. Naumovski, T. Crook, and X. Lu. 2001. ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell 8:781-794. [DOI] [PubMed] [Google Scholar]
- 53.Schedlich, L. J., and L. D. Graham. 2002. Role of insulin-like growth factor binding protein-3 in breast cancer cell growth. Microsc. Res. Tech. 59:12-22. [DOI] [PubMed] [Google Scholar]
- 54.Sedarous, M., E. Keramaris, M. O'Hare, E. Melloni, R. S. Slack, J. S. Elce, P. A. Greer, and D. S. Park. 2003. Calpains mediate p53 activation and neuronal death evoked by DNA damage. J. Biol. Chem. 278:26031-26038. [DOI] [PubMed] [Google Scholar]
- 55.Selivanova, G., V. Iotsova, I. Okan, M. Fritsche, M. Strom, B. Groner, R. C. Grafstrom, and K. G. Wiman. 1997. Restoration of the growth suppression function of mutant p53 by a synthetic peptide derived from the p53 C-terminal domain. Nat. Med. 3:632-638. [DOI] [PubMed] [Google Scholar]
- 56.Selivanova, G., L. Ryabchenko, E. Jansson, V. Iotsova, and K. G. Wiman. 1999. Reactivation of mutant p53 through interaction of a C-terminal peptide with the core domain. Mol. Cell. Biol. 19:3395-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw, P., J. Freeman, R. Bovey, and R. Iggo. 1996. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene 12:921-930. [PubMed] [Google Scholar]
- 58.Tokino, T., S. Thiagalingam, W. S. el-Deiry, T. Waldman, K. W. Kinzler, and B. Vogelstein. 1994. p53 tagged sites from human genomic DNA. Hum. Mol. Genet. 3:1537-1542. [DOI] [PubMed] [Google Scholar]
- 59.Trink, B., K. Okami, L. Wu, V. Sriuranpong, J. Jen, and D. Sidransky. 1998. A new human p53 homologue. Nat. Med. 4:747-748. [DOI] [PubMed] [Google Scholar]
- 60.Venot, C., M. Maratrat, C. Dureuil, E. Conseiller, L. Bracco, and L. Debussche. 1998. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 17:4668-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venot, C., M. Maratrat, V. Sierra, E. Conseiller, and L. Debussche. 1999. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene 18:2405-2410. [DOI] [PubMed] [Google Scholar]
- 62.Walker, K. K., and A. J. Levine. 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc. Natl. Acad. Sci. USA 93:15335-15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, A., M. Kaghad, Y. Wang, E. Gillett, M. D. Fleming, V. Dotsch, N. C. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2:305-316. [DOI] [PubMed] [Google Scholar]
- 64.Yang, A., and F. McKeon. 2000. p63 and p73: p53 mimics, menaces and more. Nat. Rev. Mol. Cell Biol. 1:199-207. [DOI] [PubMed] [Google Scholar]
- 65.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]
- 66.Zheng, X., and X. Chen. 2001. Aquaporin 3, a glycerol and water transporter, is regulated by p73 of the p53 family. FEBS Lett. 489:4-7. [DOI] [PubMed] [Google Scholar]
- 67.Zhu, J., J. Jiang, W. Zhou, K. Zhu, and X. Chen. 1999. Differential regulation of cellular target genes by p53 devoid of the PXXP motifs with impaired apoptotic activity. Oncogene 18:2149-2155. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, J., S. Nozell, J. Wang, J. Jiang, W. Zhou, and X. Chen. 2001. p73 cooperates with DNA damage agents to induce apoptosis in MCF7 cells in a p53-dependent manner. Oncogene 20:4050-4057. [DOI] [PubMed] [Google Scholar]
- 69.Zhu, J., S. Zhang, J. Jiang, and X. Chen. 2000. Definition of the p53 functional domains necessary for inducing apoptosis. J. Biol. Chem. 275:39927-39934. [DOI] [PubMed] [Google Scholar]
- 70.Zhu, J., W. Zhou, J. Jiang, and X. Chen. 1998. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J. Biol. Chem. 273:13030-13036. [DOI] [PubMed] [Google Scholar]
- 71.Zilfou, J. T., W. H. Hoffman, M. Sank, D. L. George, and M. Murphy. 2001. The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol. Cell. Biol. 21:3974-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]